Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2
Emelissa J. Mendoza, Emelissa J. Mendoza, Kathy Manguiat, Kathy Manguiat, Heidi Wood, Heidi Wood, Michael Drebot, Michael Drebot
Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has been identified as the causal agent of COronaVIrus Disease-19 (COVID-19), an atypical pneumonia-like syndrome that emerged in December 2019. While SARS-CoV-2 titers can be measured by detection of viral nucleic acid, this method is unable to quantitate infectious virions. Measurement of infectious SARS-CoV-2 can be achieved by tissue culture infectious dose−50 (TCID50), which detects the presence or absence of cytopathic effect in cells infected with serial dilutions of a virus specimen. However, this method only provides a qualitative infectious virus titer. Plaque assays are a quantitative method of measuring infectious SARS-CoV-2 by quantifying the plaques formed in cell culture upon infection with serial dilutions of a virus specimen. As such, plaque assays remain the gold standard in quantifying concentrations of replication-competent lytic virions. Here, we describe two detailed plaque assay protocols to quantify infectious SARS-CoV-2 using different overlay and staining methods. Both methods have several advantages and disadvantages, which can be considered when choosing the procedure best suited for each laboratory. These assays can be used for several research purposes, including titration of virus stocks produced from infected cell supernatant and, with further optimization, quantification of SARS-CoV-2 in specimens collected from infected animals. © 2019 The Authors.
Basic Protocol : SARS-CoV-2 plaque assay using a solid double overlay method
Alternate Protocol : SARS-CoV-2 plaque assay using a liquid overlay and fixation-staining method
INTRODUCTION
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), formerly known as 2019-nCoV, was identified as the causal agent of COronaVIrus Disease-19 (COVID-19), a pneumonia-like syndrome that emerged in Wuhan, Hubei, China in December 2019 (WHO, 2020c; Wu et al., 2020; Zhu et al., 2020). Since then, SARS-CoV-2 has spread rapidly, and the pandemic was deemed a public health emergency of international concern in January 2020 (WHO, 2020d). As of May 1, 2020, over 3 million cases and 216,000 fatalities have been reported worldwide (WHO, 2020a). To date, there remains no therapeutic or vaccine effective for the treatment or prevention of SARS-CoV-2 infection (Sanders, Monogue, Jodlowski, & Cutrell, 2020). Thus, clinical management of critically ill patients with COVID-19 relies heavily upon supportive measures, such as mechanical ventilation and hemodynamic support (Poston, Patel, & Davis, 2020). The enormous number of patients and limited resources have overwhelmed health care systems in their efforts to provide such support. Ongoing research involving infectious SARS-CoV-2 will be crucial in understanding viral-associated pathogenesis and conducting pre-clinical testing of medical countermeasures for the pathogen causing the COVID-19 pandemic.
Virus quantification assays are key tools for research involving SARS-CoV-2. Molecular tests detecting viral RNA can be used to quantify viral loads in clinical samples and virus titers of SARS-CoV-2 stocks prepared from cell culture supernatants (Chu et al., 2020; Corman et al., 2020). While this method is highly sensitive and specific, it is incapable of quantifying infectious SARS-CoV-2, particularly when a high proportion of non-infectious virions are produced during a lytic cycle. Tissue culture infectious dose−50 (TCID50) measures infectious SARS-CoV-2 by detecting the presence or absence of cytopathic effect in cell culture upon infection with serial dilutions of a virus specimen (van Doremalen et al., 2020). However, this only provides a qualitative measurement of infectious virus in TCID50 units, which describe the amount of virus needed to induce 50% CPE in susceptible cells. Plaque assays are a quantitative method of measuring infectious SARS-CoV-2 by quantifying the plaques formed in cell culture upon infection with serial dilutions of a virus specimen (Harcourt et al., 2020). Infectious virus titers are measured in plaque-forming units (PFU). As such, plaque assays remain the gold standard in quantifying concentrations of replication-competent lytic virions (Cooper, 1961; Juarez, Long, Aguilar, Kochel, & Halsey, 2013).
In this article, we describe two detailed procedures to conduct SARS-CoV-2 plaque assays. In both plaque assays, a confluent monolayer of host cells is infected with serial dilutions of SARS-CoV-2 of unknown starting concentration. After adsorption, an immobilizing overlay is used to cover the infected monolayer, to prevent virus spread and restrict virus growth to foci of cells at the sites of initial infection. During incubation, zones of cell death develop as viral infection and replication are restricted to the surrounding monolayer, leading to plaque formation. After incubation, cells are stained to enhance the contrast between plaques and the uninfected monolayer. Plaques are then enumerated and used to calculate the titer of infectious virus in the specimen. The first plaque assay method that we describe (Basic Protocol) uses Noble agar as the matrix in a solid overlay and neutral red as the stain to enhance plaque visualization. The second plaque assay method that we describe (Alternate Protocol) uses carboxymethylcellulose (CMC) as the matrix in a liquid overlay and crystal violet as the stain to enhance plaque visualization. An overview of these protocols is shown in Figure 1. We believe that both of these protocols would be useful for quantifying infectious SARS-CoV-2 in viral stocks produced from infected cell supernatant. We also believe that either method can be further optimized for other research purposes, such as quantifying infectious SARS-CoV-2 in specimens from animals experimentally infected with SARS-CoV-2.
Basic Protocol: SARS-CoV-2 PLAQUE ASSAY USING A SOLID DOUBLE OVERLAY METHOD
This protocol outlines a plaque assay method that can be used for the quantification of SARS-CoV-2 plaque-forming units (PFUs) in virus specimens, including viral stocks prepared from infected cell culture supernatants, and with further optimization, bodily fluids from animals infected with SARS-CoV-2.In brief, 10-fold serial dilutions of specimens containing an unknown amount of SARS-CoV-2 are adsorbed onto a monolayer of susceptible cells. After adsorption, a solid overlay medium (S-OM) is applied to the cell monolayer to restrict virus growth to the originally infected foci of cells. These foci develop into plaques, which are visualized after the addition of a secondary S-OM containing neutral red, which is incorporated and bound by lysosomes in viable cells (Repetto, del Peso, & Zurita, 2008). As a result, clear plaques can be distinguished from the brownish-red uninfected monolayer. Plaques are enumerated and used for the calculation of virus titers in PFU/ml.
Materials
-
Vero E6 cells (ATCC® CRL-1586™)
-
Cell maintenance medium (see recipe)
-
Gibco™ Trypsin-EDTA (0.25%), with phenol red (ThermoFisher Scientific #25200072)
-
Infection medium (see recipe)
-
Specimens to be assayed for virus
-
Gibco™ DPBS, no calcium, no magnesium (ThermoFisher Scientific #14190144)
-
Overlay diluent (see recipe)
-
2% Noble agar (NA; solid matrix, see recipe)
-
0.33% neutral red (NR; see recipe)
-
Micro-Chem Plus Disinfectant Detergent (National Chemical Laboratories #C849T34)
-
Class II biological safety cabinet
-
150-cm2 (T-150) culture flasks (Corning™ #430825 or equivalent)
-
CO2 incubator (Heracell™ 150i CO2 incubator with stainless-steel chamber, or equivalent)
-
Light microscope
-
Falcon 50-ml conical centrifuge tubes (Corning™ #C352098 or equivalent)
-
Countess II automated cell counter (or equivalent)
-
Costar® 6-well plates (Corning™ #3506 or equivalent)
-
2-ml microcentrifuge tubes (ThermoFisher Scientific #50809242PK or equivalent)
-
Vacuum source
-
Serological pipettes, sterile:
- 5-ml (Corning™ #C4487 or equivalent)
- 10-ml (Corning™ #C4488 or equivalent)
- 25-ml (Corning™ #C4489 or equivalent)
- 50-ml (Corning™ #C4490 or equivalent)
-
Pipet-Aid (or equivalent)
-
Repeat pipettor (e.g., Rainin, or equivalent from ThermoFisher Scientific)
-
Tips for repeat pipettor:
- 20-µl barrier tips (ThermoFisher Scientific #2149 or equivalent)
- 200-µl barrier tips (ThermoFisher Scientific #2770 or equivalent)
- 1000-µl barrier tips (ThermoFisher Scientific #2380 or equivalent)
-
44°C water bath
-
Timer
-
Microwave oven
-
VWR Scientific TW-26 White Light Transilluminator (or equivalent)
-
Additional reagents and equipment for basic cell culture techniques including trypsinization (see Current Protocols article: Phelan & May, 2015)
Cell culture (Day 0)
1.Cell maintenance : Maintain Vero E6 cells in T-150 flasks containing cell maintenance medium in an incubator at 37°C with 5% CO2. See Current Protocols article Phelan and May (2015) for basic cell culture techniques.
2.Seed plates : Trypsinize (see Current Protocols article: Phelan & May, 2015) a confluent T-150 flask of Vero E6 cells and bring volume to 20 ml with cell maintenance medium. Use an automated cell counter to count cells and prepare a suspension at a density of 3 × 105 cells/ml in cell maintenance medium. For each specimen to be titrated, seed four 6-well plates with 6 × 105 cells per well by adding 2 ml of the suspension to each well. Incubate at 37°C in a 5% CO2 incubator overnight to achieve 100% confluence the following day.
Specimen dilution, infection of cells, and primary overlay (Day 1, 0 days post-infection, dpi)
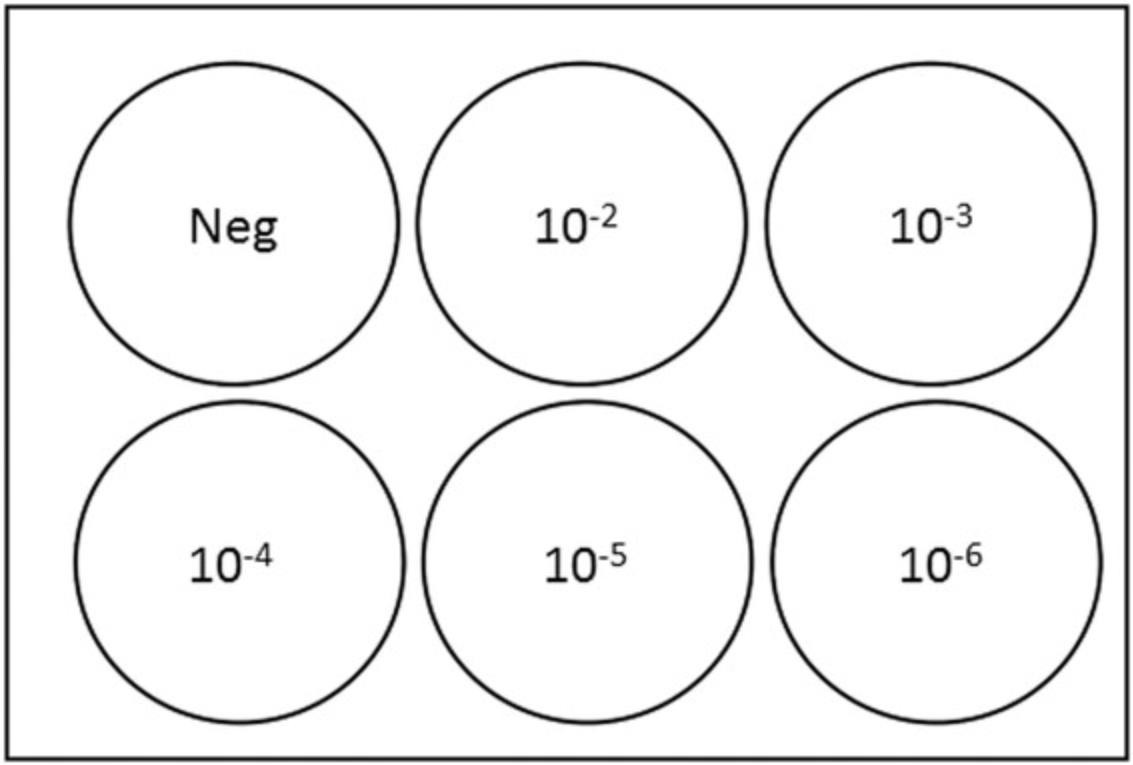
3.Specimen dilution : For each specimen to be titrated, label 2-ml microcentrifuge tubes as follows: Neg, 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6. Add 900 μl of infection medium to each tube. Serially dilute specimens 10-fold by transferring 100 μl of each specimen to the appropriate tube labeled 10−2, vortexing, and transferring 100 µl to the subsequent tube, down to 10−6.
4.Infection and adsorption : Label 6-well plates containing cells (see step 2) according to Figure 2. Aspirate medium from cell monolayers under vacuum, leaving ∼100 µl of medium to prevent monolayer from drying. Using a Pipet-Aid and a serological pipette, wash cells with 1 ml of DPBS. Leave ∼100 µl DPBS to prevent monolayer from drying out. Dispense 100 µl of the infection medium from the microcentrifuge tube labeled “N” into designated wells, as depicted in Figure 1.Dispense 100 µl of diluted specimens (10−2, 10−3, 10−4, 10−5, and 10−6) into designated wells, changing pipette tips every time (Figure 2). Incubate at 37°C in a 5% CO2 incubator for 1 hr, rocking every 15 min.
5.Preparation of primary S-OM : Incubate overlay diluent at 44°C in a water bath. Microwave to dissolve 2% NA (solid matrix) and incubate at 44°C in a water bath. After rocking plates for the last time (∼10 min before incubation is over), prepare primary S-OM by combining equal amounts (1:1) of overlay diluent and 2% NA (solid matrix). Use Formula 1 to calculate the total volume of S-OM needed, followed by Formula 2 to calculate the overlay diluent and 2% NA (solid matrix) needed to prepare the primary S-OM.
Formula 1 : Total volume of primary S-OM needed (in ml) = Number of specimens × 4 plates per specimen × 6 wells per plate × 3 ml primary S-OM per well × 1.2 (to account for pipetting error)
Formula 2 : Volume of overlay diluent or 2% NA (solid matrix) needed for primary S-OM (in ml) = total volume of primary S-OM needed ÷ 2
6.Primary solid overlay : Add 3 ml of primary S-OM to each well of the titration plates (step 4). Allow primary S-OM to solidify and incubate at 37°C in a 5% CO2 incubator for 2 days.
Secondary overlay and staining (Day 3, 2 dpi)
7.Preparation : Incubate overlay diluent at 44°C in a water bath. Microwave to dissolve 2% NA (solid matrix) and incubate at 44°C in a water bath. Prepare secondary S-OM (composition: 1:1 overlay diluent:2% NA with 0.01% neutral red) by using Formula 3 to calculate the total volume of secondary S-OM needed. Use Formula 4 to calculate the volume of 0.33% NR needed, Formula 5 to calculate the volume of overlay diluent needed, and Formula 6 to calculate the volume of 2% NA (solid matrix) needed to prepare the secondary S-OM.
Formula 3 : Total volume of secondary S-OM needed (in ml) = Number of specimens × 4 plates per specimen × 6 wells per plate × 2 ml secondary S-OM per well × 1.2 (to account for pipetting error)
Formula 4 : Volume of 0.33% NR needed for secondary S-OM (in ml) = total volume of secondary S-OM needed × 0.03
Formula 5 : Volume of overlay diluent needed for secondary S-OM (in ml) = (total volume of secondary solid overlay medium needed ÷ 2) − volume of 0.33% NR needed for secondary S-OM
Formula 6 : Volume of 2% NA (solid matrix) needed for secondary S-OM = total volume of secondary S-OM needed ÷ 2
8.Secondary solid overlay : Add 2 ml of secondary S-OM to each well of the titration plates. Allow secondary S-OM to solidify and incubate at 37°C in a 5% CO2 incubator overnight.
Enumeration of plaques and titer calculation (Day 4, 3 dpi)
9.Enumerate plaques : Use a white-light transilluminator (light box) to aid in visualizing the plaques, which will appear as peach-colored circles on a brownish-red monolayer of cells (Figure 3A). The negative control should have a uniform monolayer, which can be used as a reference. Record the number of plaques observed per well at each virus dilution.
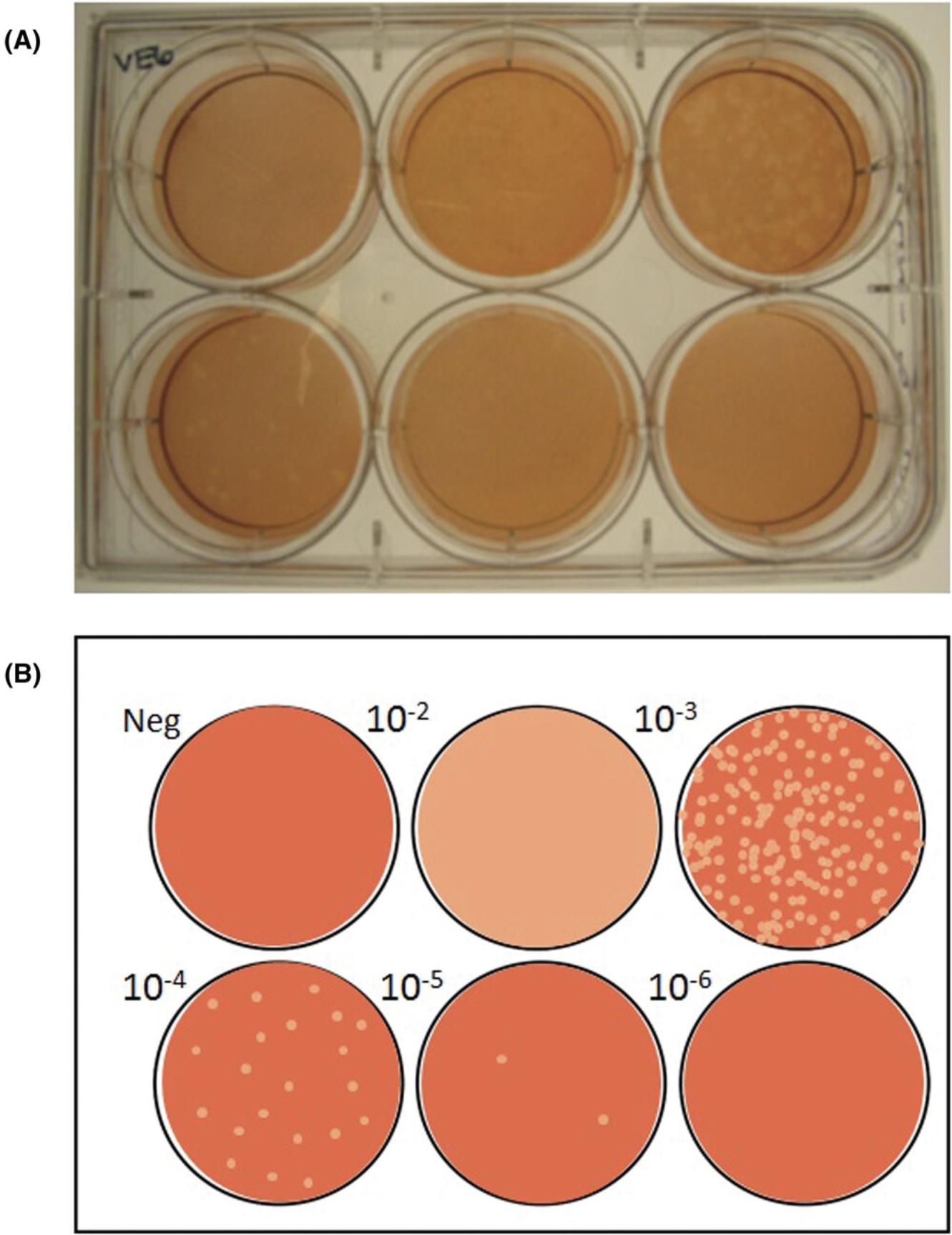
10.Titer calculation : Identify the virus dilution factor that yields 5-100 plaques per well (Baer & Kehn-Hall, 2014). Use Formula 7 to calculate the average number of plaques at this dilution. Use Formula 8 to calculate the titer of SARS-CoV-2 in the specimen using the identified dilution factor and the inoculum volume of 0.1 ml. Refer to Figure 3B for an example plaque assay plate and interpretation.
Formula 7 : Average number of plaques given by the identified virus dilution = Sum of plaques from 4 replicate wells ÷ 4
Formula 8 : Titer of SARS-CoV-2 (in PFU/ml) = Average number of plaques given by the identified virus dilution ÷ (dilution factor × 0.1 ml)
11.Waste disposal : Storage of plates prior to enumeration is not recommended because the borders of the plaques become less distinct over time. Wells of each plate can be topped up with an appropriate disinfectant (such as a 5% Micro-Chem Plus solution) and incubated at room temperature overnight prior to disposal in biohazardous waste. All biohazardous waste should be autoclaved at 121°C for 1 hr.
Alternate Protocol: SARS-CoV-2 PLAQUE ASSAY USING A LIQUID OVERLAY AND FIXATION-STAINING METHOD
This protocol can be used as an alternative to the Basic Protocol for the quantification of SARS-CoV-2 by plaque assay. The first steps in the protocol are identical to the Basic Protocol. However, after adsorption, a liquid overlay medium (L-OM) is applied to the cell monolayer to restrict virus growth to the originally infected foci of cells. In addition, instead of incorporating a vital stain into the secondary overlay (S-OM), the L-OM is removed from the monolayer, fixed, and stained with crystal violet, which binds to proteins and DNA within cells (Feoktistova, Geserick, & Leverkus, 2016). As a result, clear plaques can be distinguished from the purple monolayer. Plaques are enumerated and used for the calculation of virus titers in PFU/ml. For a comparison of the two protocols, see Commentary.
Additional Materials (also see Basic Protocol)
- 3% carboxymethylcellulose (CMC; (liquid matrix, see recipe)
- 10% neutral-buffered formalin (Sigma-Aldrich #HT501128-4L); Thermo Scientific™ Paraformaldehyde Solution, 4% in PBS (ThermoFisher Scientific #AAJ19943K2) can also be used.
- Formalex GREEN Formalin Neutralizer (Jones Scientific # H-FORMG-CB)
- 0.5% crystal violet (CV, see recipe)
Cell culture (Day 0)
1.Perform steps 1-2 of the Basic Protocol.
Specimen dilution, infection of cells, and primary overlay (Day 1, 0 dpi)
2.Perform steps 3-4 of the Basic Protocol
3.Preparation of L-OM : Incubate overlay diluent at 44°C in a water bath. After rocking plates for the last time (approximately 10 min before incubation is over), prepare L-OM by adding equal amounts (1:1) of overlay diluent and 3% CMC (liquid matrix). Use Formula 1 to calculate the total volume of L-OM needed, followed by Formula 2 to calculate the overlay diluent and 3% CMC (liquid matrix) needed to prepare the L-OM.
Formula 1 : Total volume of L-OM needed (in ml) = Number of specimens × 4 plates per specimen × 6 wells per plate × 3 ml L-OM per well × 1.2 (to account for pipetting error)
Formula 2 : Volume of overlay diluent or 3% CMC (liquid matrix) needed for L-OM (in ml) = total volume of L-OM needed ÷ 2
4.Liquid overlay : Add 3 ml of L-OM to each well of the titration plates (see Basic Protocol, step 4). Incubate at 37°C in a 5% CO2 incubator for 3 days.
Fixation, staining, enumeration of plaques, and titer calculation (Day 4, 3 dpi)
5.Fixation : Carefully aspirate L-OM from each well and discard in a waste bottle containing an appropriate disinfectant (such as a 5% Micro-Chem Plus solution). Gently wash cells with DPBS and fill wells with 10% buffered formalin or 4% paraformaldehyde. Incubate at room temperature for 1 hr.
6.Staining : Aspirate fixative from wells and discard into a waste bottle containing the appropriate amount of Formalex. Add 200 μl of 0.5% CV to each well and incubate at room temperature for 5-15 minutes.
7.Wash : Wash cells 2-4 times with distilled water. Blot dry.
8.Enumerate plaques : Plaques will appear as clear circles on a purple monolayer of cells (Figure 4A). The negative control should have a uniform monolayer, which can be used as a reference. Record the number of plaques observed per well at each virus dilution.
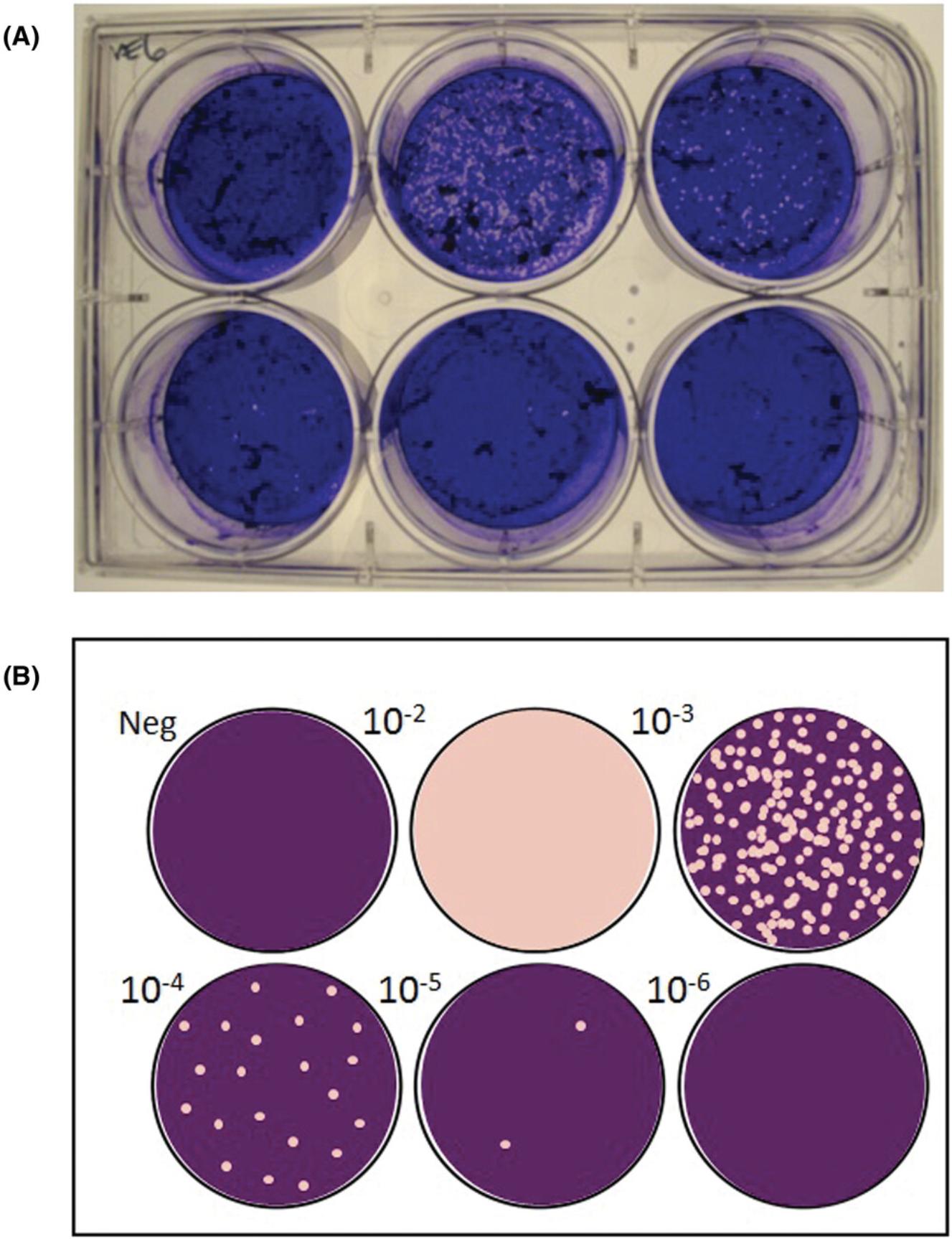
9.Titer calculation : Identify the virus dilution factor demonstrating 5-100 plaques per well. Use Formula 7 (see Basic Protocol) to calculate the average number of plaques at this dilution. Use Formula 8 (see Basic Protocol) to calculate the titer of SARS-CoV-2 in the specimen using the identified dilution factor and the inoculum volume of 0.1 ml. Refer to Figure 4B for an example plaque assay plate and interpretation.
10.Waste disposal : Dispose of plates in biohazardous waste and autoclave at 121°C for 1 hr. Liquid waste containing disinfectant should be incubated at room temperature overnight prior to disposal.
REAGENTS AND SOLUTIONS
Carboxymethylcellulose, 3% (w/v)
- 6 g carboxymethylcellulose, medium viscosity (solid; Sigma-Aldrich #C4888-500G)
- 200 ml distilled water
- Sterilize by autoclaving at 121°C for 15 min
- Store at room temperature up to 2 months
Cell maintenance medium
- 500 ml HyClone™ Dulbecco's Modified Eagles Medium, High Glucose with l-glutamine (ThermoFisher Scientific #SH30022LS)
- 50 ml FBS (ThermoFisher Scientific #16140071)
- Store at 4°C up to 2 months
Crystal violet, 1% (w/v)
- 1 g crystal violet (solid; Certified Biological Stain; ThermoFisher Scientific #C581-25)
- 20 ml absolute ethanol
- 80 ml distilled water
- Filter sterilize
- Store at room temperature up to 4 months protected from light
Infection medium
- 500 ml HyClone™ Dulbecco's Modified Eagles Medium, High Glucose, with l-glutamine (ThermoFisher Scientific #SH30022LS)
- 10 ml FBS (ThermoFisher Scientific #16140071)
- Store at 4°C up to 2 months
Neutral red, 0.33% (w/v)
- 0.33 g neutral red (solid; ACROS Organics™; ThermoFisher Scientific #AC415491000)
- 100 ml distilled water
- Filter sterilize
- Store at room temperature up to 4 months protected from light
Noble agar, 2% (w/v)
- 4 g Noble agar (solid; Ultrapure; ThermoFisher Scientific #AAJ1090736)
- 200 ml distilled water
- Sterilize by autoclaving at 121°C for 15 min
- Store at room temperature up to 2 months
Overlay diluent
- 500 ml Gibco™ MEM (2×), no phenol red (ThermoFisher Scientific #11935046)
- 40 ml FBS (ThermoFisher Scientific #16140071)
- 10 ml 200 mM l-glutamine (ThermoFisher Scientific #25030149)
- 10 ml 100× nonessential amino acids (NEAA; ThermoFisher Scientific #11140050)
- 7.5 ml 100× sodium bicarbonate (ThermoFisher Scientific #25080094)
- Store at 4°C up to 2 months
COMMENTARY
Background Information
The accurate quantification of infectious virus specimens is crucial for a multitude of applications in virology. Plaque assays were established in 1952 as an adaptation of phage assays, which were used to calculate bacteriophage titers in plant biology (Cooper, 1961; Dulbecco & Vogt, 1953). Although new techniques for viral titration continue to evolve, plaque assays continue to be the gold standard for the quantification of infectious virus (Baer & Kehn-Hall, 2014).
The plaque assay using the double overlay method (Basic Protocol) described in this article has been adapted for SARS-CoV-2 from previously characterized procedures (Berry et al., 2004; Harcourt et al., 2020; Ströher et al., 2004). Noble agar, the solid matrix used in the overlay in the Basic Protocol, is considered a traditional overlay matrix for plaque assays. However, it comes with disadvantages. Heating is required to liquefy the matrix immediately prior to application, which warrants the need to work quickly when applying this type of overlay since it can re-solidify as it cools. The secondary overlay in this protocol uses neutral red to stain the monolayer and enhance the visualization of plaques. As a vital stain, neutral red has the advantage of being applied during incubation, enabling live monitoring of plaque formation. However, the contrast between plaques and viable cells stained with neutral red is not as distinct as that produced by crystal violet and fixation in the Alternate Protocol, and requires a light box to further enhance visualization.
The plaque assay using the fixation and staining method (Alternate Protocol) described in this article has been adapted for SARS-CoV-2 from previously characterized procedures (Schneider et al., 2012; Vicenzi et al., 2004; Wang, Sakhatskyy, Chou, & Lu, 2005). In contrast to the Basic Protocol, this method uses a liquid overlay matrix, which confers benefits over solid/semi-solid overlay matrices, including the ease of removal prior to fixation. In addition, liquid overlays can be applied to cell monolayers at room temperature, eliminating issues of re-solidification during the application process. However, the disadvantage of the liquid overlay matrix is that disruption of the liquid overlays can result in smeared plaques, and thus great care must be taken to prevent movement of plates during the incubation period. The fixation and subsequent staining with crystal violet used to enhance plaque visualization in the protocol has several advantages, including increased contrast between plaques from the purple monolayers, as well as the inactivation of the virus. In addition, fixation enables plates to be stored and plaques to be enumerated at a later time. However, some disadvantages of this method includes the generation of formaldehyde waste and the additional need to properly inactivate that chemical.
As summarized in Table 1, each protocol has advantages and disadvantages that can be assessed to select the method best suited to each laboratory's resources and preferences. A comparison conducted in our laboratory demonstrated that there were no significant differences in viral titers calculated using either method.
| Characteristic | Basic Protocol | Alternate Protocol |
|---|---|---|
| Completion time | Completed in 4 days (cell culture excluded) | Completed in 4 days (cell culture excluded) |
| Number of steps | Advantage: Neutral red applied in secondary overlay bypasses need for additional washes, fixation, and staining steps | Disadvantage: Additional washing, fixation, and staining steps needed |
| Work in BSL-3 lab | Disadvantage: Secondary overlay to stain cells requires manipulation of plates the day before enumeration (assay needs 3 days working in BSL-3) | Advantage: Fixation, staining, and enumeration on same day instead of different day (assay only needs 2 days working in BSL-3 lab) |
| Overlay application | Disadvantage: Requires speed to apply solid overlay to monolayers to avoid re-solidifying |
Advantage: Liquid overlay easily applied |
| Incubation | Advantage: Plaques in solid overlay not prone to distortion if plates are moved | Disadvantage: Movement of plates during incubation can disrupt liquid overlay, causing plaques to appear as streaks |
| Additional chemicals needed | Advantage: Does not require fixation step | Disadvantage: Fixation step requires formaldehyde fixatives and chemicals used for inactivation of formaldehyde waste |
| Additional equipment needed | Disadvantage: Requires a microwave oven to dissolve agar and light box to enhance visualization of plaques | Advantage: Overlay in liquid form |
| Visualization | Disadvantage: Peach-colored plaques less distinct from brownish-red monolayer | Advantage: Clear plaques very distinct from purple monolayer |
| Storage of plates | Disadvantage: Plaques must be enumerated within a few days of secondary overlay | Advantage: Plates can be stored at 4°C to read plaques up to a month |
| Cost | Advantage: Less reagents and consumables used per assay | Disadvantage: Additional reagents and consumables used per assay |
| Safety precautions during enumeration | Disadvantage: Virus is still viable and infectious while reading plates | Advantage: Virus is inactivated during fixation step |
Critical Parameters
Since SARS-CoV-2 is a BSL-3 pathogen, procedures using SARS-CoV-2 specimens require BSL-3 facilities, equipment, and operational practices. Additional personal protective equipment (PPE), including respiratory protection, is required for working with SARS-CoV-2 (WHO, 2020b). It is crucial to consult with on-site biosafety officers, occupational hygienists, and other related personnel to conduct risk assessments and provide guidance regarding protective equipment, effective disinfectants, and practices to mitigate risks when working with the pathogen.
The monolayer is susceptible to damage from forceful pipetting, which can then be mistaken for plaques. Instead of adding medium directly on top of the monolayer, slowly add it by aiming for the walls of each well.
Troubleshooting
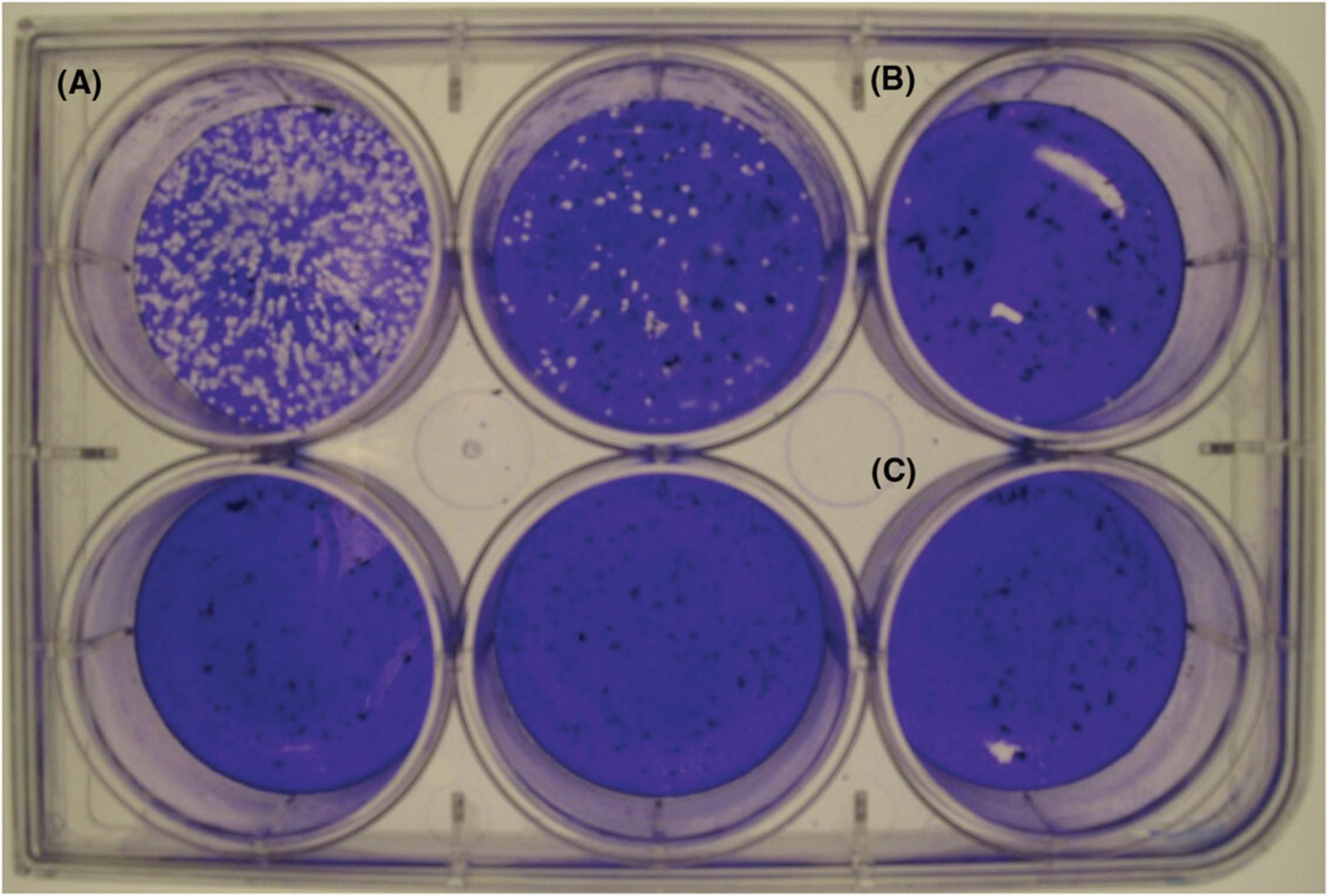
See Table 2 for commonly encountered problems, causes, and solutions. A plaque assay plate exhibiting commonly encountered problems is depicted in Figure 5.
| Problem | Possible cause | Solution |
|---|---|---|
| Plaque numbers >100 at all dilutions | Virus titers of the specimen may be greater than 107 PFU/ml | Conduct another plaque assay with further serial dilutions of the specimen (i.e., 10−7, 10−8, 10−9, etc.) |
| Improper technique for serial dilutions | Make sure to change pipet tips, vortex well, and briefly centrifuge between dilutions | |
| Plaque numbers <5 at all dilutions | Virus titers of the specimen may be less than 100 PFU/ml | Conduct another plaque assay and infect cells with the undiluted sample, as well as the sample diluted 10−1 |
| Heat from the S-OM (Basic Protocol) could possibly affect infectivity of SARS-CoV-2 | Make sure S-OM cools slightly from 44°C before applying it to cell monolayer | |
| Plaques clustered around edges of wells | Swirling the inoculum during the adsorption stage | Use a rocking motion from front to back and side to side to distribute the inoculum evenly on the cell monolayer |
| Large area of missing cells on monolayer | Overlay medium or other reagents added quickly and directly on top of monolayer | Add medium slowly and aim for edge of well |
| Cells not confluent prior to start of assay | Use a light microscope to check that cells are at least 95% confluence before starting assay | |
| “Shooting star” or smear-like appearance of plaques (Alternate Protocol) | Liquid overlay disrupted during incubation | Minimize movement of plates during incubation by leaving plates untouched throughout the incubation and reducing the number of times the incubator door is opened and closed. |
| “Crescent moon” along edge of well | Drying of cell monolayer | (a) Leave ∼50-100 µl of liquid in each well when aspirating medium and DPBS; (b) rock plates every 10-15 min during adsorption incubation; (c) work in small batches of plates to prevent the monolayer from drying out between each aspiration and medium addition |
| Large plaque-like spot | Scratching of the monolayer by pipette tip during medium addition or removal | Avoid making touching the monolayer with pipette tip by hovering above the well and aiming along the edges when adding medium. Minimize the force used when using a serological pipette tip to remove medium. |
| Medium added directly to monolayer at high speed | Aim medium along the edges at low speed |
Understanding Results
Refer to Figure 3 for an example interpretation of a schematic 6-well SARS-CoV-2 plaque assay plate. In our laboratory, we have found that the titers of most of our SARS-CoV-2 stocks passaged in Vero E6 and titrated by plaque assays using Vero E6 range from 105 to 106 PFU/ml.
Time Considerations
The Basic Protocol and Alternate Protocol can both be completed in 5-7 days, including the time needed to seed plates for the plaque assay. However, if starting Vero E6 cells from cryostocks, it would be necessary to passage the cell line at least three times prior to use in either assay to remove excess cryopreservative and allow cells to return to normal growth, extending the duration by a few days; this is not taken into account in the protocols. If plates are seeded as described in the protocols on a Monday, the plaque assay can begin on Tuesday and end with plaque enumeration on Friday. Using this schedule, the secondary overlay would be applied on Thursday for the Basic Protocol.
Acknowledgements
Development of this protocol was supported by the Public Health Agency of Canada. We thank Dr. Darwyn Kobasa, Dr. Amrit Boese, and Anders Leung for providing the SARS-CoV-2 stock, and for their constructive commentary (Public Health Agency of Canada). We would also like to thank Kristina Dimitrova (Public Health Agency of Canada) and Dr. Yohannes Berhane (Canadian Food Inspection Agency) for their constructive commentary regarding the optimization of the protocols.
Literature Cited
- Baer, A., & Kehn-Hall, K. (2014). Viral concentration determination through plaque assays: Using traditional and novel overlay systems. Journal of Visualized Experiments , 93, e52065. doi: 10.3791/52065.
- Berry, J. D., Jones, S., Drebot, M. A., Andonov, A., Sabara, M., Yuan, X. Y., … Plummer, F. (2004). Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus.Journal of Virological Methods , 120(1), 87–96. doi: 10.1016/j.jviromet.2004.04.009.
- Burnett, L. C., Lunn, G., & Coico, R. (2009). Biosafety: Guidelines for working with pathogenic and infectious microorganisms. Current Protocols in Microbiology , 13, 1A.1.1-1A.1.14. doi: 10.1002/9780471729259.mc01a01s13.
- Chu, D. K. W., Pan, Y., Cheng, S. M. S., Hui, K. P. Y., Krishnan, P., Liu, Y., … Poon, L. L. M. (2020). Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clinical Chemistry , 66(4), 549–555. doi: 10.1093/clinchem/hvaa029.
- Cooper, P. D. (1961). The plaque assay of animal viruses. Advances in Virus Research , 8, 319–78. doi: 10.1016/s0065-3527(08)60689-2.
- Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., … Drosten, C. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance , 25(3), 2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045.
- Dulbecco, R., & Vogt, M. (1953). Some problems of animal virology as studied by the plaque technique. Cold Spring Harbor Symposia on Quantitative Biology , 18, 273–9. doi: 10.1101/sqb.1953.018.01.039.
- Feoktistova, M., Geserick, P., & Leverkus, M. (2016). Crystal violet assay for determining viability of cultured cells. Cold Spring Harbor Protocols , 2016(4), pdb.prot087379. doi: 10.1101/pdb.prot087379.
- Harcourt, J., Tamin, A., Lu, X., Kamili, S., Sakthivel, S. K., Murray, J., … Thornburg, N. J. (2020). Severe acute respiratory syndrome coronavirus 2 from patient with 2019 novel coronavirus disease, United States. Emerging Infectious Diseases , 26(6). doi: 10.3201/eid2606.200516.
- Juarez, D., Long, K. C., Aguilar, P., Kochel, T. J., & Halsey, E. S. (2013). Assessment of plaque assay methods for alphaviruses. Journal of Virological Methods , 187(1), 185–189. doi: 10.1016/j.jviromet.2012.09.026.
- Phelan, K., & May, K. M. (2015). Basic techniques in mammalian cell culture. Current Protocols in Cell Biology , 66, 1.1.1–1.1.22. doi: 10.1002/0471143030.cb0101s66.
- Poston, J. T., Patel, B. K., & Davis, A. M. (2020). Management of critically ill adults with COVID-19. Journal of the American Medical Association , 323 ( 18 ) , 1839–1841. doi: 10.1001/jama.2020.4914.
- Repetto, G., del Peso, A., & Zurita, J. L. (2008). Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nature Protocols , 3(7), 1125–1131. doi: 10.1038/nprot.2008.75.
- Sanders, J. M., Monogue, M. L., Jodlowski, T. Z., & Cutrell, J. B. (2020). Pharmacologic treatments for coronavirus disease 2019 (COVID-19). Journal of the American Medical Association , 323(18), 1824–1836. doi: 10.1001/jama.2020.6019.
- Schneider, M., Ackermann, K., Stuart, M., Wex, C., Protzer, U., Schätzl, H. M., & Gilch, S. (2012). Severe acute respiratory syndrome coronavirus replication is severely impaired by MG132 due to proteasome-independent inhibition of M-calpain. Journal of Virology , 86(18), 10112–22. doi: 10.1128/JVI.01001-12.
- Ströher, U., DiCaro, A., Li, Y., Strong, J. E., Aoki, F., Plummer, F., … Feldmann, H. (2004). Severe acute respiratory syndrome–related coronavirus is inhibited by interferon-α. The Journal of Infectious Diseases , 189(7), 1164–1167. doi: 10.1086/382597.
- van Doremalen, N., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., … Munster, V. J. (2020). Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1 (Supplementary Appendix). New England Journal of Medicine , 382(16): 1564–1567. doi: 10.1056/NEJMc2004973.
- Vicenzi, E., Canducci, F., Pinna, D., Mancini, N., Carletti, S., Lazzarin, A., … Clementi, M. (2004). Coronaviridae and SARS-associated coronavirus strain HSR1. Emerging Infectious Diseases , 10(3), 413–8. doi: 10.3201/eid1003.030683.
- Wang, S., Sakhatskyy, P., Chou, T.-H. W., & Lu, S. (2005). Assays for the assessment of neutralizing antibody activities against Severe Acute Respiratory Syndrome (SARS) associated coronavirus (SCV). Journal of Immunological Methods , 301(1–2), 21–30. doi: 10.1016/j.jim.2005.03.008.
- WHO. (2020a). Coronavirus disease 2019 (COVID-19) Situation Report—99. Retrieved April 28, 2020, from https://static.yanyin.tech/literature/current_protocol/10.1002/cpmc.105/attachments/20200428-sitrep-99-covid-19.pdf.
- WHO. (2020b). Laboratory biosafety guidance related to the novel coronavirus (2019-nCoV). Retrieved April 24, 2020, from https://static.yanyin.tech/literature/current_protocol/10.1002/cpmc.105/attachments/laboratory-biosafety-novel-coronavirus-version-1-1.pdf.
- WHO. (2020c). Novel Coronavirus—China. Retrieved March 27, 2020, from https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/.
- WHO. (2020d). Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). Retrieved March 29, 2020, from https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov).
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., … Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature , 579(7798), 265–269. doi: 10.1038/s41586-020-2008-3.
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., … China Novel Coronavirus Investigating and Research Team. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine , 382(8), 727–733. doi: 10.1056/NEJMoa2001017.
Citing Literature
Number of times cited according to CrossRef: 162
- Natalia E Ketaren, Fred D Mast, Peter C Fridy, Jean Paul Olivier, Tanmoy Sanyal, Andrej Sali, Brian T Chait, Michael P Rout, John D Aitchison, Nanobody repertoire generated against the spike protein of ancestral SARS-CoV-2 remains efficacious against the rapidly evolving virus, eLife, 10.7554/eLife.89423.3, 12 , (2024).
- Natalia E Ketaren, Fred D Mast, Peter C Fridy, Jean Paul Olivier, Tanmoy Sanyal, Andrej Sali, Brian T Chait, Michael P Rout, John D Aitchison, Nanobody repertoire generated against the spike protein of ancestral SARS-CoV-2 remains efficacious against the rapidly evolving virus, eLife, 10.7554/eLife.89423, 12 , (2024).
- Anoop Kumar, Prajna Tripathi, Prashant Kumar, Ritu Shekhar, Rajiv Pathak, From Detection to Protection: Antibodies and Their Crucial Role in Diagnosing and Combatting SARS-CoV-2, Vaccines, 10.3390/vaccines12050459, 12 , 5, (459), (2024).
- Einat Toister, Lilach Cherry, Edith Lupu, Arik Monash, Eyal Dor, Lilach Levin, Meni Girshengorn, Niva Natan, Shira Chapman, Shlomo Shmaya, Eyal Epstein, Yaakov Adar, Ran Zichel, Yakir Ophir, Eran Diamant, Development and Validation of a Plaque Assay to Determine the Titer of a Recombinant Live-Attenuated Viral Vaccine for SARS-CoV-2, Vaccines, 10.3390/vaccines12040374, 12 , 4, (374), (2024).
- Vinícius Pinto Costa Rocha, Bruna Aparecida Souza Machado, Helenita Costa Quadros, Antônio Márcio Santana Fernandes, Bianca Sampaio Dotto Fiuza, Cássio Santana Meira, Vitória Torres Barbosa da Silva, Afrânio Ferreira Evangelista, Larissa Moraes dos Santos Fonseca, Roberto José da Silva Badaró, Milena Botelho Pereira Soares, High-Content Imaging-Based Assay for SARS-CoV-2-Neutralizing Antibodies, Vaccines, 10.3390/vaccines12030236, 12 , 3, (236), (2024).
- Seung-Min Hong, Eun-Jin Ha, Ho-Won Kim, Seung-Ji Kim, Sun-Min Ahn, Se-Hee An, Gun Kim, Suji Kim, Hyuk-Joon Kwon, Kang-Seuk Choi, Effects of G and SH Truncation on the Replication, Virulence, and Immunogenicity of Avian Metapneumovirus, Vaccines, 10.3390/vaccines12010106, 12 , 1, (106), (2024).
- Nadine Alvarez, Gregory C. Adam, John A. Howe, Vijeta Sharma, Matthew D. Zimmerman, Enriko Dolgov, Risha Rasheed, Fatima Nizar, Khushboo Sahay, Andrew M. Nelson, Steven Park, Xiaoyan Zhou, Christine Burlein, John F. Fay, Daniel V. Iwamoto, Carolyn M. Bahnck-Teets, Krista L. Getty, Shih Lin Goh, Imad Salhab, Keith Smith, Christopher W. Boyce, Tamara D. Cabalu, Nicholas Murgolo, Nicholas G. Fox, Todd W. Mayhood, Valerie W. Shurtleff, Mark E. Layton, Craig A. Parish, John A. McCauley, David B. Olsen, David S. Perlin, Novel Pan-Coronavirus 3CL Protease Inhibitor MK-7845: Biological and Pharmacological Profiling, Viruses, 10.3390/v16071158, 16 , 7, (1158), (2024).
- Sankar Prasad Chaki, Melissa M. Kahl-McDonagh, Benjamin W. Neuman, Kurt A. Zuelke, Validating the inactivation of viral pathogens with a focus on SARS-CoV-2 to safely transfer samples from high-containment laboratories, Frontiers in Cellular and Infection Microbiology, 10.3389/fcimb.2024.1292467, 14 , (2024).
- Saba Farooq, undefined Atia-tul-Wahab, Muhamamd Iqbal Choudhary, undefined Atta-ur-Rahman, Preventive Potential of Extracts of Some Dietary Plants against SARSCoV- 2 Infection, Current Traditional Medicine, 10.2174/2215083809666230116141143, 10 , 1, (2024).
- Jérémie Le Pen, Gabrielle Paniccia, Volker Kinast, Marcela Moncada-Velez, Alison W. Ashbrook, Michael Bauer, H.-Heinrich Hoffmann, Ana Pinharanda, Inna Ricardo-Lax, Ansgar F. Stenzel, Edwin A. Rosado-Olivieri, Kenneth H. Dinnon, William C. Doyle, Catherine A. Freije, Seon-Hui Hong, Danyel Lee, Tyler Lewy, Joseph M. Luna, Avery Peace, Carltin Schmidt, William M. Schneider, Roni Winkler, Elaine Z. Yip, Chloe Larson, Timothy McGinn, Miriam-Rose Menezes, Lavoisier Ramos-Espiritu, Priyam Banerjee, John T. Poirier, Francisco J. Sànchez-Rivera, Aurélie Cobat, Qian Zhang, Jean-Laurent Casanova, Thomas S. Carroll, J. Fraser Glickman, Eleftherios Michailidis, Brandon Razooky, Margaret R. MacDonald, Charles M. Rice, A genome-wide arrayed CRISPR screen identifies PLSCR1 as an intrinsic barrier to SARS-CoV-2 entry that recent virus variants have evolved to resist, PLOS Biology, 10.1371/journal.pbio.3002767, 22 , 9, (e3002767), (2024).
- Abigail D. Solstad, Parker J. Denz, Adam D. Kenney, Najmus S. Mahfooz, Samuel Speaks, Qiaoke Gong, Richard T. Robinson, Matthew E. Long, Adriana Forero, Jacob S. Yount, Emily A. Hemann, IFN-λ uniquely promotes CD8 T cell immunity against SARS-CoV-2 relative to type I IFN, JCI Insight, 10.1172/jci.insight.171830, 9 , 13, (2024).
- Judith M. J. Veugen, Tom Schoenmakers, Inge H. M. van Loo, Bart L. Haagmans, Mathie P. G. Leers, Mart M. Lamers, Mayk Lucchesi, Bas C. T. van Bussel, Walther N. K. A. van Mook, Rudy M. M. A. Nuijts, Paul H. M. Savelkoul, Mor M. Dickman, Petra F. G. Wolffs, Advancing COVID-19 diagnostics: rapid detection of intact SARS-CoV-2 using viability RT-PCR assay, Microbiology Spectrum, 10.1128/spectrum.00160-24, 12 , 9, (2024).
- Dominic Wooding, Kate Buist, Alessandra Romero-Ramirez, Helen Savage, Rachel Watkins, Daisy Bengey, Caitlin Greenland-Bews, Caitlin R. Thompson, Nadia Kontogianni, Richard Body, Gail Hayward, Rachel L. Byrne, Susan Gould, Christopher Myerscough, Barry Atkinson, Victoria Shaw, Bill Greenhalf, Emily Adams, Ana Cubas-Atienzar, Saye Khoo, Tom Fletcher, Thomas Edwards, A. Joy Allen, Julian Braybrook, Peter Buckle, Paul Dark, Kerrie Davis, Adam Gordon, Anna Halstead, Charlotte Harden, Colette Inkson, Naoko Jones, William Jones, Dan Lasserson, Joseph Lee, Clare Lendrem, Andrew Lewington, Mary Logan, Massimo Micocci, Brian Nicholson, Rafael Perera-Salazar, Graham Prestwich, D. Ashley Price, Charles Reynard, Beverley Riley, A. J. Simpson, Valerie Tate, Philip Turner, Mark Wilcox, Melody Zhifang, Optimization of SARS-CoV-2 culture from clinical samples for clinical trial applications, mSphere, 10.1128/msphere.00304-24, (2024).
- Harrison Dulin, Ramya S. Barre, Duo Xu, Arrmund Neal, Edward Vizcarra, Jerald Chavez, Arzu Ulu, Myeon-Sik Yang, Siddiqur Rahman Khan, Keidy Wuang, Nikhil Bhakta, Chanvoraboth Chea, Emma H. Wilson, Luis Martinez-Sobrido, Rong Hai, Harnessing preexisting influenza virus-specific immunity increases antibody responses against SARS-CoV-2, Journal of Virology, 10.1128/jvi.01571-23, 98 , 2, (2024).
- Yi-Hao Chan, Vanja Lundberg, Jérémie Le Pen, Jiayi Yuan, Danyel Lee, Francesca Pinci, Stefano Volpi, Koji Nakajima, Vincent Bondet, Sanna Åkesson, Noopur V. Khobrekar, Aaron Bodansky, Likun Du, Tina Melander, Alice-Andrée Mariaggi, Yoann Seeleuthner, Tariq Shikh Saleh, Debanjana Chakravarty, Per Marits, Kerry Dobbs, Sofie Vonlanthen, Viktoria Hennings, Karolina Thörn, Darawan Rinchai, Lucy Bizien, Matthieu Chaldebas, Ali Sobh, Tayfun Özçelik, Sevgi Keles, Suzan A. AlKhater, Carolina Prando, Isabelle Meyts, Paul Bastard, Alessandro Borghesi, Aziz Bousfiha, Oksana Boyarchuk, Petter Brodin, Jacinta Bustamante, Giorgio Casari, Rémi Chevalier, John Christodoulou, Roger Colobran, Antonio Condino-Neto, Juan Carlos Aldave Becerra, Lisa Arkin, Evangelos Andreakos, Christian W. Thorball, Sara Espinosa, Carlos Flores, Amyrath Geraldo, Rabih Halwani, Nevin Hatipoğlu, Brahim Melaiki, Jacques Fellay, Alenka Gagro, Yuval Itan, Chandima Jeewandara, Filomeen Haerynck, Davood Mansouri, Leslie Naesens, Lisa F.P. Ng, Keisuke Okamoto, Pere Soler-Palacin, Laurent Renia, Aurora Pujol Onofre, Igor B. Resnick, José Luis Franco Restrepo, Jacques G. Rivière, Anna Scherbina, Anna Šedivá, Mikko R.J. Seppänen, Helen Su, Stuart G Tangye, Sehime G. Temel, Ahmad Abou Tayoun, Stuart Turvey, K.M. Furkan Uddin, Diederik van de Beek, Tom Le Voyer, Donald C. Vinh, Shen-Ying Zhang, Jean-Laurent Casanova, Michael R. Wilson, Jérémie Rosain, Emmanuelle Jouanguy, Mélodie Aubart, Laurent Abel, Trine H. Mogensen, Qiang Pan-Hammarström, Daxing Gao, Darragh Duffy, Aurélie Cobat, Stefan Berg, Luigi D. Notarangelo, Oliver Harschnitz, Charles M. Rice, Lorenz Studer, Jean-Laurent Casanova, Olov Ekwall, Shen-Ying Zhang, SARS-CoV-2 brainstem encephalitis in human inherited DBR1 deficiency, Journal of Experimental Medicine, 10.1084/jem.20231725, 221 , 9, (2024).
- Uday Chintapula, Shazeed-Ul Karim, Priyanka Raghunathan Iyer, Haritha Asokan-Sheeja, Biswas Neupane, Farzana Nazneen, He Dong, Fengwei Bai, Kytai T. Nguyen, A novel nanocomposite drug delivery system for SARS-CoV-2 infections, Nanoscale Advances, 10.1039/D4NA00361F, 6 , 15, (3747-3758), (2024).
- Shashi Gujar, Jonathan G. Pol, Vishnupriyan Kumar, Manuela Lizarralde-Guerrero, Prathyusha Konda, Guido Kroemer, John C. Bell, Tutorial: design, production and testing of oncolytic viruses for cancer immunotherapy, Nature Protocols, 10.1038/s41596-024-00985-1, 19 , 9, (2540-2570), (2024).
- Jingjing Zhang, Yanmei Li, Fengyuan Zeng, Changyong Mu, Change Liu, Lichun Wang, Xiaowu Peng, Liping He, Yanrui Su, Hongbing Li, An Wang, Lin Feng, Dongxiu Gao, Zhixiao Zhang, Gang Xu, Yixuan Wang, Rong Yue, Junbo Si, Lichun Zheng, Xiong Zhang, Fuyun He, Hongkun Yi, Zhongshu Tang, Gaocan Li, Kaili Ma, Qihan Li, Virus-like structures for combination antigen protein mRNA vaccination, Nature Nanotechnology, 10.1038/s41565-024-01679-1, 19 , 8, (1224-1233), (2024).
- Rúbens Prince dos Santos Alves, Julia Timis, Robyn Miller, Kristen Valentine, Paolla Beatriz Almeida Pinto, Andrew Gonzalez, Jose Angel Regla-Nava, Erin Maule, Michael N. Nguyen, Norazizah Shafee, Sara Landeras-Bueno, Eduardo Olmedillas, Brett Laffey, Katarzyna Dobaczewska, Zbigniew Mikulski, Sara McArdle, Sarah R. Leist, Kenneth Kim, Ralph S. Baric, Erica Ollmann Saphire, Annie Elong Ngono, Sujan Shresta, Human coronavirus OC43-elicited CD4+ T cells protect against SARS-CoV-2 in HLA transgenic mice, Nature Communications, 10.1038/s41467-024-45043-2, 15 , 1, (2024).
- Jinseung Bae, Petr Bednar, Rong Zhu, Cheolwoo Bong, Moon Soo Bak, Sarah Stainer, Kyoungjun Kim, Junghun Lee, Chulsoo Yoon, Yugyeong Lee, Omobolaji Taye Ojowa, Maximilian Lehner, Peter Hinterdorfer, Daniel Ruzek, Sungsu Park, Yoo Jin Oh, Mechanisms of Plasma Ozone and UV-C Sterilization of SARS-CoV-2 Explored through Atomic Force Microscopy, ACS Applied Materials & Interfaces, 10.1021/acsami.4c11057, 16 , 37, (49176-49185), (2024).
- Agnieszka Nagorska, Ruben M. F. Tomás, Afifah Tasnim, Nicole C. Robb, Matthew I. Gibson, Cryopreserved Kidney Epithelial (Vero) Cell Monolayers for Rapid Viral Quantification, Enabled by a Combination of Macromolecular Cryoprotectants, Biomacromolecules, 10.1021/acs.biomac.4c00760, 25 , 8, (5352-5358), (2024).
- Manar E. Khalifa, Muhammad Munir, Protocol for constructing and characterizing recombinant vectored vaccines for rabies virus, STAR Protocols, 10.1016/j.xpro.2024.103392, 5 , 4, (103392), (2024).
- Asja Korajkic, Brian R. McMinn, Adin C. Pemberton, Julie Kelleher, Warish Ahmed, The comparison of decay rates of infectious SARS-CoV-2 and viral RNA in environmental waters and wastewater, Science of The Total Environment, 10.1016/j.scitotenv.2024.174379, 946 , (174379), (2024).
- Gabriela Leite, Sepideh Mehravar, Mark Pimentel, Ruchi Mathur, Gil Y Melmed, Volha Teagle, Gillian M Barlow, Ali Rezaie, Extracellular and intracellular antiviral effects of ultraviolet A against severe acute respiratory syndrome coronavirus-2 are variant-independent, Photodiagnosis and Photodynamic Therapy, 10.1016/j.pdpdt.2024.104097, 47 , (104097), (2024).
- Line L. Bang, Ditte R. Tornby, Stephanie T.D. Pham, Kristian Assing, Sören Möller, Yaseelan Palarasah, Lone W. Madsen, Karina G. Thomsen, Isik S. Johansen, Rune M. Pedersen, Thomas E. Andersen, Culturing of SARS-CoV-2 from patient samples: Protocol for optimal virus recovery and assessment of infectious viral load, Journal of Virological Methods, 10.1016/j.jviromet.2024.114912, 326 , (114912), (2024).
- Rajesh Rajaiah, Kabita Pandey, Arpan Acharya, Anoop Ambikan, Narendra Kumar, Reema Guda, Sean N. Avedissian, Luis J. Montaner, Samuel M. Cohen, Ujjwal Neogi, Siddappa N. Byrareddy, Differential immunometabolic responses to Delta and Omicron SARS-CoV-2 variants in golden syrian hamsters, iScience, 10.1016/j.isci.2024.110501, 27 , 8, (110501), (2024).
- Seunghoon Choi, Jusung Lee, Suhyeon Kim, Youn Woo Lee, Gi-Cheon Kim, Seung-Min Hong, Se-Hee An, Hyuna Noh, Kyung Eun Kim, Dain On, Sang Gyu Lee, Hui Jeong Jang, Sung-Hee Kim, Jiseon Kim, Jung Seon Seo, Jeong Jin Kim, In Ho Park, Jooyeon Oh, Da-Jung Kim, Jong-Hwi Yoon, Sang-Hyuk Seok, Yu Jin Lee, Seo Yeon Kim, Young Been Kim, Ji-Yeon Hwang, Hyo-Jung Lee, Hong Bin Kim, Jun Won Park, Jun-Won Yun, Jeon-Soo Shin, Jun-Young Seo, Ki Taek Nam, Kang-Seuk Choi, Ho-Keun Kwon, Ho-Young Lee, Jong Kyoung Kim, Je Kyung Seong, A longitudinal molecular and cellular lung atlas of lethal SARS-CoV-2 infection in K18-hACE2 transgenic mice, eBioMedicine, 10.1016/j.ebiom.2023.104932, 99 , (104932), (2024).
- Anne Sophie Rufyikiri, Rebecca Martinez, Philip W. Addo, Bo-Sen Wu, Mitra Yousefi, Danielle Malo, Valérie Orsat, Silvia M. Vidal, Jörg H. Fritz, Sarah MacPherson, Mark Lefsrud, Germicidal efficacy of continuous and pulsed ultraviolet-C radiation on pathogen models and SARS-CoV-2, Photochemical & Photobiological Sciences, 10.1007/s43630-023-00521-2, 23 , 2, (339-354), (2024).
- Valentina Gentili, Silvia Beltrami, Doretta Cuffaro, Giorgia Cianci, Gloria Maini, Roberta Rizzo, Marco Macchia, Armando Rossello, Daria Bortolotti, Elisa Nuti, JG26 attenuates ADAM17 metalloproteinase-mediated ACE2 receptor processing and SARS-CoV-2 infection in vitro, Pharmacological Reports, 10.1007/s43440-024-00650-0, (2024).
- Gui-Juan Zhao, Min Yang, Yan-Qun Zhang, Gui-Feng Li, Gui Pu, Sui Yao, Dong Miao, Yin-Ke Li, Qiu-fen Hu, Xin-Zhou Yang, Prenylated Flavonoids from Desmodium caudatum and as Potential Activators for SARS-CoV-2, Chemistry of Natural Compounds, 10.1007/s10600-024-04342-2, 60 , 3, (423-427), (2024).
- Ping Zhang, Wen-Yu Liu, Hua-Yin Liu, Yu-Ping Wu, Yin-Ke Li, Guang-Hui Kong, Gao-Kun Zhao, Wei-Guang Wang, Qiu-Fen Hu, Guang-Hai Zhang, Two New Anti-SARS-CoV-2 Naphthoquinones from Cigar-Tobacco-Derived Endophytic Fusarium oxysporum, Chemistry of Natural Compounds, 10.1007/s10600-024-04339-x, 60 , 3, (410-414), (2024).
- Gui-Feng Li, Yan-Qun Zhang, Gui-Juan Zhao, Gui Pu, Yue-Yu Ma, Min Zhou, Yin-Ke Li, Dong Miao, Qiu-fen Hu, Xin-Zhou Yang, Two New Stilbenes from the Whole Plant of Arundina graminifolia and Their Anti-SARS-CoV-2 Activity, Chemistry of Natural Compounds, 10.1007/s10600-024-04335-1, 60 , 3, (393-397), (2024).
- Yan-Qun Zhang, Gui-Feng Li, Gui-Juan Zhao, Gui Pu, Yue-Yu Ma, Min Zhou, Yin-Ke Li, Dong Miao, Qiu-fen Hu, Xin-Zhou Yang, Two New Stilbenes from the Leaves and Stems of Bletilla striata and Their Anti-SARS-CoV-2 Activity, Chemistry of Natural Compounds, 10.1007/s10600-024-04290-x, 60 , 2, (211-214), (2024).
- Gaurav Kumar Sharma, Sanketkumar Nehul, Shweta Choudhary, Sonalika Mahajan, In Vitro and in Vivo Evaluation Tools of SARS-CoV-2 Antiviral Drugs, Advances in Antiviral Research, 10.1007/978-981-99-9195-2_11, (287-305), (2024).
- Adriano Souza Carolino, Xaiane Martins Silva Freitas, Célio Matias Airone Macalia, Juliana Coatrini Soares, Andrey Coatrini Soares, Camila Costa Pinto, Aguyda Rayany Cavalcante Barbosa, Jaqueline Araújo Bezerra, Pedro Henrique Campelo, Marcos Marques Silva Paula, Pritesh Jaychand Lalwani, Natalia Mayumi Inada, Ștefan Țãlu, Adriana Malheiro, Edgar Aparecido Sanches, Virus adsorbent systems based on Amazon holocellulose and nanomaterials, Microscopy Research and Technique, 10.1002/jemt.24566, 87 , 8, (1933-1954), (2024).
- Byeongseok Kim, Ki Hoon Park, Ok-Hee Lee, Giwan Lee, Hyukjung Kim, Siyoung Lee, Semi Hwang, Young Bong Kim, Youngsok Choi, Effect of severe acute respiratory syndrome coronavirus 2 infection during pregnancy in K18-hACE2 transgenic mice, Animal Bioscience, 10.5713/ab.22.0143, 36 , 1, (43-52), (2023).
- Samuel P. Smith, Rebecca Shipley, Pascal Drake, Anthony R. Fooks, Julian Ma, Ashley C. Banyard, Characterisation of a Live-Attenuated Rabies Virus Expressing a Secreted scFv for the Treatment of Rabies, Viruses, 10.3390/v15081674, 15 , 8, (1674), (2023).
- Vinícius Pinto Costa Rocha, Helenita Costa Quadros, Antônio Márcio Santana Fernandes, Luana Pereira Gonçalves, Roberto José da Silva Badaró, Milena Botelho Pereira Soares, Bruna Aparecida Souza Machado, An Overview of the Conventional and Novel Methods Employed for SARS-CoV-2 Neutralizing Antibody Measurement, Viruses, 10.3390/v15071504, 15 , 7, (1504), (2023).
- Keven Lothert, Elena Bagrin, Michael W. Wolff, Evaluating Novel Quantification Methods for Infectious Baculoviruses, Viruses, 10.3390/v15040998, 15 , 4, (998), (2023).
- Miaomiao Guo, Li Deng, Hongyang Liang, Yuyao Du, Wenrui Gao, Na Tian, Ying Bi, Jinghua Li, Tiancong Ma, Yuntao Zhang, Hui Wang, Development and Preliminary Application of a Droplet Digital PCR Assay for Quantifying the Oncolytic Herpes Simplex Virus Type 1 in the Clinical-Grade Production, Viruses, 10.3390/v15010178, 15 , 1, (178), (2023).
- Reem Binsuwaidan, Walaa A. Negm, Engy Elekhnawy, Nashwah G. M. Attallah, Eman Ahmed, Sameh Magdeldin, Ehssan Moglad, Sally Abdallah Mostafa, Suzy A. El-Sherbeni, In Vitro Antiviral Effect and Potential Neuroprotection of Salvadora persica L. Stem Bark Extract against Lipopolysaccharides-Induced Neuroinflammation in Mice: LC-ESI-MS/MS Analysis of the Methanol Extract, Pharmaceuticals, 10.3390/ph16030398, 16 , 3, (398), (2023).
- Nashwah G. M. Attallah, Amal Kabbash, Walaa A. Negm, Engy Elekhnawy, Reem Binsuwaidan, Omnia Momtaz Al-Fakhrany, Moataz A. Shaldam, Ehssan Moglad, Marwa Tarek, Nehal Samir, Heba M. Fawzy, Protective Potential of Saussurea costus (Falc.) Lipsch. Roots against Cyclophosphamide-Induced Pulmonary Injury in Rats and Its In Vitro Antiviral Effect, Pharmaceuticals, 10.3390/ph16020318, 16 , 2, (318), (2023).
- Rebeca Gonzalez-Pastor, Saskya E. Carrera-Pacheco, Johana Zúñiga-Miranda, Cristina Rodríguez-Pólit, Arianna Mayorga-Ramos, Linda P. Guamán, Carlos Barba-Ostria, Current Landscape of Methods to Evaluate Antimicrobial Activity of Natural Extracts, Molecules, 10.3390/molecules28031068, 28 , 3, (1068), (2023).
- Raghad Khaleafi, Jelena Zeleznjak, Sapir Cordela, Shani Drucker, Tihana Lenac Rovis, Stipan Jonjic, Yotam Bar-On, Reovirus infection of tumor cells reduces the expression of NKG2D ligands, leading to impaired NK-cell cytotoxicity and functionality, Frontiers in Immunology, 10.3389/fimmu.2023.1231782, 14 , (2023).
- Endeshaw Chekol Abebe, Tadesse Asmamaw Dejenie, Protective roles and protective mechanisms of neutralizing antibodies against SARS-CoV-2 infection and their potential clinical implications, Frontiers in Immunology, 10.3389/fimmu.2023.1055457, 14 , (2023).
- Austin Featherstone, Amanda Claire Brown, Sapna Chitlapilly Dass, Understanding how different surfaces and environmental biofilms found in food processing plants affect the spread of COVID-19, PLOS ONE, 10.1371/journal.pone.0286659, 18 , 6, (e0286659), (2023).
- Brittany Humphrey, Matthew Tezak, Mia Lobitz, Anastasia Hendricks, Andres Sanchez, Jake Zenker, Steven Storch, Ryan D. Davis, Bryce Ricken, Jesse Cahill, Viral Preservation with Protein-Supplemented Nebulizing Media in Aerosols, Applied and Environmental Microbiology, 10.1128/aem.01545-22, 89 , 3, (2023).
- Danyel Lee, Jérémie Le Pen, Ahmad Yatim, Beihua Dong, Yann Aquino, Masato Ogishi, Rémi Pescarmona, Estelle Talouarn, Darawan Rinchai, Peng Zhang, Magali Perret, Zhiyong Liu, Iolanda Jordan, Sefika Elmas Bozdemir, Gulsum Iclal Bayhan, Camille Beaufils, Lucy Bizien, Aurelie Bisiaux, Weite Lei, Milena Hasan, Jie Chen, Christina Gaughan, Abhishek Asthana, Valentina Libri, Joseph M. Luna, Fabrice Jaffré, H.-Heinrich Hoffmann, Eleftherios Michailidis, Marion Moreews, Yoann Seeleuthner, Kaya Bilguvar, Shrikant Mane, Carlos Flores, Yu Zhang, Andrés A. Arias, Rasheed Bailey, Agatha Schlüter, Baptiste Milisavljevic, Benedetta Bigio, Tom Le Voyer, Marie Materna, Adrian Gervais, Marcela Moncada-Velez, Francesca Pala, Tomi Lazarov, Romain Levy, Anna-Lena Neehus, Jérémie Rosain, Jessica Peel, Yi-Hao Chan, Marie-Paule Morin, Rosa Maria Pino-Ramirez, Serkan Belkaya, Lazaro Lorenzo, Jordi Anton, Selket Delafontaine, Julie Toubiana, Fanny Bajolle, Victoria Fumadó, Marta L. DeDiego, Nadhira Fidouh, Flore Rozenberg, Jordi Pérez-Tur, Shuibing Chen, Todd Evans, Frédéric Geissmann, Pierre Lebon, Susan R. Weiss, Damien Bonnet, Xavier Duval, Qiang Pan-Hammarström, Anna M. Planas, Isabelle Meyts, Filomeen Haerynck, Aurora Pujol, Vanessa Sancho-Shimizu, Clifford L. Dalgard, Jacinta Bustamante, Anne Puel, Stéphanie Boisson-Dupuis, Bertrand Boisson, Tom Maniatis, Qian Zhang, Paul Bastard, Luigi Notarangelo, Vivien Béziat, Rebeca Perez de Diego, Carlos Rodriguez-Gallego, Helen C. Su, Richard P. Lifton, Emmanuelle Jouanguy, Aurélie Cobat, Laia Alsina, Sevgi Keles, Elie Haddad, Laurent Abel, Alexandre Belot, Lluis Quintana-Murci, Charles M. Rice, Robert H. Silverman, Shen-Ying Zhang, Jean-Laurent Casanova, Loubna Alavoine, Sylvie Behillil, Charles Burdet, Charlotte Charpentier, Aline Dechanet, Diane Descamps, Xavier Duval, Jean-Luc Ecobichon, Vincent Enouf, Wahiba Frezouls, Nadhira Houhou, Ouifiya Kafif, Jonathan Lehacaut, Sophie Letrou, Bruno Lina, Jean-Christophe Lucet, Pauline Manchon, Mariama Nouroudine, Valentine Piquard, Caroline Quintin, Michael Thy, Sarah Tubiana, Sylvie van der Werf, Valérie Vignali, Benoit Visseaux, Yazdan Yazdanpanah, Abir Chahine, Nawal Waucquier, Maria-Claire Migaud, Dominique Deplanque, Félix Djossou, Mayka Mergeay-Fabre, Aude Lucarelli, Magalie Demar, Léa Bruneau, Patrick Gérardin, Adrien Maillot, Christine Payet, Bruno Laviolle, Fabrice Laine, Christophe Paris, Mireille Desille-Dugast, Julie Fouchard, Denis Malvy, Duc Nguyen, Thierry Pistone, Pauline Perreau, Valérie Gissot, Carole Le Goas, Samatha Montagne, Lucie Richard, Catherine Chirouze, Kévin Bouiller, Maxime Desmarets, Alexandre Meunier, Benjamin Lefèvre, Hélène Jeulin, Karine Legrand, Sandra Lomazzi, Bernard Tardy, Amandine Gagneux-Brunon, Frédérique Bertholon, Elisabeth Botelho-Nevers, Kouakam Christelle, Leturque Nicolas, Layidé Roufai, Karine Amat, Sandrine Couffin-Cadiergues, Hélène Espérou, Samia Hendou, Laurent Abel, Hassan Abolhassani, Sergio Aguilera-Albesa, Alessandro Aiuti, Ozge Metin Akcan, Nihal Akcay, Gulsum Alkan, Suzan A. Alkhater, Luis Miguel Allende, Yosunkaya Alper, Naima Amenzoui, Mark S. Anderson, Lisa Arkin, Melodie Aubart, Iryna Avramenko, Şehnaz Aydemir, Zeynep Gökçe Gayretli Aydin, Caner Aytekin, Gökhan Aytekin, Selma Erol Aytekin, Silvia Yumi Bando, Kathie Beland, Serkan Belkaya, Catherine M. Biggs, Agurtzane Bilbao Aburto, Geraldine Blanchard-Rohner, Daniel Blázquez-Gamero, Marketa Bloomfield, Dusan Bogunovic, Anastasia Bondarenko, Alessandro Borghesi, Amed Aziz Bousfiha, Oksana Boyarchuk, Petter Brodin, Yenan Bryceson, Giorgia Bucciol, Valeria Calcaterra, Giorgio Casari, Andre Cavalcanti, Jale Bengi Celik, George P. Chrousos, Roger Colobran, Antonio Condino-Neto, Francesca Conti, Megan Cooper, Taner Coskuner, Cyril Cyrus, Enza D’Auria, Selket Delafontaine, Beth A. Drolet, Burcu Bursal Duramaz, Loubna El Zein, Marwa H. Elnagdy, Melike Emiroglu, Emine Hafize Erdeniz, Marianna Fabi, Hagit Baris Feldman, Jacques Fellay, Filip Fencl, Filippos Filippatos, Julie Freiss, Jiri Fremuth, Alenka Gagro, Blanca Garcia-Solis, Gianluca Vergine, Rafaela González-Montelongo, Yahya Gul, Belgin Gülhan, Sara Sebnem Kilic Gultekin, Marta Gut, Rabih Halwani, Lennart Hammarström, Nevin Hatipoğlu, James Heath, Sarah E. Henrickson, Elisa Hernandez-Brito, Ilse Hoffman, Levi Hoste, Elena Hsieh, Antonio Íñigo-Campos, Yuval Itan, Petr Jabandziev, Bahar Kandemir, Saliha Kanık-Yüksek, Hasan Kapakli, Adem Karbuz, Ozgur Kasapcopur, Robin Kechiche, Yasemin Kendir Demirkol, Omer Kilic, Stella Kim Hansen, Adam Klocperk, Yu-Lung Lau, Jan Lebl, José M. Lorenzo-Salazar, Carrie L. Lucas, Majistor Maglorius, Laura Marque, Yeray Novoa Medina, Abián Montesdeoca Melián, Alexios-Fotios A. Mentis, Michele T. Pato, Athanasios Michos, Joshua D. Milner, Trine H. Mogensen, Adrián Muñoz-Barrera, Serdar Nepesov, João Farela Neves, Ashley Ng, Lisa F. P. Ng, Antonio Novelli, Giuseppe Novelli, Fatma Nur Oz, J. Gonzalo Ocejo-Viñals, Satoshi Okada, Zerrin Orbak, Ahmet Osman Kilic, Hind Ouair, Şadiye Kübra Tüter Öz, Tayfun Özçelik, Esra Akyüz Özkan, Aslınur Özkaya Parlakay, Carlos N. Pato, Estela Paz-Artal, Simon Pelham, Isabelle Pellier, Quentin Philippot, Laura Planas-Serra, Samira Plassart, Petra Pokorna, Meltem Polat, Cecilia Poli, Carolina Prando, Laurent Renia, Jacques G. Rivière, Agustí Rodríguez-Palmero, Lucie Roussel, Luis A. Rubio-Rodriguez, Moro Salifu, Lumir Sasek, Laura Sasia, Anna Scherbina, Erica Schmitt, Anna Sediva, Esra Sevketoglu, Katerina Slaba, Ondrej Slaby, Ali Sobh, Jordi Solé-Violán, Pere Soler-Palacin, Lien De Somer, Betül Sözeri, András N. Spaan, Yuriy Stepanovskiy, Stuart G. Tangye, Gonul Tanir, Elizabeth Barbara Tatsi, Christian W. Thorball, Selda Hancerli Torun, Stuart Turvey, Mohammed J. Uddin, Emel Uyar, Juan Valencia-Ramos, Ana Maria Van Den Rym, Hulya Vatansev, Martín Castillo de Vera, François Vermeulen, Donald C. Vinh, Alla Volokha, Horst von Bernuth, Carine Wouters, Aysun Yahşi, Volkan Yarar, Osman Yesilbas, Mehmet Yıldız, Mayana Zatz, Pawel Zawadzki, Gianvincenzo Zuccotti, Shen-Ying Zhang, Jean-Laurent Casanova, Inborn errors of OAS–RNase L in SARS-CoV-2–related multisystem inflammatory syndrome in children, Science, 10.1126/science.abo3627, 379 , 6632, (2023).
- Asmaa Saleh, Dalia H. Abdelkader, Thanaa A. El-Masry, Duaa Eliwa, Badriyah Alotaibi, Walaa A. Negm, Engy Elekhnawy, Antiviral and antibacterial potential of electrosprayed PVA/PLGA nanoparticles loaded with chlorogenic acid for the management of coronavirus and Pseudomonas aeruginosa lung infection , Artificial Cells, Nanomedicine, and Biotechnology, 10.1080/21691401.2023.2207606, 51 , 1, (255-267), (2023).
- Tairan Liu, Yuzhu Li, Hatice Ceylan Koydemir, Yijie Zhang, Ethan Yang, Merve Eryilmaz, Hongda Wang, Jingxi Li, Bijie Bai, Guangdong Ma, Aydogan Ozcan, Rapid and stain-free quantification of viral plaque via lens-free holography and deep learning, Nature Biomedical Engineering, 10.1038/s41551-023-01057-7, 7 , 8, (1040-1052), (2023).
- Yifang Liu, Jacob L. Potts, Dylan Bloch, Keqing Nian, Caroline A. McCormick, Oleksandra Fanari, Sara H. Rouhanifard, Paired Capture and FISH Detection of Individual Virions Enable Cell-Free Determination of Infectious Titers, ACS Sensors, 10.1021/acssensors.3c00239, 8 , 7, (2563-2571), (2023).
- Nur Syafiqah Mohamad Ishak, Tomoe Numaguchi, Kazuto Ikemoto, Antiviral Effects of Pyrroloquinoline Quinone through Redox Catalysis To Prevent Coronavirus Infection, ACS Omega, 10.1021/acsomega.3c06040, 8 , 47, (44839-44849), (2023).
- Samson W. L. Mah, Denver P. Linklater, Vassil Tzanov, Phuc H. Le, Chaitali Dekiwadia, Edwin Mayes, Ranya Simons, Daniel J. Eyckens, Graeme Moad, Soichiro Saita, Saulius Joudkazis, David A. Jans, Vladimir A. Baulin, Natalie A. Borg, Elena P. Ivanova, Piercing of the Human Parainfluenza Virus by Nanostructured Surfaces, ACS Nano, 10.1021/acsnano.3c07099, 18 , 2, (1404-1419), (2023).
- Colleen R. McCollum, Colleen M. Courtney, Nolan J. O’Connor, Thomas R. Aunins, Tristan X. Jordan, Keegan L. Rogers, Stephen Brindley, Jared M. Brown, Prashant Nagpal, Anushree Chatterjee, Safety and Biodistribution of Nanoligomers Targeting the SARS-CoV-2 Genome for the Treatment of COVID-19, ACS Biomaterials Science & Engineering, 10.1021/acsbiomaterials.2c00669, 9 , 3, (1656-1671), (2023).
- Junxia Cao, Hongjing Gu, Xueting Zhang, Hongfang Yun, Jiarong Li, Chuan-Yimu Si, Jiyan Zhang, Hui Wang, Intranasal inoculation of female BALB/c mice with replication-deficient human adenovirus type 5 expressing SARS-CoV‐2 nucleocapsid protein aggravates lung pathology upon re-encountering the antigen, Virus Research, 10.1016/j.virusres.2023.199201, 335 , (199201), (2023).
- Brian R. McMinn, Asja Korajkic, Adin C. Pemberton, Julie Kelleher, Warish Ahmed, Eric N. Villegas, Kevin Oshima, Assessment of two volumetrically different concentration approaches to improve sensitivities for SARS-CoV-2 detection during wastewater monitoring, Journal of Virological Methods, 10.1016/j.jviromet.2022.114645, 311 , (114645), (2023).
- Kiwamu Nakamura, Masahiro Sugiyama, Hikari Ishizuka, Tomomi Sasajima, Yoko Minakawa, Hiroko Sato, Masatsugu Miyazawa, Kazuhiro Kitakawa, Shohei Fujita, Nozomi Saito, Naoko Kashiwabara, Hironobu Kohata, Yasuka Hara, Yumiko Kanari, Toshikatsu Shinka, Keiji Kanemitsu, Prolonged infective SARS-CoV-2 omicron variant shedding in a patient with diffuse large B cell lymphoma successfully cleared after three courses of remdesivir, Journal of Infection and Chemotherapy, 10.1016/j.jiac.2023.05.003, 29 , 8, (820-824), (2023).
- Rofiqul A'la, Andi Yasmin Wijaya, Helen Susilowati, Suryo Kuncorojakti, undefined Diyantoro, Jola Rahmahani, Fedik Abdul Rantam, Inactivated SARS-CoV-2 vaccine candidate immunization on non-human primate animal model: B-cell and T-cell responses immune evaluation, Heliyon, 10.1016/j.heliyon.2023.e18039, 9 , 7, (e18039), (2023).
- Voddu Suresh, Padmanava Behera, Deepti Parida, Amlan Priyadarshee Mohapatra, Suraja Kumar Das, Sneha Kumari, Kiran Avula, Amruta Mohapatra, Gulam Hussain Syed, Shantibhusan Senapati, Therapeutic role of N-acetyl cysteine (NAC) for the treatment and/or management of SARS-CoV-2-induced lung damage in hamster model, European Journal of Pharmacology, 10.1016/j.ejphar.2022.175392, 938 , (175392), (2023).
- Ishani Wickramage, Jeffrey VanWye, Klaas Max, John H. Lockhart, Ismet Hortu, Ezinne F. Mong, John Canfield, Hiran M. Lamabadu Warnakulasuriya Patabendige, Ozlem Guzeloglu-Kayisli, Kimiko Inoue, Atsuo Ogura, Charles J. Lockwood, Kemal M. Akat, Thomas Tuschl, Umit A. Kayisli, Hana Totary-Jain, SINE RNA of the imprinted miRNA clusters mediates constitutive type III interferon expression and antiviral protection in hemochorial placentas, Cell Host & Microbe, 10.1016/j.chom.2023.05.018, 31 , 7, (1185-1199.e10), (2023).
- Jérémie Rosain, Anna-Lena Neehus, Jérémy Manry, Rui Yang, Jérémie Le Pen, Wassim Daher, Zhiyong Liu, Yi-Hao Chan, Natalia Tahuil, Özden Türel, Mathieu Bourgey, Masato Ogishi, Jean-Marc Doisne, Helena M. Izquierdo, Takayoshi Shirasaki, Tom Le Voyer, Antoine Guérin, Paul Bastard, Marcela Moncada-Vélez, Ji Eun Han, Taushif Khan, Franck Rapaport, Seon-Hui Hong, Andrew Cheung, Kathrin Haake, Barbara C. Mindt, Laura Pérez, Quentin Philippot, Danyel Lee, Peng Zhang, Darawan Rinchai, Fatima Al Ali, Manar Mahmoud Ahmad Ata, Mahbuba Rahman, Jessica N. Peel, Søren Heissel, Henrik Molina, Yasemin Kendir-Demirkol, Rasheed Bailey, Shuxiang Zhao, Jonathan Bohlen, Mathieu Mancini, Yoann Seeleuthner, Marie Roelens, Lazaro Lorenzo, Camille Soudée, María Elvira Josefina Paz, María Laura González, Mohamed Jeljeli, Jean Soulier, Serge Romana, Anne-Sophie L’Honneur, Marie Materna, Rubén Martínez-Barricarte, Mathieu Pochon, Carmen Oleaga-Quintas, Alexandre Michev, Mélanie Migaud, Romain Lévy, Marie-Alexandra Alyanakian, Flore Rozenberg, Carys A. Croft, Guillaume Vogt, Jean-François Emile, Laurent Kremer, Cindy S. Ma, Jörg H. Fritz, Stanley M. Lemon, András N. Spaan, Nicolas Manel, Laurent Abel, Margaret R. MacDonald, Stéphanie Boisson-Dupuis, Nico Marr, Stuart G. Tangye, James P. Di Santo, Qian Zhang, Shen-Ying Zhang, Charles M. Rice, Vivien Béziat, Nico Lachmann, David Langlais, Jean-Laurent Casanova, Philippe Gros, Jacinta Bustamante, Human IRF1 governs macrophagic IFN-γ immunity to mycobacteria, Cell, 10.1016/j.cell.2022.12.038, 186 , 3, (621-645.e33), (2023).
- Balamurugan Shanmugaraj, Narach Khorattanakulchai, Weena Paungpin, Yada Akkhawattanangkul, Suwimon Manopwisedjaroen, Arunee Thitithanyanont, Waranyoo Phoolcharoen, Immunogenicity and efficacy of recombinant subunit SARS-CoV-2 vaccine candidate in the Syrian hamster model, Biotechnology Reports, 10.1016/j.btre.2022.e00779, 37 , (e00779), (2023).
- Dong-Gun Kim, Uijin Kim, In Ho Park, Bumhan Ryu, Youngki Yoo, Jeong Seok Cha, Ga-Yeon Yoon, Sung-Hee Kim, Heeju Oh, Jun-Young Seo, Ki Taek Nam, Je Kyung Seong, Jeon-Soo Shin, Hyun-Soo Cho, Hak-Sung Kim, A bivalent form of a RBD-specific synthetic antibody effectively neutralizes SARS-CoV-2 variants, Antiviral Research, 10.1016/j.antiviral.2023.105738, 220 , (105738), (2023).
- Farah Shahjin, Milankumar Patel, Jatin Machhi, Jacob D. Cohen, Mohammad Ullah Nayan, Pravin Yeapuri, Chen Zhang, Emiko Waight, Mahmudul Hasan, Mai Mohamed Abdelmoaty, Prasanta K. Dash, You Zhou, Irene Andreu, Howard E. Gendelman, Bhavesh D. Kevadiya, Multipolymer microsphere delivery of SARS-CoV-2 antigens, Acta Biomaterialia, 10.1016/j.actbio.2022.12.043, 158 , (493-509), (2023).
- Ram Gopal Nitharwal, In vitro and in vivo approaches for evaluating antiviral efficacy, Viral Infections and Antiviral Therapies, 10.1016/B978-0-323-91814-5.00016-7, (693-707), (2023).
- Thamby Rajah Mahendran, Binsin Cynthia, Ganapaty Manickavasagam, John J. Thambirajah, Maheswaran Solayappan, Thirumalai Komala, Isolation and evaluation of antiviral plant compounds against respiratory disease-causing viruses: a review, Advances in Traditional Medicine, 10.1007/s13596-023-00723-0, 24 , 3, (725-753), (2023).
- Mariana Marques, Ana Rita Ferreira, Daniela Ribeiro, Determining the Importance of Peroxisomal Proteins for Viral Infections in Cultured Mammalian Cells, Peroxisomes, 10.1007/978-1-0716-3048-8_21, (309-319), (2023).
- Anne K. Haudenschild, Blaine A. Christiansen, Sophie Orr, Erin E. Ball, Christopher M. Weiss, Hongwei Liu, David P. Fyhrie, Jasper H. N. Yik, Lark L. Coffey, Dominik R. Haudenschild, Acute bone loss following SARS‐CoV‐2 infection in mice, Journal of Orthopaedic Research, 10.1002/jor.25537, 41 , 9, (1945-1952), (2023).
- Sushil Kumar Sharma, Paban Kumar Dash, Ram Govind Yadav, Ambuj Shrivastava, Rohit Menon, Jyoti S. Kumar, Shashi Sharma, Suman Dhankher, Sunil Dhiman, Divya Kumari, Manisha Shukla, Vineet Relhan, Suresh Kumar, Manmohan Parida, Isolation and characterization of emerging Mpox virus from India, Journal of Medical Virology, 10.1002/jmv.28911, 95 , 7, (2023).
- Qasem Ramadan, Rana Hazaymeh, Mohammed Zourob, Immunity‐on‐a‐Chip: Integration of Immune Components into the Scheme of Organ‐on‐a‐Chip Systems, Advanced Biology, 10.1002/adbi.202200312, 7 , 12, (2023).
- Zaigham Abbas Rizvi, Rajdeep Dalal, Srikanth Sadhu, Akshay Binayke, Jyotsna Dandotiya, Yashwant Kumar, Tripti Shrivastava, Sonu Kumar Gupta, Suruchi Aggarwal, Manas Ranjan Tripathy, Deepak Kumar Rathore, Amit Kumar Yadav, Guruprasad R Medigeshi, Amit Kumar Pandey, Sweety Samal, Shailendra Asthana, Amit Awasthi, Golden Syrian hamster as a model to study cardiovascular complications associated with SARS-CoV-2 infection, eLife, 10.7554/eLife.73522, 11 , (2022).
- Aleksandra Nowakowska, Hanul Choi, Kihoon Park, Jinha Kim, Yuyeon Jang, Jungmin Chu, Young Bong Kim, Hee-Jung Lee, In vitro Antiviral Activity of Remdesivir Against SARS-CoV-2 and its Variants, Journal of Bacteriology and Virology, 10.4167/jbv.2022.52.4.149, 52 , 4, (149-159), (2022).
- Lise Lamoureux, Babu Sajesh, Jessy A. Slota, Sarah J. Medina, Matthew Mayor, Kathy L. Frost, Bryce Warner, Kathy Manguiat, Heidi Wood, Darwyn Kobasa, Stephanie A. Booth, Non-Productive Infection of Glial Cells with SARS-CoV-2 in Hamster Organotypic Cerebellar Slice Cultures, Viruses, 10.3390/v14061218, 14 , 6, (1218), (2022).
- Sunghoon Jung, Jun-Young Yang, Donghwan Jang, Taeyoon Kim, Ki Ho Baek, Hyunkyung Yoon, Joo Young Park, Sang Kwon Kim, Jinhyuk Hong, Sungweon Ryoo, Ho Won Jang, Seunghun Lee, Sustainable Antibacterial and Antiviral High-Performance Copper-Coated Filter Produced via Ion Beam Treatment, Polymers, 10.3390/polym14051007, 14 , 5, (1007), (2022).
- Fatemah A. Alherz, Walaa A. Negm, Engy Elekhnawy, Thanaa A. El-Masry, Eman M. Haggag, Moneerah J. Alqahtani, Ismail A. Hussein, Silver Nanoparticles Prepared Using Encephalartos laurentianus De Wild Leaf Extract Have Inhibitory Activity against Candida albicans Clinical Isolates, Journal of Fungi, 10.3390/jof8101005, 8 , 10, (1005), (2022).
- Marian O. Pacho, Dylan Deeney, Emily A. Johnson, Bryanna N. Bravo, Kishen Patel, Mark A. Latta, Michael A. Belshan, Stephen M. Gross, Characterization of Ag-Ion Releasing Zeolite Filled 3D Printed Resins, Journal of Functional Biomaterials, 10.3390/jfb14010007, 14 , 1, (7), (2022).
- Richard Kuan-Lin Lee, Tian-Neng Li, Sui-Yuan Chang, Tai-Ling Chao, Chun-Hsien Kuo, Max Yu-Chen Pan, Yu-Ting Chiou, Kuan-Ju Liao, Yi Yang, Yi-Hsuan Wu, Chen-Hao Huang, Hsueh-Fen Juan, Hsing-Pang Hsieh, Lily Hui-Ching Wang, Identification of Entry Inhibitors against Delta and Omicron Variants of SARS-CoV-2, International Journal of Molecular Sciences, 10.3390/ijms23074050, 23 , 7, (4050), (2022).
- Sabrina Semeraro, Anastasia Serena Gaetano, Luisa Zupin, Carlo Poloni, Elvio Merlach, Enrico Greco, Sabina Licen, Francesco Fontana, Silvana Leo, Alessandro Miani, Francesco Broccolo, Pierluigi Barbieri, Operative Protocol for Testing the Efficacy of Nasal Filters in Preventing Airborne Transmission of SARS-CoV-2, International Journal of Environmental Research and Public Health, 10.3390/ijerph192113790, 19 , 21, (13790), (2022).
- Craig Westover, Savlatjon Rahmatulloev, David Danko, Evan E. Afshin, Niamh B. O’Hara, Rachid Ounit, Daniela Bezdan, Christopher E. Mason, Ozone Disinfection for Elimination of Bacteria and Degradation of SARS-CoV2 RNA for Medical Environments, Genes, 10.3390/genes14010085, 14 , 1, (85), (2022).
- Santhamani Ramasamy, Afsal Kolloli, Ranjeet Kumar, Seema Husain, Patricia Soteropoulos, Theresa L. Chang, Selvakumar Subbian, Comprehensive Analysis of Disease Pathology in Immunocompetent and Immunocompromised Hosts following Pulmonary SARS-CoV-2 Infection, Biomedicines, 10.3390/biomedicines10061343, 10 , 6, (1343), (2022).
- Mohga E. Hamza, Muhammad A. Othman, Mohamed A. Swillam, Plasmonic Biosensors: Review, Biology, 10.3390/biology11050621, 11 , 5, (621), (2022).
- Farhad Vesuna, Ivan Akhrymuk, Amy Smith, Paul T. Winnard, Shih-Chao Lin, Lauren Panny, Robert Scharpf, Kylene Kehn-Hall, Venu Raman, RK-33, a small molecule inhibitor of host RNA helicase DDX3, suppresses multiple variants of SARS-CoV-2, Frontiers in Microbiology, 10.3389/fmicb.2022.959577, 13 , (2022).
- María I. Zapata-Cardona, Lizdany Flórez-Álvarez, Wildeman Zapata-Builes, Ariadna L. Guerra-Sandoval, Carlos M. Guerra-Almonacid, Jaime Hincapié-García, María T. Rugeles, Juan C. Hernandez, Atorvastatin Effectively Inhibits Ancestral and Two Emerging Variants of SARS-CoV-2 in vitro, Frontiers in Microbiology, 10.3389/fmicb.2022.721103, 13 , (2022).
- Stephen T. Yeung, Thomas A. Premeaux, Li Du, Toshiro Niki, Satish K. Pillai, Kamal M. Khanna, Lishomwa C. Ndhlovu, Galectin-9 protects humanized-ACE2 immunocompetent mice from SARS-CoV-2 infection, Frontiers in Immunology, 10.3389/fimmu.2022.1011185, 13 , (2022).
- Antonia Dibernardo, Nikki PL Toledo, Alyssia Robinson, Carla Osiowy, Elizabeth Giles, Jacqueline Day, L Robbin Lindsay, Michael A Drebot, Timothy F Booth, Tamara Pidduck, Ashley Baily, Carmen L Charlton, Graham Tipples, Jamil N Kanji, Gino Brochu, Amanda Lang, Christian Therrien, Mélina Bélanger-Collard, Sylvie-Nancy Beaulac, Brian M Gilfix, Guy Boivin, Marie-Ève Hamelin, Julie Carbonneau, Simon Lévesque, Philippe Martin, Andrés Finzi, Gabrielle Gendron-Lepage, Guillaume Goyette, Mehdi Benlarbi, Romain Gasser, Claude Fortin, Valérie Martel-Lafferrière, Myriam Lavoie, Renée Guérin, Louis-Patrick Haraoui, Christian Renaud, Craig Jenkins, Sheila F O'Brien, Steven J Drews, Valerie Conrod, Vanessa Tran, Bill Awrey, Robert Scheuermann, Alan DuPuis, Anne Payne, Casey Warszycki, Roxie Girardin, William Lee, George Zahariadis, Lei Jiao, Robert Needle, James Cordenbach, Jerry Zaharatos, Kellee Taylor, Marty Teltscher, Matthew Miller, May Elsherif, Peter Robertson, Jason L Robinson, Evaluation of the performance of multiple immunoassay diagnostic platforms on the National Microbiology Laboratory SARS-CoV-2 National Serology Panel, Official Journal of the Association of Medical Microbiology and Infectious Disease Canada, 10.3138/jammi-2021-0026, 7 , 3, (186-195), (2022).
- Waleed Ezzat Madboly, Ahmed M. Abu-Dief, Viral Infectivity Inhibition and Viral Biological Elements Destruction using Safe and Low Power Electrons Generated by Life Restoration Device (LRD): An In Vitro Study, Current Biotechnology, 10.2174/2211550111666220830123424, 11 , 2, (158-171), (2022).
- Eric S. Pringle, Brett A. Duguay, Maxwell P. Bui-Marinos, Rory P. Mulloy, Shelby L. Landreth, Krishna Swaroop Desireddy, Stacia M. Dolliver, Shan Ying, Taylor Caddell, Trinity H. Tooley, Patrick D. Slaine, Stephen L. Bearne, Darryl Falzarano, Jennifer A. Corcoran, Denys A. Khaperskyy, Craig McCormick, Thiopurines inhibit coronavirus Spike protein processing and incorporation into progeny virions, PLOS Pathogens, 10.1371/journal.ppat.1010832, 18 , 9, (e1010832), (2022).
- Mariel Kleer, Rory P. Mulloy, Carolyn-Ann Robinson, Danyel Evseev, Maxwell P. Bui-Marinos, Elizabeth L. Castle, Arinjay Banerjee, Samira Mubareka, Karen Mossman, Jennifer A. Corcoran, Human coronaviruses disassemble processing bodies, PLOS Pathogens, 10.1371/journal.ppat.1010724, 18 , 8, (e1010724), (2022).
- Ashok Chakraborty, Anil Diwan, Vijetha Chiniga, Vinod Arora, Preetam Holkar, Yogesh Thakur, Jay Tatake, Randall Barton, Neelam Holkar, Rajesh Pandey, Bethany Pond, Dual effects of NV-CoV-2 biomimetic polymer: An antiviral regimen against COVID-19, PLOS ONE, 10.1371/journal.pone.0278963, 17 , 12, (e0278963), (2022).
- Ki Ho Baek, Donghwan Jang, Taeyoon Kim, Joo Young Park, Dojoon Kim, Sungweon Ryoo, Seunghun Lee, Instant inactivation of aerosolized SARS-CoV-2 by dielectric filter discharge, PLOS ONE, 10.1371/journal.pone.0268049, 17 , 5, (e0268049), (2022).
- Luan D Vu, Shonta Wallace, Anh TQ Phan, Rebecca C Christofferson, Erik Turner, Sean Parker, Karen Elkind-Hirsch, Darrell Landry, Austin Stansbury, Rebecca Rose, David J Nolan, Susanna L Lamers, Michael Hirezi, Beverly Ogden, Stephania A Cormier, Absence of antibody responses to SARS-CoV-2 N protein in COVID-19 vaccine breakthrough cases, Experimental Biology and Medicine, 10.1177/15353702221134097, 247 , 21, (1923-1936), (2022).
- Shambhunath Choudhary, Isis Kanevsky, Soner Yildiz, Rani S. Sellers, Kena A. Swanson, Tania Franks, Raveen Rathnasinghe, Raquel Munoz-Moreno, Sonia Jangra, Olga Gonzalez, Philip Meade, Timothy Coskran, Jessie Qian, Thomas A. Lanz, Jillian G. Johnson, Cassandra A Tierney, Justin D. Smith, Kristin Tompkins, Arthur Illenberger, Paula Corts, Tara Ciolino, Philip R. Dormitzer, Edward J. Dick, Vinay Shivanna, Shannan Hall-Ursone, Journey Cole, Deepak Kaushal, Jane A. Fontenot, Carles Martinez-Romero, Meagan McMahon, Florian Krammer, Michael Schotsaert, Adolfo García-Sastre, Modeling SARS-CoV-2: Comparative Pathology in Rhesus Macaque and Golden Syrian Hamster Models, Toxicologic Pathology, 10.1177/01926233211072767, 50 , 3, (280-293), (2022).
- Concha Ortiz-Cartagena, Laura Fernández-García, Lucia Blasco, Olga Pacios, Inés Bleriot, María López, Rafael Cantón, María Tomás, Reverse Transcription-Loop-Mediated Isothermal Amplification-CRISPR-Cas13a Technology as a Promising Diagnostic Tool for SARS-CoV-2, Microbiology Spectrum, 10.1128/spectrum.02398-22, 10 , 5, (2022).
- Austin B. Featherstone, Amanda Claire Brown, Sapna Chitlapilly Dass, Murine Hepatitis Virus, a Biosafety Level 2 Model for SARS-CoV-2, Can Remain Viable on Meat and Meat Packaging Materials for at Least 48 Hours, Microbiology Spectrum, 10.1128/spectrum.01862-22, 10 , 5, (2022).
- Sebastian J. Conte, Are Intuitions Treated as Evidence? Cases from Political Philosophy*, Journal of Political Philosophy, 10.1111/jopp.12277, (2022).
- Donghwan Jang, Dagyum Lee, Young Chul Shin, Joong Su Lee, Jihee Jung, Sungweon Ryoo, Low molecular weight chitooligosaccharide inhibits infection of SARS‐CoV‐2 in vitro, Journal of Applied Microbiology, 10.1111/jam.15618, 133 , 2, (1089-1098), (2022).
- Zeynep Ece Kuloğlu, Rojbin El, Gulen Guney‐Esken, Yeşim Tok, Zeynep Gülçe Talay, Tayfun Barlas, Mert Ahmet Kuskucu, Özgür Albayrak, Özlem Doğan, Serap Şimşek Yavuz, Kenan Midilli, Önder Ergönül, Füsun Can, Effect of BTN162b2 and CoronaVac boosters on humoral and cellular immunity of individuals previously fully vaccinated with CoronaVac against SARS‐CoV‐2: A longitudinal study, Allergy, 10.1111/all.15316, 77 , 8, (2459-2467), (2022).
- Richard M. Mariita, Amy C. Wilson Miller, Rajul V. Randive, Evaluation of the virucidal efficacy of Klaran UVC LEDs against surface-dried norovirus, Access Microbiology, 10.1099/acmi.0.000323, 4 , 1, (2022).
- Qian Zhang, Daniela Matuozzo, Jérémie Le Pen, Danyel Lee, Leen Moens, Takaki Asano, Jonathan Bohlen, Zhiyong Liu, Marcela Moncada-Velez, Yasemin Kendir-Demirkol, Huie Jing, Lucy Bizien, Astrid Marchal, Hassan Abolhassani, Selket Delafontaine, Giorgia Bucciol, Laurent Abel, Hassan Abolhassani, Alessandro Aiuti, Ozge Metin Akcan, Saleh Al-Muhsen, Fahd Al-Mulla, Gulsum Alkan, Mark S. Anderson, Evangelos Andreakos, Andrés A. Arias, Jalila El Bakkouri, Hagit Baris Feldman, Alexandre Belot, Catherine M. Biggs, Dusan Bogunovic, Alexandre Bolze, Anastasiia Bondarenko, Ahmed A. Bousfiha, Sefika Elmas Bozdemir, Petter Brodin, Yenan Bryceson, Carlos D. Bustamante, Manish J. Butte, Giorgio Casari, John Christodoulou, Roger Colobran, Antonio Condino-Neto, Stefan N. Constantinescu, Megan A. Cooper, Clifton L. Dalgard, Murkesh Desai, Beth A. Drolet, Jamila El Baghdadi, Melike Emiroglu, Emine Hafize Erdeniz, Sara Espinosa-Padilla, Jacques Fellay, Carlos Flores, José Luis Franco, Antoine Froidure, Peter K. Gregersen, Bodo Grimbacher, Belgin Gulhan, Filomeen Haerynck, David Hagin, Rabih Halwani, Lennart Hammarström, James R. Heath, Sarah E. Henrickson, Elena W.Y. Hsieh, Eystein Husebye, Kohsuke Imai, Yuval Itan, Petr Jabandziev, Erich D. Jarvis, Timokratis Karamitros, Adem Karbuz, Kai Kisand, Cheng-Lung Ku, Yu-Lung Lau, Yun Ling, Carrie L. Lucas, Tom Maniatis, Davood Mansouri, László Maródi, Ayse Metin, Isabelle Meyts, Joshua D. Milner, Kristina Mironska, Trine H. Mogensen, Tomohiro Morio, Lisa F.P. Ng, Luigi D. Notarangelo, Antonio Novelli, Giuseppe Novelli, Cliona O'Farrelly, Satoshi Okada, Keisuke Okamoto, Şadiye Kübra Tüter Öz, Tayfun Ozcelik, Qiang Pan-Hammarström, Maria Papadaki, Jean W. Pape, Aslinur Ozkaya Parlakay, Rebeca Perez de Diego, David S. Perlin, Graziano Pesole, Anna M. Planas, Petra Pokorna, Carolina Prando, Aurora Pujol, Lluis Quintana-Murci, Sathishkumar Ramaswamy, Laurent Renia, Igor Resnick, Jacques G. Rivière, Carlos Rodríguez-Gallego, Vanessa Sancho-Shimizu, Anna Sediva, Mikko R.J. Seppänen, Mohammed Shahrooei, Anna Shcherbina, Katerina Slaba, Ondrej Slaby, Andrew L. Snow, Pere Soler-Palacín, Lien De Somer, András N. Spaan, Ivan Tancevski, Stuart G. Tangye, Ahmad Abou Tayoun, Dimitris Thanos, Stuart E. Turvey, K M Furkan Uddin, Mohammed J. Uddin, Diederik van de Beek, François Vermeulen, Donald C. Vinh, Horst von Bernuth, Joost Wauters, Carine Wouters, Aysun Yahsi, Saliha Kanik Yuksek, Mayana Zatz, Pawel Zawadzki, Helen C. Su, Jean-Laurent Casanova, Gulsum Ical Bayhan, Sevgi Keles, Ayca Kiykim, Selda Hancerli, Filomeen Haerynck, Benoit Florkin, Nevin Hatipoglu, Tayfun Ozcelik, Guillaume Morelle, Mayana Zatz, Lisa F.P. Ng, David Chien Lye, Barnaby Edward Young, Yee-Sin Leo, Clifton L. Dalgard, Richard P. Lifton, Laurent Renia, Isabelle Meyts, Emmanuelle Jouanguy, Lennart Hammarström, Qiang Pan-Hammarström, Bertrand Boisson, Paul Bastard, Helen C. Su, Stéphanie Boisson-Dupuis, Laurent Abel, Charles M. Rice, Shen-Ying Zhang, Aurélie Cobat, Jean-Laurent Casanova, Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia, Journal of Experimental Medicine, 10.1084/jem.20220131, 219 , 8, (2022).
- Keven Lothert, Friederike Eilts, Michael W. Wolff, Quantification methods for viruses and virus-like particles applied in biopharmaceutical production processes, Expert Review of Vaccines, 10.1080/14760584.2022.2072302, 21 , 8, (1029-1044), (2022).
- See more

