Section 3: Libraries quality control (QC)
Ester Kalef-Ezra, Christos Proukakis, Ben Harvey, Katherine Roper
Abstract
This protocol details quality control of libraries and should be performed after Section 2: NGS library preparation for sequencing.
Attachments
Steps
Libraries quality control (QC)
Thaw the libraries On ice.
Centrifuge briefly.
Quantify the libraries using BR or HS Qubit (depending on the starting material and the amplification cycles).
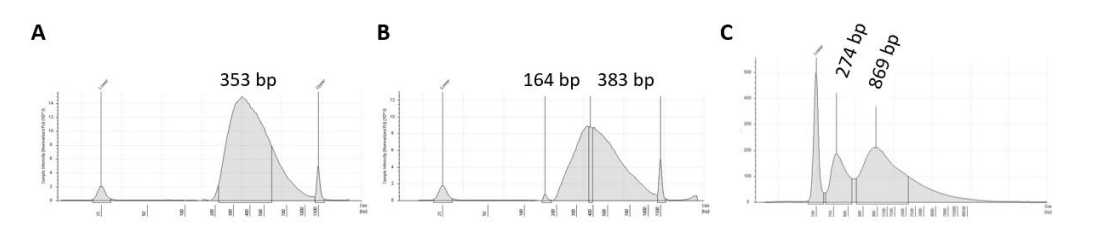
Analyse libraries using TapeStation with HS D1000 or D1000 Screen tapes and reagents (depending on the starting material and the amplification cycles), according to manufacturer guidelines. Examples of these are presented in Figure 8. Other methods can be used such as Agilent 2100 Bioanalyzer system with DNA 1000 kit.
Dilute the libraries to concentration needed and pool them together. The QC the pooled library using both HS Qubit and HS D1000 tapes.

