Nutil Data Integration
Michael X. Henderson
Abstract
This protocol describes Nutil data integration from segmentation and registration of mouse brain data.
Attachments
Steps
Nutil Data Quantification
Find the 3 folders you created at the beginning of this workflow and populated during the workflow and populate them into their corresponding list above. (segmentation is your input folder, brain atlas map folder is your atlas folder, output folder is output and should be empty still XML or JSON anchoring file is the QuickN JSON file that you created earlier – you can also use visualign json file it doesn’t matter the anchor is set in quickNII and nonlinear adjustments are made in reference to it. Object color Is black because that’s what the staining of our segmentations look like if it were a different color we would need to look for another object color.
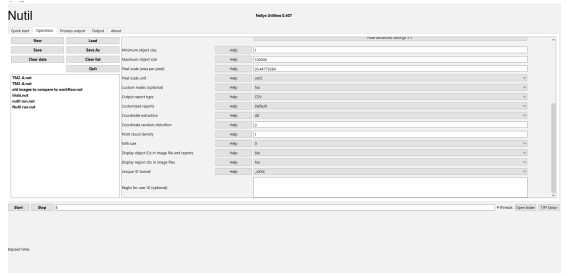
Navigate to advanced settings In the row labeled Global Pixel Scale/Unit, set it to 36.44778384 um2. (6.0 is new value for van andel scans) The NEW images that are scanned here at Van Andel are resolved at 0.5031 microns per pixel, so if we downsized by 12 we get 6.0372 then this number is squared to account for um2. the final value is 36.44778384.