hissPCR: A simple, single-tube overlapping amplicon-targeted Illumina sequencing assay.
Jason D Limberis, Joel Ernst, Alina Nalyvayko, john.metcalfe
Abstract
Targeted amplicon sequencing to identify pathogens, resistance-conferring mutations, and strain types is an important tool in diagnosing and treating infections. However, due to the short read limitations of Illumina sequencing, many applications require the splitting of limited clinical samples between two reactions. Here, we outline hairpin Illumina single-tube sequencing PCR ( hiss PCR) which allows for the generation of overlapping amplicons containing Illumina indexes and adapters in a single tube, effectively extending the Illumina read length while maintaining reagent and sample input requirements.
Attachments
Steps
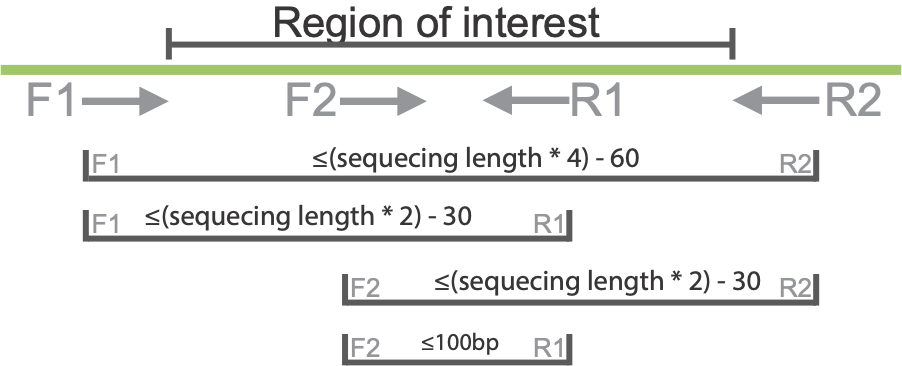
Primer design
Primers must be designed as shown in the figure below, if any amplicon is larger than the 2 x sequencing length, the inner portion of the amplicon will not be covered. We have developed a pipeline to assist in creating primers, which is available here: hisPCR primer design, and is used as follows.
hisPCR_primer_designer.sh \
--name "rpob_demo" \
--seq_cycles 300 \
--start 100 \
--end 800 \
--template "ttgaccgatgaccccggttcaggcttcaccacagtgtggaacgcggtcgtctccgaacttaacggcgaccctaaggttgacgacggacccagcagtgatgctaatctcagcgctccgctgacccctcagcaaagggcttggctcaatctcgtccagccattgaccatcgtcgaggggtttgctctgttatccgtgccgagcagctttgtccaaaacgaaatcgagcgccatctgcgggccccgattaccgacgctctcagccgccgactcggacatcagatccaactcggggtccgcatcgctccgccggcgaccgacgaagccgacgacactaccgtgccgccttccgaaaatcctgctaccacatcgccagacaccacaaccgacaacgacgagattgatgacagcgctgcggcacggggcgataaccagcacagttggccaagttacttcaccgagcgcccgcacaataccgattccgctaccgctggcgtaaccagccttaaccgtcgctacacctttgatacgttcgttatcggcgcctccaaccggttcgcgcacgccgccgccttggcgatcgcagaagcacccgcccgcgcttacaaccccctgttcatctggggcgagtccggtctcggcaagacacacctgctacacgcggcaggcaactatgcccaacggttgttcccgggaatgcgggtcaaatatgtctccaccgaggaattcaccaacgacttcattaactcgctccgcgatgaccgcaaggtcgcattcaaacgcagctaccgcgacgtagacgtgctgttggtcgacgacatccaattcattgaaggcaaagagggtattcaagaggagttcttccacaccttcaacaccttgcacaatgccaacaagcaaatcgtcatctcatctgaccgcccacccaagcagctcgccaccctcgaggaccggctgagaacccgctttgagtgggggctgatcactgacgtacaaccacccgagctggagacccgcatcgccatcttgcgcaagaaagcacagatggaacggctcgcggtccccgacgatgtcctcgaactcatcgccagcagtatcgaacgcaatatccgtgaactcgagggcgcgctgatccgggtcaccgcgttcgcctcattgaacaaaacaccaatcgacaaagcgctggccgagattgtgcttcgcgatctgatcgccgacgccaacaccatgcaaatcagcgcggcgacgatcatggctgccaccgccgaatacttcgacactaccgtcgaagagcttcgcgggcccggcaagacccgagcactggcccagtcacgacagattgcgatgtacctgtgtcgtgagctcaccgatctttcgttgcccaaaatcggccaagcgttcggccgtgatcacacaaccgtcatgtacgcccaacgcaagatcctgtccgagatggccgagcgccgtgaggtctttgatcacgtcaaagaactcaccactcgcatccgtcagcgctccaagcgctag"
Stage 1 PCR
| A | B |
|---|---|
| COMPONENT | Volume (µl) |
| 5X Q5 Reaction Buffer | 10 |
| 5X Q5 High GC Buffer | 10 |
| 10 mM dNTPs | 1 |
| Q5 High-Fidelity DNA Polymerase | 0.5 |
| 10µM Forward primer 1 | 1 |
| 10µM Reverse primer 2 | 1 |
| 20 mg/ml BSA | 5 |
| Template DNA (~1ng/ul) | 5 |
| Nuclease-Free Water | 18.5 |
The total volume is 50ul at this stage
| A | B | C | D |
|---|---|---|---|
| Step | Temp (C) | Time (s) | Cycles |
| Denaturation | 98 | 120 | 1 |
| Denaturation | 98 | 10 | 10 |
| Annealing | 64 | 15 | |
| Extension | 72 | 45 | |
| Extension | 4 | Forever | 1 |
Cycle parameters
As soon as the sample hits 4°C, proceed to step 2.
Stage 2 PCR
Add the below into the reaction while at 4°C and proceed to the second PCR cycling
| A | B |
|---|---|
| COMPONENT | Volume (µl) |
| 10µM Forward primer 2 | 0.15 |
| 10µM Reverse primer 1 | 0.15 |
| 10µM Forward Illumina adapter primer | 0.35 |
| 10µM Reverse Illumina adapter primer | 0.35 |
The total volume is 51ul at this stage
| A | B | C | D |
|---|---|---|---|
| Step | Temp (C) | Time (s) | Cycles |
| Denaturation | 98 | 10 | 1 |
| Denaturation | 98 | 10 | 20 |
| Annealing | 65 | 15 | |
| Extension | 72 | 30 | |
| Extension | 72 | 120 | 1 |
Cycle parameters
Bead cleanup
Add 40µL of resuspended AMPure XP beads to each reaction in a PCR tube
Mix by pipetting 10x
Incubate 0h 5m 0s at 65Room temperature
Place on magnet
Wash 2x with 200µL freshly-prepared 70% (v/v) ethanol
Air dry for0h 0m 30s, don't allow the beads to become cracked
Remove the tubes from the magnetic rack
Add 50µL 10 mM Tris-HCl pH 8.0 with 50 mM NaCl
NOTE: The BSA in the reaction causes the beads to clump.
Flick the tubes to partially resuspend the beads
Mix by pipetting 10x
Incubate 0h 5m 0s at 65Room temperature
Place on the magnet, aspirate 50µL of the eluant into a new tube
Run 10µL on a 0.8% agarose gel
Optional: Determine the proportion of amplicons with both Illumina adapters via qPCR
Library quantification can be done using a commercial kit such as the KAPA Library Quantification Kits or using the below custom protocol. KAPA Library Quantification Kits or using the below custom protocol.
Dilute the cleaned up amplicons 1:100.
Make the master mix below using Illumina_quant_Inner primer set and a second master mix for Illumina_quant_Outer primer set.
| A | B |
|---|---|
| COMPONENT | Volume (µl) |
| PowerUp SYBR Green Master Mix (2X) | 5 |
| 10µM Forward primer | 0.5 |
| 10µM Reverse primer | 0.5 |
| Diluted amplicon | 2 |
| Nuclease-Free Water | 2 |
qPCR master mix
Aliquot 8µL to each well and add in 2µL of the diluted amplicon.
| A | B | C | D | E |
|---|---|---|---|---|
| Step | Temp (C) | Time (s) | Cycles | Ramp Rate (C/s) |
| UDG activation | 50 | 120 | 1 | 2.73 |
| Denaturation | 95 | 120 | 1 | 2.73 |
| Denaturation | 95 | 1 | 2.73 | |
| Amplification | 60 | 30 | 2.11 | |
| Capture | 60 | 0 | – | |
| Melt Curve | 95 | 1 | 1 | 2.73 |
| 60 | 20 | 1 | 2.11 | |
| 95 | – | 1 | 0.15 | |
| Capture | – | 1 | – |
Cycle parameters for QuantStudio 3
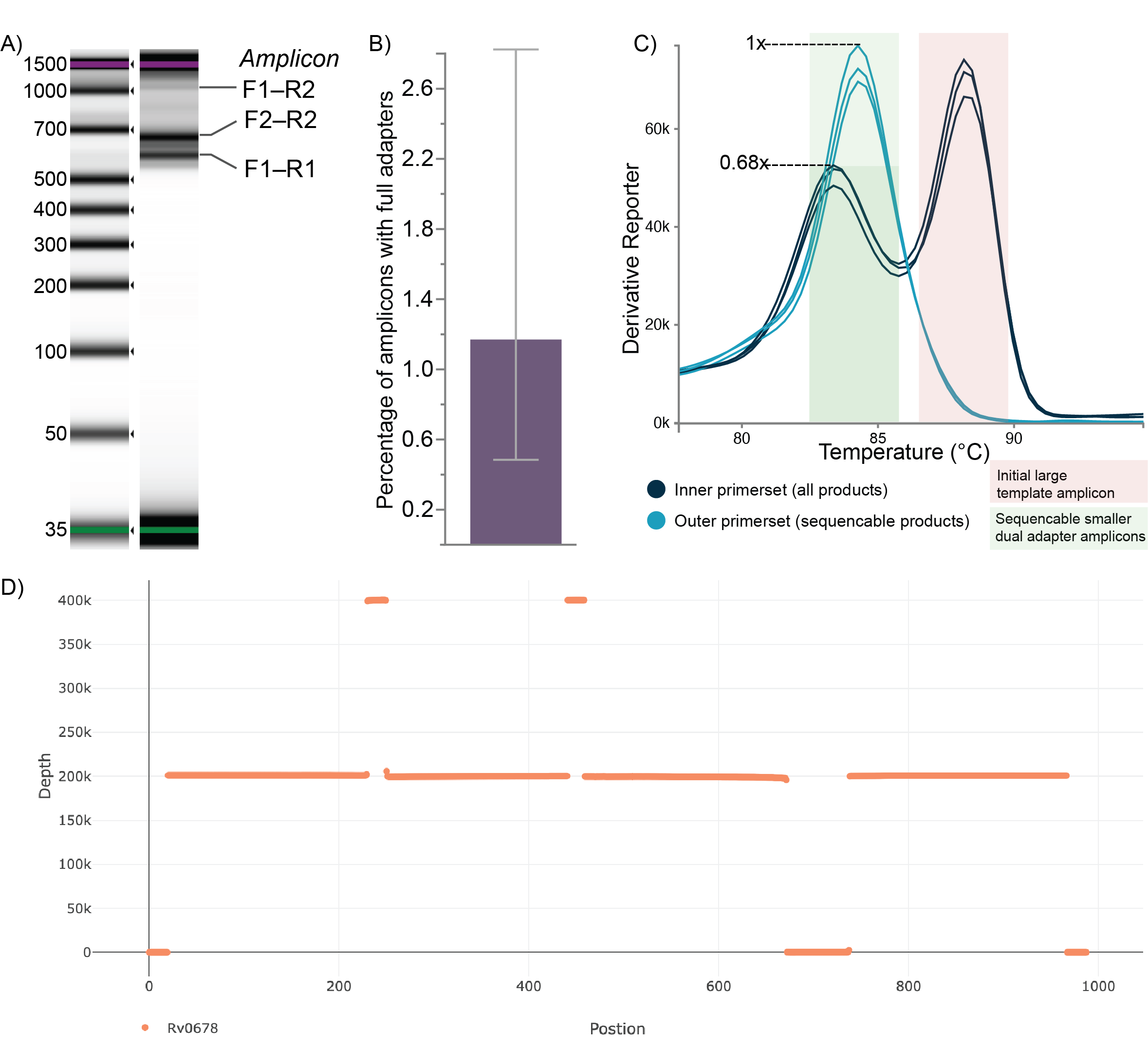
The ratio of the Illumina _quant_Inner primer set to the Illumina _quant_Outer primer set will give the proportion of the DNA in the tube that contain both Illumina adapters and is sequencable (use this for pooling). While the melt curve will provide information on the proportion of each amplicon in the sample.
Expected Result
Depth and Pooling Calculations
The amount of amplicon to be pooled and loaded can be calculated using the Illumina Sequencing Coverage Calculator by selecting DNA input then custom content and
depends on numerous factors, including the amplicon size and number, and the kit and device used. An example output is shown below.
| A | B |
|---|---|
| Application or product: | Custom Content |
| Genome or region size (Mbases) | 0.001 |
| Read length | 600 |
| On target (%) | 95 |
| Coverage (x) | 50000 |
| Duplicates (%) | 0 |
| Instrument | MiSeq |
| Run type | v3 Reagents |
| Clusters | 25,000,000 per flow cell |
| Output per unit (flow cell or lane) | 15,000,000,000 per flow cell |
| Exceeds maximum read length? | Does not exceed maximum (2x300) |
| Number of units per sample (flow cell or lane) | 0.004 flow cells |
| Samples per unit (flow cell or lane) | 285/flow cell Most Illumina library prep kits enable up to 96 indexes; Nextera XT up to 384 indexes. |
| Comments | Upgraded software; MCS v2.3 or later; MiSeq Reagent Kit v3 (150/600) |
| Product | MiSeq Reagent Kit v3 |
| Link | http://www.illumina.com/products/miseq-reagent-kit-v3.html |
Illumina coverage estimator output.
Data Analysis
The sequence data can be performed using most amplicon processing pipelines; however, the primer sequences must be trimmed from the reads to eliminate their effect on mutation identification. We have developed a pipeline to process data which is available at hissPCR analysis with further details, below is its basic usage command.
bash hissPCR.sh \
--R1 "test_data/read_R1_001.fastq.gz" \
--R2 "test_data/read_R2_001.fastq.gz" \
--ref "refs/BDQ_duplex.fasta" \
--primers "refs/primers.bed" \

