Primer design using NIH's Primer-BLAST tool.
Pedro C. Díaz, Francisco Franco García Calderon
Published: 2024-04-23 DOI: 10.17504/protocols.io.rm7vzj8j5lx1/v1
Abstract
This protocol demonstrates the basic usage of the Primer-BLAST tool in order to design PCR primers from a given FASTA sequence.
The results should show the sequence of at least one pair of primers (forward and reverse) within the chosen parameters.
Steps
Go to NIH's Primer-BLAST tool
1.
Open your internet browser (Google Chrome recommended).
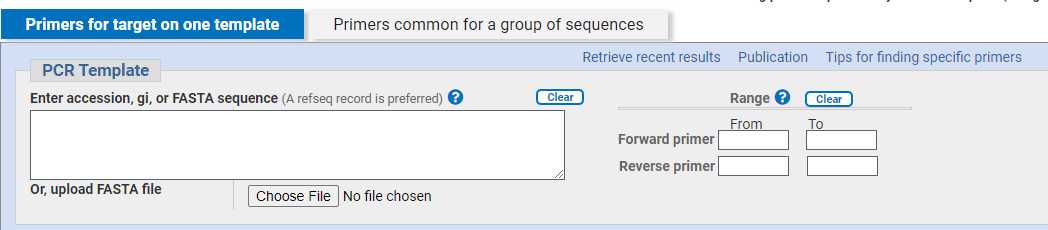
Enter FASTA sequence
3.
4.
You can select a custom range (i.e. the positional value of the first and last nucleotide of your ROI) for the forward and reverse primers.
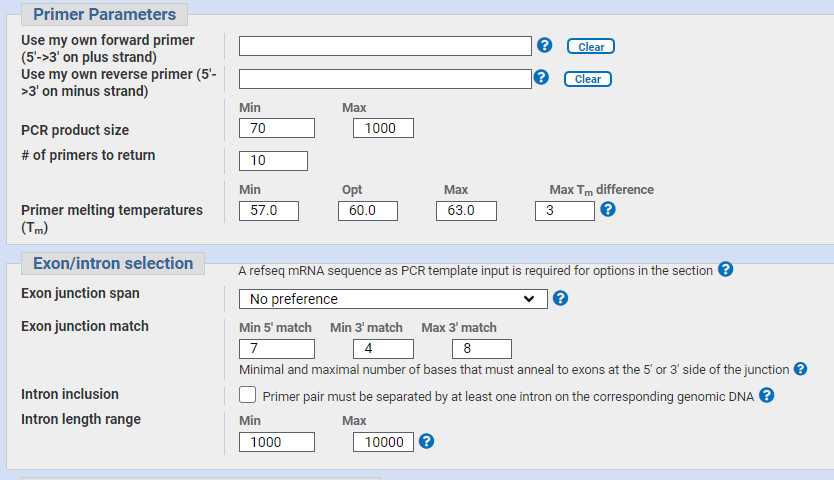
Enter primer parameters
5.