Towards Development, Maintenance, and Standardized Phenotypic Characterization of Single-Seed-Descent Genetic Resources for Lupins
Saleh Alseekh, Saleh Alseekh, Alisdair R. Fernie, Magdalena Kroc, Magdalena Kroc, Magdalena Tomaszewska, Magdalena Tomaszewska, Katarzyna Czepiel, Katarzyna Czepiel, Elena Bitocchi, Elena Bitocchi, Markus Oppermann, Markus Oppermann, Kerstin Neumann, Kerstin Neumann, Luis Guasch, Luis Guasch, Elisa Bellucci, Elisa Bellucci, Andreas Graner, Andreas Graner, Roberto Papa, Karolina Susek
germplasm
intelligent collections
INCREASE
genetic resources
lupin
plant standardized phenotyping
single-seed descent line
Abstract
Well-characterized genetic resources are fundamental to maintain and provide the various genotypes for pre-breeding programs for the production of new cultivars (e.g., wild relatives, unimproved material, landraces). The aim of the current article is to provide protocols for the characterization of the genetic resources of two lupin crop species: the European Lupinus albus and the American Lupinus mutabilis. Intelligent nested collections of lupins derived from homozygous lines (single-seed descent) are being developed, established, and exploited using cutting-edge approaches for genotyping, phenotyping, data management, and data analysis within the INCREASE project (EU Horizon 2020). This will allow us to predict the phenotypic performance of genotyped lines, and will further boost research and development in lupins. Lupins stand out due to their high-quality seed protein (∼40% of seed dry weight) and other primary components in the seeds, which include fatty acids, dietary fiber, and minerals. The potential of lupins as a crop is highlighted by the multiple benefits of plant-based food in terms of food security, nutrition, human health, and sustainable production. The use of lupins in foods, along with other well-studied and widely used food legumes, will also provide a greatly diversified plant-based food palette to meet the Global Goals for Sustainable Development to improve people's lives by 2030. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Lupin seed phenotypic descriptors
Basic Protocol 2 : Lupin seed imaging
Basic Protocol 3 : Standardized phenotypic characterization of lupin genetic resources grown towards primary seed increase (development of single-seed descent genetic resources)
INTRODUCTION
The biodiversity of lupins (genus Lupinus) is reflected in their complex genome structure (Hufnagel et al., 2020a; Susek et al., 2019), the phenotypic variations of their agricultural traits [e.g., seed variations: (Clements, Dracup, Buirchell, & Smith, 2005)], their biochemical complexity and versatility [e.g., conglutins (Foley et al., 2015) and alkaloids (Święcicki et al., 2019; Wink, Meißner, & Witte, 1995)], and their adaptive potential [e.g., drought tolerance (Annicchiarico, Romani, & Pecetti, 2018) and vernalization (Adhikari, Buirchell, & Sweetingham, 2012)]. Their striking diversity is associated with complex evolutionary events, and it has been significantly shaped by their polyploidy, which might also have promoted the adaptation of lupins to challenging environments. Not surprisingly, lupins also serve as a valuable repository of agrobiodiversity and as novel genetic resources in breeding programs. Moreover, they represent a very attractive food source for humans.
Lupins belong to the basally branching Genistoid clade of papilionoid legumes (Cardoso et al., 2013; Lavin, Herendeen, & Wojciechowski, 2005). Lupinus is a large and diverse genus within the legume family that comprises ∼280 species (Gladstones, 1998), although this number has been extended even further more recently, to up to 1000 species (Kurlovich, 2002). Lupin species are widespread across various climatic zones—which range from subarctic regions (e.g., Alaska), through the Mediterranean and the mountain regions of East Africa and Mexico as well as the Andes and the High Rockies, to subtropical regions of eastern South America—thus also highlighting their adaptation to varied agroclimatic conditions (Gladstones, 1988).
The lupin genus has been geographically separated into two centers of diversity. The species distributed throughout the Mediterranean basin and in North and East Africa form the group of ‘Old World’ lupins, while the species of North and South America constitute the ‘New World’ lupins (Gladstones, 1998). The Old World lupins include 13 to 15 species that are all annual and herbaceous, and these are mostly autogamous species (Gladstones, 1998; Pascual, 2004; Święcicki, Święcicki, & Wolko, 1996). In contrast, the majority of lupins are distributed throughout the New World. These species show diverse growth forms, which include annuals and herbaceous and woody perennials, with compound or unifoliolate leaves, and are well adapted to various ecosystems and climates across wide altitudinal ranges, e.g., coastal dunes, grasslands, and mountains (Aïnouche et al., 2004; Drummond, Eastwood, Miotto, & Hughes, 2012).
Four lupin species have achieved agricultural importance (for review, see Wolko, Clements, Naganowska, Nelson, and Yang, 2011). Among these, there are three Old World lupins that are considered novel foods by the European Commission (https://ec.europa.eu/food/safety/novel_food/catalogue_en): Lupinus albus L. (white lupin); Lupinus luteus L. (yellow lupin); and Lupinus angustifolius L. (narrow-leafed lupin). Among the New World lupins, only one species, Lupinus mutablis (tarwi), is an important food crop.
EVOLUTION, DOMESTICATION, AND USE FOR HUMAN FOOD
Recently, it was confirmed that the genus Lupinus originated in the Old World, and was subsequently dispersed throughout the New World (Drummond et al., 2012). Lupins have been divided into three basic lineages (Drummond et al., 2012): the Mediterranean and North African species (4.6–12.5 million years; Ma), which are sisters to the unifoliolate species from eastern North America (0.1–2.4 Ma); the eastern South American species (2.3–7.1 Ma), which are sisters to the Texas bluebonnets from eastern North America (0.1–2.3 Ma); and the western New World species (5.0–13.2 Ma), which include the Andean and Mexican species (1.19–3.50 Ma) that are derived from a paraphyletic assemblage of western North American species (2.1–5.5 Ma).
Whole-genome triplication appears to have been the main process involved in lupin evolution (Hane et al., 2016; Kroc, Koczyk, Swiecicki, Kilian, & Nelson, 2014; Xu et al., 2020), which has included multiple chromosome rearrangements (Susek et al., 2019; Susek, Bielski, Hasterok, Naganowska, & Wolko, 2016) and epigenetic changes (Susek et al., 2017). However, of particular note, the western New World lupins show an exceptionally high rate of diversification, with no evidence that polyploidy has had any role in the diversification of the species in this clade (Nevado, Atchison, Hughes, & Filatov, 2016). For this group of lupins, a rapid rate of speciation is believed to have been strongly associated with their transition from an annual to a perennial life history, along with their colonization of higher altitudes. This also enabled exploitation of novel ecological opportunities (Drummond et al., 2012; Nevado et al., 2016).
Old World and New World lupins were domesticated independently, with the incorporation of key traits of the domestication syndrome, such as non-shattering pods, permeable seed coats, and large seeds (Atchison et al., 2016). Lupinus albus (white lupin; annual, 2 n =50; genome size, ∼580 Mbp) dates back to the times of the ancient Greeks and Romans. At that time, in 1000-800 BC, farmers used white lupin for soil improvement and crop rotation while also using the seeds for food and animal feed. They were the first who selected for large permeable seeds and non-shattering pods. Greece and the Balkan Peninsula are considered to have the greatest diversity of L. albus and its wild subspecies (subsp. graecus , subsp. termis , subsp. albus) and cultivated types (i.e., landraces) (Gladstones, 1998). Lupinus mutabilis (tarwi, Andean lupin, pearl lupin; annual, 2 n =48; genome size, ∼930 Mbp) is considered to have been primarily domesticated in the Andes between 1800 and 2600 BC (Atchison et al., 2016). The latest data suggest that tarwi was domesticated in the highlands of northern Peru (most likely the Cajamarca region), and that Lupinus piurensis is the likely progenitor of tarwi (Atchison et al., 2016). The history of tarwi domestication inferred from demographic analyses also suggests that after the split from its progenitor, it went through a classical domestication bottleneck, with subsequent rapid population expansion as a widely cultivated species across the Andes (Atchison et al., 2016). Nowadays, L. mutabilis is only known in cultivation, with no wild populations discovered to date. The South American Indians used L. mutabilis as food (after de-bittering), and as green manure and medicine (e.g., for cardiac disease, rheumatism, malaria) (Bebeli et al., 2020). Interestingly, the seed size increased two-fold (i.e., doubled) during the domestication of L. mutabilis (Atchison et al., 2016).
Lupinus albus and L. mutabilis have therefore been traditional food legumes in the Mediterranean regions and the Andes, respectively, for thousands of years (Cowling, Buirchell, & Tapia, 1998). The seeds of L. albus and L. mutabilis are relatively large, and across lupins in general, they are characterized by the highest protein content (up to 38%, 41%-51% of seed dry weight, respectively) and oil content (9%-13%, 14%-24% of seed dry weight, respectively), and can thus be used as dual-purpose, providing both protein and oil for human consumption (Gresta et al., 2017; Gulisano, Alves, Martins, & Trindade, 2019). Moreover, the oil from L. albus and L. mutabilis seeds is of high quality, with a high proportion of unsaturated fatty acids, low erucic acid, and a long shelf life (Cowling et al., 1998; Rybiński et al., 2018). The main anti-nutritional factor of lupin seeds is the alkaloid content (Wink et al., 2010). However, owing to intensive breeding efforts, the seed alkaloid contents of modern lupin cultivars are often lower than the accepted industry threshold (Kamel, Święcicki, Kaczmarek, & Barzyk, 2016; Kroc et al., 2017).
Many potential health benefits have been defined for the consumption of lupin seeds, including the cholesterol-lowering activity of the fiber, the protection against cardiovascular disease by the tocopherols, and the activity of the conglutin-γ protein fraction (Arnoldi, Boschin, Zanoni, & Lammi, 2015; Boschin & Arnoldi, 2011; Fontanari, Batistuti, Cruz, Saldiva, & Arêas, 2012).
LUPIN GENOMICS AND GERMPLASM DIVERSITY
Lupin genetics, genomics, and germplasm resources lag far behind those of other major crops. Substantial progress has been made recently in the molecular characterization of the L. albus transcriptome (Secco, Shou, Whelan, & Berkowitz, 2014; Wang et al., 2014), with two genome assemblies of cultivar ‘Amiga’ (Hufnagel et al., 2020a; Xu et al., 2020) and a pangenome assembly (Hufnagel et al., 2020b). These provide basic resources for studies into the biology and breeding of this species, and will allow future investigations into the influence of white lupin domestication on its genomic variability (Hufnagel et al., 2020b). On the other hand, L. mutabilis remains an under-studied crop, which still lacks extensive genomics resources. Recently, a large genome-wide DNA polymorphism dataset was generated for L. mutabilis using ‘next restriction site associated DNA sequencing’ (nextRADseq; Atchison et al., 2016). L. mutabilis was also included in a chloroplast genome investigation that provided comparative analysis and further resolution of phylogenies within Lupinus (Keller et al., 2017). The genetic diversity within L. mutabilis and its relation to other lupin species was illustrated with aid of molecular markers (Chirinos-Arias, Jiménez, & Vilca-Machaca, 2015; Olczak, Rurek, Jańska, Augustyniak, & Sawicka-Sienkiewicz, 2001; Talhinhas, Neves-Martins, & Leitao, 2003).
Data on the variations within the genetic resources for white lupin and tarwi are scarce. A set of target phenotypic traits and phenology showed variations in both wild and domesticated white lupins that were grouped into seven clusters (Berger, Shrestha, & Ludwig, 2017). Here, the number of days to flowering varied from 65 days for an average Mediterranean climate (cluster 5) to 70 days for the cooler long-season Iberian climate with higher rainfall (cluster 2). White lupins have been defined in the context of large seeds, high early vigor, rapid growth rates, plant heights, harvest indices, and seed and biological yields (Berger et al., 2017), along with their characterization according to differences in the quality of their seed contents.
A valuable source of Ethiopian white lupin variations has been described, where about 500 white lupin genotypes were collected and are conserved at the Ethiopia Biodiversity Institute (Beyene, 2020). Molecular analyses of 212 Ethiopian landraces revealed the high genetic diversity, and highlighted distinct gene pools (Atnaf et al., 2017). A local, high-alkaloid, white lupin variety has also been described that is grown in north-western Ethiopia, which is also partially non-shattering, high-yielding, and most importantly, resistant to lupin anthracnose disease (Yeheyis, Kijora, Melaku, Girma, & Peters, 2010). In addition, 25 landraces in north-western and southern Ethiopia were characterized by their differences in days after sowing for emergence time, first flowering, 50% flowering, and maturity, along with plant height (Beyene, 2020). Variations in landraces of white lupin have also been reported to include their yield characteristics, as numbers of pods per plant, numbers of seeds per pod, and pod length, and also in terms of their protein content. Some other seed traits are not significantly different across these Ethiopian landraces (e.g., 100-seed weight, seed length, and width; Beyene, 2020).
The genetic diversity of L. mutabilis is shown by the phenotypic variations in its flower, stem, and seed colors, and its indeterminate growth. The variations in seed traits relate to their shape (from lenticulate to spherical) and primary color (from pearly white to dark, with some intermediates), along with various patterns of pigmentation. However, most L. mutabilis cultivar accessions of Andean germplasm collections have a pearl white color (i.e., 95%). Lupins with a dark seed color also have darker flowers, which suggests that the white color is recessive (Gulisano et al., 2019). Seed protein and oil content were shown to vary among 149 Peruvian tarwi accessions, which suggested that the seeds of the taller and larger plants tended to be richer in protein, while those of the smaller plants were richer in oil (Neves Martins, Talhinhas, & De Sousa, 2016). Of note, an analysis of the variations across 23 L. mutabilis accessions in terms of their combined genetic and genomic characteristics and morphological traits revealed variations in many traits, such as time to flowering, plant height, seed weight, and number of pods (Guilengue, Alves, Talhinhas, & Neves-Martins, 2020). It has been suggested that indeterminate and determinate plants are adaptive to interannual meteorological variations under southern (Mediterranean) and northern and central European conditions, respectively (Neves Martins et al., 2016).
The white and tarwi lupin germplasm collections that are conserved in genebanks across the world generally include both cultivated material and wild populations. Nonetheless, these materials have not been well characterized. Most accessions indicate the country of origin only, with the other ‘passport’ data limited or missing. According to the European Search Catalogue for Plant Genetic Resources database (EURISCO Catalogue; http://eurisco.ecpgr.org, 2020-12-02), the greatest number of white lupin genetic resources are conserved in Portugal (927 accessions), Ukraine (750), Spain (743), the Russian Federation (501), Germany (352), Poland (298), and Italy (211). Among the countries outside Europe, the largest white lupin collections are held in Australia (890) and the U.S.A. (457) (Global Portal on Plant Genetic Resources; https://www.genesys-pgr.org, 2020-12-02).
For L. mutabilis , the most numerous genetic resources in Europe are conserved in Germany (695 accessions), Portugal (150), and the Russian Federation (132) (http://eurisco.ecpgr.org, 2020-12-02). The largest and most relevant germplasm collections of tarwi outside Europe are held mainly in the U.S.A. (102) and Australia (74) (https://www.genesys-pgr.org, 2020-12-02), and also Peru, Ecuador, and Bolivia. However, reports suggest that much of the L. mutabilis diversity remains uncollected (Gulisano et al., 2019; Jacobsen & Mujica, 2008), and little is known about the genetic variability of these collections (Guilengue et al., 2020).
DEVELOPMENT AND MAINTENANCE OF THE LUPIN INTELLIGENT COLLECTION: INCREASE
The need to meet the global challenges for the management and use of genetics resources was very recently, and urgently, emphasized by Mccouch et al. (2020). They highlighted that joint efforts are crucial for efficient and expeditious characterization and use of agrodiversity, through the establishment of the necessary platforms to empower genebank managers, researchers, breeders, and farmers to more effectively use genetic variation not just for research but also for accelerated crop improvement and sustainable production. The progress and shift in research strategies from traditional plant phenotyping to combined phenotypic and genotypic studies (e.g., association studies, using the whole genome) now pave the way for the accurate and comprehensive definition of this diversity.
To date, there has been limited information available on lupin phenotypic descriptors, and even when available, the data have generally only provided basic descriptive information. The exception here is the Agricultural Research Service of the U.S. Department of Agriculture, where some images of seeds are already included in the collection, although without any protocol to define how to image and analyze the seed traits (https://npgsweb.ars-grin.gov/gringlobal/taxon/taxonomydetail?id=22802). According to the International Treaty on Plant Genetic Resources for Food and Agriculture, only 644 white lupin and 706 tarwi accessions have digital object identifiers (DOIs), with these mainly registered by genebanks, e.g., the Leibniz Institute of Plant Genetics and Crop Plant Research in Germany (https://ssl.fao.org/glis).
The INCREASE project gets its name from “In telligent C ollections of Food Legume Genetic R esources for E uropean A grofood S ystems,” and is funded by the European Union as part of Horizon 2020 (Bellucci et al., 2021). This has provided the unique opportunity to develop phenotyping data and to integrate these with genotypic information for thousands of lupin accessions from different sources (e.g., genebanks, research institutes, private companies, project stakeholders). The protocols for exploring the variations within lupins and other food legumes studied within INCREASE (i.e., also chickpea, common bean, lentil; see Current Protocols articles: Guerra-García, Gioia, Von Wettberg, Logozzo, & Bett, in preparation; Kumar et al., in preparation; and Cortinovis et al., 2021) are being provided to enhance the standardization and data handling of the available genetic resources of these legumes and to increase the effectiveness of their conservation and use (https://www.pulsesincrease.eu).
Thus, a set of lupin nested core collections is being established within the INCREASE project, as the Reference-core (R-core), Training-core (T-core), and Hyper-core (H-core) collections. These will be multiplied, characterized genetically and/or phenotypically, and conserved for further use. The wide range of phenotypic traits will be defined and analyzed to provide the set of genotypes that show the phenotypic diversity, to group genotypes according to their similarities and dissimilarities, and to integrate these with various types of genetic and genomic data.
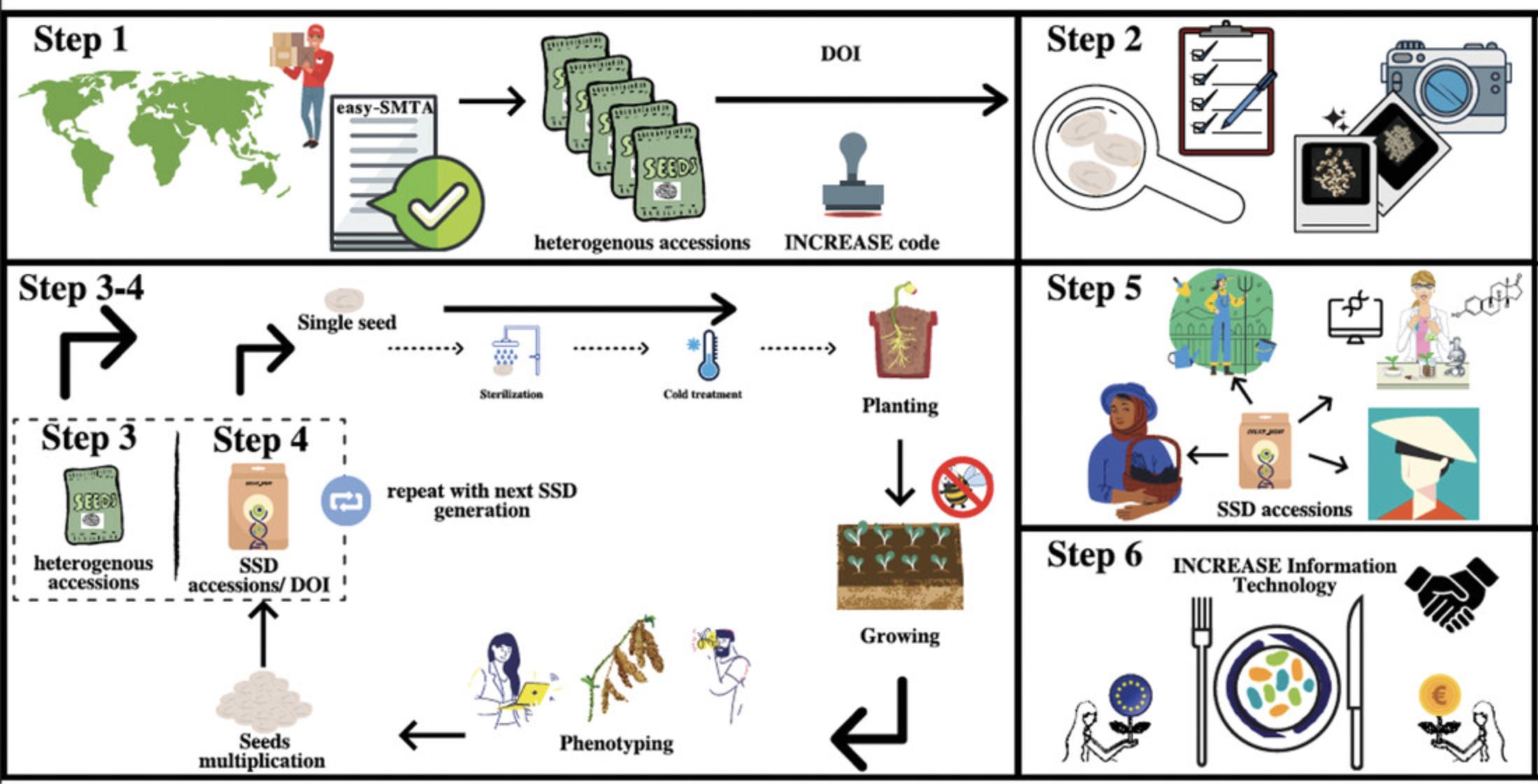
In the present study, we describe the procedures that are established and implemented in the INCREASE project to develop lupin Intelligent Collections. These protocols are also recommended for use in genebanks and research institutions and are aimed at the development of single-seed descent (SSD) lines for conservation and maintenance of seeds. The protocols will facilitate the characterization of lupin genetic resources and the integration of the data obtained into both centralized and decentralized systems, which will eventually be accessible to each end user. We have developed the basic protocols for the lupin primary seed increase based on Lupin Descriptors (International Board for Plant Genetic Resources, 1981; https://www.bioversityinternational.org/fileadmin/bioversity/publications/Web_version/103/ch1.htm) and Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability (International Union for the Protection of New Varieties of Plants, 2004; https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.191/attachments/tg066.pdf). Our basic protocol for lupin seed imaging is based on Current Protocols article Cortinovis et al. (2021). The scheme for the development of the SSD lines and seed multiplication cycles, as well as for the sharing and management of the genetic resources within the INCREASE project, is illustrated in Figure 1. This workflow includes the following steps:
- Step 1 : List of genetic resources. The basic initial step is the preparation of the list of accessions obtained from various providers, and to order these with their unique INCREASE codes, so as to be able to manage the accessions properly and avoid any mistakes while sharing seeds. Moreover, DOIs will be assigned to the accessions that are selected for the R-core, by registering them on the Global Information System of the International Treaty on Plant Genetic Resources for Food and Agriculture (https://ssl.fao.org/glis). It is essential to share the seeds under easy Standard Material Transfer Agreements or policies.
- Step 2 : Characteristics of the seeds from heterogeneous accessions. The seeds indicated for the development the SSD lines are characterized according to Basic Protocol 1 (Lupin seed phenotypic descriptors), with their image analysis using Basic Protocol 2 (Lupin seed imaging).
Of importance: Pictures of the original seeds (heterogeneous material) used to develop these SSD lines, as well as the SSD seeds at the subsequent selfing cycles, should be archived for further comparison between the originals and the next generations of SSD genotypes.
- Step 3 : Development of SSD lines. One single seed of each heterogeneous accession is chosen at random to be grown under insect-free conditions (i.e., greenhouse).
Recommended: Seeds are surface-sterilized to avoid potential development of pathogen-caused diseases, and are cold-treated for 21 days at 6° to 8°C to induce flowering in genotypes responsive to vernalization. Of importance: The phenotypic observations for each genotype are carried out for each cycle of the SSD line development to compare the phenotypic traits among the SSD generations, using Basic Protocol 3 (Standardized phenotypic characterization of lupin genetic resources grown for primary seed increase—development of SSD genetic resources).
- Step 4 : The multiplication cycles. One single seed from the first SSD line is chosen at random to develop the second selfing cycle, according to Step 3, above (same approach should apply in the subsequent selfing cycle; e.g., to produce a third cycle of SSD lines, the single seed from the second round of selfing is chosen at random). Seed characterization and imaging are carried out as described for Step 2.
- Step 5 : SSD line sharing and conservation. The SSD lines developed are shared among different genebanks (and stakeholders) to multiply the seeds themselves, using the INCREASE lupin protocols, and to integrate and compare the data provided in the reciprocal approaches.
- Step 6 : Data integration. A large amount of phenotypic information from each genotype for each particular seed selfing cycle will be obtained. These data will be integrated with large-scale genotypic data into the INCREASE information technology system, so that the data can be stored, shared, and explored by multiple users through an open and publicly available web portal.
Basic Protocol 1: LUPIN SEED PHENOTYPIC DESCRIPTORS
This protocol describes the assessment of the seed morphological traits prior to each seed multiplication cycle. The seed traits are recorded at the beginning of each seed increase cycle, starting from the first cycle of SSD line development using the heterogeneous genotypes. These assessments will enable comprehensive characterization of seeds from different lupin genetic resources, as well as detection and elimination of errors that might occur during the SSD cycles.
Materials
- Seeds of lupin genetic resources
- Validated measuring devices, such as a ruler
- A standardized color scale (i.e., ColorChecker Classic Nano, X-Rite)
- Template file (spreadsheet)
NOTE : The seed traits listed below should be recorded at the beginning of each primary seed multiplication cycle.
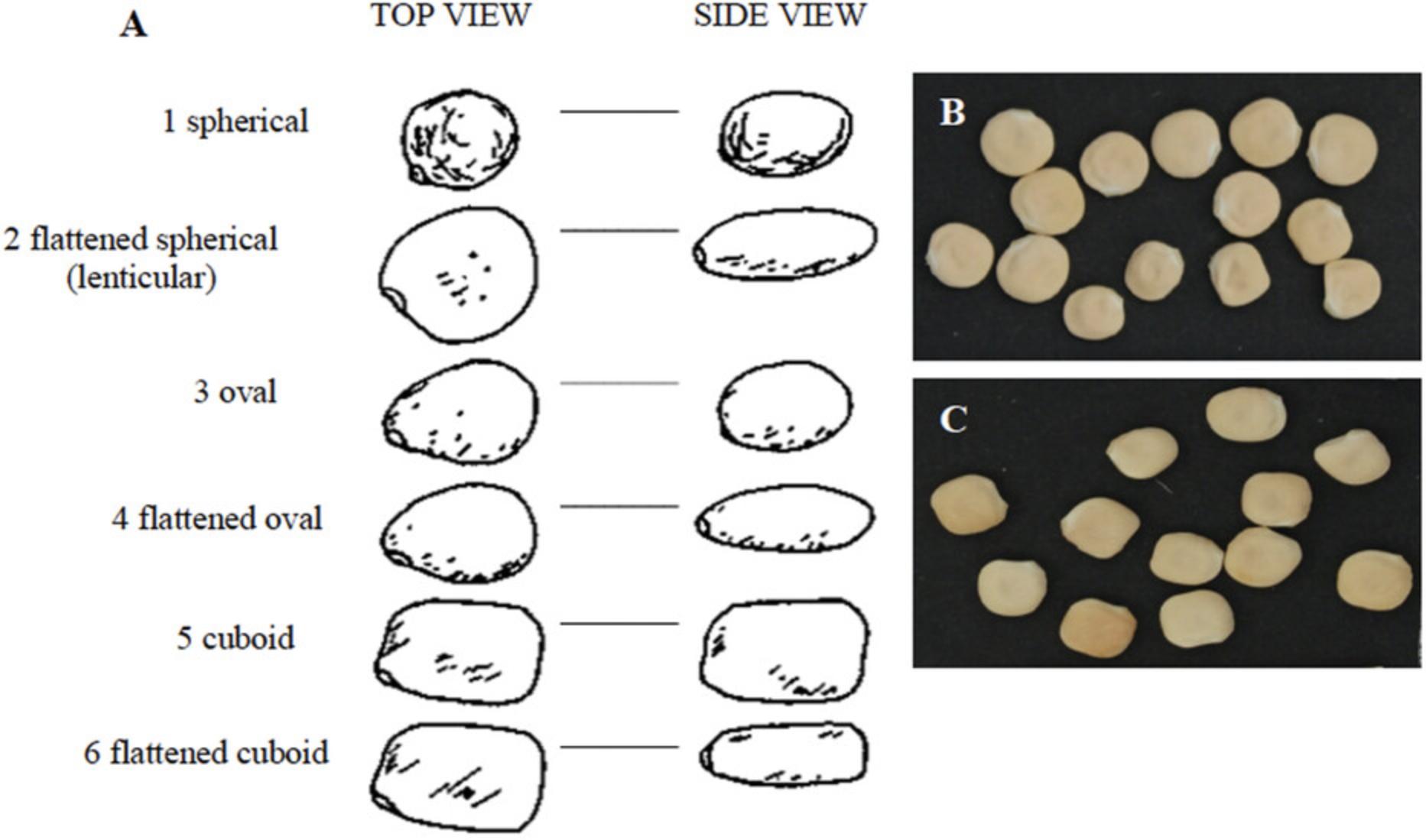
1.Take at least five seeds from each accession (heterogeneous materials or SSD lines), and through visual assessment record the Seed Shape , according to the following categories (Fig. 2):
- 1 = spherical
- 2 = flattened spherical (lenticular)
- 3 = oval
- 4 = flattened oval
- 5 = cuboid
- 6 = flattened cuboid
2.Take at least five seeds from each accession (heterogeneous materials or SSD lines), and through visual assessment record the Seed Primary Color , according to the following categories:
- 1 = white
- 2 = brown
- 3 = other
The seed primary color is the dominating basal color of the seed.
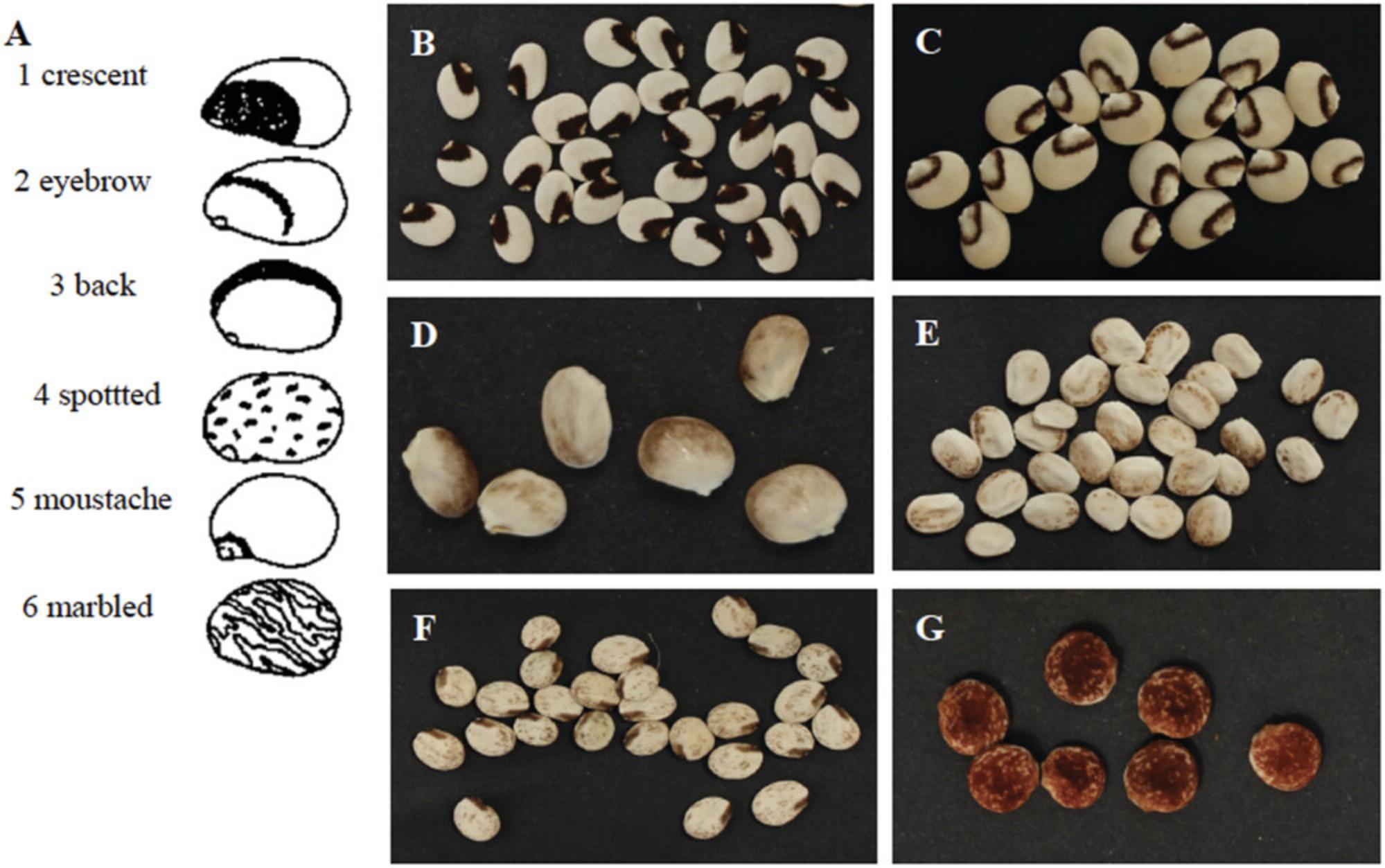
3.Take at least five seeds from each accession (heterogeneous materials or SSD lines), and through visual assessment record the Seed Ornamentation , according to the following categories (for example of lupin seed ornamentation, see Fig. 3):
- 1 = crescent
- 2 = eyebrow
- 3 = back
- 4 = spotted
- 5 = moustache
- 6 = marbled
- 7 = marbled crescent
- 8 = marbled plus eyebrow
- 9 = spotted plus eyebrow
- 10 = spotted plus moustache
- 11 = other
Ornamentation refers to well-defined seed coat pattern that is different from the primary color. It should be assessed at full maturity of the seeds.
4.Take at least five seeds from each accession (heterogeneous materials or SSD lines), and through visual assessment record the Seed Ornamentation Color , according to the following categories:
- 1 = gray
- 2 = light brown
- 3 = dark brown
- 4 = black
- 5 = other
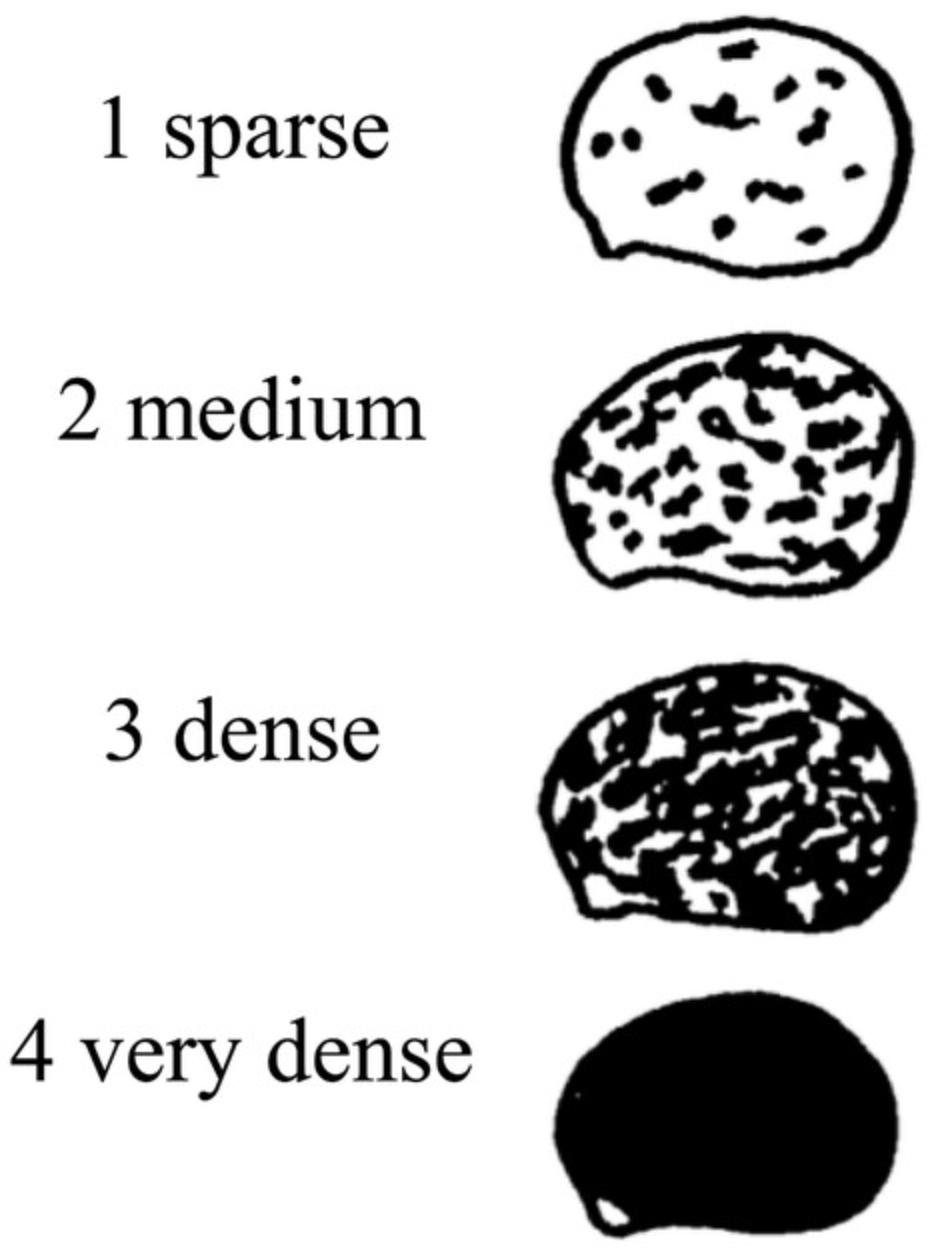
Density of seed ornamentation
5.Take at least five seeds from each accession (heterogeneous materials or SSD lines), and through visual assessment record the Density of Seed Ornamentation , according to the following categories (Fig. 4):
- 1 = sparse
- 2 = medium
- 3 = dense
- 4 = very dense
6.Take at least five seeds from each accession (heterogeneous materials or SSD lines), and through visual assessment record the Seed Quality , i.e., visibly healthy seeds, not affected by any pest or disease.
Basic Protocol 2: LUPIN SEED IMAGING
This protocol describes the best-practice standards for seed imaging within the INCREASE project. The images of the seeds are archived prior to each seed multiplication cycle, starting from the first cycle of SSD line development using the heterogeneous genotypes. Such records will serve as a documentation of seed quality, as well as shape, color, and size of seeds, and might be an input for automated image analysis.
Materials
- Seeds of lupin genetic resources
- Validated measuring devices, such as a ruler
- A standardized color scale (i.e., ColorChecker Classic Nano, X-Rite)
- Uniform background
- Digital camera (≥20 MP resolution)
1.Take at least five seeds from each accession (heterogeneous materials or SSD line).
2.Place the seeds on the uniform background.
3.Place the seeds close to the ruler.
4.Make sure that the seeds are well separated from each other.
5.Place the standardized color scale to provide color quality control and enable automated image post-processing.
6.Add accession label or leave enough space for the subsequent insertion of the label information and the barcode (QR code).
7.Take a picture of each accession.
8.Transfer the finalized images to a central repository for the project.
Basic Protocol 3: STANDARDIZED PHENOTYPIC CHARACTERIZATION OF LUPIN GENETIC RESOURCES GROWN FOR PRIMARY SEED INCREASE (DEVELOPMENT OF SSD GENETIC RESOURCES)
Here we describe the protocol to phenotypically characterize the lupin genetic material grown under controlled conditions, for primary seed increase. The protocol should be applied at the stage of SSD development from heterogeneous material, as well as for further selfing cycles. Collected data will be uploaded into the database of the project and integrated with the results obtained for the same line in other experiments (e.g., field trials, molecular characterization), thus serving as a comprehensive information resource.
Materials
- L. albus and L. mutabilis plants grown in insect-free conditions
- The following conditions are offered for guidance in providing optimal growing conditions for lupins: pod capacity, 7.5 L; potting medium, peat and vermiculite mixture (1:1 by volume); day and night temperatures of 22°C and 18°C, respectively; at last 14- to 16-hr photoperiod; air humidity, 60%–65%; plant watering as required, average, 200 ml per pot, twice a week; the commercial rhizobia inoculum treatment might be applied prior to sowing (to the seeds or soil); during the flowering period plants to be fertilized once a week; biological pest controls as well as protective biological products (e.g., mites, microorganisms) to be used through the whole experimental period; chemical pest control to be used when other attempts have failed.
- Validated measuring devices, such as a meter stick or a ruler
- Data collection template file (spreadsheet)
NOTE : Prior to sowing, it is recommended to surface-sterilize the seeds to avoid potential development of pathogen diseases. As this approach weakens the seed coat, it might serve as an scarification treatment to stimulate germination. Soak the seeds in 70% ethanol (30 s for L. albus , 20 s for L. mutabilis); rinse the seeds under tap water; soak the seeds in 1.5% sodium hypochlorite (4 min for L. albus , 1 min for L. mutabilis); rinse the seeds under sterile water; and put the seeds into sterile petri dishes and then in the refrigerator.
1.Prior to protocol implementation, collect the following information on the experimental site:
- Insect-free conditions: greenhouse/tunnel/ growth chamber
- Growing conditions (day/night temperature, photoperiod, humidity, watering)
- Location of experimental trial
- Latitude of experimental trial
- Longitude of experimental trial
- Altitude of experimental trial
- Data collector name
- Time of collection of the data (year/season)
- Number of generations from collection of the original seeds
- Type of INCREASE Intelligent Collections
The following mandatory traits need to be recorded. These traits are essential to the lupin INCREASE project, and the data should be collected for all accessions of the INCREASE Intelligent Collections (i.e., R-core, T-core, H-core).
2.Record the Dates of Cold Treatment (dd/mm/yyyy).
3.Record the Date of Sowing the seeds (dd/mm/yyyy).
4.Record the Date of Emergence , when the cotyledons emerge from the soil and open with their leave visible (dd/mm/yyyy; Fig. 5).

5.Record the Date of First Bud Occurrence on the Main Stem , when bud becomes clearly visible (dd/mm/yyyy ; Fig. 6A).

6.Record the Date of First Bud Occurrence on the Lateral Branches , when bud becomes clearly visible (dd/mm/yyyy).
7.Record the Date of First Petal Occurrence on the Main Stem (dd/mm/yyyy ; Fig. 6B).
8.Record the Date of First Petal Occurrence on the Later Branches (dd/mm/yyyy).
9.Record the Date of First Flower Opened on the Main Stem (dd/mm/yyyy).
10.Record the Date of First Flower Opened on the Lateral Branches , at any node (dd/mm/yyyy).
11.Through visual assessment, record the dominant Color of Wing Petal in the Just-Opened Flower on the Main Stem , according to the following categories:
- 1 = white
- 2 = white/blue; white color is more expanded than blue
- 3 = blue
- 4 = blue/white; blue color is more expanded than white
- 5 = white/violet; white color is more expanded than violet
- 6 = violet
- 7 = violet/white; violet color is more expanded than white
- 8 = white/pink; white color is more expanded than pink
- 9 = pink
- 10 = pink/white; pink color is more expanded than white
12.Through visual assessment, record the dominant Color of Standard Petal in the Just-Opened Flower of the Main Stem , according to the following categories:
- 1 = white
- 2 = white/blue; white color is more expanded than blue
- 3 = blue
- 4 = blue/white; blue color is more expanded than white
- 5 = white/violet; white color is more expanded than violet
- 6 = violet
- 7 = violet/white; violet color is more expanded than white
- 8 = white/pink; white color is more expanded than pink
- 9 = pink
- 10 = pink/white; pink color is more expanded than white
13.Through visual assessment, record the Color of the Keel in Just-Opened Flower on the Main Stem (observed in L. mutabilis only) , according to the following categories:
- 1 = white
- 2 = white/blue; white color is more expanded than blue
- 3 = blue/white; blue color is more expanded than white
14.Record the Date of Full Flowering on the Main Stem , when 98% to 100% of the flowers of the inflorescences have opened (dd/mm/yyyy ; Fig. 6D).
15.Through visual assessment, record the dominant Color of Wing Petals at Full Flowering on the Main Stem , according to the following categories:
- 1 = white
- 2 = white/blue
- 3 = blue
- 4 = blue/white
- 5 = white/violet
- 6 = violet
- 7 = violet/white
- 8 = white/pink
- 9 = pink
- 10 = pink/white
16.Through visual assessment, record the dominant Color of Standard Petal at Full Flowering on the Main Stem , according to the following categories:
- 1 = white
- 2 = white/blue
- 3 = blue
- 4 = blue/white
- 5 = white/violet
- 6 = violet
- 7 = violet/white
- 8 = white/pink
- 9 = pink
- 10 = pink/white
17.Through visual assessment, record the Color of the Keel Edge at Full Flowering on the Main Stem , according to the following categories:
- 1 = transparent
- 2 = white
- 3 = white/ blue
- 4 = slight blue hue
- 5 = intensely colored—black
18.Through visual assessment, record the Type of Keel Edge on the Main Stem , according to the following categories:
- 1 = curved
- 2 = straight
- 3 = enclosed
19.Record the Date of First Flower Withering on the Main Stem (dd/mm/yyyy).
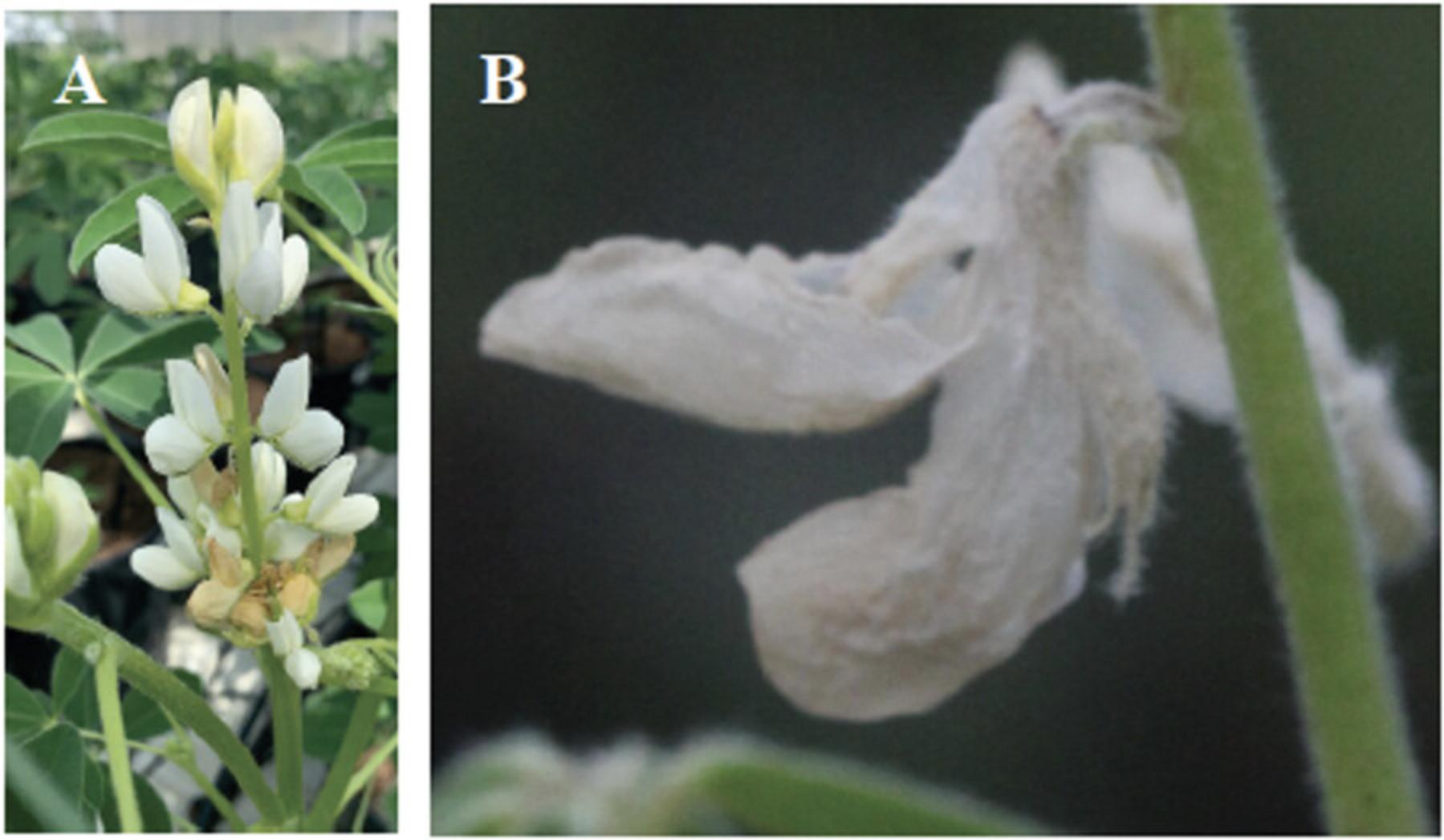
20.Through visual assessment, record the Color of Keel Edge at Flower Withering on the Main Stem , according to the following categories:
- 1 = transparent
- 2 = white
- 3 = white/ blue
- 4 = slight blue hue
- 5 = intensely colored—black
21.Through visual assessment, record the Purple Pigmentation of the Main Stem at 2 months and 4 months after emergence, according to the following categories:
- 1 = none
- 2 = slightly
- 3 = medium intensity—more than half of the stem is purple
- 4 = very intense—stem and internodes are purple
- 5 = no pigment in stem plus purple internodes
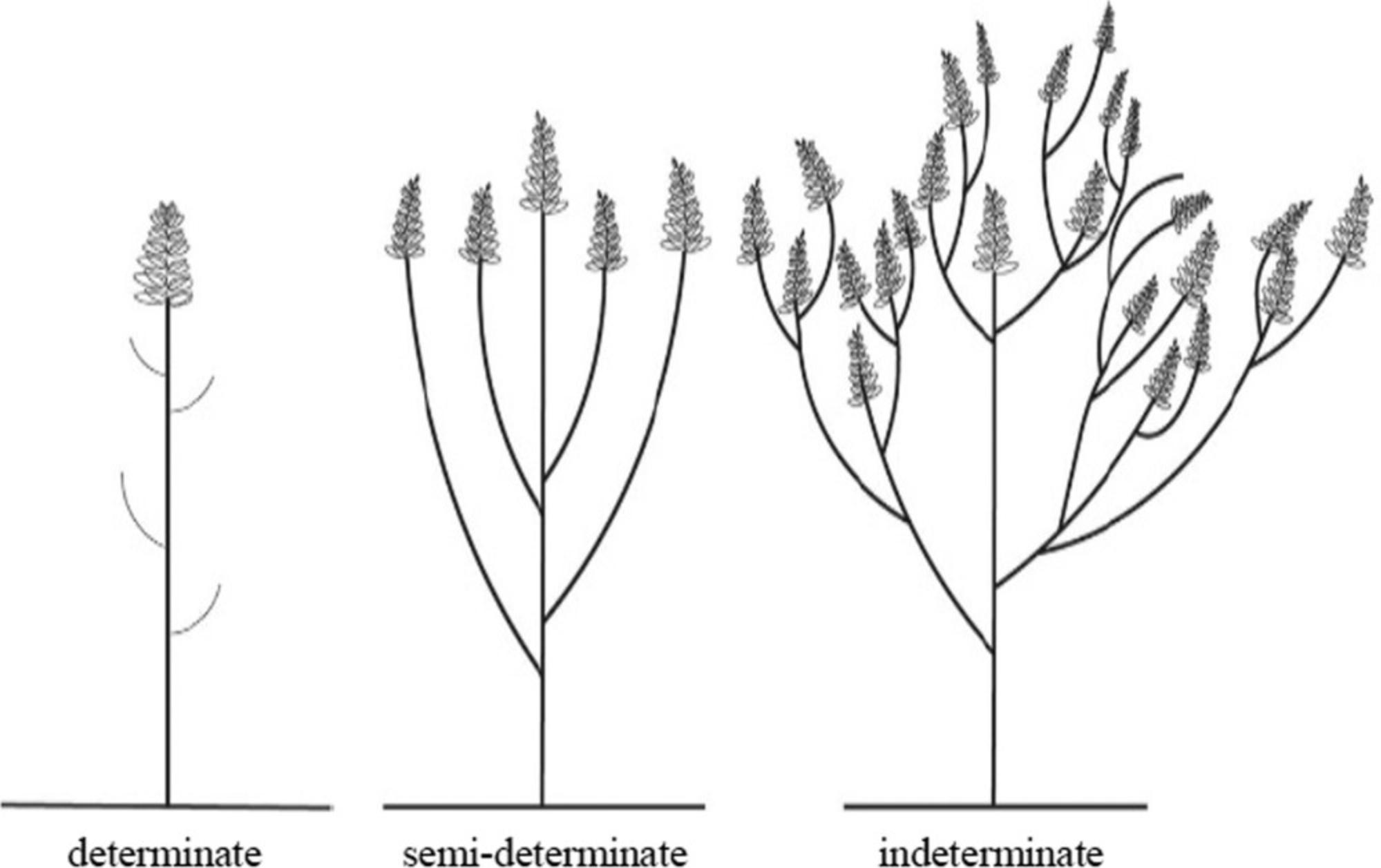
22.At flower withering, record the Growth Habit , according to the following categories (Fig. 8):
1.determinate 2.semi-determinate 3.indeterminate
Determinate means determinate inflorescence on the main stem. Indeterminate means lateral branches develop along the main stem with further branching. The secondary lateral branches grow higher than the primary lateral branches, and so on. Flowers develop in compact inflorescences at the tops of branches. The expression is the same in the spring type and winter type.
23.Record the Date of First Pod Set on the Main Stem (dd/mm/yyyy).
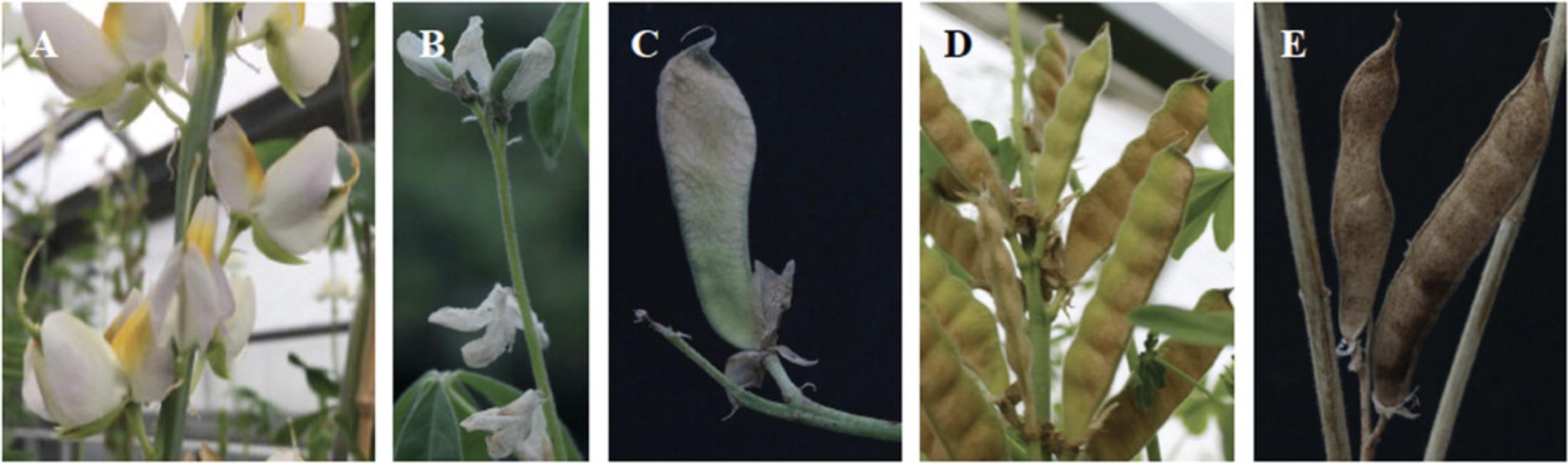
24.Record the Date of First Pod set on the Lateral Branches (dd/mm/yyyy).
25.Record the Date of First Pod Color Changes on the Main Stem (dd/mm/yyyy).
26.Record the Date of First Pod Color Changes on the Lateral Branches (dd/mm/yyyy).
27.At the stage of harvesting, through visual assessment, record the Pod Shattering on the Main Stem , according to the following categories:
- 1 = none
- 2 = slight
- 3 = moderate
- 4 = complete
28.At the stage of harvesting, through visual assessment, record the Pod Shattering on the Lateral Branches , according to the following categories:
- 1 = none
- 2 = slight
- 3 = moderate
- 4 = complete
29.Record the Date of Harvesting Pods on the Main Stem (dd/mm/yyyy), i.e., when the pods have dried and turned a golden-brown color.
30.Record the Date of Harvesting Pods on the Lateral Branches (dd/mm/yyyy), i.e., when the pods have dried and turned a golden-brown color.
31.Record the Plant Height of the Main Stem (in cm) from the ground level to the lowest end of the first ripe pod, at the stage of harvesting.
32.Record the Total Plant Height (in cm) from the ground level to the highest point of the plant, including lateral branches, at the stage of harvesting.
33.Record the Number of Pods per Plant on the Main Stem of each plant.
34.Record the Number of Pods per Plant on the Lateral Branches of each plant.
35.Record the Number of Qualified Seeds on the Main Stem of each plant, i.e., healthy, well-developed, non-infected.
36.Record the Number of Qualified Seeds on the Lateral Branches of each plant, i.e., healthy, well-developed, non-infected.
37.Record the Total Number of Qualified Seeds from each plant from both the main stem and the lateral branches, i.e., healthy, well-developed, non-infected.
38.Record the Total Mass of the Qualified Seeds harvested from each plant.
39.Through visual assessment, record the Presence of any Disease and/or Pathogen , and if possible, describe it or make notes according to the following categories:
- 1 = thrips
- 2 = powdery mildew
- 3 = aphids
- 4 = anthracnose
- 5 = fusarium
- 6 = virus
- 7 = other
Based on the mandatory data recorded in Basic Protocol 3 (steps 1-39), the following traits can be estimated
40.Based on the recorded dates, estimate the Days to Emergence based on the recorded date of sowing.
41.Based on the recorded dates, estimate the Length of Vegetative Phase /Days to Floral Time based upon the recorded date of sowing and the first bud occurrence on the main stem.
42.Based on the recorded dates, estimate the Length of the Generative Phase on the Main Stem :
- Count from the first bud occurrence to the first flower wither
- Count from date of pod set to pod color changes
The generative phase begins when the flower bud becomes visible and ends when the first flower has begun to wither and the pod color changes to khaki on the main stem.
43.Based on the recorded dates, estimate the Length of the Pod and Seed Growth Phase on the Main Stem (physiological maturity):
- Count from date of pod set to harvest
- Count from date of pod color change to harvest
The pod and seed growth phase begins when a pod is set (reaches 8-10 mm) and ends when the pod changes color (from green, through khaki, to light brown).
44.Based on the recorded dates, estimate the Length of the Pod and Seed Growth Phase on the Lateral Branches (physiological maturity):
- Count from date of pod set to harvest
- Count from date of pod color change to harvest
The pod and seed growth phase begins when a pod is set (reaches 8-10 mm) and ends when the pod changes color (from green, through khaki, to light brown).
COMMENTARY
Conclusions and Future Prospects
Comprehensive characterization, management, and use of the genetic resources is crucial for enhancing lupin agrobiodiversity and efficient exploitation of these species. Worldwide germplasm collections of lupin, which is one of the most important food legumes, are not well characterized. Within the INCREASE project, intelligent nested collections of the white and tarwi lupins will be established and exploited using cutting-edge approaches to genotyping, phenotyping, and data management and sharing. Here, we provide ready-to-use protocols to develop and describe SSD lines, comprising the broad set of phenotypic traits that can be used. These protocols will facilitate the screening and maintenance of lupin germplasm variations for agricultural improvement.
These protocols are valuable templates for a wide range of users, such as genebank managers, researchers, breeders, and farmers. Their use will not only reduce commonly seen misunderstandings, but more importantly will provide the gold standard for lupin genetic resource characterization and use when targeting specific traits. The approach to germplasm characterization and management that is being developed within the INCREASE project represents a fundamental basis for integration of the increasingly accessible genomic resources, along with the advanced ‘-omics’ analyses, to accelerate the discovery of improved lupin agricultural diversity and to determine valuable sources for breeding programs.
Acknowledgments
This study was conducted within the INCREASE project and is funded by the European Union Horizon 2020 Research and Innovation Programme under grant agreement No 862862. We thank Prof Wojciech K. Święcicki (Institute of Plant Genetics, Polish Academy of Sciences, Poznan) for comments and suggestions on the manuscript and Prof Łukasz Stępień (Institute of Plant Genetics, Polish Academy of Sciences, Poznan) who took pictures of particular growth stages of lupins.
Author Contributions
Magdalena Kroc : conceptualization, investigation, methodology, supervision, visualization, writing original draft, writing review and editing; Magdalena Tomaszewska : conceptualization, investigation, methodology; Katarzyna Czepiel : investigation, methodology, visualization, writing review and editing; Elena Bitocchi : conceptualization, writing review and editing; Markus Oppermann : formal analysis, writing review and editing; Kerstin Neumann : resources, writing review and editing; Luis Guasch Pereira : resources; Elisa Bellucci : writing review and editing; Saleh Alseekh : writing review and editing; Andreas Graner : resources, writing review and editing; Alisdair Fernie : writing review and editing; Roberto Papa : conceptualization, funding acquisition, writing review & editing; Karolina Susek : conceptualization, data curation, investigation, methodology, supervision, visualization, writing original draft, writing review and editing.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Open Research
Data Availability Statement
Data sharing not applicable—no new data generated.
Literature Cited
- Adhikari, K. N., Buirchell, B. J., & Sweetingham, M. W. (2012). Length of vernalization period affects flowering time in three lupin species. Plant Breeding , 131, 631–636. doi: 10.1111/j.1439-0523.2012.01996.x.
- Annicchiarico, P., Romani, M., & Pecetti, L. (2018). White lupin (Lupinus albus) variation for adaptation to severe drought stress. Plant Breeding , 137, 782–789. doi: 10.1111/pbr.12642.
- Arnoldi, A., Boschin, G., Zanoni, C., & Lammi, C. (2015). The health benefits of sweet lupin seed flours and isolated proteins. Journal of Functional Foods , 18, 550–563. doi: 10.1016/j.jff.2015.08.012.
- Atchison, G. W., Nevado, B., Eastwood, R. J., Contreras-Ortiz, N., Reynel, C., Madriñán, S., … Hughes, C. E. (2016). Lost crops of the Incas: Origins of domestication of the Andean pulse crop tarwi, Lupinus mutabilis. American Journal of Botany , 103, 1592–1606. doi: 10.3732/ajb.1600171.
- Atnaf, M., Yao, N., Martina, K., Dagne, K., Wegary, D., & Tesfaye, K. (2017). Molecular genetic diversity and population structure of Ethiopian white lupin landraces: Implications for breeding and conservation. PloS One , 12, e0188696–e0188696. doi: 10.1371/journal.pone.0188696.
- Bebeli, P. J., Lazaridi, E., Chatzigeorgiou, T., Suso, M.-J., Hein, W., Alexopoulos, A. A., … Van Den Berg, M. (2020). State and progress of Andean lupin cultivation in Europe: A review. Agronomy , 10, 1038. doi: 10.3390/agronomy10071038.
- Bellucci, E., Agrawal, S. K., Aguilar, O. M., Alseekh, S., Bett, K., Brezeanu, C., … R, P. The INCREASE project: Intelligent collections of food-legume genetic resources for European agrofood systems. Plant Journal , Submitted for publication.
- Berger, J. D., Shrestha, D., & Ludwig, C. (2017). Reproductive strategies in mediterranean legumes: Trade-Offs between phenology, seed size and vigor within and between wild and domesticated Lupinus species collected along aridity gradients. Frontiers in Plant Science , 8, 548. doi: 10.3389/fpls.2017.00548.
- Bellucci, E., Aguilar, M., Alseekh, S., Bett, K., Brezeanu, C., Cook, D., … Papa, R. (2021). The INCREASE project: Intelligent collections of food-legume genetic resources for European agrofood systems. Plant Journal , In press.
- Beyene, C. (2020). Genetic variation among white lupin (Lupinus albus L.) landraces from Northwestern and Southern Ethiopia for agronomic traits and nutrient contents of grain. Journal of Plant Breeding and Crop Science , 122, 159–169.
- Boschin, G., & Arnoldi, A. (2011). Legumes are valuable sources of tocopherols. Food Chemistry , 127, 1199–1203. doi: 10.1016/j.foodchem.2011.01.124.
- Cardoso, D., Pennington, R. T., De Queiroz, L. P., Boatwright, J. S., Van Wyk, B. E., Wojciechowski, M. F., & Lavin, M. (2013). Reconstructing the deep-branching relationships of the papilionoid legumes. South African Journal of Botany , 89, 58–75. doi: 10.1016/j.sajb.2013.05.001.
- Chirinos-Arias, M. C., Jiménez, J. E., & Vilca-Machaca, L. S. (2015). Analysis of genetic variability among thirty accessions of Andean Lupin (Lupinus mutabilis Sweet) using ISSR molecular markers. Scientia Agropecuaria , 6(1), 17–30. doi: 10.17268/sci.agropecu.2015.01.02.
- Clements, J. C., Dracup, M., Buirchell, B. J., & Smith, C. G. (2005). Variation for seed coat and pod wall percentage and other traits in a germplasm collection and historical cultivars of lupins. Australian Journal of Agricultural , 56, 75–83. doi: 10.1071/AR03114.
- Cortinovis, G., Oppermann, M., Neumann, K., Graner, A., Gioia, T., Marsella, M., Alseekh, S., … Bitocchi, E. (2021). Towards development, maintenance and standardized phenotypic characterization of single-seed-descent genetic resources for common bean. Current Protocols , 1, e133. doi: 10.1002/cpz1.133.
- Cowling, W. A., Buirchell, B. J., & Tapia, M. E. (1998). Lupin. Lupinus L. Promoting the conservation and use of underutilized and neglected crops. 23. Rome, Italy: International Board for Plant Genetic Resources (IBPGR).
- Drummond, C. S., Eastwood, R. J., Miotto, S. T. S., & Hughes, C. E. (2012). Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): Testing for key innovation with incomplete taxon sampling. Systematic Biology , 61, 443–460. doi: 10.1093/sysbio/syr126.
- Foley, R. C., Jimenez-Lopez, J. C., Kamphuis, L. G., Hane, J. K., Melser, S., & Singh, K. B. (2015). Analysis of conglutin seed storage proteins across lupin species using transcriptomic, protein and comparative genomic approaches. BMC Plant Biology , 15, 106–106. doi: 10.1186/s12870-015-0485-6.
- Fontanari, G. G., Batistuti, J. P., Cruz, R. J. D., Saldiva, P. H. N., & Arêas, J.a.G. (2012). Cholesterol-lowering effect of whole lupin (Lupinus albus) seed and its protein isolate. Food Chemistry , 132, 1521–1526. doi: 10.1016/j.foodchem.2011.11.145.
- Gladstones, J. S. (1998). Distribution, origin, taxonomy, history and importance. In J. S. Gladstones, C. A. Atkins, & J. Hamblin (Eds.). Lupins as crop plants: Biology, production, and utilization (pp. 1–36), Wallingford, UK: CAB International.
- Gresta, F., Wink, M., Prins, U., Abberton, M., Capraro, J., Scarafoni, A., & Hill, G. (2017). Lupins in European cropping systems. In D. Murphy-Bokern, F. Stoddard, & C. Watson (Eds.), Legumes in Cropping Systems (pp. 88–108). Wallingford, Oxfordshire, UK: CABI.
- Guerra-García, A., Gioia, T., Von Wettberg, E., Logozzo, G., & Bett, K. E. INCREASEing lentil genetic resources: Evolutionary history, recent genomic characterization of germplasm, and the need for well characterized collections. Current Protocols in Plant Biology , Manuscript in preparation.
- Guilengue, N., Alves, S., Talhinhas, P., & Neves-Martins, J. (2020). Genetic and genomic diversity in a tarwi (Lupinus mutabilis Sweet) germplasm collection and adaptability to mediterranean climate conditions. Agronomy , 10, 21. doi: 10.3390/agronomy10010021.
- Gulisano, A., Alves, S., Martins, J. N., & Trindade, L. M. (2019). Genetics and breeding of Lupinus mutabilis : An emerging protein crop. Frontiers in Plant Science , 10, 1385. doi: 10.3389/fpls.2019.01385.
- Hane, J. K., Ming, Y., Kamphuis, L. G., Nelson, M. N., Garg, G., Atkins, C. A., … Singh, K. B. (2016). A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: Insights into plant-microbe interactions and legume evolution. Plant Biotechnology Journal , 2016, 1–13.
- Hufnagel, B., Marques, A., Soriano, A., Marquès, L., Divol, F., Doumas, P., … Péret, B. (2020a). High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nature Communication , 11, 492. doi: 10.1038/s41467-019-14197-9.
- Hufnagel, B., Soriano, A., Taylor, J., Divol, F., Kroc, M., Sanders, H., … Péret, B. (2020b). Pangenome of white lupin provides insights into the diversity of the species. bioRxiv , 2020.2006.2021.163378.
- International Board for Plant Genetic Resources. (1981). Lupin Descriptors. Available at https://www.bioversityinternational.org/fileadmin/bioversity/publications/Web_version/103/ch1.htm.
- International Union for the Protection of New Varieties of Plants. (2004). Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability. Available at https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.191/attachments/tg066.pdf.
- Jacobsen, S.-E., & Mujica, A. (2008). Geographical distribution of the Andean lupin (Lupinus mutabilis Sweet). Plant Genetic Resources Newsletter , 155, 1–8.
- Kamel, K. A., Święcicki, W., Kaczmarek, Z., & Barzyk, P. (2016). Quantitative and qualitative content of alkaloids in seeds of a narrow-leafed lupin (Lupinus angustifolius L.) collection. Genetic Resources and Crop Evolution , 63, 711–719. doi: 10.1007/s10722-015-0278-7.
- Keller, J., Rousseau-Gueutin, M., Martin, G. E., Morice, J., Boutte, J., Coissac, E., … Aïnouche, A. (2017). The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses of four novel legume chloroplast genomes from Lupinus. DNA Research , 24, 343–358. doi: 10.1093/dnares/dsx006.
- Kroc, M., Koczyk, G., Swiecicki, W., Kilian, A., & Nelson, M. N. (2014). New evidence of ancestral polyploidy in the Genistoid legume Lupinus angustifolius L. (narrow-leafed lupin). Theoretical and Applied Genetics , 127, 1237–1249. doi: 10.1007/s00122-014-2294-y.
- Kroc, M., Rybiński, W., Wilczura, P., Kamel, K., Kaczmarek, Z., Barzyk, P., & Święcicki, W. (2017). Quantitative and qualitative analysis of alkaloids composition in the seeds of a white lupin (Lupinus albus L.) collection. Genetic Resources and Crop Evolution , 64, 1853–1860. doi: 10.1007/s10722-016-0473-1.
- Kumar, A., et al. Chickpea INCREASE Intelligent Collections: Characterization and development of single seed descent genetic resources. Current Protocols in Plant Biology , Manuscript in preparation.
- Kurlovich, B. S. (2002). Geography, classification, genetic resources and breeding. St. Petersburg, Russia and Pellosniemi, Finland: OY International North Express.
- Lavin, M., Herendeen, P. S., & Wojciechowski, M. F. (2005). Evolutionary rates analysis of leguminosae implicates a rapid diversification of lineages during the Tertiary. Systems Biology , 54, 575–594. doi: 10.1080/10635150590947131.
- Mccouch, S., Navabi, Z. K., Abberton, M., Anglin, N. L., Barbieri, R. L., Baum, M., … Rieseberg, L. H. (2020). Mobilizing crop biodiversity. Molecular Plant , 13, 1341–1344. doi: 10.1016/j.molp.2020.08.011.
- Nevado, B., Atchison, G. W., Hughes, C. E., & Filatov, D. A. (2016). Widespread adaptive evolution during repeated evolutionary radiations in New World lupins. Nature Communications , 7, 12384. doi: 10.1038/ncomms12384.
- Neves Martins, J. M., Talhinhas, P., & De Sousa, R. P. (2016). Yield and seed chemical composition of Lupinus mutabilis in Portugal. Revista de Ciências Agrárias , 2016, 39(4), 518–525. doi: 10.19084/RCA16079.
- Olczak, T., Rurek, M., Jańska, H., Augustyniak, H., & Sawicka-Sienkiewicz, E. J. (2001). Screening of cytoplasmic DNA diversity between and within Lupinus mutabilis Sweet and Lupinus albus sensu lato by restriction fragment length polymorphism (RFLP). Journal of Applied Genetics , 42, 127–137.
- Pascual, H. (2004). Lupinus mariae-josephi (Fabaceae), nueva y sorprendente especie descubierta en España. Anales del Jardín Botánico de Madrid , 61, 69–72.
- Rybiński, W., Święcicki, W., Bocianowski, J., Börner, A., Starzycka-Korbas, E., & Starzycki, M. (2018). Variability of fat content and fatty acids profiles in seeds of a Polish white lupin (Lupinus albus L.) collection. Genetic Resources and Crop Evolution , 65, 417–431. doi: 10.1007/s10722-017-0542-0.
- Secco, D., Shou, H., Whelan, J., & Berkowitz, O. (2014). RNA-seq analysis identifies an intricate regulatory network controlling cluster root development in white lupin. BMC Genomics , 15, 1471–2164. doi: 10.1186/1471-2164-15-230.
- Susek, K., Bielski, W., Czyz, K. B., Hasterok, R., Jackson, S. A., Wolko, B., & Naganowska, B. (2019). Impact of chromosomal rearrangements on the interpretation of lupin karyotype evolution. Genes (Basel) , 10, 259. doi: 10.3390/genes10040259.
- Susek, K., Bielski, W. K., Hasterok, R., Naganowska, B., & Wolko, B. (2016). A first glimpse of wild lupin karyotype variation as revealed by comparative cytogenetic mapping. Frontiers in Plant Science , 7, 1152. doi: 10.3389/fpls.2016.01152.
- Susek, K., Braszewska-Zalewska, A., Bewick, A. J., Hasterok, R., Schmitz, R. J., & Naganowska, B. (2017). Epigenomic diversification within the genus Lupinus. PLoS One , 12, e0179821. doi: 10.1371/journal.pone.0179821.
- Święcicki, W., Czepiel, K., Wilczura, P., Barzyk, P., Kaczmarek, Z., & Kroc, M. (2019). Chromatographic fingerprinting of the Old World lupins seed alkaloids: A supplemental tool in species discrimination. Plants (Basel) , 8(12), 548. doi: 10.3390/plants8120548.
- Święcicki, W., Święcicki, W. K., & Wolko, W. (1996). Lupinus anatolicus —a new lupin species of the Old World. Genetic Resources and Crop Evolution , 43, 109–117. doi: 10.1007/BF00126753.
- Talhinhas, P., Neves-Martins, J., & Leitao, J. (2003). AFLP, ISSR and RAPD markers reveal high levels of genetic diversity among Lupinus spp. Plant Breeding , 122, 507–510. doi: 10.1111/j.1439-0523.2003.00892.x.
- Wang, Z., Straub, D., Yang, H., Kania, A., Shen, J., Ludewig, U., & Neumann, G. (2014). The regulatory network of cluster-root function and development in phosphate-deficient white lupin (Lupinus albus) identified by transcriptome sequencing. Physiologia Plantarum , 151, 323–338. doi: 10.1111/ppl.12187.
- Wink, M., Botschen, F., Gosmann, C., Schäfer, H., & Waterman, P. (2010). Chemotaxonomy Seen from a Phylogenetic Perspective and Evolution of Secondary Metabolism. In M. Wink (Ed.), Biochemistry of Plant Secondary Metabolism, Annual Plant Reviews, vol. 40, Wiley-Blackwell, Oxford, pp 364–433.
- Wink, M., Meißner, C., & Witte, L. (1995). Patterns of quinolizidine alkaloids in 56 species of the genus Lupinus. Phytochemistry , 38, 139–153. doi: 10.1016/0031-9422(95)91890-D.
- Wolko, B., Clements, J. C., Naganowska, B., Nelson, M. N., & Yang, H. (2011). Lupinus. In C. Kole (Ed.), Wild Crop Relatives: Genomic and Breeding Resources (pp. 153–206). Heidelberg, Germany: Springer-Verlag.
- Xu, W., Zhang, Q., Yuan, W., Xu, F., Muhammad Aslam, M., Miao, R., … Cheng, F. (2020). The genome evolution and low-phosphorus adaptation in white lupin. Nature Communications , 11, 1069.
- Yeheyis, L., Kijora, C., Melaku, S., Girma, A., & Peters, K. J. (2010). White lupin (Lupinus albus L.), the neglected multipurpose crop: Its production and utilization in the mixed crop-livestock farming system of Ethiopia. Livestock Research for Rural Development , 22(4), 2010.
Internet Resources
The Novel Food Catalogue list.
European Search Catalogue for Plant Genetic Resources database.
Global Portal on Plant Genetic Resources.
Agricultural Research Service of the U.S. Department of Agriculture.
INCREASE project.
Global Information System of the International Treaty on Plant Genetic Resources for Food and Agriculture.
Lupin descriptors: The International Board for Plant Genetic Resources (IBPGR), IBPGR Executive Secretariat Plant Production and Protection Division Food and Agriculture Organization of the United Nations, Via delle Terme di Caracalla, 00100 Rome, Italy (1981).
Guidelines for the conduct of tests for distinctness, uniformity, and stability (Lupinus albus L., Lupinus angustifolius L., Lupinus luteus L.). The International Union for the Protection of New Varieties of Plants (UPOV), TG/66/4, Geneva, Switzerland (2004).
Citing Literature
Number of times cited according to CrossRef: 5
- Athanasios Mavromatis, Irini Nianiou-Obeidat, Alexios Polidoros, Zoi Parissi, Eleni Tani, Maria Irakli, Konstantinos A. Aliferis, Ioannis Zafeiriou, Photini V. Mylona, Efi Sarri, Evgenia-Anna Papadopoulou, Rafail Tagiakas, Leonidas Kougiteas, Stavroula Kostoula, Eleni M. Abraham, Characterization of Lupin Cultivars Based on Phenotypical, Molecular and Metabolomic Analyses, Agronomy, 10.3390/agronomy13020370, 13 , 2, (370), (2023).
- Mustafa Bulut, Regina Wendenburg, Elena Bitocchi, Elisa Bellucci, Magdalena Kroc, Tania Gioia, Karolina Susek, Roberto Papa, Alisdair R. Fernie, Saleh Alseekh, A comprehensive metabolomics and lipidomics atlas for the legumes common bean, chickpea, lentil and lupin, The Plant Journal, 10.1111/tpj.16329, 116 , 4, (1152-1171), (2023).
- Lorenzo Rocchetti, Tania Gioia, Giuseppina Logozzo, Creola Brezeanu, Luis Guasch Pereira, Lucía De Rosa, Stefania Marzario, Alice Pieri, Alisdair R. Fernie, Saleh Alseekh, Karolina Susek, Douglas R. Cook, Rajeev K. Varshney, Shiv Kumar Agrawal, Aladdin Hamwieh, Elena Bitocchi, Roberto Papa, Towards the Development, Maintenance and Standardized Phenotypic Characterization of Single‐Seed‐Descent Genetic Resources for Chickpea, Current Protocols, 10.1002/cpz1.371, 2 , 2, (2022).
- Ioannis Zafeiriou, Alexios N. Polidoros, Eirini Baira, Konstantinos M. Kasiotis, Kyriaki Machera, Photini V. Mylona, Mediterranean White Lupin Landraces as a Valuable Genetic Reserve for Breeding, Plants, 10.3390/plants10112403, 10 , 11, (2403), (2021).
- Elisa Bellucci, Orlando Mario Aguilar, Saleh Alseekh, Kirstin Bett, Creola Brezeanu, Douglas Cook, Lucía De la Rosa, Massimo Delledonne, Denise F. Dostatny, Juan J. Ferreira, Valérie Geffroy, Sofia Ghitarrini, Magdalena Kroc, Shiv Kumar Agrawal, Giuseppina Logozzo, Mario Marino, Tristan Mary‐Huard, Phil McClean, Vladimir Meglič, Tamara Messer, Frédéric Muel, Laura Nanni, Kerstin Neumann, Filippo Servalli, Silvia Străjeru, Rajeev K. Varshney, Marta W. Vasconcelos, Massimo Zaccardelli, Aleksei Zavarzin, Elena Bitocchi, Emanuele Frontoni, Alisdair R. Fernie, Tania Gioia, Andreas Graner, Luis Guasch, Lena Prochnow, Markus Oppermann, Karolina Susek, Maud Tenaillon, Roberto Papa, The INCREASE project: Intelligent Collections of food‐legume genetic resources for European agrofood systems, The Plant Journal, 10.1111/tpj.15472, 108 , 3, (646-660), (2021).

