Precision genome editing using cytosine and adenine base editors in mammalian cells
Tony P. Huang, Gregory A. Newby, David R. Liu





























Supplementary information
Supplementary Information
Supplementary Fig. 1.
Reporting Summary
Supplementary Tables 1–6
Additional information on base editor components and a calculator for determining HTS library concentrations.
Supplementary Data 1
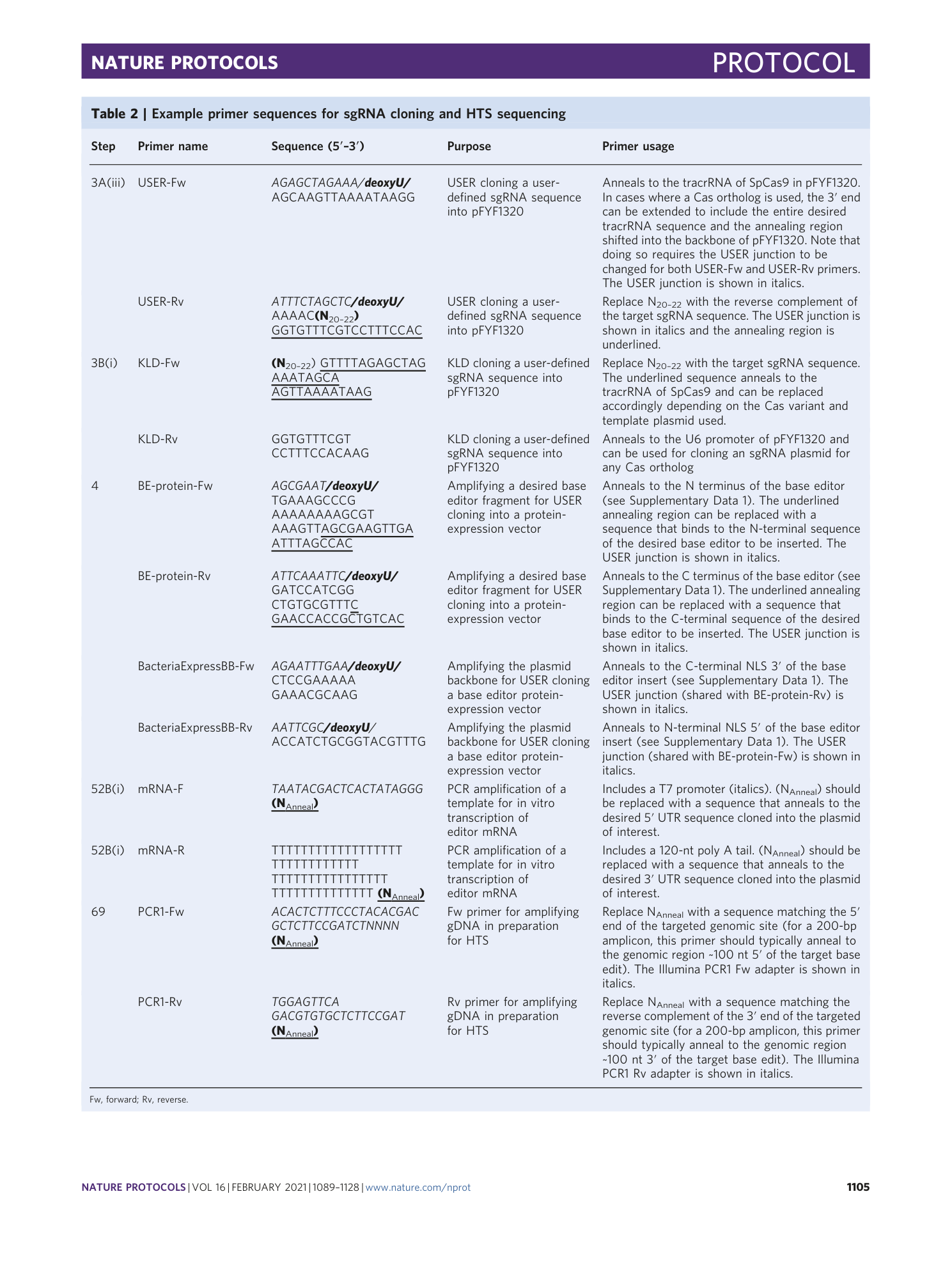
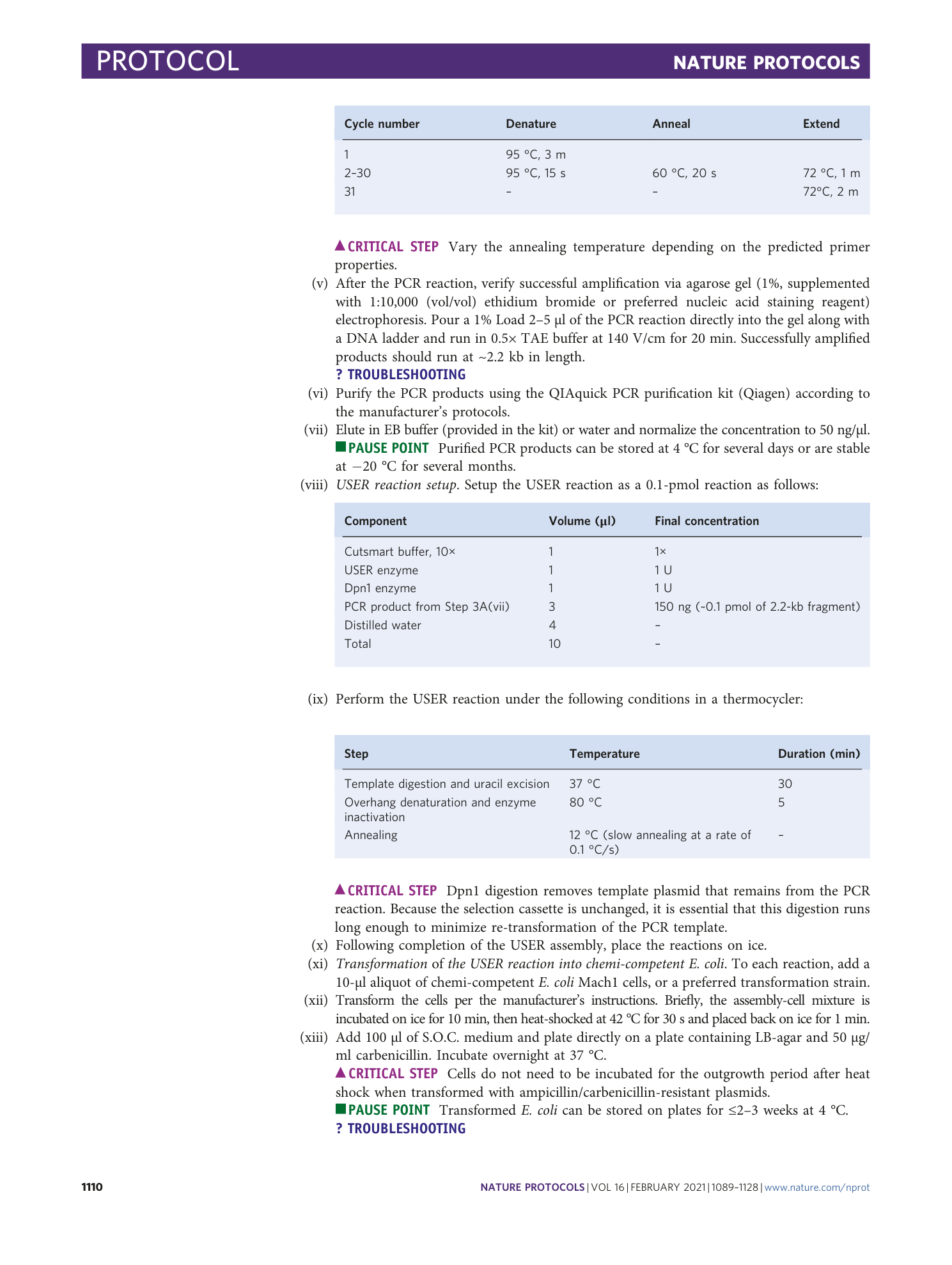
A plasmid map annotated with example primer sequences (Table 2) for cloning a base editor into a protein expression vector
Supplementary Data 2
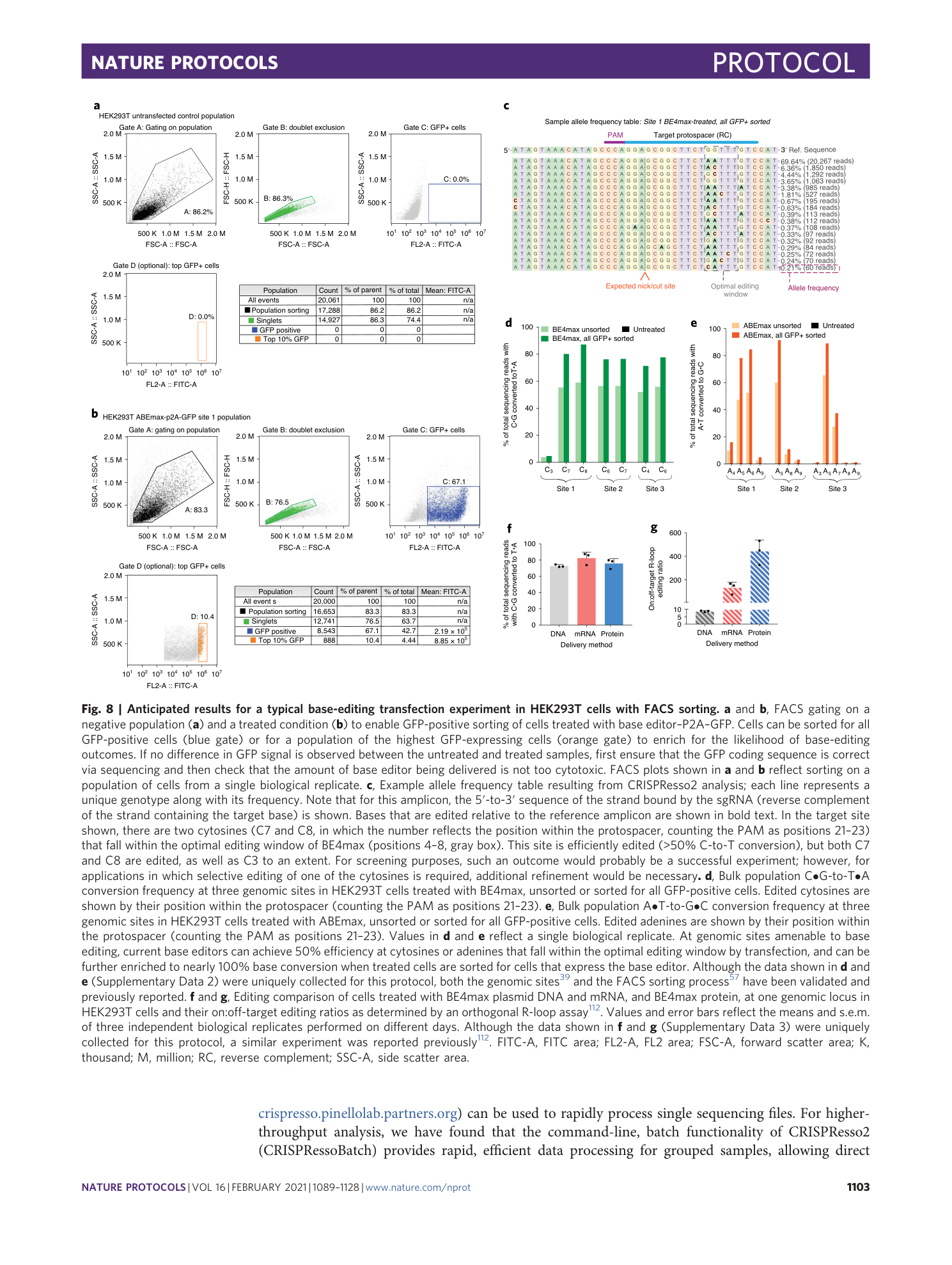
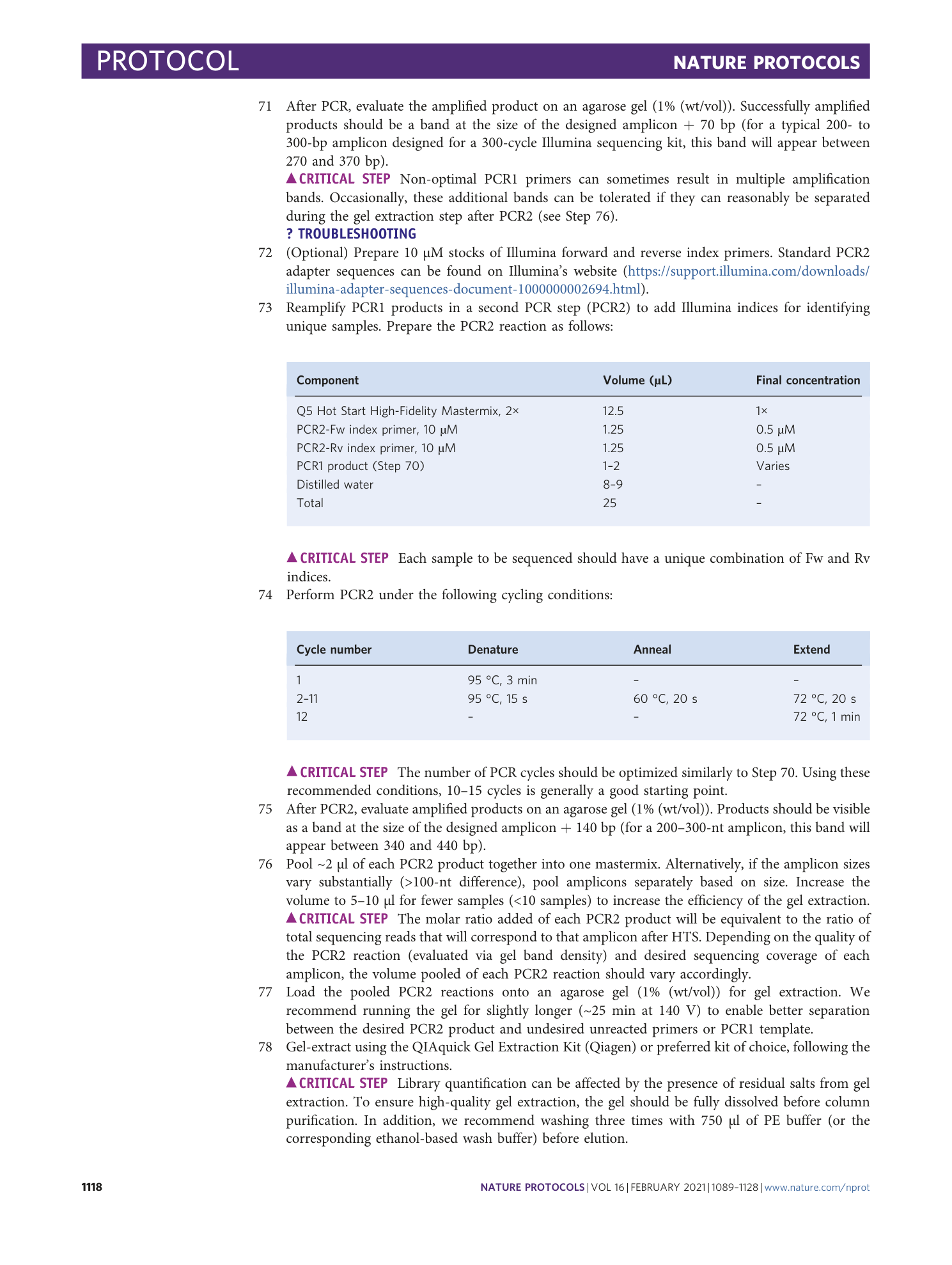
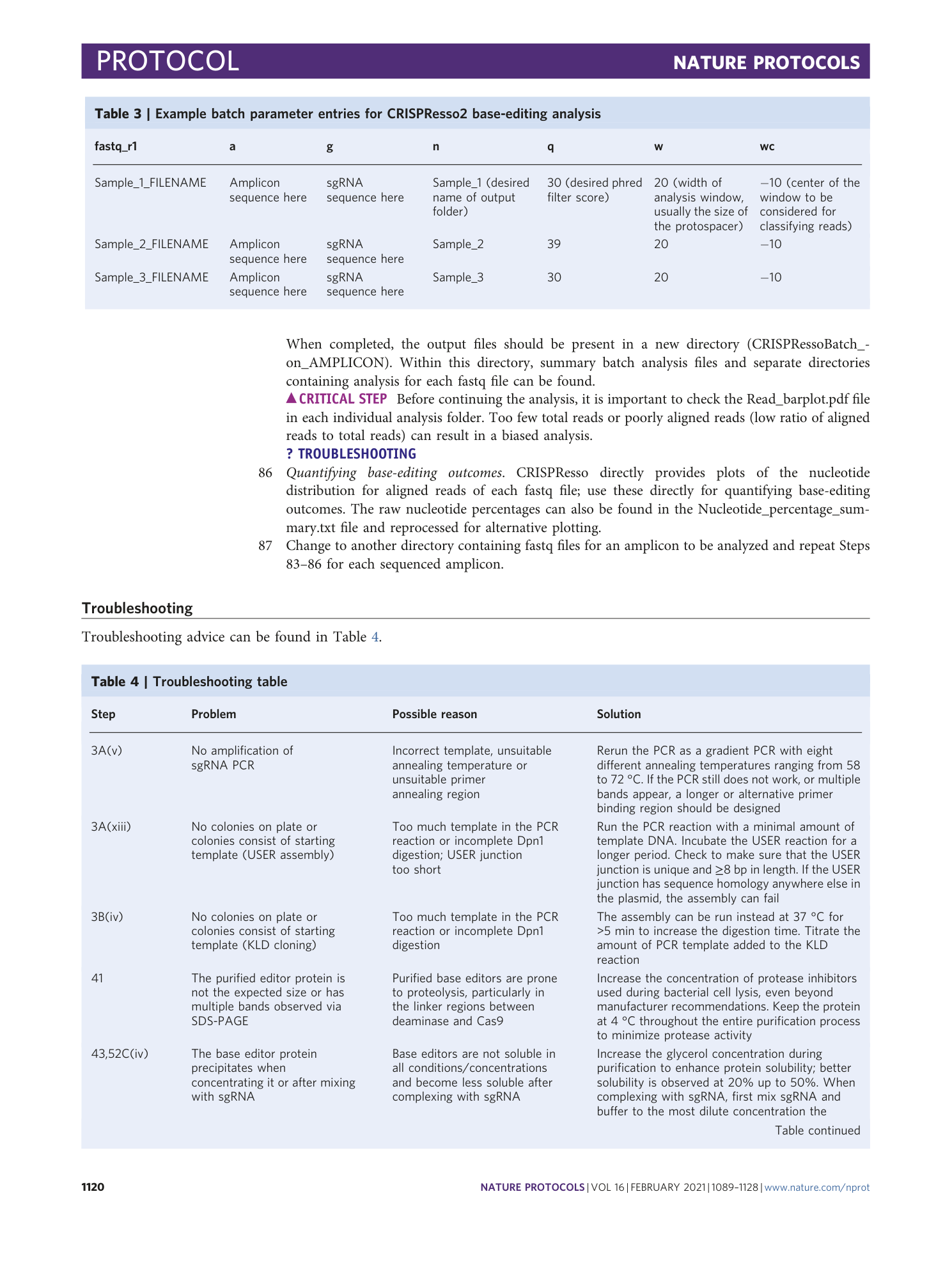
Sequencing files associated with the data shown in Fig. 8, d and e. The raw fastq files are provided, along with a CRISPRessoBatch parameter file for each amplicon in the pre-processing folder. Successfully analyzed files can be found in the post-processing folder.
Supplementary Data 3
Sequencing files associated with the data shown in Fig. 8, f and g. The raw fastq files are provided, along with a CRISPRessoBatch parameter file for each amplicon. Fastq files are named by the delivery method used.

