Primary Data Analysis - Basecalling, Demultiplexing, and Consensus Building for ONT Fungal Barcodes
Stephen Douglas Russell
Abstract
This protocol assumes that your MinION run has been completed and the data from the run has been saved. It should take you from raw data to usable FASTA files containing consensus sequences for each of your fungal barcodes.
Note: This protocol assumes you are using 10.4.1 flowcells with V14 chemistry, so there is a process to filter out and combine your duplex reads with your simplex reads. It will also show the average quality scores from each simplex and duplex component.
Steps
Initial Post-Run Preparation
This protocol assumes the experiment name is "FirstRun."
Create a new working folder on the desktop. Ex - FirstRun. Within that create a new folder called "fast5," another called "Programs," and a final one called "NGSpeciesID."
I will start by copying all of the fast5 files from:
/var/lib/minknow/data/./FirstRun/CellName/long_unique_name/fast5 to the newly created fast5 folder on the desktop
Create an index file from your extraction template papers. This will allow you to link all of your reads with the individual specimens. A template for 10 plates (960 specimens) can be found here:
NANOPORE TEMPLATE SEVENTH RUN.xlsx
This .xlsx is formatted to utilize the Lab Code and iNaturalist # columns as the only inputs. It will combine these and all of the other columns into a single cell - concatenating them all into the final file name. For the Lab Code, I will typically put these into the iNaturalist "Voucher Number(s)" Observational Field, and then export them all at once into a .csv from iNat. This allows me to simply copy and paste many iNat numbers at once, without ever needing to input any of the numbers manually.
After editing, save as a tab-delimited text file in the NGSpeciesID folder. You will need to remove most of the final columns from the template. The final output should be saved like this:
Copy these Python scripts into the Programs folder you just created.
My shorthand "quick" guide for all of the following commands can be found in the file below. I use this file whenever working through this protocol. It is saved on my desktop. Just copy-and-paste the commands sequentially into the terminal.
SUP Basecalling with Guppy
Run the basecalling command. The command below uses Super-accuracy mode with Guppy.
guppy_basecaller -x "cuda:all" -i ~/Desktop/FirstRun/fast5 -s ~/Desktop/FirstRun/simplex_calls --config dna_r10.4.1_e8.2_400bps_sup.cfg --records_per_fastq 0 --trim_adapters --trim_strategy dna --chunks_per_runner 256
For a Flongle cell with 1.15Gb of bases and 700 - 1.18M reads, this command takes about 2h 0m 0s to run. Example output:
Sometimes after the run I need to restart the CPU before this command runs successfully.
This command will install the duplex tools you will need for filtering them out and combining them with the simplex reads.
python -m venv venv --prompt duplex
. venv/bin/activate
pip install duplex_tools
Around 20-30% of the reads in your final results will be duplex reads - the single fragment is read twice - once in the forward direction and one in the reverse direction. These reads need to be identified with the command below.
duplex_tools pairs_from_summary ~/Desktop/FirstRun/simplex_calls/sequencing_summary.txt ~/Desktop/FirstRun/simplex_calls/pairs
duplex_tools filter_pairs ~/Desktop/FirstRun/simplex_calls/pairs/pair_ids.txt ~/Desktop/FirstRun/simplex_calls/pass
Rerun Guppy basecalling with the duplex reads alone that were parsed out in the previous step. This step may take about 1h 15m 0s.
guppy_basecaller_duplex \
-i ~/Desktop/FirstRun/fast5 \
-r -s ~/Desktop/FirstRun/duplex_calls \
-x 'cuda:all' -c dna_r10.4.1_e8.2_260bps_sup.cfg \
--chunks_per_runner 256 \
--duplex_pairing_mode from_pair_list \
--duplex_pairing_file ~/Desktop/FirstRun/simplex_calls/pairs/pair_ids_filtered.txt
Combine all FASTQ files into a single file for simplex and duplex calls, show the read counts, and zip them.
cat ~/Desktop/FirstRun/simplex_calls/pass/*runid*.fastq > ~/Desktop/FirstRun/simplex_calls/pass/basecall.fastq
cat ~/Desktop/FirstRun/simplex_calls/pass/basecall.fastq | wc -l | awk '{print $1/4}'
gzip ~/Desktop/FirstRun/simplex_calls/pass/basecall.fastq
cat ~/Desktop/FirstRun/duplex_calls/pass/*runid*.fastq > ~/Desktop/FirstRun/duplex_calls/pass/basecall.fastq
cat ~/Desktop/FirstRun/duplex_calls/pass/basecall.fastq | wc -l | awk '{print $1/4}'
gzip ~/Desktop/FirstRun/duplex_calls/pass/basecall.fastq
gzip ~/Desktop/FirstRun/duplex_calls/pass/*.fastq
Remove the uncombined FASTQ files.
rm ~/Desktop/FirstRun/simplex_calls/pass/*runid*.fastq
Validate the QC of the Run
Compile quality control summary charts for the simplex and duplex reads.
cd ~/Desktop/FirstRun/Programs
Rscript MinIONQC.R -i ~/Desktop/FirstRun/simplex_calls/sequencing_summary.txt -o ~/Desktop/FirstRun/QC-Reports-simplex
Rscript MinIONQC.R -i ~/Desktop/FirstRun/duplex_calls/sequencing_summary.txt -o ~/Desktop/FirstRun/QC-Reports-duplex
Review the images that are generated. Ensure the quality scores of your run are in an appropriate range. For a 10.4.1 Flongle with "Q20+" V14 chemistry, I typically get a peak in the 15-16 range.
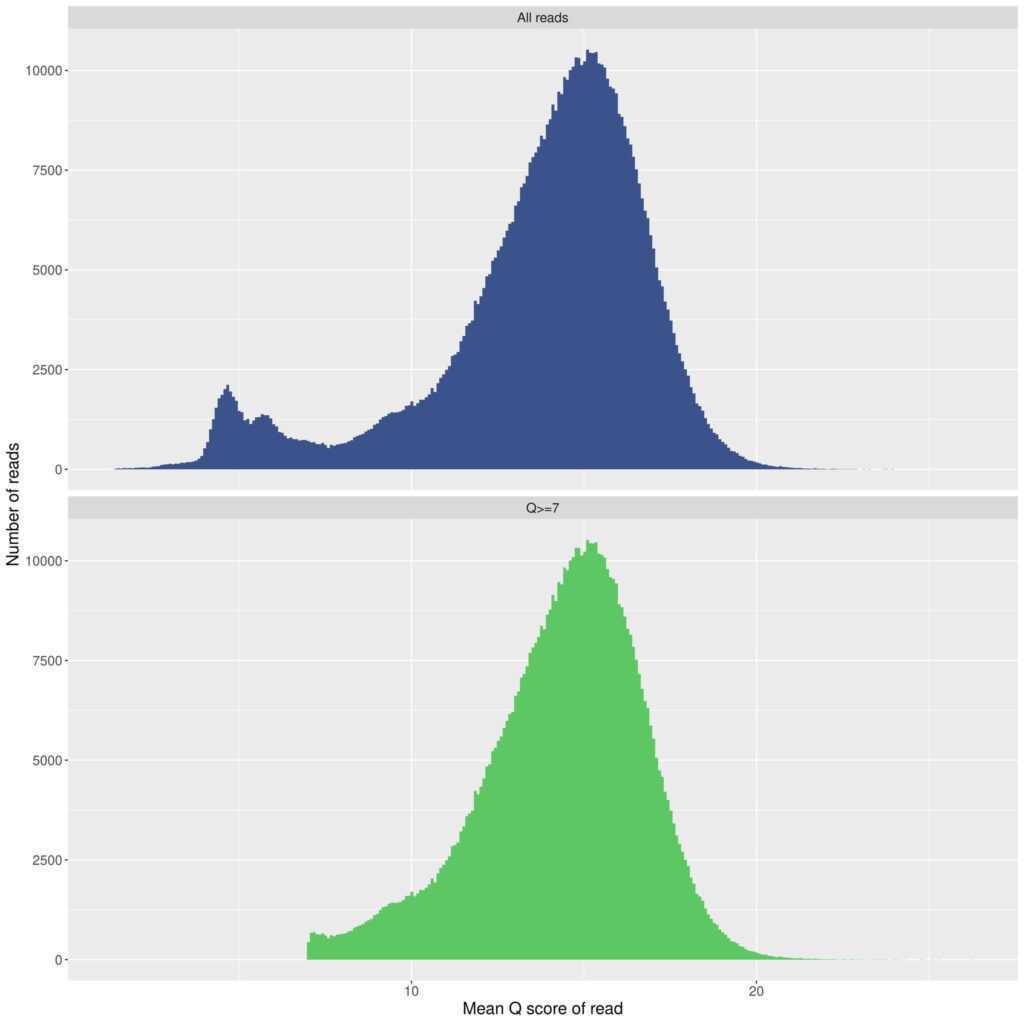
Example of all outputs from this command: MinIONQC.zip
Merge the Simplex and Duplex Reads
Use seqkit to extract all the filenames of reads from the simplex run.
seqkit seq --name ~/Desktop/FirstRun/simplex_calls/pass/basecall.fastq.gz > ~/Desktop/FirstRun/simplex_calls/pass/simplex_ids_txt
Create additional file structure and move around necessary files.
mkdir ~/Desktop/FirstRun/combined_bases/
cd ~/Desktop/FirstRun/combined_bases/
cp ~/Desktop/FirstRun/simplex_calls/pass/basecall.fastq.gz ~/Desktop/FirstRun/combined_bases/simplex_basecall.fastq.gz
cp ~/Desktop/FirstRun/duplex_calls/pass/basecall.fastq.gz ~/Desktop/FirstRun/combined_bases/duplex_basecall.fastq.gz
cp ~/Desktop/FirstRun/simplex_calls/pass/simplex_ids_txt ~/Desktop/FirstRun/combined_bases/simplex_ids_txt
cp ~/Desktop/FirstRun/simplex_calls/pairs/pair_ids_filtered.txt ~/Desktop/FirstRun/combined_bases/pair_ids_filtered.txt
Replace the reads in the simplex with the duplex reads.
{ sed 's/ /\n/' ~/Desktop/FirstRun/combined_bases/pair_ids_filtered.txt | \
seqkit grep -v -f - ~/Desktop/FirstRun/combined_bases/simplex_basecall.fastq.gz ; \
zcat ~/Desktop/FirstRun/duplex_calls/pass/*.fastq.gz ; } \
| gzip - > combined.fastq.gz
Note: This code was derived from a post in the ONT Community Forums. It may be in need of some future revision.
Manually unzip the file here: ~/Desktop/FirstRun/combined_bases/combined.fastq.gz
Review the total number of reads in your final file and move some files for additional analysis.
cat ~/Desktop/FirstRun/combined_bases/combined.fastq | wc -l | awk '{print $1/4}'
cp ~/Desktop/FirstRun/combined_bases/combined.fastq ~/Desktop/FirstRun/NGSpeciesID/combined.fastq
cp ~/Desktop/FirstRun/Programs/minibar.py ~/Desktop/FirstRun/NGSpeciesID/minibar.py
cp ~/Desktop/FirstRun/Programs/primers.txt ~/Desktop/FirstRun/NGSpeciesID/primers.txt
cp ~/Desktop/FirstRun/Programs/Index.txt ~/Desktop/FirstRun/NGSpeciesID/Index.txt
cp ~/Desktop/FirstRun/Programs/summarize.py ~/Desktop/FirstRun/NGSpeciesID/summarize.py
Demultiplex the Reads
Demultiplex your samples using MiniBar. This should take less than 0h 5m 0s.
cd ~/Desktop/FirstRun/NGSpeciesID
./minibar.py -F Index.txt combined.fastq
Remove several large files that are not necessary for most use cases and will just make your final analysis take longer.
rm combined.fastq
rm sample_Multiple_Matches.fastq
rm sample_unk.fastq
Create the Final Consensus Sequences
Utilize NGSpecies ID to generate your final consensus sequences from your demultiplexed samples. This can take 8h 0m 0s. It would be great if someone could get this to run through the GPU.
conda activate NGSpeciesID
for file in *.fastq; do
bn=`basename $file .fastq`
NGSpeciesID --ont --consensus --sample_size 500 --m 730 --s 400 --medaka --primer_file primers.txt --fastq $file --outfolder ${bn}
done
Summarize the Data
Create a summary file for your results.
python summarize.py ~/Desktop/FirstRun/NGSpeciesID

