Protein Isolation from 3D Hydrogel Scaffolds
Gabriela Da Silva André, Gabriela Da Silva André, Lorenza Garau Paganella, Lorenza Garau Paganella, Asia Badolato, Asia Badolato, Sibilla Sander, Sibilla Sander, Costanza Giampietro, Costanza Giampietro, Mark W Tibbitt, Mark W Tibbitt, Jörn Dengjel, Jörn Dengjel, Céline Labouesse, Céline Labouesse
Abstract
Protein isolation is an essential tool in cell biology to characterize protein abundance under various experimental conditions. Several protocols exist, tailored to cell culture or tissue sections, and have been adapted to particular downstream analyses (e.g., western blotting or mass spectrometry). An increasing trend in bioengineering and cell biology is to use three-dimensional (3D) hydrogel-based scaffolds for cell culture. In principle, the same protocols can be used to extract protein from hydrogel-based cell and tissue constructs. However, in practice the yield and quality of the recovered protein pellet is often substantially lower when using standard protocols and requires tuning of multiple steps, including the selected lysis buffer and the scaffold homogenization strategy, as well as the methods for protein purification and reconstitution. We present here specific protocols tailored to common 3D hydrogels to help researchers using hydrogel-based 3D cell culture improve the quantity and quality of their extracted protein. We focus on three materials: protease-degradable PEG-based hydrogels, collagen hydrogels, and alginate hydrogels. We discuss how the protein extraction procedure can be adapted to the scaffold of interest (degradable or non-degradable gels), proteins of interests (soluble, matrix-bound, or phosphoproteins), and downstream biochemical assays (western blotting or mass spectrometry). With the growing interest in 3D cell culture, the protocols presented should be useful to many researchers in cell biology, protein science, biomaterials, and bioengineering communities. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Isolating proteins from PEG-based hydrogels
Basic Protocol 2 : Isolating proteins from collagen hydrogels
Basic Protocol 3 : Isolating proteins from alginate hydrogels
Alternate Protocol : Isolating protein from alginate gels using EDTA to dissolve the gel
Support Protocol : Isolating protein and RNA simultaneously from the same samples
INTRODUCTION
Protein isolation from cell cultures has been a mainstay of cell and molecular biology for decades. It is an essential step in the process of identifying changes in protein abundance, post-translational modifications, and interaction partners (Walker, 2009). Protein-based assays are dependent on the quality and the yield of the protein lysate. In the last decade, cell and tissue biologists have embraced 3D multicellular constructs to engineer more complex in vitro models. Such 3D models include cell encapsulation into various types of hydrogels, spheroid formation, and organoids (free-floating or encapsulated; Caliari & Burdick, 2016; Gjorevski et al., 2016). Many researchers will acknowledge that protein isolation from 3D scaffolds is more challenging than that from standard 2D plastic- or glass-based cultures. The challenges lie in the ability to isolate enough protein with high quality and a similar throughput as from 2D assays. However, these challenges are rarely discussed in the experimental section of research articles, and few published protocols directly address them. Here we address the specific challenges associated with isolating protein from 3D cell cultures. We borrowed some techniques used to isolate proteins from tissue samples but with a specific focus on isolating cells from 3D-hydrogel-based scaffolds.
The general steps of all protein isolation protocols are (i) cell lysis, (ii) purification and precipitation, and (iii) resuspension of the pellet in native or denaturing conditions. The three main issues encountered in the process are protein yield, efficiency, and maintenance of post-translational modifications (PTMs), such as phosphorylated groups. The yield will depend both on the number of cells and on the isolation protocol, and tends to be a limiting factor in the success of downstream assays. Various protocols for isolation differ in the choice of lysis buffer, purification method, and resuspension buffer. Lower efficiency can arise when some steps do not work as well in the presence of a gel phase swollen in an aqueous solvent. Non-dissolved gel phase can clump and compromise downstream processing of the samples. The third challenge is to be able to work fast enough to preserve PTMs, e.g., phosphorylated groups that are highly labile, as processes in 3D, such as breaking up or degrading the scaffold, typically take longer than standard 2D protocols. We discuss all of these challenges in the protocols presented here.
In addressing these challenges, the properties of the hydrogel scaffold are important. First, for the best results, the scaffold must be broken up or degraded to ensure complete cell lysis and protein recovery. Scaffolds can be either mechanically degraded or chemically dissolved. Some scaffolds are also designed to be degradable by light (e.g., Kloxin et al., 2010). Second, the swelling properties of the polymers in the aqueous and organic solvents used must be considered. Phase separation is one method of choice to purify protein content from whole cell lysates. It relies on the separation of an organic phase, often based on phenol/chloroform, containing the protein content from an aqueous phase containing RNA. The DNA normally found in the interphase can be precipitated with ethanol. How the gel phase interacts with the solvents used will influence the quality of the purification process. Increasing the volume of organic solvent or the number of incubation steps can be useful to ensure proper purification. Strategies can be adapted to the type of scaffold and its degradability. Finally, the physical properties of the scaffold are important, namely its volume and its protein content. Natural-based hydrogels, such as polysaccharide (e.g., alginate, hyaluronic acid)- or protein-based hydrogels (e.g., collagen), can be more difficult to fully eliminate compared with synthetic hydrogels, such as poly(ethylene glycol) (PEG).
Having a good understanding of the scaffold that is used, as well as the intended protein target(s) to be analyzed and the methodology of analysis is essential to ensure suitable purity and yield for the downstream assays. When using 3D scaffolds, cell number and scaffold size can be a limiting factor, and therefore the protocols are less forgiving if a specific amount of protein is needed for downstream assays. Western blots typically require 10 µg of protein per sample, and up to 20 to 40 µg of protein for phosphorylated targets. For classical bottom-up liquid crystallography–tandem mass spectrometry (LC-MS/MS)-based proteomics, similar amounts of protein per sample are typically used, whereas for co-immunoprecipitation (co-IP), milligrams of protein are needed. These numbers also depend on the abundance of the target proteins and depth of analysis. To obtain sufficient yield, it is essential to tailor the isolation protocols for each scaffold type and cell type used. In addition, for any protein that is secreted or extracellular, and thus potentially interacts with the scaffold, the investigator must ensure that the scaffold degradation method does not interfere with protein integrity or detection (Sawicki et al., 2018).
Our aim is to guide the investigator in adapting standard protein isolation protocols for their own specific type of 3D scaffolds. Here we give detailed protocols for three types of scaffolds, a synthetic scaffold (PEG), a natural fibrous protein scaffold (collagen), and a polysaccharide-based scaffold (alginate), addressing specific challenges linked to their different degradation mechanism and swelling properties. These three basic protocols focus on the isolation of proteins from cells grown in hydrogels using TRIzol™-based lysis and phase separation. We give one alternate protocol that uses a different scaffold degradation strategy, and one support protocol for simultaneously isolating RNA and protein from the same sample, which applies to all types of scaffolds. We discuss in the Commentary section our rationale, and the downstream applications that are possible from the isolated protein solutions (western blotting, mass spectrometry, immunoprecipitation).
CAUTION : All protocols given here are done using the phenol/chloroform phase separation method with TRIzol™ reagent. TRIzol™ and chloroform are toxic to inhale and must be handled in a fume hood with adequate ventilation. Please follow the safety procedures outlined on the safety data sheets of each product. If one uses a nontoxic lysis buffer (e.g., radioimmunoprecipitation assay [RIPA] lysis buffer), there is no need to perform these steps under a fume hood.
STRATEGIC PLANNING
We describe in the protocols below the result of protocol refinement for PEG, collagen, and alginate scaffolds. These protocols should be further adapted to each new scaffold. To guide the investigator in this process, we detail here the main aspects to consider before starting any protein isolation process. These are summed up in the two tables below: Know Your Scaffold (Table 1) and Know Your Target (Table 2).
| Hydrogel degradation strategy | Type | Time per sample | Comment |
|---|---|---|---|
| Manual breakage | 5-10 min/sample | Take care to avoid sample cross-contamination | |
| Homogenization | 1-5 min/sample | Take care to avoid sample cross-contamination | |
| Digestion (enzymatic or with EDTA) | 7-15 min | Possible if protein abundance not expected to change over 15-min time frame | |
| Light-based degradation | Minutes | ||
| Hydrogel swelling properties | Solvent | Swelling ratio or swelling behavior | Comment |
| Aqueous buffer | How much water is retained in the gel phase and transferred to the lysate? (will depend on the degradation strategy) | Increase number of washing steps prior to lysing and add more chloroform to the phase separation step | |
| Ethanol | If some gel phase is still in the supernatant after washing with ethanol, repeat the ethanol washing step once more |
- a Examples of hydrogel degradation strategies and swelling behaviors in different solvents. EDTA, ethylenediaminetetraacetic acid.
| Type of target protein(s) | Required cell numbers for western blotting | Required cell number for mass spectrometry | |
|---|---|---|---|
| Non-phosphorylated | High abundance | 100,000 cells/sample | 10,000-100,000 cells/sample |
| Low abundance | 400,000 cells/sample | ||
| Phosphorylated protein | High abundance | 200,000 cells/sample | |
| Low abundance | >500,000 cells/sample | ||
| ECM protein (native) | No interaction with scaffold | The scaffold can be dissolved/degraded with any method | |
| Interacts/entangles with the scaffold | Do not use collagenase to digest the hydrogel. Other methods can be used |
- a Here we provide rough estimates of cell numbers. Note that in all cases, the detection level will depend on the sensitivity of the antibody used for the total or the phosphorylated proteins. The antibodies should always be tested first.
Know your scaffold
The polymer part of the scaffold must be broken up for efficient protein isolation. Either the polymer scaffold is intrinsically degradable (by light, by enzymes, or by chelating an ionic cross-linker, not to be confused with cell-directed degradability), or else it must be mechanically disrupted. The scaffold degradation step can be done before or after adding the lysis buffer (see Fig. 1). If done before cell lysis, it is important that this step is fast to avoid any changes or loss in the protein content. Mechanical disruption can be done manually by grinding the gel in a mortar or in a tube with a spatula, with optional use of a needle (gauge 23G to 25G) to remove small clumps. Alternatively, mechanical disruption with a tissue homogenizer or a bead mill homogenizer (also called a bead beater) can be more efficient. Mechanical disruption yields better results when freezing the hydrogel before degradation. Finally, some scaffolds are, intrinsically or by design, degradable by peptidases or proteases (e.g., sortases, collagenases or other matrix metalloproteinases; Bretherton et al., 2023; Lutolf & Hubbell, 2005; Valdez et al., 2017). In principle, collagenase or an appropriate protease could be used in these cases to enzymatically digest the gel. However, enzymatic digestion can be long (>15 min, depending on the gel volume and protease concentration), which could affect the abundance and post-translational modifications of the protein(s) of interest. Therefore, this method should be used only if no change in protein abundance is expected on these time scales. For hydrogels that are difficult to break up fast, combining chemical dissolution with mechanical degradation can help to obtain fast scaffold degradation and efficient cell lysis.
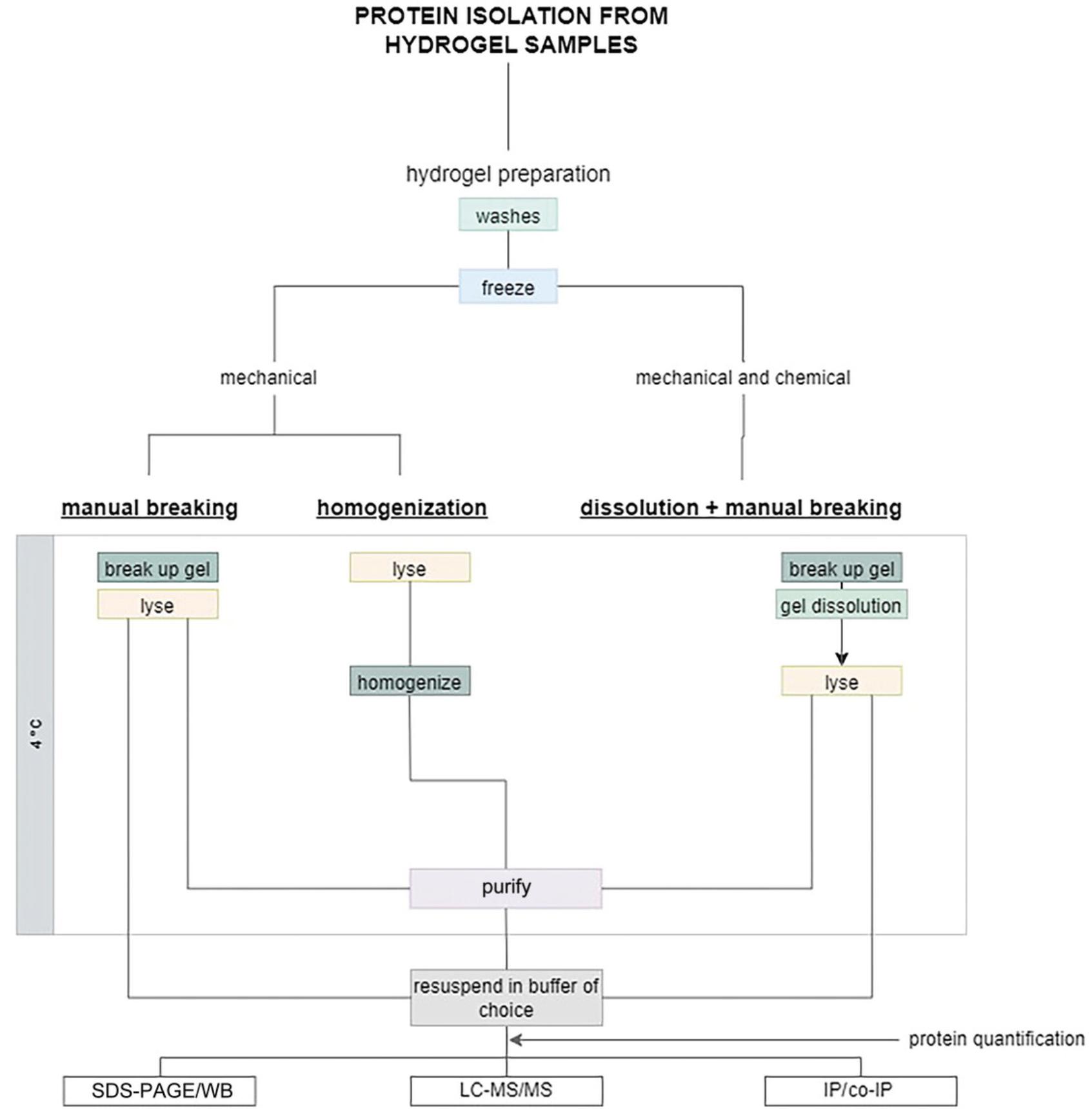
Efficient hydrogel degradation and cell lysis usually requires immersing the hydrogel into 1-5 times its volume of lysis buffer. Although this could be done directly in a lysis buffer, such as the RIPA lysis buffer traditionally used for many protein isolation protocols, it would entail using large volumes of RIPA buffer and correspondingly diluting the protein content. To avoid this problem, we chose to use a phase-separation technique using TRIzol™ and chloroform followed by protein precipitation and purification. Following this approach, the protein pellet can be resuspended using a buffer and volume of choice to adjust the final protein concentration as required by the planned downstream application. Note that TRIzol™ does denature proteins to some extent, and thus this procedure is not compatible with subsequent applications that require native protein conformation (co-immunoprecipitation, native gel electrophoresis); in such cases, using a mild, non-denaturing lysis buffer is recommended followed by dialysis to re-concentrate the protein if needed (Savorani et al., 2021).
Hydrogels swell
We recommend measuring the swelling ratio and final water content of the scaffold. For comparison, tissues are 70% water, whereas hydrogels are often 95% or more water. In addition, the cell density is often substantially lower than tissue. Furthermore, some hydrogels can swell in aqueous solutions, increasing the relative water content. Hydrogels that swell a lot in aqueous media, such as PEG, may need multiple washing steps to be fully flushed of the medium components that could contain proteases. Moreover, protein precipitation steps are done in ethanol or isopropanol. However, the gel phase may have different solubility in these solvents, and this could affect the efficiency of the processes. Again here, the number of washing steps should be adjusted to the polymer scaffold used.
Know your target
Protein isolation protocols must be aligned with the downstream application and the desired target. The number of cells can be controlled by cell seeding density or scaffold volume. As a general point, as cells proliferate slower in 3D hydrogels than on 2D plastic or glass, one should aim to seed cells in hydrogels at high density. If quantification of phosphoproteins is desired, a larger amount of protein is needed than for assays looking at total proteins irrespective of their phosphorylation state. In addition, one must work fast and at low temperatures (keeping samples on ice) to avoid loss of phosphorylated groups due to phosphatase activity. The use of phosphatase inhibitors is also recommended in this case.
Basic Protocol 1: ISOLATING PROTEINS FROM PEG-BASED HYDROGELS
PEG-based hydrogels have become a prominent biomaterial to encapsulate cells and create 3D in vitro tissue models (Caliari & Burdick, 2016; Lutolf & Hubbell, 2005). PEG chains can interfere with downstream applications, so it is important to separate and remove the PEG from the protein content. This protocol details how to break up PEG gels, isolate protein using TRIzol™ and chloroform phase separation, and purify protein.
NOTE : Volumes here are given assuming a 100-µl starting gel volume (before swelling). Adjust the volumes of lysis buffer as needed to suit your sample volume.
Materials
-
Starting sample: cells or microtissues encapsulated in a PEG-based hydrogel (see Strategic Planning in regard to cell numbers)
-
Phosphate-buffered saline (PBS), 4°C
-
Pierce™ Protease Inhibitor Mini Tablets, EDTA-free (cat. no. A32955, Sigma-Aldrich)
-
Optional : Pierce™ Phosphatase Inhibitor Mini Tablets (cat. no. A32957, Sigma-Aldrich)
-
TRIzol™ (cat. no. 15596018, Invitrogen; or equivalent monophasic solution of phenol and guanidine isothiocyanate), 4°C
-
Chloroform
-
70% (v/v) ethanol, diluted from 100% ethanol in nuclease- and protease-free water (e.g., UltraPure™ Distilled Water, cat. no. 10977-035, Invitrogen)
-
100% ethanol, BioUltra for molecular biology (cat. no. 51976–500ML-F, Sigma-Aldrich)
-
100% isopropanol, BioUltra for molecular biology (cat. no. 59304, Sigma-Aldrich)
-
0.3 M guanidine hydrochloride
-
Resuspension buffer: e.g., Hot SDS buffer
-
1-ml and 200-µl pipets
-
Fume hood
-
Metal spatula
-
5- and 2-ml PCR clean tubes (cat. nos. 0030119460 and 022431048, Eppendorf)
-
Liquid nitrogen or dry ice
-
Ice
-
Tissue homogenizer: T 25 digital ULTRA-TURRAX homogenizing with dispersing tool (cat. no. S25 N-8G-ST, IKA)
-
Centrifuge, cooled, with maximum speed of at least 13,000 rcf
-
Freezer, −20°C
-
Hot plate, capable of heating up to 75°C
1.Wash the PEG gel samples twice, each time using 1 ml PBS containing protease inhibitors (and phosphatase inhibitors, if required), precooled to 4°C.
2.Pre-fill a 5-ml tube with 1 ml TRIzol™ and label it.
3.Using a spatula, transfer your gel to the TRIzol™-filled tube and immediately snap-freeze it in liquid nitrogen.
4.Place the samples on ice.
5.Homogenize the sample until the hydrogel is broken up into small flakes, typically <1 mm in size. You can use the homogenizer at the maximum speed (∼15,000 rpm).
6.Between samples, wash your homogenizer probe twice in cold 70% ethanol using sufficient volume to immerse the probe, to avoid sample cross-contamination. Change the washing solution after each wash.
7.Incubate samples with TRIzol™ for 15 min at room temperature.
8.Add chloroform to each sample, 200 µl to every 1 ml of TRIzol™. Incubate 10 min at room temperature. Mix the tubes by vigorously inverting them.
9.Centrifuge the samples for 15 min at 13,000 rcf, 4°C, to induce phase separation. The samples should separate into a top transparent phase (aqueous phase), an opaque interphase (usually thin), and a bottom pink organic phase containing the protein content.
10.Carefully discard the top, aqueous phase using a pipet. If you wish to keep the RNA, save the aqueous phase (see Support Protocol).
11.Add 100% ethanol, 300 µl for each 1 ml of TRIzol™, to precipitate DNA and the remainder of the gel phase. Incubate 2-3 min at room temperature.
12.Spin down for 5 min at 2500 rcf, 4°C.
13.Recover the supernatant into a fresh 2-ml tube using a pipet. Discard any pellet.
14.Repeat steps 11-13.
15.Add 100% isopropanol, 1.5 ml for each 1 ml of TRIzol™ used.
16.Incubate in isopropanol overnight at −20°C to precipitate the protein.
17.Spin down for 10 min at 12,000 rcf, 4°C. A protein pellet should form and be visible.
18.Discard the supernatant using a pipet.
19.Add 2 ml guanidine hydrochloride. Mix gently by inversion to detach the pellet from the tube wall. Incubate 20 min at room temperature.
20.Spin down for 5 min at 7500 rcf, 4°C.
21.Remove the supernatant using a pipet, and add another 2 ml of guanidine hydrochloride. Mix gently by inversion to detach the pellet from the tube wall.
22.Spin down for 5 min at 7500 rcf, 4°C.
23.Discard the supernatant using a pipet, and add 2 ml of 100% ethanol.
24.Spin down for 7 min at 7500 rcf, 4°C.
25.Discard the supernatant using a pipet.
26.Allow the ethanol to evaporate by leaving the samples (open tubes) at room temperature for 5-10 min.
27.Resuspend the pellet in your buffer of choice. Typically, 100 µl per sample yields high enough concentrations.
Basic Protocol 2: ISOLATING PROTEIN FROM COLLAGEN HYDROGELS
Collagen gels are also used routinely to encapsulate cells for 3D cell culture (Rhee, 2009). Collagen does not swell much, which makes it easier to estimate the total volume of gel phase and aqueous phase. If not properly broken down, collagen fibers can create problems in SDS-PAGE band migration and will also contribute to high signal in mass spectrometry, given that it is a protein. Therefore, it is important to try to fully remove the collagen component from the protein pellet. Here we give a protocol based on manual mechanical disruption of collagen gels. This has been the most efficient way in our hands to break up collagen gels. Note that collagen gels can be digested with collagenase; however, this process takes 20-30 min and must be done at 37°C. In these conditions, the cellular protein content could be degraded or altered, especially in the case of phosphorylated targets. Collagenase is therefore not the preferred method.
NOTE : Volumes here are given assuming a 1.5-ml starting gel volume (before swelling). Adjust the volumes of lysis buffer as needed to suit your sample volume.
Materials
-
Starting sample: cells or microtissues encapsulated in a collagen hydrogel (see Strategic Planning about cell numbers)
-
Phosphate-buffered saline (PBS), 4°C
-
Pierce™ Protease Inhibitor Mini Tablets, EDTA-free, (cat. no. A32955, Sigma-Aldrich)
-
Optional : Pierce™ Phosphatase Inhibitor Mini Tablets (cat. no. A32957, Sigma-Aldrich)
-
TRIzol™ (cat. no. 15596018, Invitrogen; or equivalent monophasic solution of phenol and guanidine isothiocyanate), 4°C
-
Chloroform
-
100% ethanol, BioUltra for molecular biology (cat. no. 51976-500ML-F, Sigma-Aldrich)
-
Isopropanol, BioUltra for molecular biology (cat. no. 59304, Sigma Aldrich)
-
0.3 M guanidine hydrochloride
-
Resuspension buffer, e.g., Hot SDS buffer
-
1-ml and 200-µl pipets
-
Metal spatula
-
5- and 2-ml PCR clean tubes (cat. nos. 0030119460 and 022431048, Eppendorf)
-
Fume hood
-
Liquid nitrogen or dry ice
-
Ice
-
Centrifuge, cooled, with maximum speed of at least 13,000 rcf
-
Freezer, −20°C
-
Hot plate, heating up to 75°C
1.Wash the collagen gel samples twice, each time using with 3 ml PBS containing protease inhibitors (and phosphatase inhibitors, if required), precooled to 4°C.
2.Transfer your gel using a spatula to a 5-ml tube and immediately snap-freeze it in liquid nitrogen.
3.Add 300 µl cold TRIzol™ to each sample.
4.Mechanically break up the gel with a sharp spatula or a grinder. You should be able to break the gel up into small pieces, not more than a few millimeters wide.
5.Add an additional 1 ml of TRIzol™.
6.Place the samples on ice.
7.Incubate samples with TRIzol™ for 15 min at room temperature.
8.Add chloroform to each sample. Phase separation is best achieved by adding 280 µl of chloroform for the 1.3 ml TRIzol™ added. Incubate 10 min at room temperature. Mix the tubes by vigorously inverting them.
9.Centrifuge the samples for 15 min at 13,000 rcf, 4°C, to induce phase separation. The samples should separate into a top transparent phase (aqueous phase), an opaque interphase (usually thin), and a bottom pink organic phase containing the protein content.
10.Carefully discard the top, aqueous phase using a pipet. If you wish to keep the RNA, save the aqueous phase (see Support Protocol).
11.Add 100% ethanol, 300 µl for each 1 ml of TRIzol™, to precipitate DNA and the remainder of the gel phase. Incubate 2-3 min at room temperature.
12.Spin down for 5 min at 2500 rcf, 4°C. Repeat steps 11-13.
13.Collect the supernatant into a fresh 2-ml tube using a pipet. Discard any pellet.
14.Add 1.5 ml of 100% isopropanol per sample.
15.Incubate in isopropanol overnight at −20°C to precipitate the protein.
16.Spin down for 10 min at 12,000 rcf, 4°C. A visible protein pellet should form.
17.Discard the supernatant using a pipet.
18.Add 2 ml guanidine hydrochloride solution. Mix gently by inversion to detach the pellet from the tube wall. Incubate 20 min at room temperature.
19.Spin down for 10 min at 12,000 rcf, 4°C.
20.Remove the supernatant using a pipet and add another 2 ml of guanidine hydrochloride solution. Mix gently by inversion to detach the pellet from the tube wall.
21.Spin down for 5 min at 7500 rcf, 4°C.
22.Discard the supernatant using a pipet and add 2 ml of 100% ethanol.
23.Spin down for 7 min at 7500 rcf, 4°C.
24.Discard the supernatant using a pipet.
25.Allow the ethanol to evaporate by leaving the samples (open tubes) for 5-10 min at room temperature.
26.Resuspend the pellet in your buffer of choice. Typically, 100 µl per sample yields high enough concentrations.
Basic Protocol 3: ISOLATING PROTEIN FROM ALGINATE HYDROGELS
Alginate gels are emerging as a common scaffold for the 3D culture of mammalian and microbial cells alike (Balasubramanian et al., 2021; Chaudhuri et al., 2016). Alginate gelation can be induced by cross-linking the polysaccharide chains with divalent cations, such as Ca2+. The alginate molecular weight, the polymer density, and the divalent ion concentration all influence how easy it is to break up the formed gel. Mechanical degradation remains the best way to break the gels, but this can be combined with chemical treatment, namely using a calcium chelator, such as ethylenediaminetetraacetic acid (EDTA). For investigators who do not have a tissue homogenizer or grinder, the combined chemical and mechanical degradation is a suitable option; however, this process takes ∼15 min, so it is only useful for protein content that is not expected to change over this time period. The protocol using EDTA is provided in Alternate Protocol.
NOTE : Volumes here are given assuming a 100-µl starting gel volume (before swelling). Adjust the volumes of lysis buffer as needed to suit your sample volume.
Materials
-
Starting sample: cells or microtissues encapsulated in an alginate hydrogel (see Strategic Planning about cell numbers)
-
Phosphate-buffered saline (PBS), 4°C
-
Pierce™ Protease Inhibitor Mini Tablets, EDTA-free, (cat. no. A32955, Sigma-Aldrich)
-
Optional : Pierce™ Phosphatase Inhibitor Mini Tablets (cat. no. A32957, Sigma-Aldrich)
-
TRIzol™ (cat. no. 15596018, Invitrogen; or equivalent monophasic solution of phenol and guanidine isothiocyanate), 4°C
-
70% (v/v) ethanol, diluted from 100% ethanol in nuclease- and protease-free water (e.g., UltraPure™ Distilled Water, cat. no. 10977-035, Invitrogen), cold
-
Chloroform
-
100% ethanol, BioUltra for molecular biology (cat. no. 51976–500ML-F, Sigma-Aldrich)
-
Isopropanol, BioUltra for molecular biology (cat. no. 59304, Sigma Aldrich)
-
0.3 M guanidine hydrochloride
-
Resuspension buffer, e.g., Hot SDS buffer
-
1-ml and 200-µl pipets
-
Fume hood
-
Metal spatula
-
5- and 2-ml PCR clean tubes (cat. nos. 0030119460 and 022431048, Eppendorf)
-
Liquid nitrogen or dry ice
-
Ice
-
Tissue homogenizer T 25 digital ULTRA-TURRAX, with dispersing tool (cat. no. S25 N-8G-ST, IKA)
-
Centrifuge, cooled, with maximum speed of at least 13,000 rcf
-
Freezer, −20°C
-
Hot plate, with heating up to 75°C
1.Wash the alginate samples twice, each time using 1 ml PBS containing protease inhibitors (and phosphatase inhibitors if required), precooled to 4°C.
2.Pre-fill a 5-ml tube with 1 ml TRIzol™ and label it.
3.Transfer your gel using a spatula to the TRIzol™-filled tube and immediately snap-freeze it in liquid nitrogen.
4.Place the samples on ice.
5.Homogenize the sample three to five times until no pieces of hydrogel are visible (the solution should look clear). You can use the homogenizer at the maximum speed (∼15,000 rpm).
6.Between samples, wash your homogenizer probe in cold 70% ethanol twice in sufficient volume to immerse the probe to avoid sample cross-contamination. Change the washing solution after each wash.
7.Incubate samples with TRIzol™ for 15 min at room temperature.
8.Add chloroform to each sample, 200 µl for every 1 ml of TRIzol™. Incubate 10 min at room temperature. Mix the tubes by vigorously inverting them.
9.Centrifuge the samples for 15 min at 13,000 rcf, 4°C, to induce phase separation. The samples should separate into a top transparent phase (aqueous phase), an opaque interphase (usually thin), and a bottom pink organic phase containing the protein content.
10.Discard the top, aqueous phase using a pipet. If you wish to keep the RNA, save the aqueous phase (see Support Protocol).
11.Add 100% ethanol, 300 µl for each 1 ml of TRIzol™, to precipitate DNA and the remainder of the gel phase. Incubate 2-3 min at room temperature.
12.Spin down for 15 min at 13,000 rcf, 4°C.
13.Recover the supernatant into a fresh 2-ml tube using a pipet. Discard any pellet.
14.Repeat steps 12 and 13 to ensure residue-free supernatant.
15.Add 100% isopropanol, 1.5 ml for each 1 ml of TRIzol™ used.
16.Incubate in isopropanol overnight at −20°C to precipitate the protein.
17.Spin down for 10 min at 12,000 rcf, 4°C.
18.Discard the supernatant using a pipet.
19.Add 2 ml guanidine hydrochloride. Mix gently by inversion to detach the pellet from the tube wall. Incubate 20 min at room temperature.
20.Once the protein forms a nice pellet, remove the supernatant and add another 2 ml guanidine hydrochloride.
21.Spin down for 5 min at 7500 rcf, 4°C.
22.Discard the supernatant using a pipet and add 2 ml of 100% ethanol.
23.Spin down for 7 min at 7500 rcf, 4°C.
24.Discard the supernatant using a pipet.
25.Allow the ethanol to evaporate by leaving the samples (open tubes) at room temperature for 5-10 min.
26.Resuspend the pellet in your buffer of choice. Typically, 100 µl per sample yields high enough concentrations.
Alternate Protocol: ISOLATING PROTEIN FROM ALGINATE GELS USING EDTA TO DISSOLVE THE GEL
This procedure requires the same list of reagents and materials as for Basic Protocol 3 (see above), except that tissue homogenizer is not needed.
Additional Materials (see Basic Protocol 3)
- 50 mM EDTA solution
- Vortex
1.Wash the alginate samples twice, each time using 1 ml of PBS containing protease inhibitors (and phosphatase inhibitors, if required), precooled to 4°C.
2.Transfer your gel using a spatula to a tube and immediately snap-freeze it in liquid nitrogen.
3.Crush the gel using a spatula in the tube.
4.Incubate the samples with 1 ml ice-cold 50 mM EDTA for 5-15 min or until the gel is fully dissolved. Vortex in between to help with gel dissolution.
5.Spin down for 5 min at 7500 rcf, 4°C.
6.Discard the supernatant using a pipet. The cell pellet should be visible.
7.Add 500 µl TRIzol™ to the sample.
8.Continue with Basic Protocol 3 starting from step 7.
Support Protocol: ISOLATING PROTEIN AND RNA SIMULTANEOUSLY FROM THE SAME SAMPLES
The protocol presented here describes how to isolate both proteins and RNA from the same samples, enabling quantification of both gene expression and protein abundance. We give detailed steps on how to isolate and purify the RNA. When isolating RNA, attention must be given to avoiding ribonucleases (RNases), which are ubiquitous, and thus surfaces and materials must be thoroughly cleaned. Always keep samples on ice, unless otherwise stated. This protocol starts from the phase-separation steps above (steps 9 in Basic Protocols 1, 2, and 3)
Materials
-
Phosphate-buffered saline (PBS)
-
Pierce™ Protease Inhibitor Mini Tablets, EDTA-free (cat. no. A32955, Sigma-Aldrich)
-
Optional : Pierce™ Phosphatase Inhibitor Mini Tablets (cat. no. A32957, Sigma-Aldrich)
-
TRIzol™ (cat. no. 15596018, Invitrogen; or equivalent monophasic solution of phenol and guanidine isothiocyanate), 4°C
-
100% ethanol, BioUltra for molecular biology (cat. no. 51976–500ML-F, Sigma-Aldrich)
-
Nuclease- and protease-free water (e.g., UltraPure™ Distilled Water, cat. no. 10977-035, Invitrogen)
-
70% (v/v) and 75% (v/v) ethanol, diluted from 100% ethanol in nuclease-free water
-
Chloroform
-
Isopropanol (cat. no. 59304, Sigma Aldrich)
-
GlycoBlue (cat. no. AM9515, Thermofisher)
-
Centrifuge, cooled, with maximum speed of at least 13,000 rcf
-
1-ml, 200-µl, 20-µl, and 10-µl pipets
-
Fume hood
-
Liquid nitrogen or dry ice
-
Metal spatula
-
Ice
-
5- and 2-ml PCR clean tubes (cat. nos. 0030119460 and 022431048, Eppendorf)
-
Tissue homogenizer T 25 digital ULTRA-TURRAX with dispersing tool (cat. no. S25 N-8G-ST; for PEG-based hydrogels and alginate hydrogels)
-
Vortex
-
Freezer, −20°C
1.Perform steps 1 to 8 of Basic Protocol 1, 2, or 3, as appropriate.
2.Centrifuge the samples for 15 min at 13,000 rcf, 4°C, to induce phase separation. The samples should separate into a top transparent phase (aqueous phase), an opaque interphase (usually thin), and a bottom pink organic phase containing the protein.
3.Carefully recover the top, aqueous phase (containing the RNA) using a 200-µl pipet and transfer it to a fresh labeled 2-ml tube, being sure not to carry over any trace of the interphase or organic phase so as to ensure the purity of the RNA.
4.Add an equal volume of precooled isopropanol to the aqueous phase (e.g., if 500 µl of aqueous phase was recovered, add 500 µl isopropanol).
5.Add 1.5 µl of GlycoBlue co-precipitant. This will help visualize the RNA pellet in later steps.
6.Vortex briefly.
7.Incubate overnight at −20°C.
8.Spin down for 8 min at 12,000 rcf, 4°C.
9.Discard the supernatant using a pipet.
10.Wash with 1 ml of 75% ethanol. Mix gently by inversion to detach the pellet from the tube wall.
11.Spin down for 5 min at 7500 rcf, 4°C.
12.Discard the supernatant using a pipet.
13.Repeat steps 9-11 three more times.
14.Aspirate any remaining ethanol with a small pipet until the pellet is completely dry.
15.Once the pellet is completely dry, resuspend it in 20-50 µl nuclease-free water (or alternatively 0.1 mM EDTA or 0.5% SDS). Adjust the final volume depending on the required RNA concentration.
REAGENTS AND SOLUTIONS
EDTA for alginate gel dissolution, 50 mM
- Dissolve 9.3 g EDTA (cat. no. 03677, Sigma Aldrich) in 450 ml of deionized water. Stir. Add 1 M NaOH dropwise to help dissolve EDTA (EDTA will not fully dissolve until the pH reaches ∼8). Adjust the volume to 500 ml.
- Store up to 1 year at room temperature
Guanidine hydrochloride solution, 0.3 M
- Prepare a 95% ethanol solution by mixing 95 ml of 100% ethanol and 5 ml of protease-free water.
- Dissolve 2.87 g of guanidine hydrochloride in 100 ml of 95% ethanol (cat. no. G3272-25G, Sigma-Aldrich)
- Store up to 4 weeks at room temperature
Hot SDS buffer for protein resuspension, 2×
- Add 1.97 g Tris•Cl to 30 ml protease-free water (250 mM Tris•Cl final concentration)
- Add 5 g of sodium dodecyl sulfate (SDS; 10% [w/v] final)
- Add 15 ml glycerol (30% [v/v] final)
- Adjust the pH to 6.1
- Adjust the final volume to 50 ml using protease-free water
- Store at room temperature up to 4 weeks
- Dilute 1:1 in protease-free water before use
COMMENTARY
Background Information
In addition to hydrogels, there are many other 3D culture systems used in cell biology and bioengineering. The protocols detailed above were inspired by protein isolation protocols for tissue sections, formalin-fixed paraffin-embedded (FFPE) specimens, and organoids (Addis et al., 2009; Dussoyer et al., 2022; Ericsson & Nistér, 2011; Gjorevski et al., 2016). All of these 3D systems have in common that they require a matrix disaggregation step. A procedure for extracting proteins from cryopreserved tissues was proposed by Ericsson et al., combining SDS-based solubilization with frozen tissue disintegration (Ericsson & Nistér, 2011). Dussoyer et al. compared several protocols for enriching extracellular matrix proteins in mass spectrometry analysis (Dussoyer et al., 2022). Addis et al. presented a strategy to obtain high-quality protein extracts from formalin-fixed, paraffin-embedded (FFPE) tissues, which is based on antigen retrieval procedures used for immunohistochemistry (Addis et al., 2009). In this approach, the recovered proteins can then be examined further using western blotting, ELISA, protein microarrays, or mass spectrometry. Avances in proteomics analysis have led to the refinement of procedures for isolating proteins from FFPE sections over the years; however, protocol standardization in terms of buffers and parameters is still required. Garcia-Vence et al. reviewed these protocols, with the goal of establishing a process for deparaffination and protein extraction of FFPE kidney tissues for downstream proteomic analysis (García-Vence et al., 2021). Combining high incubation temperature with mechanical tissue disaggregation using RIPA lysis buffer resulted in the best protein extraction efficiency.
Protocols involving the simultaneous extraction of proteins and nucleic acids were also investigated in the context of solid tissues. Pena-Llopis et al. presented a thorough approach for extracting genomic DNA, mRNA, noncoding RNA, and proteins from tumor tissue at the same time, allowing integrative analysis of gene expression and protein levels (Peña-Llopis & Brugarolas, 2013). This was achieved by merging and adapting two different commercially available kits (Qiagen's AllPrep and Ambion's mirVana). The protocol involves homogenization of frozen tissue in alternating dry and wet ice, acid-phenol/chloroform RNA extraction, and protein precipitation in acetone. Strategies for isolating proteins from distinct cell compartments have also been proposed, as some studies require the identification of proteins from a specific location. Baghirova et al. and Schiller et al. both proposed protocols to sequentially lyse cells for protein extraction from the cytosol, membrane-bound organelles, and nucleus (Baghirova et al., 2015; Schiller et al., 2015). This method can be extended to tissue samples by adding a homogenization step.
With the advancements in organoid technologies in recent years, accurate methods for characterizing their proteomes have become critical. However, because organoids are usually cultured in soft 3D environments, isolation of intracellular proteins requires the breakdown of the surrounding matrix. Matrigel is the most common material used to support organoid culture, and its incomplete dissolution can affect proteomic analysis. Recent work from Wang et al. addresses this issue by comparing different Matrigel dissolution methods to isolate organoids (Wang et al., 2022). Their approaches involved enzymatic, non-enzymatic, and chemical strategies, with the enzymatic one being the most effective.
When using other substrates for organoid growth, the dissolution strategies vary depending on the nature of the chosen material. Several groups have used PEG-based hydrogels to encapsulate cells and organoids. Mahoney et al. encapsulated neural cells from which they analyzed the proteins and nucleic acid, and used hydrogel homogenization in lysis buffer to disrupt the polymer network (Mahoney & Anseth, 2006). Gjorevski et al. designed modular PEG-based hydrogels to define key extracellular matrix (ECM) parameters that drive organoid formation. These synthetic hydrogels were enzymatically cross-linked, and their dissociation to release the embedded cell colonies was achieved by enzymatic digestion. Others have successfully isolated proteins and/or RNA from cells embedded in agarose hydrogels (Wang et al., 2009), agarose-collagen hydrogels (Cambria et al., 2020), chitosan-based hydrogels (Yu et al., 2013), or self-assembled peptide hydrogels (Burgess et al., 2017). Finally, Sawicki et al. proposed an approach to isolate proteins secreted into the microenvironment by first decellularizing the sample and then enzymatically degrading the PEG hydrogel, cross-linked by collagenase-degradable cross-linkers.
Critical Parameters
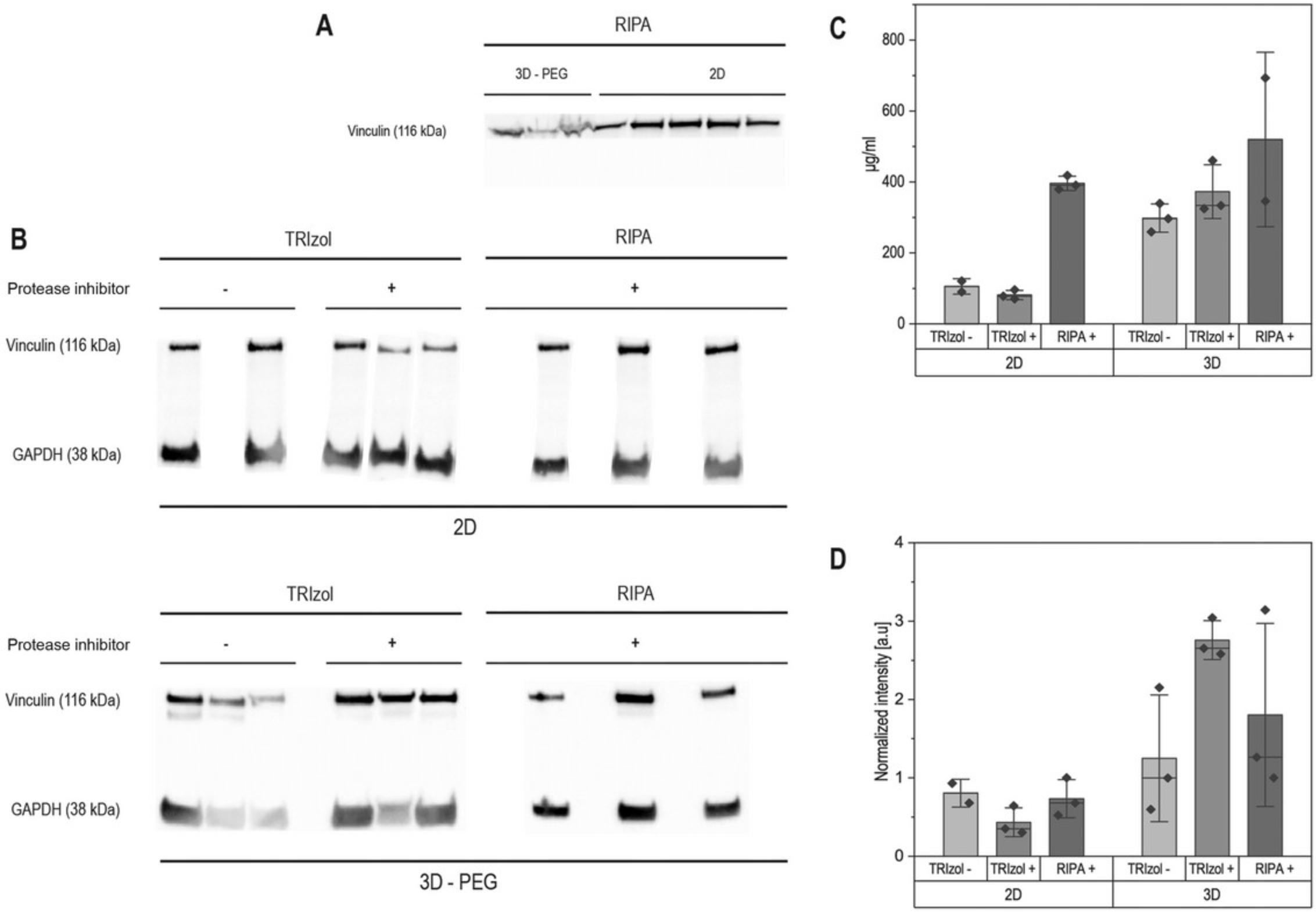
As mentioned in some of the articles cited above, the choice of lysis buffer is critical to any protein isolation process. RIPA lysis buffer is the most commonly used lysis buffer and does not require a phase-separation step. However, we found that using RIPA lysis buffer alone does not allow robust elimination of the gel phases for two reasons: the volumes of buffer typically used to obtain high protein concentration (∼100 µl per sample) are too small to allow homogenization, and manual breakage is not always sufficient to break the gels into small parts; in addition, this procedure does not include a protein purification stage, which in our hands helped to eliminate the gel phase (Fig. 2). Both these issues stem from the difficulty of fully drying a hydrogel (unless using a freeze-dryer) or of fully extracting cells from a hydrogel without a digestion step. Although digestion steps can be implemented (for example, using trypsin or collagenases), making it possible to then pellet cells, they usually require long incubation times, which can modify protein abundance. Therefore, we recommend using TRIzol™-based isolation for 3D scaffolds, as described in the protocols above. This approach allows the initial use of large volumes, fast inactivation of protease and phosphatases, and downstream pelleting of protein prior to resuspension in the desired buffer.
Note that the choice and composition of the resuspension buffer should be adapted to the target proteins and downstream application. For SDS-PAGE and western blotting, either a denaturing buffer containing high levels of SDS (“Hot SDS buffer”) or RIPA, which contains low levels of SDS (typically 0.1%), are appropriate. For mass spectrometry, SDS and strong detergents must be efficiently removed before analysis as they commonly ionize well and interfere with peptide ion identification and quantification. Native gel electrophoresis and co-immunoprecipitation applications rely on protein structure and protein-protein interactions, and therefore a mild, non-denaturing buffer (that does not contain SDS or any reducing agent) should be used (Giampietro et al., 2017; Savorani et al., 2021).
Any protein isolation protocol also requires limiting protease activity (and optionally phosphatase activity if phosphoproteins are to be investigated). This is typically done by cooling down the samples (working on ice), using denaturing agents and metal-chelating agents that block proteases action, and using protease inhibitor cocktails and phosphatase inhibitors (e.g., sodium orthovanadate, PMSF). When isolating proteins from a 3D scaffold, isolation protocols are typically longer, due to the need to break up the scaffold. Although the same principles apply, it may help to reduce batch size (process fewer samples at once) to keep total processing time short. If phospho-groups need to be maintained intact, it is recommended to add phosphatase inhibitors in the washing buffer, to have immediate inhibition of phosphatases.
Finally, the volume of the swollen hydrogel phase should be kept in mind. The fact that many hydrogels used in tissue engineering will hold 95% to 98% aqueous volume makes it difficult to wash any component out completely. Moreover, diffusible factors, including proteases, can be trapped in the scaffold pores. Typically, one needs to add several washing steps compared to standard 2D protocols and use agitation. For protein quantification using bicinchoninic acid (BCA) or Bradford assays, one must wash out the serum component of the medium before cell collection and lysis, as otherwise there is a risk of the serum components dominating the protein content. Multiple washes in serum-free medium, or a period of serum starvation of at least 12 hr or overnight, are recommended. For the phase-separation step, the volume of the scaffold and of the contained fluid phase must be accounted for, with sufficient TRIzol™ used per sample volume.
Troubleshooting
Further tips to troubleshoot results are given in Table 3.
| Problem | Possible cause | Solution |
|---|---|---|
| Hydrogel is difficult to rinse | The hydrogel might be a low-polymer fraction and hold a large volume of fluid (medium, buffer, etc.) | Increase the number of washes |
| Phase separation does not have clear distinct phases | Too high an aqueous phase content, due to volume carried by the hydrogel | Adapt the volume of chloroform to 20% of total sample volume |
| Too high a gel content relative to TRIzol™ volume | Adjust TRIzol™ volume so that it exceeds gel volume by ∼10-fold | |
| Aqueous phase is not clear but opaque or pink | TRIzol™ and chloroform were not mixed properly, or too much chloroform was added |
Repeat the phase separation Add more TRIzol™ |
| Protein pellet does not dissolve | The pellet may contain some undissolved polymer phase | Add a second ethanol wash step to remove any remaining polymer phase |
| The pellet may contain a lot of fibrous protein from the extracellular matrix | Sonicate the sample for a few minutes | |
| Low protein yield | Low cell density | Seed more cells |
| Low efficiency of the isolation process | Increase incubation times; ensure proper gel breakup/dissolution | |
| Protein quantification is not accurate | Serum contained in the medium is the dominant contribution to quantification assay, confounding readings | Starve the cells or rinse with serum-free medium multiple times before harvesting |
| Phosphoprotein marker is absent | Process was done without inactivating phosphatases | Add phosphatase inhibitors to the first buffer used to wash; work on ice or at 4°C; resuspend in a denaturing buffer to inactivate phosphatases |
| Cell density is too low | Increase cell seeding density |
Understanding Results
Results of protein isolation protocols are typically evaluated by (i) quantifying yield using a protein absorbance assays (BCA or Bradford), (ii) running an SDS-PAGE and western blot, or (iii) quantifying protein using mass spectrometry. Other downstream assays include IP or co-IP and native PAGE. We discuss here how a successful isolation procedure manifests in the downstream assays, as well as potential problems (see also Table 3).
Absorbance assays for protein quantification
We quantified our protein yield from all three protocols using the BCA. This assay is based on the reduction of Cu2+ by proteins and the colorimetric detection of Cu+. It is advisable to check the solvent compatibility chart of the BCA assay to ensure that the buffer used for protein resuspension will not interfere with the read-out. We typically obtained protein concentrations of 100 to 500 µg/ml when resuspending pellets in 100 µl of Hot SDS buffer. The BCA assay has a detection range of 5 to 2000 µg/ml and requires 25 µl of each sample (Fig. 2).
Western blotting
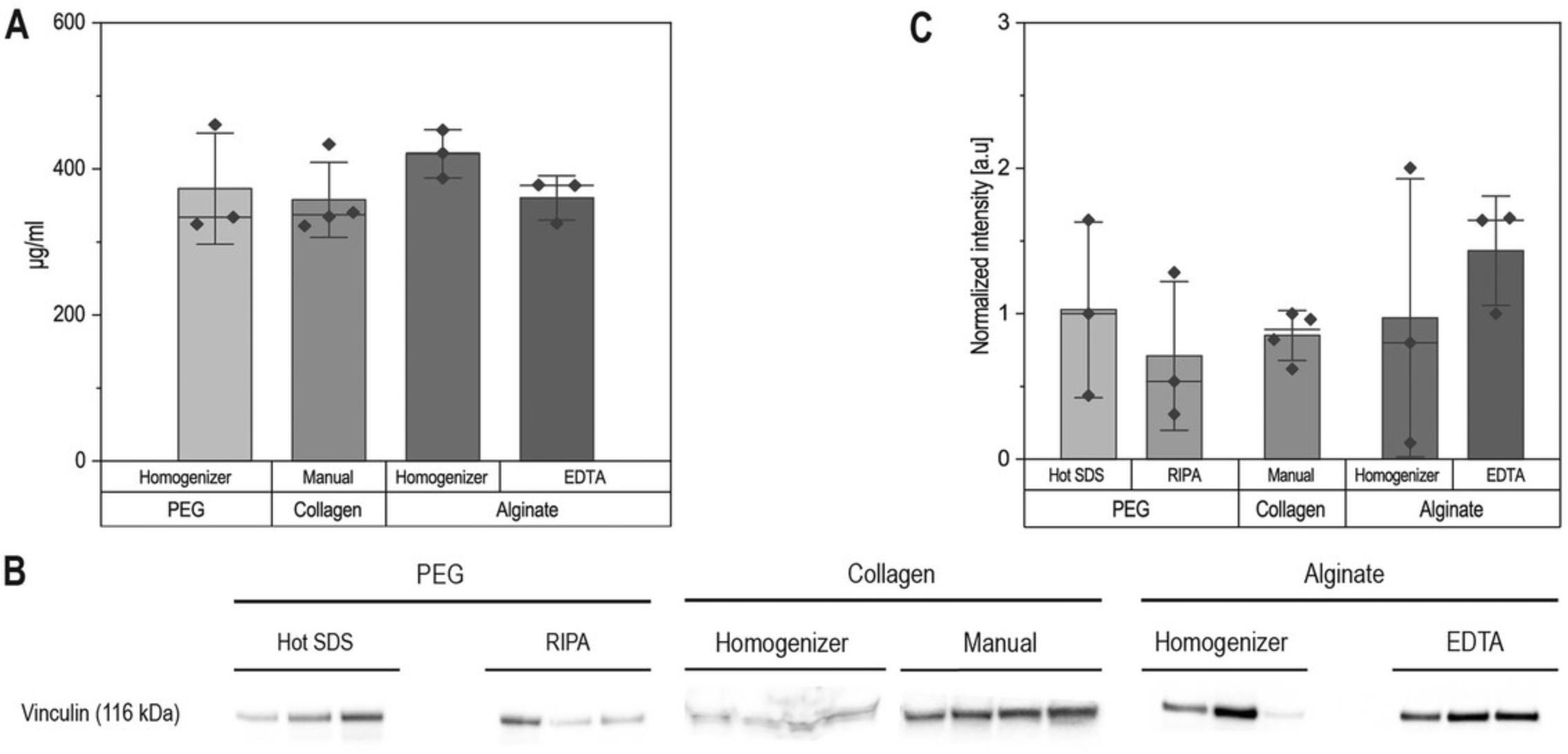
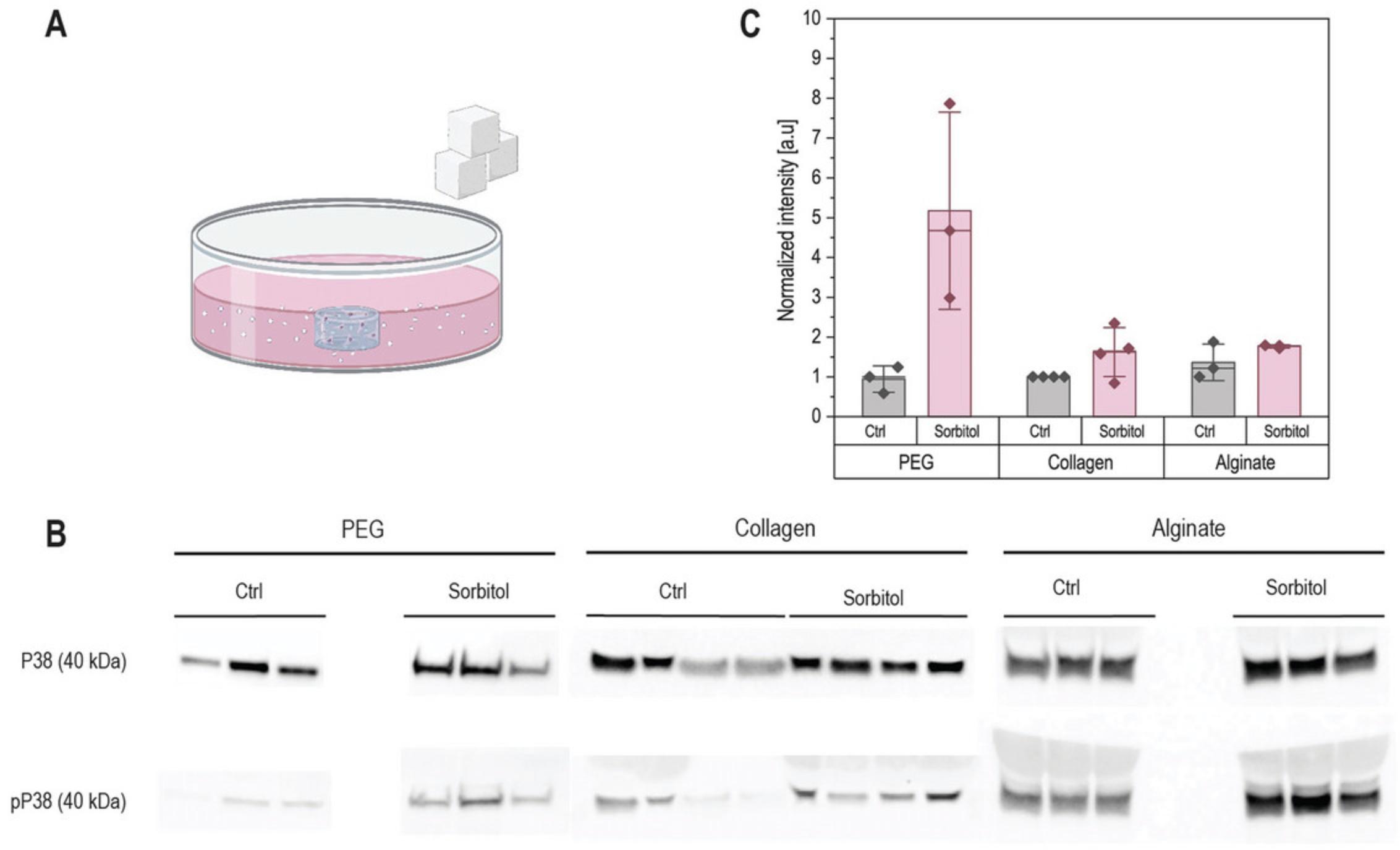
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) is typically done with at least 5 µg of protein (or 20 µg if staining for phosphoproteins). Samples in which the hydrogel polymer components have not been fully removed will show band distortions in the high-molecular-weight regions and can compromise band quantification (Fig. 2). Proper hydrogel breakup and protein purification should lead to straight bands (Fig. 3). We used Basic Protocols 1, 2, and 3 to isolate proteins from PEG-based, collagen, and alginate hydrogels, and stained for housekeeping protein (Fig. 3). We then used a hyperosmotic stress known to activate the P38 MAP kinase by phosphorylation to verify that our protocols preserve phospho-groups, showing increased levels of phospho-P38 (pP38) in treated samples (Fig. 4). Detailed methods used for western blotting are given in the Supporting Information.
Mass spectrometry
For bottom-up proteomics, liquid chromatography–tandem mass spectrometry (LC-MS/MS) requires further tryptic protein digestion into peptides. This is typically done by starting from a protein pellet diluted in a detergent-containing buffer such as RIPA buffer or 5% sodium deoxycholate. Samples can be sonicated to support resuspension. A starting material of 10 µg of protein is recommended. Any remaining scaffold polymer can also affect the LC-MS/MS analysis and may appear with a strong signature in the MS spectra. For example, PEG as a polymer of (O−CH2−CH2) units can be easily identified by ions with a characteristic 44-Da difference in mass. By using the SP3 technology for MS sample preparation (Hughes et al., 2019), we verified that we can obtain ∼5500 protein identifications using single-shot, data-independent acquisition (DIA)-LC-MS/MS starting from ∼15,000 to 20,000 cells encapsulated in alginate gels (with 18 days of culture and an average 270-hr doubling time), using Basic Protocol 3 (Fig. 5). Detailed methods used for mass spectrometry are given in the Supporting Information.
Time Considerations
Processing around six frozen samples can take up to 3 hr on each processing day (with an overnight incubation step in between), due to the number of incubation steps and washing steps. The time-limiting step is the breaking up of the hydrogel scaffold, as both mechanical breaking and homogenization must be done on each sample individually, limiting the throughput. In the subsequent processing steps (phase separation, purification, and washing), multiple samples can be processed together. Processing samples in batches of 6 to 8 makes it possible to ensure that all samples are incubated for the same amount of time in TRIzol™/chloroform after the initial homogenization or manual breaking. When isolating RNA and protein from the same samples (Support Protocol), it is important that both the RNA isolation and protein isolation be done immediately after phase separation. Therefore, here again, it is important to keep the number of samples manageable. The RNA precipitation in isopropanol can be done overnight, or also shortened to a few hours.
Acknowledgments
This work was supported by funds from the ETH Zurich (Open ETH project SKINTEGRITY.CH) and the Swiss National Science Foundation under project number SNF NRP79 (SNF_206394_NFP79). For the purpose of Open Access, a CC BY public copyright license is applied to any Author Accepted Manuscript (AAM) version arising from this submission.
Author Contributions
Gabriela Da Silva André : Conceptualization; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review and editing. Lorenza Garau Paganella : Conceptualization; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review and editing. Asia Badolato : Conceptualization; formal analysis; investigation; methodology; validation; writing—original draft; writing—review and editing. Sibilla Sander : Conceptualization; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review and editing. Costanza Giampietro : Conceptualization; methodology; supervision; visualization; writing—original draft; writing—review and editing. Mark W Tibbitt : Conceptualization; funding acquisition; methodology; supervision; visualization; writing—original draft; writing—review and editing. Jörn Dengjel : Conceptualization; funding acquisition; methodology; supervision; visualization; writing—original draft; writing—review and editing. Céline Labouesse : Conceptualization; investigation; methodology; supervision; validation; visualization; writing—original draft; writing—review and editing.
Conflict of Interest
All authors declare no conflict of interest.
Open Research
Data Availability Statement
The data, tools, and materials (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Supporting Information
| Filename | Description |
|---|---|
| cpz1966-sup-0001-SuppMat.docx3.1 MB | Additional methods for (i) fabrication of hydrogels, (ii) sorbitol treatment, (iii) western blotting, and (iv) mass spectrometry, along with Supplementary Figures S1 to S7 showing uncropped versions of the western blots in Figures 2 to 4. |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Addis, M. F., Tanca, A., Pagnozzi, D., Crobu, S., Fanciulli, G., Cossu-Rocca, P., & Uzzau, S. (2009). Generation of high-quality protein extracts from formalin-fixed, paraffin-embedded tissues. Proteomics , 9(15), 3815–3823. https://doi.org/10.1002/pmic.200800971
- Baghirova, S., Hughes, B. G., Hendzel, M. J., & Schulz, R. (2015). Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX , 2, 440–445. https://doi.org/10.1016/j.mex.2015.11.001
- Balasubramanian, S., Yu, K., Meyer, A. S., Karana, E., & Aubin-Tam, M.-E. (2021). Bioprinting of regenerative photosynthetic living materials. Advanced Functional Materials , 31(31), 2011162. https://doi.org/10.1002/adfm.202011162
- Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., Fridman, W.-H., Pagès, F., Trajanoski, Z., & Galon, J. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics , 25(8), 1091–1093. https://doi.org/10.1093/bioinformatics/btp101
- Bretherton, R. C., Haack, A. J., Kopyeva, I., Rahman, F., Kern, J. D., Bugg, D., Theberge, A. B., Davis, J., & DeForest, C. A. (2023). User-controlled 4D biomaterial degradation with substrate-selective sortase transpeptidases for single-cell biology. Advanced Materials , 35(19), 2209904. https://doi.org/10.1002/adma.202209904
- Burgess, K. A., Miller, A. F., Oceandy, D., & Saiani, A. (2017). Western blot analysis of cells encapsulated in self-assembling peptide hydrogels. BioTechniques , 63(6), 253–260. https://doi.org/10.2144/000114617
- Caliari, S. R., & Burdick, J. A. (2016). A practical guide to hydrogels for cell culture. Nature Methods , 13(5), 5. https://doi.org/10.1038/nmeth.3839
- Cambria, E., Brunner, S., Heusser, S., Fisch, P., Hitzl, W., Ferguson, S. J., & Wuertz-Kozak, K. (2020). Cell-laden agarose-collagen composite hydrogels for mechanotransduction studies. Frontiers in Bioengineering and Biotechnology , 8, 346. https://doi.org/10.3389/fbioe.2020.00346
- Chaudhuri, O., Gu, L., Klumpers, D., Darnell, M., Bencherif, S. A., Weaver, J. C., Huebsch, N., Lee, H., Lippens, E., Duda, G. N., & Mooney, D. J. (2016). Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nature Materials , 15(3), 326–334. https://doi.org/10.1038/nmat4489
- Dussoyer, M., Page, A., Delolme, F., Rousselle, P., Nyström, A., & Moali, C. (2022). Comparison of extracellular matrix enrichment protocols for the improved characterization of the skin matrisome by mass spectrometry. Journal of Proteomics , 251, 104397. https://doi.org/10.1016/j.jprot.2021.104397
- Ericsson, C., & Nistér, M. (2011). Protein extraction from solid tissue. In J. Dillner (Ed.), Methods in Biobanking (Vol. 675, pp. 307–312). Humana Press. https://doi.org/10.1007/978-1-59745-423-0_17
- García-Vence, M., Chantada-Vazquez, M. D. P., Sosa-Fajardo, A., Agra, R., Barcia de la Iglesia, A., Otero-Glez, A., García-González, M., Cameselle-Teijeiro, J. M., Nuñez, C., Bravo, J. J., & Bravo, S. B. (2021). Protein extraction from FFPE kidney tissue samples: A review of the literature and characterization of techniques. Frontiers in Medicine , 8, 657313. https://doi.org/10.3389/fmed.2021.657313
- Giampietro, C., Lionetti, M. C., Costantini, G., Mutti, F., Zapperi, S., & La Porta, C. A. M. (2017). Cholesterol impairment contributes to neuroserpin aggregation. Scientific Reports , 7(1), 1. https://doi.org/10.1038/srep43669
- Gjorevski, N., Sachs, N., Manfrin, A., Giger, S., Bragina, M. E., Ordóñez-Morán, P., Clevers, H., & Lutolf, M. P. (2016). Designer matrices for intestinal stem cell and organoid culture. Nature , 539(7630), 7630. https://doi.org/10.1038/nature20168
- Hughes, C. S., Moggridge, S., Müller, T., Sorensen, P. H., Morin, G. B., & Krijgsveld, J. (2019). Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nature Protocols , 14(1), 68–85. https://doi.org/10.1038/s41596-018-0082-x
- Kloxin, A. M., Tibbitt, M. W., & Anseth, K. S. (2010). Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nature Protocols , 5(12), 12. https://doi.org/10.1038/nprot.2010.139
- Lutolf, M. P., & Hubbell, J. A. (2005). Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature Biotechnology , 23(1), 47–55. https://doi.org/10.1038/nbt1055
- Mahoney, M. J., & Anseth, K. S. (2006). Three-dimensional growth and function of neural tissue in degradable polyethylene glycol hydrogels. Biomaterials , 27(10), 2265–2274. https://doi.org/10.1016/j.biomaterials.2005.11.007
- Peña-Llopis, S., & Brugarolas, J. (2013). Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nature Protocols , 8(11), 11. https://doi.org/10.1038/nprot.2013.141
- Rhee, S. (2009). Fibroblasts in three dimensional matrices: Cell migration and matrix remodeling. Experimental & Molecular Medicine, 41(12), 858–865. https://doi.org/10.3858/emm.2009.41.12.096
- Savorani, C., Malinverno, M., Seccia, R., Maderna, C., Giannotta, M., Terreran, L., Mastrapasqua, E., Campaner, S., Dejana, E., & Giampietro, C. (2021). A dual role of YAP in driving TGFβ-mediated endothelial-to-mesenchymal transition. Journal of Cell Science , 134(15), jcs251371. https://doi.org/10.1242/jcs.251371
- Sawicki, L. A., Choe, L. H., Wiley, K. L., Lee, K. H., & Kloxin, A. M. (2018). Isolation and identification of proteins secreted by cells cultured within synthetic hydrogel-based matrices. ACS Biomaterials Science & Engineering, 4(3), 836–845. https://doi.org/10.1021/acsbiomaterials.7b00647
- Schiller, H. B., Fernandez, I. E., Burgstaller, G., Schaab, C., Scheltema, R. A., Schwarzmayr, T., Strom, T. M., Eickelberg, O., & Mann, M. (2015). Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Molecular Systems Biology , 11(7), 819. https://doi.org/10.15252/msb.20156123
- Valdez, J., Cook, C. D., Ahrens, C. C., Wang, A. J., Brown, A., Kumar, M., Stockdale, L., Rothenberg, D., Renggli, K., Gordon, E., Lauffenburger, D., White, F., & Griffith, L. (2017). On-demand dissolution of modular, synthetic extracellular matrix reveals local epithelial-stromal communication networks. Biomaterials , 130, 90–103. https://doi.org/10.1016/j.biomaterials.2017.03.030
- J. M. Walker (Ed.). (2009). The Protein Protocols Handbook. Humana Press. https://doi.org/10.1007/978-1-59745-198-7
- Wang, C., Li, X., Yao, Y., & Wang, D.-A. (2009). Concurrent extraction of proteins and RNA from cell-laden hydrogel scaffold free of polysaccharide interference. Journal of Chromatography B , 877(29), 3762–3766. https://doi.org/10.1016/j.jchromb.2009.09.012
- Wang, M., Yu, H., Zhang, T., Cao, L., Du, Y., Xie, Y., Ji, J., & Wu, J. (2022). In-depth comparison of Matrigel dissolving methods on proteomic profiling of organoids. Molecular & Cellular Proteomics, 21(1), 100181. https://doi.org/10.1016/j.mcpro.2021.100181
- Yu, C., Young, S., Russo, V., Amsden, B. G., & Flynn, L. E. (2013). Techniques for the isolation of high-quality RNA from cells encapsulated in chitosan hydrogels. Tissue Engineering. Part C, Methods , 19(11), 829–838. https://doi.org/10.1089/ten.tec.2012.0693
Citing Literature
Number of times cited according to CrossRef: 1
- Lorenza Garau Paganella, Asia Badolato, Céline Labouesse, Gabriel Fischer, Catharina S. Sänger, Andreas Kourouklis, Costanza Giampietro, Sabine Werner, Edoardo Mazza, Mark W. Tibbitt, Variations in fluid chemical potential induce fibroblast mechano-response in 3D hydrogels, Biomaterials Advances, 10.1016/j.bioadv.2024.213933, 163 , (213933), (2024).

