Preparation and Co-Culture of iPSC-Derived Dopaminergic Neurons and Astrocytes
Aurelie de Rus Jacquet, Aurelie de Rus Jacquet
astrocytes
co-culture
dopaminergic neurons
iPSC differentiation
non-cell-autonomous
Parkinson's disease
Abstract
Induced pluripotent stem cell (iPSC)-based models are powerful tools to study neurodegenerative diseases such as Parkinson's disease. The differentiation of patient-derived neurons and astrocytes allows investigation of the molecular mechanisms responsible for disease onset and development. In particular, these two cell types can be mono- or co-cultured to study the influence of cell-autonomous and non-cell-autonomous contributors to neurodegenerative diseases. We developed a streamlined procedure to produce high-quality/high-purity cultures of dopaminergic neurons and astrocytes that originate from the same population of midbrain floor-plate progenitors. This unit describes differentiation, quality control, culture parameters, and troubleshooting tips to ensure the highest quality and reproducibility of research results. © 2019 The Authors.
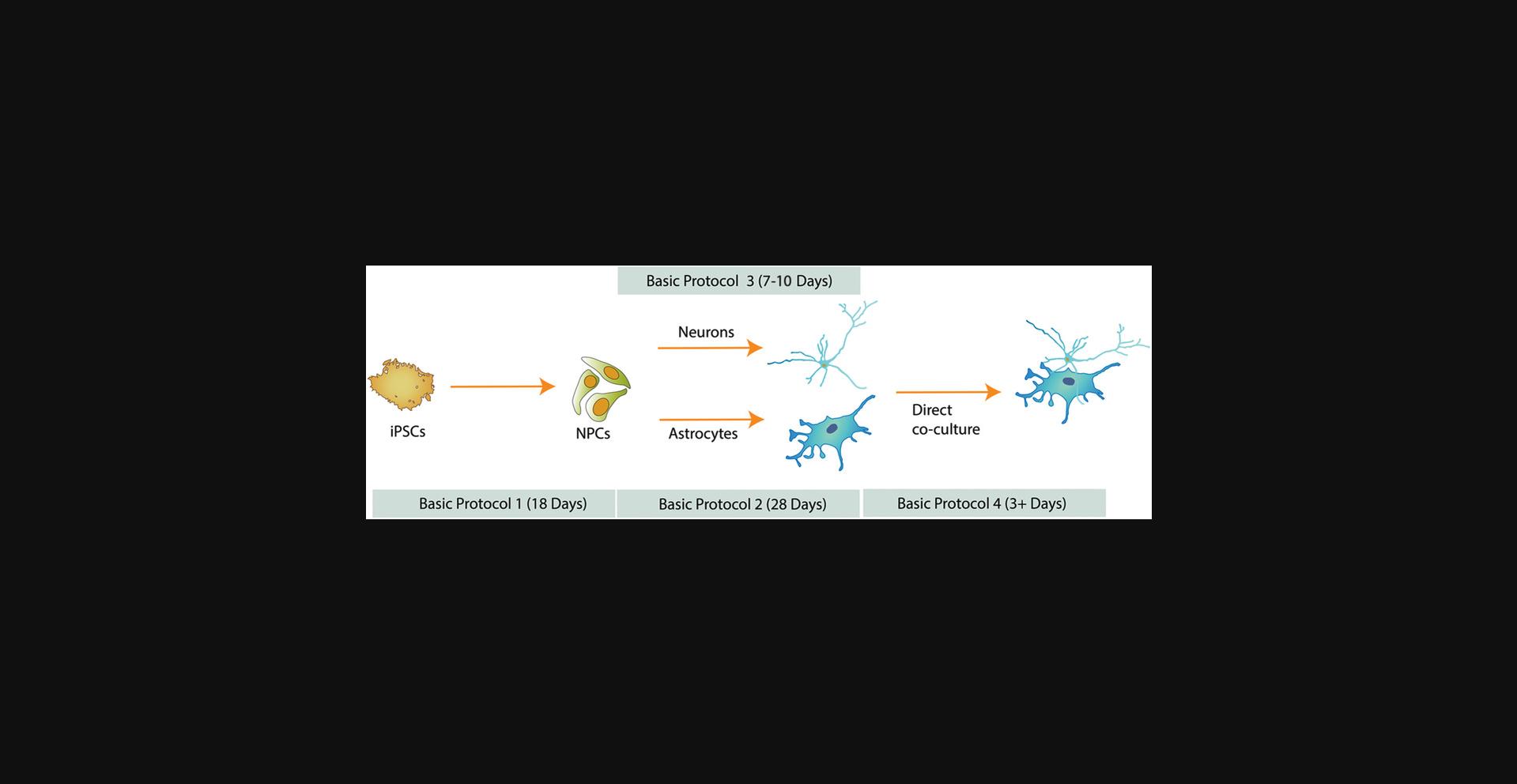
Basic Protocol 1 : Differentiation of iPSCs into midbrain-patterned neural progenitor cells
Support Protocol : Quality control of neural progenitor cells
Basic Protocol 2 : Differentiation of neural progenitor cells into astrocytes
Basic Protocol 3 : Differentiation of neural progenitor cells into dopaminergic neurons
Basic Protocol 4 : Co-culture of iPSC-derived neurons and astrocytes
INTRODUCTION
The study of neurodegenerative diseases such as Parkinson's disease has long been hampered by the lack of models that accurately reproduce human brain cells in vitro. However, in recent years, major technical advances enabled efficient differentiation of induced pluripotent stem cells (iPSCs) into different cell types, including dopaminergic neurons and astrocytes (Emdad, D'Souza, Kothari, Qadeer, & Germano, 2012; Kriks et al., 2011; TCW et al., 2017). These iPSC-differentiated cells carry the same genetic signature as the donor and allow researchers to interrogate the causes of neurodegenerative diseases. This unit describes several protocols to differentiate human iPSCs into midbrain-patterned neural progenitor cells (NPCs), astrocytes, and neurons. Importantly, it allows production of dopaminergic neurons and astrocytes originating from the same midbrain-patterned NPC population. Astrocytes possess core properties as well as specialized functions determined by their regional identity (Chai et al., 2017; Yeh, Lee, Gianino, & Gutmann, 2009), and this specialization may play a role in the development and progression of neurodegenerative diseases (Clarke et al., 2018; Kostuk, Cai, & Iacovitti, 2019). Therefore, the ability to produce astrocytes and neurons originating from the same population of midbrain-patterned NPCs is a key advantage for studying neurodegenerative diseases such as Parkinson's disease. The interaction and chemical communication between neurons and astrocytes are critical to maintaining neuronal health, and dysfunctional astrocytes have been shown to induce neurodegeneration (Di Giorgio, Boulting, Bobrowicz, & Eggan, 2008; Tyzack et al., 2017). Several cell culture systems allow study of astrocyte-neuron communication, and this unit describes establishment and maintenance of a co-culture of dopaminergic neurons and astrocytes, with four detailed protocols and one support protocol for differentiating iPSCs into midbrain-patterned NPCs (Basic Protocol 1) and performing quality control of these cells (Support Protocol), differentiating NPCs into astrocytes (Basic Protocol 2) and dopaminergic neurons (Basic Protocol 3), and co-culturing these dopaminergic neurons and astrocytes (Basic Protocol 4).
STRATEGIC PLANNING
The timing of differentiation varies depending on the cell type of interest (Fig. 1). As a consequence, it is important to anticipate and time the production of each cell type according to the desired experiment. NPCs and astrocytes can be produced in large quantities and cryopreserved to make stocks readily available to users. Successful cryopreservation of iPSC-derived neurons has also been described (Wakeman et al., 2017); however, we usually prepare fresh neurons 7 to 10 days before a co-culture experiment.
NOTE : All culture incubations are at 37°C unless otherwise specified.
NOTE : Cells should be handled according to BSL-2 regulations. Cells and reagents must be maintained under sterile conditions at all times, and biohazard waste needs to be disposed of accordingly.
Basic Protocol 1: DIFFERENTIATION OF iPSCs INTO MIDBRAIN-PATTERNED NEURAL PROGENITOR CELLS
NPCs have the ability to differentiate into neural cell types including neurons and astrocytes (Tcw et al., 2017). When these NPCs are patterned toward a specific regional identity, they can produce neurons specific to that brain region (Kirkeby et al., 2012; Kriks et al., 2011). The production of iPSC-derived dopaminergic neurons described by Kriks et al. (2011) is a two-phase protocol, with initial midbrain floor-plate patterning of neural progenitors, followed by differentiation of these progenitors into neurons (Kriks et al., 2011). The protocol described here is an adaptation of that original protocol: it adheres to Kriks’ initial 11 days of differentiation and then differs by establishing a population of self-renewing midbrain-patterned NPCs. The advantage of this method is the possibility to produce large numbers of NPCs that can be frozen and stored. These NPCs can be returned to culture and readily used to produce astrocytes (Basic Protocol 2) and neurons (Basic Protocol 3), alleviating the need for maintaining iPSCs and reducing the risk of variability between downstream differentiations.
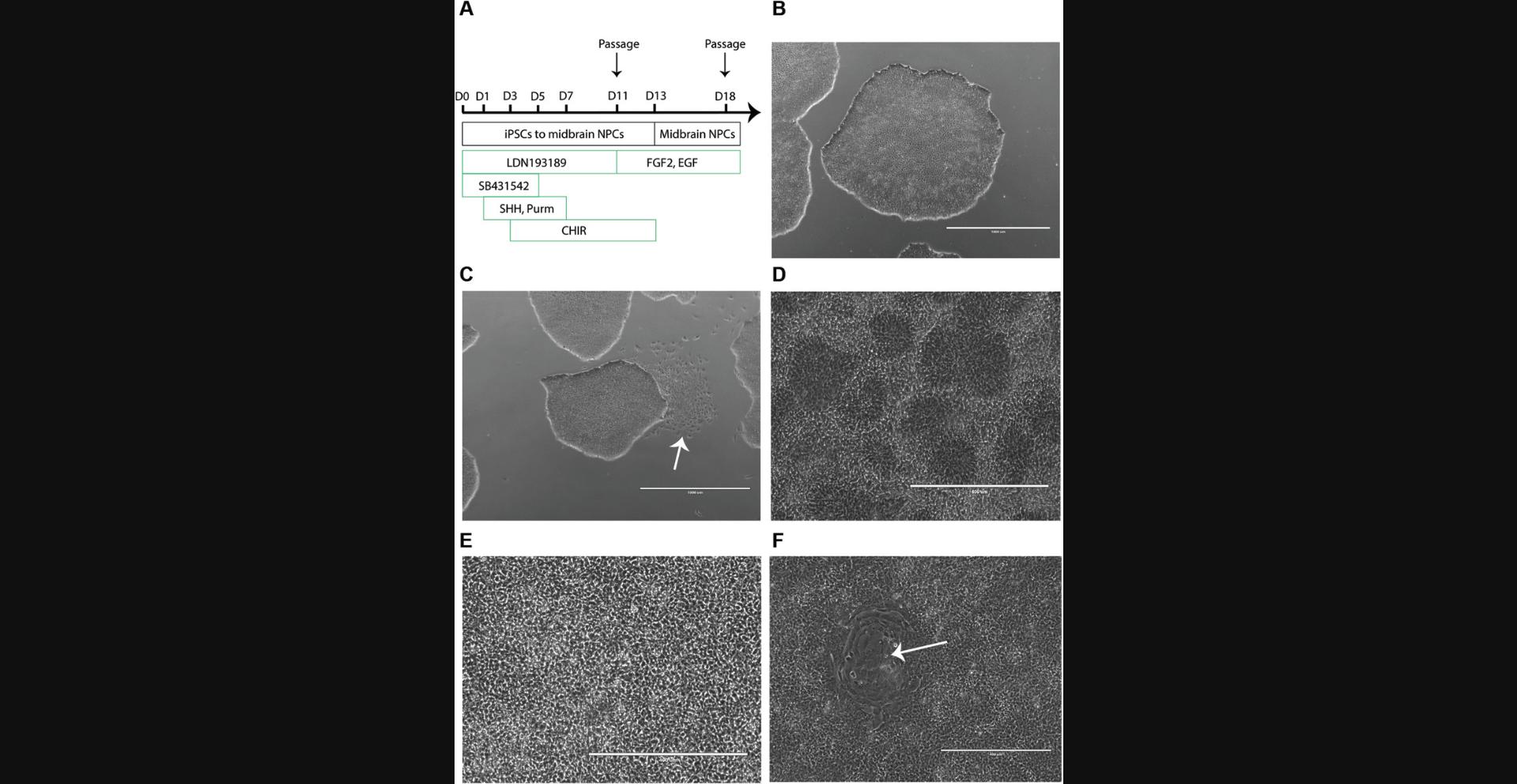
To initiate differentiation, iPSCs are plated as a monolayer and subsequently neuralized via dual SMAD inhibition (Fig. 2A). After 11 days in culture, the cells are passaged; cultured for an additional 7 days; and subsequently frozen, sub-cultured, or differentiated into regionalized astrocytes (Basic Protocol 2) and neurons (Basic Protocol 3). Newly produced NPCs need to undergo quality control (Support Protocol) to ensure expression of region-specific marker genes.
Materials
-
Confluent iPSCs in growth medium in 10-cm dish
-
mTeSR1 or mTeSR Plus medium (Stemcell Technologies, cat. no. 85870 or 05825; store in aliquots at −20°C)
-
Accumax (Innovative Cell Technologies, cat. no. AM105-500; store in aliquots at −20°C)
-
Dulbecco's phosphate-buffered saline (DPBS; Thermo Fisher, cat. no. 14190250)
-
ROCK inhibitor (Y-27632, Stemcell Technologies, cat. no. 72304)
-
Serum-free medium (SRM; see recipe), 37°C
-
LDN193189 hydrochloride (Sigma, cat. no. SML0559)
-
SB431542 (Stemcell Technologies, cat. no. 72234)
-
SHH(C25II), recombinant mouse protein (SHH; R&D Systems, cat. no. 464-SH-200)
-
Purmorphamine (Millipore, cat. no. 540223)
-
CHIR99021 (Stemcell Technologies, cat. no. 72054)
-
N2 medium (see recipe), 37°C
-
Complete neurobasal medium (see recipe), 37°C
-
NPC medium (see recipe), 37°C
-
Light microscope
-
15-ml conical centrifuge tubes
-
Standard tabletop centrifuge
-
Hemocytometer
-
Geltrex-coated 6-well plates (see recipe)
Day -1
1.Observe confluent iPSCs in growth medium in a 10-cm dish under a light microscope to monitor cell health, ensuring that colonies with tight edges and a dense center are present (Fig. 2B). Under sterile conditions, manually remove areas of spontaneous differentiation using tip of a pipet tip (Fig. 2C).
2.Pre-warm aliquots of mTeSR1 or mTeSR Plus medium and Accumax to 37°C. Equilibrate DPBS at room temperature.
3.Prepare 10 ml pre-warmed mTeSR1 medium supplemented with 10 µM ROCK inhibitor.
4.Remove growth medium from the iPSCs from step 1 and wash cells once with 5 ml DPBS. Add 3 ml pre-warmed Accumax.
5.Incubate cells at 37°C until they become rounded and detach from the dish.
6.Gently triturate cells to prepare a single-cell suspension. Transfer cell suspension to a 15-ml conical centrifuge tube.
7.Wash the 10-cm dish with 8 ml mTeSR1 medium to collect as many cells as possible and transfer to 15-ml tube containing the cell suspension.
8.Centrifuge 3 min at 300 × g. Aspirate supernatant and resuspend cells in 3 ml mTeSR1 medium supplemented with 10 µM ROCK inhibitor (see step 3).
9.Count cells with a hemocytometer and plate at a density of 1.9 × 106 cells/2 ml/well in a Geltrex-coated 6-well plate.
Day 0
10.The next day, wash cells with 2 ml DPBS per well to remove dead cells and debris.
11.Monitor cell viability under a light microscope, ensuring a confluent monolayer of iPSCs. If there are areas of the plate without cells, let culture grow for one more day in fresh mTeSR1 medium without ROCK inhibitor. If the monolayer is ready for differentiation, add 2 ml SRM freshly supplemented with 100 nM LDN193189 hydrochloride and 10 µM SB431542.
Day 1
12.Aspirate Day 0 medium and add SRM freshly supplemented with 100 nM LDN193189, 10 µM SB431542, 100 ng/ml SHH, and 2 µM purmorphamine.
Day 2
13.Aspirate Day 1 medium and add SRM freshly supplemented with 100 nM LDN193189, 10 µM SB431542, 100 ng/ml SHH, and 2 µM purmorphamine.
Day 3
14.Aspirate Day 2 medium and add SRM medium freshly supplemented with 100 nM LDN193189, 10 µM SB431542, 100 ng/ml SHH, 2 µM purmorphamine, and 3 µM CHIR99021.
Day 5
15.Combine SRM and N2 medium at a 3:1 ratio. Supplement with 100 nM LDN193189, 100 ng/ml SHH, 2 µM purmorphamine, and 3 µM CHIR99021.
16.Aspirate medium from the cells and feed cells with the medium prepared above.
Day 7
17.Combine SRM and N2 medium at a 1:1 ratio. Supplement with 100 nM LDN193189 and 3 µM CHIR99021.
18.Aspirate medium from the cells and feed cells with the medium prepared above.
Day 9
19.Combine SRM and N2 medium at a 1:3 ratio. Supplement with 100 nM LDN193189 and 3 µM CHIR99021.
20.Aspirate medium from the cells and feed cells with the medium prepared above.
Day 11
21.Passage cells as follows:
-
Wash cells once with 2 ml DPBS per well and add 1 ml pre-warmed Accumax per well.
-
Incubate at 37°C for 3 min. After 3 min, monitor cells under a light microscope.
If the cells are rounded and detached from the plate, they are ready to harvest. If not, incubate 2 to 3 min longer in the incubator. Do not over-incubate because it may reduce the viability of the cells.
-
Transfer cell suspension to a 15-ml conical centrifuge tube and add 5 ml complete neurobasal medium. Wash wells with 1 ml complete neurobasal medium to collect the remaining cells and transfer to 15-ml tube containing the cell suspension.
-
Centrifuge cell suspension for 3 min at 300 ×g. Aspirate supernatant and resuspend pellet in NPC medium freshly supplemented with 3 µM CHIR99021 and 10 µM ROCK inhibitor.
The resuspension volume varies depending on the number of wells collected. It is recommended to resuspend in 1 ml per collected well.
- Count cells and plate at a density of 7 × 106cells/2 ml/well in a Geltrex-coated 6-well plate. Label plate with the passage of the iPSCs at the time of plating and the NPC passage number.
For example, if the iPSCs were passage 20 when plated at Day -1, label the plate P20N0. This step is passage 0 of the NPCs. The NPCs are not fully patterned yet and require an additional 2 days of patterning with CHIR99021.
Day 12
22.The next day, monitor cell viability under a light microscope.
23.Aspirate medium and wash cells once with 2 ml DPBS to remove the dead cells and debris.
24.Feed cells with NPC medium freshly supplemented with 3 µM CHIR99021.
Days 13 to 16
25.Each day, aspirate medium and feed cells with NPC medium.
Day 18
26.Passage cells on Day 18 and then every 7 days at 7 × 106 cells/well in NPC medium freshly supplemented with 10 µM ROCK inhibitor in a Geltrex-coated 6-well plate.
Support Protocol: QUALITY CONTROL OF NEURAL PROGENITOR CELLS
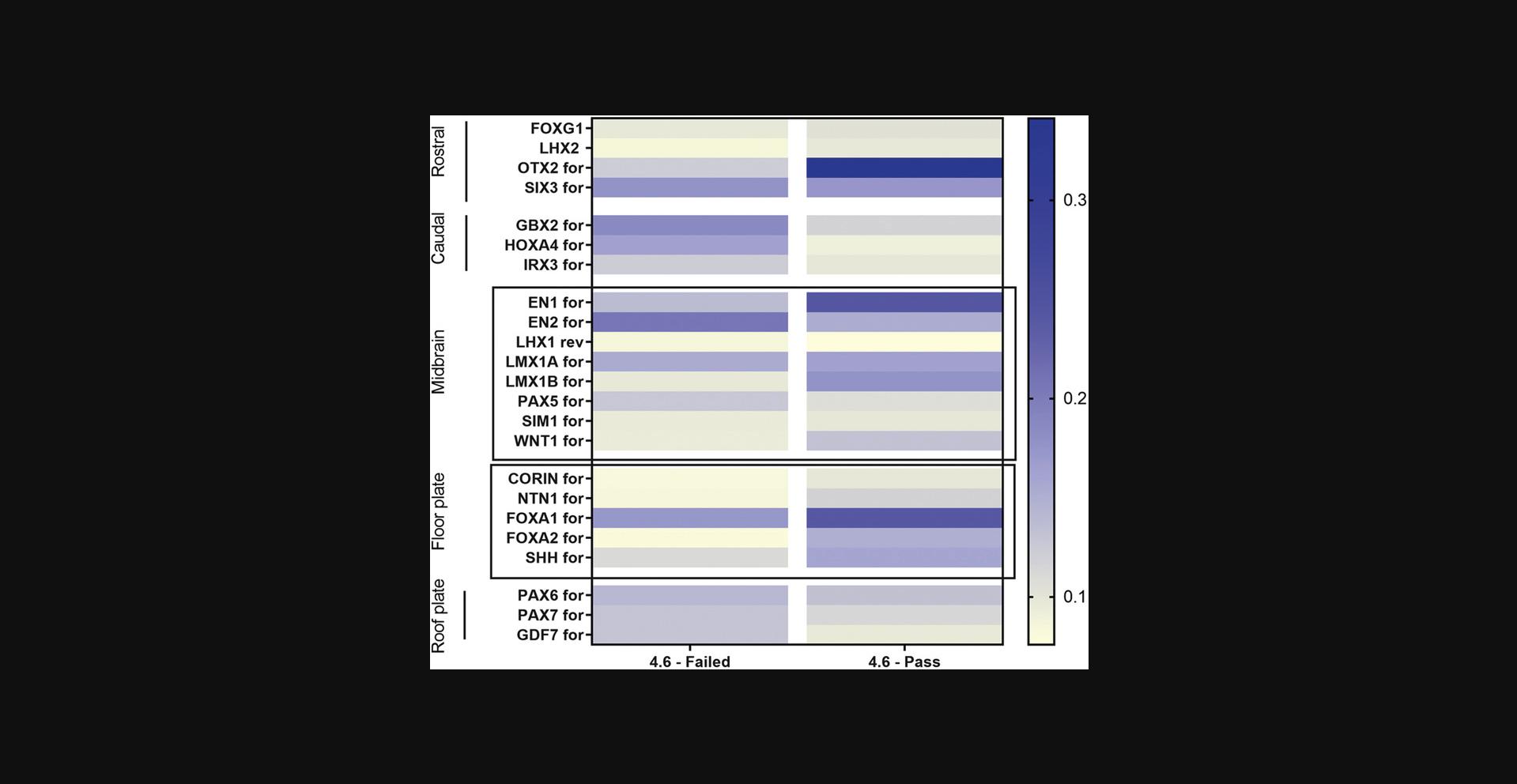
This protocol describes quality control of the NPCs obtained in Basic Protocol 1.It is very important to perform and document this quality control to ensure that the NPCs express gene markers of a midbrain and floor-plate fate (Fig. 3). It is suggested to perform this quality control once, early after differentiation, such as at passage 1 or 2.If the NPCs do not express satisfactory levels of marker genes, it is likely that they will not produce desirable yields of dopaminergic neurons (Basic Protocol 3). In such a case, new NPCs will need to be produced. To confirm NPC identity (rather than their brain-region patterning), additional techniques such as immunofluorescence can be used to check for expression of NPC markers (not described here; Cheng et al., 2015).
Materials
-
NPCs (see Basic Protocol 1)
-
NPC medium (see recipe), 37°C
-
DPBS (Thermo Fisher, cat. no. 14190250)
-
TRIzol reagent (Thermo Fisher, cat. no. 15596026)
-
RNA purification kit (Qiagen, cat. no. 74104)
-
cDNA synthesis kit (Thermo Fisher Scientific, cat. no. K1622)
-
qPCR kit (Roche, cat. no. 07959583001)
-
Geltrex-coated 12-well plates (see recipe)
-
1.5-ml Eppendorf tubes
-
Additional reagents and equipment for RNA purification, cDNA synthesis, and qPCR (see manufacturer's guidance)
CAUTION : TRIzol is a toxic reagent. To prevent inhalation of vapors, always work in a chemical hood. Use appropriate personal protective equipment, such as gloves and a laboratory coat. Set up a waste container to collect TRIzol-contaminated material (e.g., pipet tips, tubes).
1.Plate NPCs into individual wells in a Geltrex-coated 12-well plate at a density of 3 × 106 cells/well. Feed NPCs with 1 ml NPC medium daily for 48 hr.
2.After 48 hr, transfer plate to a chemical hood. Wash cells once with 1 ml DPBS and add 500 µl TRIzol reagent.
3.After 5 min, triturate contents of the well and collect TRIzol extraction into a 1.5-ml Eppendorf tube. Process sample for RNA extraction and cDNA synthesis immediately using RNA purification and cDNA synthesis kits, respectively, or store at −80°C.
4.Perform qPCR using a qPCR kit and NPC gene panel available in Table 1 and calculate 1/delta Ct value for each gene.
| Gene namea | Forward primer | Reverse primer |
|---|---|---|
| FOXG1 | TGGCCCATGTCGCCCTTCCT | GCCGACGTGGTGCCGTTGTA |
| LHX2 | GGGCGACCACTTCGGCATGAA | CGTCGGCATGGTTGAAGTGTGC |
| OTX2 | ACAAGTGGCCAATTCACTCC | GAGGTGGACAAGGGATCTGA |
| SIX3 | ACCGGCCTCACTCCCACACA | CGCTCGGTCCAATGGCCTGG |
| GBX2 | GTTCCCGCCGTCGCTGATGAT | GCCGGTGTAGACGAAATGGCCG |
| HOXA4 | ACGCTCTGTTTGTCTGAGCGCC | AGAGGCCGAGGCCGAATTGGA |
| IRX3 | GGCTTGCGCCCCGTAGAAATGT | AGGAGCCAGGTCAGGTCCGAAC |
| EN1 | CGTGGCTTACTCCCCATTTA | TCTCGCTGTCTCTCCCTCTC |
| EN2 | CCTCCTGCTCCTCCTTTCTT | GACGCAGACGATGTATGCAC |
| LHX1 | AGGTGAAACACTTTGCTCCG | CTCCAGGGAAGGCAAACTCT |
| LMX1A | CGCATCGTTTCTTCTCCTCT | CAGACAGACTTGGGGCTCAC |
| LMX1B | CTTAACCAGCCTCAGCGACT | TCAGGAGGCGAAGTAGGAAC |
| PAX5 | CCCCATTGTGACAGGCCGTGAC | TCAGCGTCGGTGCTGAGTAGCT |
| SIM1 | AAAGGGGGCCAAATCCCGGC | TCCGCCCCACTGGCTGTCAT |
| WNT1 | GAGCCACGAGTTTGGATGTT | TGCAGGGAGAAAGGAGAGAA |
| CORIN | CATATCTCCATCGCCTCAGTTG | GGCAGGAGTCCATGACTGT |
| NTN1 | GCATGCAGGTTGCAGTTACA | GCTGCAAGCCCTTCCACTA |
| FOXA1 | GGGCAGGGTGGCTCCAGGAT | TGCTGACCGGGACGGAGGAG |
| FOXA2 | CCGTTCTCCATCAACAACCT | GGGGTAGTGCATCACCTGTT |
| SHH | CCAATTACAACCCCGACATC | AGTTTCACTCCTGGCCACTG |
| PAX6 | TGGTATTCTCTCCCCCTCCT | TAAGGATGTTGAACGGGCAG |
| PAX7 | CTTCAGTGGGAGGTCAGGTT | CAAACACAGCATCGACGG |
| GDF7 | GACGCTGCTCAACTCCATGGCA | TTGGCGGCGTCGATGTAGAGGA |
- a
This list of primers was published by Kirkeby and colleagues and is used to establish a quality-control gene panel (Kirkeby et al., 2012).
Basic Protocol 2: DIFFERENTIATION OF NEURAL PROGENITOR CELLS INTO ASTROCYTES
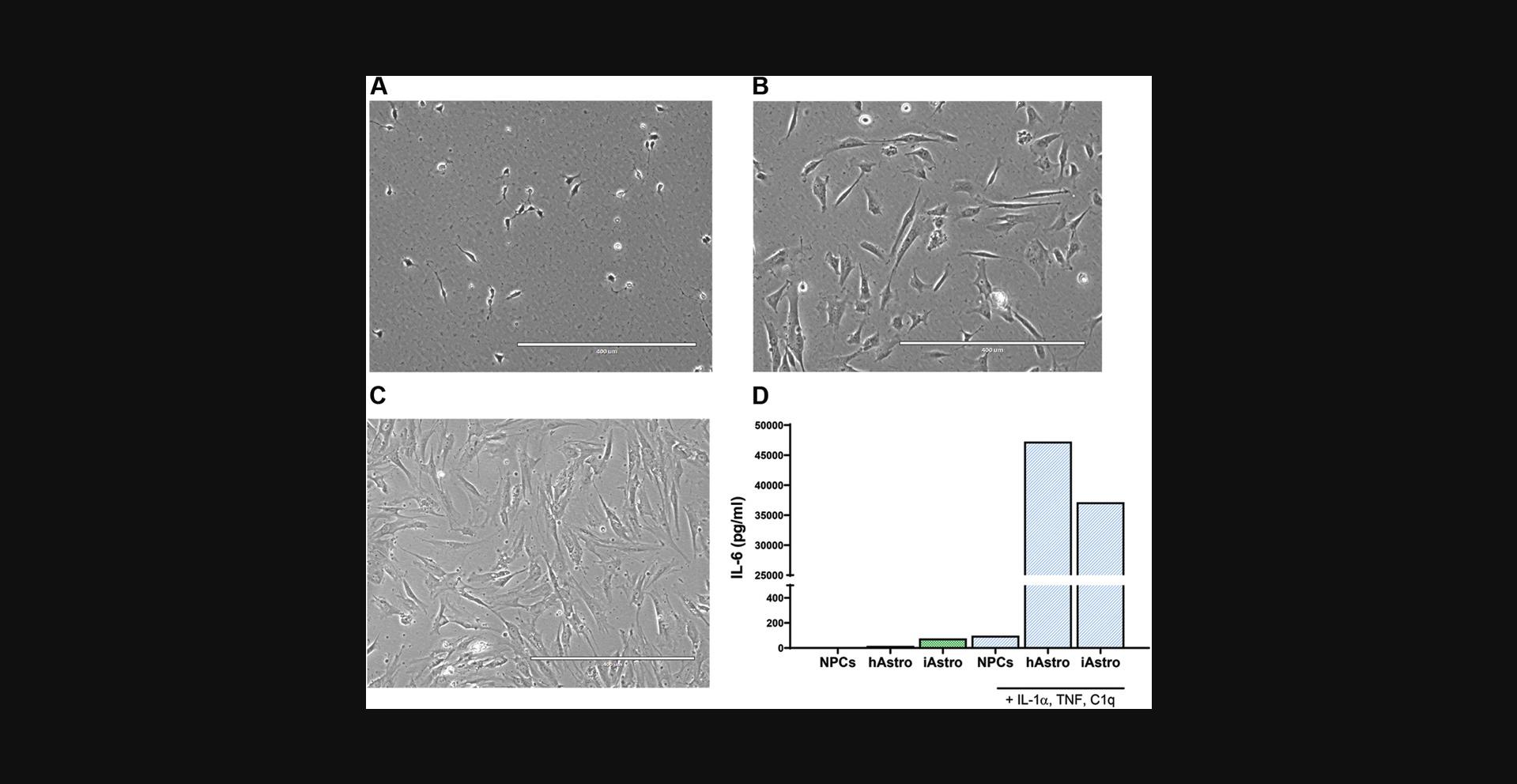
Differentiation of astrocytes can be achieved by different methods, using either iPSCs or neuralized progenitors as starting material (Chandrasekaran, Avci, Leist, Kobolak, & Dinnyes, 2016). TCW and colleagues described production of astrocytes from forebrain NPCs using a commercial medium produced by ScienCell Research Laboratories (TCW et al., 2017). This method has many advantages: it requires a single type of medium, involves little hands-on time, and produces functioning astrocytes in a relatively short period of time (28 days). However, it requires supplementation of medium with 2% fetal bovine serum (FBS), which may affect downstream applications. Basic Protocol 2 describes production of astrocytes from a population of midbrain-patterned NPCs (Basic Protocol 1) using ScienCell astrocyte medium. To initiate differentiation, NPCs are plated at low density, continuously maintained in astrocyte differentiation medium, and passaged every 7 days until Day 28.The growth rate of different NPC lines may vary, especially in the first 14 days of differentiation. Therefore, the timing of cell passaging should be adjusted according to the NPC line of interest. Cell morphology is a visual indicator of differentiation and progressively changes, from small NPCs to large, flat astrocytes (Fig. 4A-C).
Materials
-
NPCs (see Basic Protocol 1)
-
DPBS (Thermo Fisher, cat. no. 14190250)
-
Accumax (Innovative Cell Technologies, cat. no. AM105-500; store in aliquots at −20°C), 37°C
-
Complete neurobasal medium (see recipe), 37°C
-
NPC medium (see recipe), 37°C
-
ROCK inhibitor (Y-27632; Stemcell Technologies, cat. no. 72304)
-
Astrocyte medium (ScienCell Research Laboratories, cat. no. 1801), 37°C
-
Astrocyte freezing medium: astrocyte medium supplemented with 10% (v/v) dimethylsulfoxide (DMSO)
-
Geltrex-coated 6-well plates and 10- and 15-cm dishes (see recipe)
-
15- and 50-ml conical centrifuge tubes
-
Standard tabletop centrifuge
-
Hemocytometer
Day 1
1.Maintain NPCs in one well of a Geltrex-coated 6-well plate (see Basic Protocol 1, step 26). Seven days after the previous passage, split cells:
-
Coat one well of a 6-well plate with Geltrex (see Reagents and Solutions) ≥1 hr prior to plating.
-
Wash NPCs once with 2 ml DPBS. Aspirate DPBS and add 1 ml pre-warmed Accumax per well. Incubate at 37°C until cells detach (3 to 6 min).
-
Transfer cell suspension to a 15-ml conical centrifuge tube and add 5 ml complete neurobasal medium.
-
Centrifuge cell suspension for 3 min at 300 ×g. Aspirate supernatant and resuspend pellet in 2 ml NPC medium freshly supplemented with 10 µM ROCK inhibitor. Count cells with a hemocytometer and plate at a density of 15,000 cells/cm2in one well of Geltrex-coated 6-well plate. To keep track of the history of the cells, label plate with the passage number of the iPSCs, followed by the passage number of the NPCs and the passage number of the astrocytes.
For example, the label P30N4A0means that the iPSCs were at passage 30 when they were differentiated into NPCs. These NPCs were at passage 4 when they were plated to differentiate into astrocytes. The astrocytes are labeled A0in their first week of differentiation, resulting in the final label P30N4A0.
Day 0
2.Wash -cells once with 2 ml DPBS and add 2 ml astrocyte medium.
Days 2, 4, and 6
3.Change medium on Day 2 and then every other day until Day 7.
Day 7
4.Passage cells as in step 1, with slight modifications:
-
Coat all wells of 6-well plate with Geltrex.
-
Centrifuge cell suspension in astrocyte medium instead of complete neurobasal medium.
-
Plate 15,000 cells/cm2in up to six wells of Geltrex-coated 6-well plate.
After this first passage, the astrocytes are labeled A1.
Continuing from the previous example (see step 1), the culture would now be labeled P30N4A1.
Day 9
5.Change medium on Day 9 and then every other day until Day 14.
Day 14
6.Passage cells as in step 4, with slight modifications:
-
Coat three 10-cm dishes with Geltrex.
-
Plate dishes at 15,000 cells/cm2.
After this second passage, the astrocytes are labeled A2.
Continuing from the previous example (see step 4), the culture would now be labeled P30N4A2.
Day 16
7.Change medium on Day 16 and then every other day until Day 21.
Day 21
8.Passage cells as in step 4, with slight modifications:
-
Coat three 15-cm dishes with Geltrex.
-
Add 3 ml pre-warmed Accumax to each dish.
-
Transfer cell suspension to a 50-ml conical centrifuge tube and add 15 ml astrocyte medium.
-
Resuspend pellet in 5 ml astrocyte medium. Plate dishes at 15,000 cells/cm2.
After this third passage, the astrocytes are labeled A3.
Continuing from the previous example (see step 6), the culture would now be labeled P30N4A3.
Day 23
9.Change medium on Day 23 and then every other day until Day 28.
Day 28
10.Repeat step 4, with slight modifications:
-
If astrocytes will be plated for experiments, prepare Geltrex-coated plates as necessary.
-
Add 5 ml pre-warmed Accumax to each 15-cm dish.
-
Transfer cell suspension to a 50-ml conical centrifuge tube and add 20 ml astrocyte medium.
-
Resuspend pellet in 10 ml astrocyte medium. Plate at desired density for experiments in Geltrex-coated plates.
After this fourth passage, the astrocytes are labeled A4.
- Freeze remaining cells at the desired density in astrocyte freezing medium, including ≥30% additional cells to account for cell loss during handling and thawing.
Day 28 is the final astrocyte differentiation stage. The astrocytes can be frozen and/or plated for experiments.
Depending on the cell line, the rate of astrocyte growth can decrease after the final differentiation stage.
Appropriate quality control of the astrocytes (not described here) should be performed. This includes at least immunofluorescence staining of astrocytic markers (e.g., GFAP, CD44, vimentin; TCW et al.,2017; di Domenico et al.,2019) and a functional assay such as an IL-6 cytokine secretion assay (Fig.4D).
Basic Protocol 3: DIFFERENTIATION OF NEURAL PROGENITOR CELLS INTO DOPAMINERGIC NEURONS
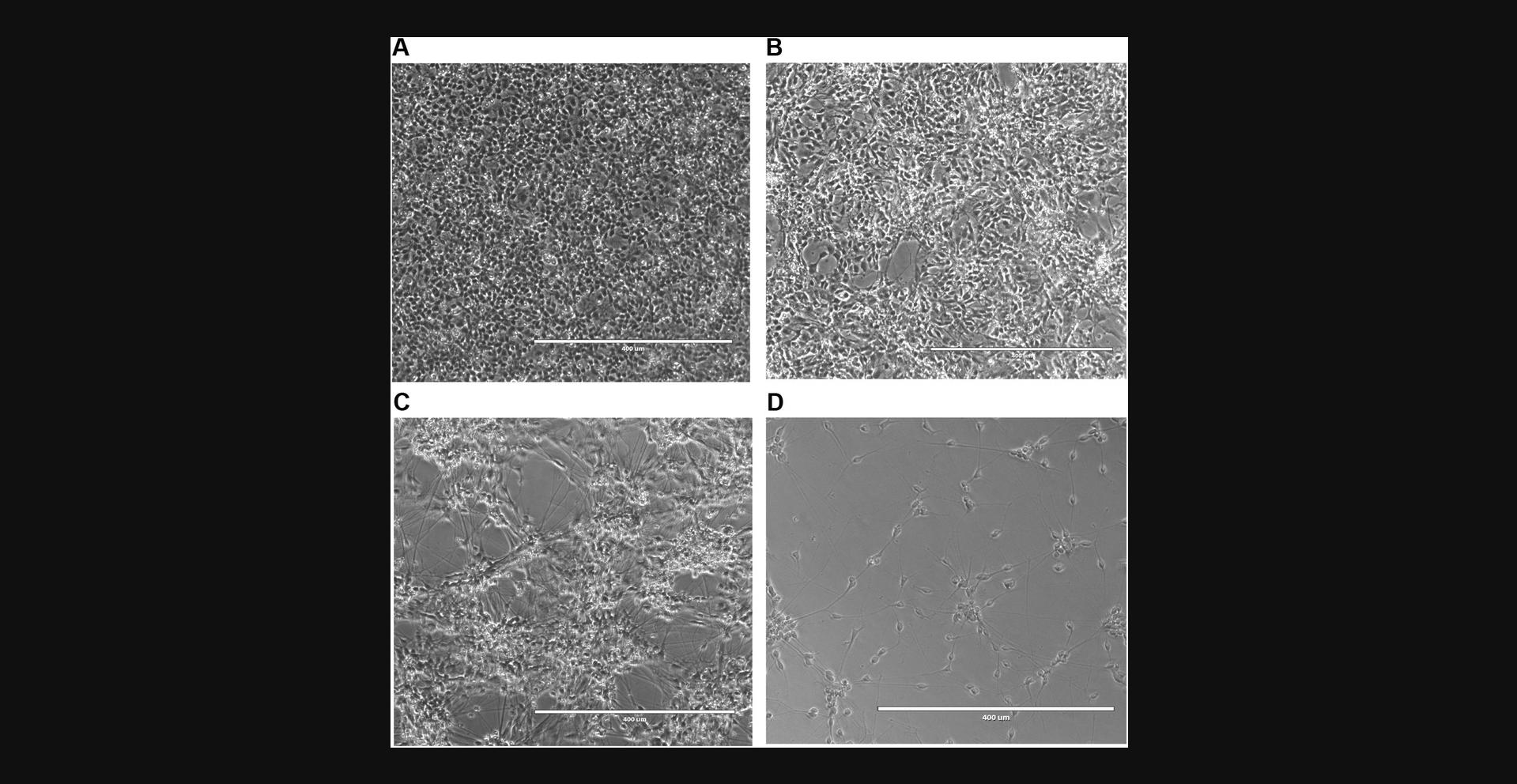
The following protocol describes differentiation of regionalized NPCs (Basic Protocol 1) into dopaminergic neurons. For differentiation, NPCs are plated at a high density and subsequently incubated in dopaminergic neuron differentiation medium (Fig. 5A-C). Between Day 7 and Day 10, most cells extend long neurites (Fig. 5C), whereas others fail to differentiate. The culture is then depleted of these undifferentiated CD133+ NPCs by magnetic-activated cell sorting (MACS), resulting in a highly enriched neuronal culture ready to be plated for experiments (Fig. 5D).
Materials
-
NPCs (see Basic Protocol 1)
-
Geltrex-coated 12-well and other plates (see recipe)
-
DPBS (Thermo Fisher, cat. no. 14190250)
-
Accumax (Innovative Cell Technologies, cat. no. AM105-500; store in aliquots at −20°C), 37°C
-
Complete neurobasal medium (see recipe), 37°C
-
NPC medium (see recipe), 37°C
-
ROCK inhibitor (Y-27632; Stemcell Technologies, cat. no. 72304)
-
Dopaminergic neuron differentiation medium (see recipe), 37°C
-
MACS buffer (see recipe)
-
APC mouse anti-human CD133 (BD Biosciences, cat. no. 566596)
-
APC-conjugated magnetic beads (Anti-APC MicroBeads, Miltenyi, cat. no. 130-090-855)
-
Astrocytes (see Basic Protocols 2 and 4; optional)
-
15-ml conical centrifuge tubes
-
Standard tabletop centrifuge, room temperature or 4°C
-
Hemocytometer
-
Strainer tube (round-bottom polystyrene test tube with cell-strainer snap cap, Corning, cat. no. 352235)
-
1.5-ml Eppendorf tubes
-
LD column (Miltenyi, cat. no. 130-042-901)
-
Multistand (MACS MultiStand, Miltenyi, cat. no. 130-042-303)
-
Separator (MidiMACS Separator, Miltenyi, cat. no. 130-042-302)
1.Maintain NPCs in one well of a Geltrex-coated 6-well plate (see Basic Protocol 1, step 26). Seven days after the previous passage, split cells:
-
Coat one well of a 12-well plate with Geltrex (see Reagents and Solutions) ≥1 hr prior to plating.
-
Wash NPCs once with 2 ml DPBS and add 1 ml pre-warmed Accumax. Incubate at 37°C until cells detach (3 to 6 min).
-
Transfer cell suspension to a 15-ml conical centrifuge tube and add 5 ml complete neurobasal medium.
-
Centrifuge cell suspension for 3 min at 300 ×g. Aspirate supernatant and resuspend pellet in 2 ml NPC medium freshly supplemented with 10 µM ROCK inhibitor. Count cells with a hemocytometer and plate 3 × 106cells in 1 ml in one well of Geltrex-coated 12-well plate.
2.The next day, wash cells once with 1 ml DPBS and add 1 ml dopaminergic neuron differentiation medium. Feed cells every other day with dopaminergic neuron differentiation medium.
3.After 7 to 10 days, deplete neuronal culture of undifferentiated NPCs by MACS and plate cells for experiments:
- Coat plates with Geltrex (see Reagents and Solutions).
The type of plate depends on the purpose of the experiment. For example, 96- or 48-well plates with optical bottoms or coverslips are useful for imaging experiments. For biochemical experiments, it is recommended to use larger plates, such as 12- or 6-well plates.
-
Wash cells once with 1 ml DPBS and add 0.5 ml pre-warmed Accumax. Incubate at 37°C for 10 min.
-
Transfer cell suspension to a 15-ml conical centrifuge tube and add 5 ml complete neurobasal medium. Centrifuge cell suspension for 3 min at 300 ×g. Aspirate supernatant and resuspend pellet in 500 µl complete neurobasal medium.
-
Prepare a strainer tube by passing 300 µl complete neurobasal medium through it. Filter cell suspension to remove large aggregates and collect single cells. Wash membrane with 400 µl complete neurobasal medium. Discard filter, transfer cell suspension to a 1.5-ml Eppendorf tube, and centrifuge 3 min at 300 ×g.
-
Resuspend pellet in 1 ml MACS buffer and count total number of cells. Centrifuge 3 min at 300 ×g.
-
Resuspend every 1 × 106cells in 95 µl MACS buffer supplemented with 5 µl APC mouse anti-human CD133. Incubate at 4°C for 10 min.
-
Add 1 ml MACS buffer and centrifuge 3 min at 300 ×g, 4°C.
-
Resuspend pellet in 80 µl MACS buffer supplemented with 20 µl APC-conjugated magnetic beads (≤108cells per 100 µl final volume). Incubate at 4°C for 10 min.
-
To magnetically deplete undifferentiated NPCs, set up MACS multistand and separator according to the manufacturer's instructions and prepare the LD column by applying 2 ml MACS buffer to the column and discarding flow-through. When the column is ready, set up a 1.5-ml Eppendorf tube to collect flow-through containing the neurons.
-
Add 400 µl MACS buffer to cell suspension to dilute it and filter through column as described in the manufacturer's instructions. Collect flow-through in the 1.5-ml tube.
The flow-through contains the unlabeled neurons. The magnetically labeled CD133+NPCs are retained in the column. Do not remove the column from the magnetic field (MACS multistand and separator) until the cell sorting is completed.
-
Wash column by applying 500 µl MACS buffer and collect cells in the flow-through in the same 1.5-ml tube. Repeat wash once.
-
Centrifuge 1.5-ml tube for 3 min at 300 ×g, 4°C. Discard LD column.
-
Gently resuspend cell pellet in 0.5 ml dopaminergic neuron differentiation medium, count cells, and plate on Geltrex-coated plates at 55,000 to 70,000 cells/well (96-well plate) or on astrocytes (see Basic Protocol4).
4.To avoid disturbing the neurons, replace only 50% of the dopaminergic neuron differentiation medium every 2 days until cells are ready for analysis.
Basic Protocol 4: CO-CULTURE OF iPSC-DERIVED NEURONS AND ASTROCYTES
Communication between neurons and astrocytes regulates a variety of processes that can be recapitulated in vitro (di Domenico et al., 2019; Hasel et al., 2017; Ioannou et al., 2019). To investigate these interactions, different culture systems can be implemented, such as direct or indirect co-culture as well as culture in conditioned medium prepared from neurons or astrocytes. The following protocol describes the steps needed to establish a direct iPSC-derived neuron/astrocyte co-culture that ensures optimal viability of both neurons and astrocytes for ≥2 weeks.
Materials
-
iPSC-derived astrocytes (see Basic Protocol 2)
-
DPBS (Thermo Fisher, cat. no. 14190250)
-
Accumax (Innovative Cell Technologies, cat. no. AM105-500; store in aliquots at −20°C), 37°C
-
Astrocyte medium (ScienCell Research Laboratories, cat. no. 1801), 37°C
-
Geltrex-coated 96-well plate (see recipe)
-
2× AraC (Sigma, cat. no. C1768) concentrated solution (i.e., 10 to 16 µM)
-
Complete neurobasal medium (see recipe) supplemented with 0.5% (v/v) FBS (Thermo Fisher, cat. no. A3160601), 37°C
-
15-ml conical centrifuge tubes
-
Standard tabletop centrifuge
-
Hemocytometer
-
Additional reagents and equipment for preparation of iPSC-derived dopaminergic neurons (see Basic Protocol 3)
CAUTION : AraC is an antimitotic agent and must be handled with caution.
1.Prepare iPSC-derived astrocytes as described in Basic Protocol 2 or thaw a vial of frozen astrocytes. Culture cells until confluent.
2.Wash cells once with 1 or 3 ml DPBS and incubate in 1 or 3 ml pre-warmed Accumax at 37°C until cells detach from the plate or dish. Transfer cell suspension to a 15-ml conical centrifuge tube and add 5 ml astrocyte medium.
3.Centrifuge cell suspension for 3 min at 300 × g. Aspirate supernatant and resuspend pellet in 1 ml astrocyte medium. Count cells with a hemocytometer and plate 9600 cells/100 µl/well in a Geltrex-coated 96-well plate.
4.The next morning, supplement astrocyte medium with 5 to 8 µM AraC by adding 100 µl of 2× AraC concentrated solution (i.e., 10 to 16 µM) directly to each well (1× final).
5.After 24 hr, remove AraC-containing medium and replace it with 200 µl fresh astrocyte medium per well.
6.Prepare iPSC-derived dopaminergic neurons as described in Basic Protocol 3.After MACS depletion of the culture, resuspend CD133- neurons in 300 µl complete neurobasal medium supplemented with 0.5% FBS. Count cells with a hemocytometer and plate 10,000 to 13,000 neurons/200 µl/well into 96-well plates pre-plated with AraC-treated, iPSC-derived astrocytes (see step 5).
7.Maintain the co-culture in complete neurobasal medium supplemented with 0.5% FBS and only change 50% of the medium every 2 to 3 days.
REAGENTS AND SOLUTIONS
Complete neurobasal medium
- Neurobasal medium (Thermo Fisher, cat. no. 21103-049)
- 2% (v/v) B27 supplement (minus vitamin A; Thermo Fisher, cat. no. 12587010)
- 100 U/ml penicillin-streptomycin (10,000 U/ml stock; Thermo Fisher, cat. no. 15140122)
- 1% (v/v) GlutaMAX (Thermo Fisher, cat. no. 35050061)
- Filter sterilize using filter bottle (e.g., Millipore, cat. no. SCGVU02RE)
- Store ≤3 weeks at 4°C
The storage time is given assuming that users do not warm up the entire bottle, but instead warm up aliquots prepared from the stock medium.
Dopaminergic neuron differentiation medium
- Complete neurobasal medium (see recipe)
- 0.2 mM ascorbic acid (Sigma, cat. no. A92902)
- 0.5 mM dibutyryl cAMP (Millipore Sigma, cat. no. 28745)
- 10 µM DAPT (Tocris Bioscience, cat. no. 2634)
- 20 ng/ml recombinant human BDNF (PeproTech, cat. no. 450-02)
- 20 ng/ml recombinant human GDNF (PeproTech, cat. no. 450-10)
- 1 ng/ml recombinant human TGF-β3 (R&D Systems, cat. no. 8420-B3-025)
- Store ≤4 days at 4°C
Geltrex-coated plates or dishes
To aliquot Geltrex:
- 1.Thaw Geltrex (Fisher Scientific, cat. no. A1413302) overnight on ice at 4°C (e.g., in a small Styrofoam box filled with ice in a laboratory refrigerator).
- 2.The next morning, pre-chill 1.5-ml Eppendorf tubes, tube rack, and pipet tips.
- 3.Mix thawed Geltrex by inversion to homogenize. Place pre-chilled tubes in the cold tube rack to maintain a low temperature while aliquoting. Prepare single-use aliquots of 60 and 120 µl. Immediately store aliquots at −20°C.
To coat plates or dishes with Geltrex:
Thaw an aliquot of Geltrex on ice.
Thaw Geltrex aliquots only once.
Prepare a 1:100 dilution of Geltrex in cold DMEM/F-12, HEPES (Fisher Scientific, cat. no. 11330057) basal medium. Dispense solution into tissue culture–treated, sterile plates or dishes immediately and incubate for ≥1 hr at 37°C.
To coat a 6-well plate, supplement 6 ml cold DMEM/F-12/HEPES with 60 µl Geltrex. Add 1 ml of this solution to each well.
When the plates or dishes are ready, aspirate basal medium and directly add cell suspension.
It is not necessary to wash the plates/dishes before plating the cells.
Do not let the Geltrex-coated plates/dishes dry; aspirate the basal medium just before plating.
Use plates or dishes within 24 hr or add DMEM/F-12, HEPES basal medium to prevent drying.
Geltrex-coated plates/dishes can be used after ≤1 week of incubation at 37°C or after ≤2 weeks if stored at 4°C. Before using Geltrex-coated plates/dishes stored at 4°C, equilibrate them in the incubator for ≥10 min prior to plating.
MACS buffer
- HBSS (with calcium and magnesium and without phenol red; Thermo Fisher, cat. no. 14025092)
- 1% (v/v) sodium pyruvate (Thermo Fisher, cat. no. 11360070)
- 1% (v/v) GlutaMAX (Thermo Fisher, cat. no. 35050061)
- 100 U/ml penicillin-streptomycin (10,000 U/ml stock; Thermo Fisher, cat. no. 15140122)
- 1% (v/v) HEPES (Thermo Fisher, cat. no. 15630106)
- 0.5% (w/v) bovine serum albumin (BSA; Fisher Scientific, cat. no. BP1600-100)
- Filter sterilize using filter bottle (e.g., Millipore, cat. no. SCGVU02RE)
- Store ≤1 month at 4°C
N2 medium
- Neurobasal medium (Thermo Fisher, cat. no. 21103-049)
- 1% (v/v) N2 supplement (Thermo Fisher, cat. no. 17502048)
- 2% (v/v) B27 supplement (minus vitamin A; Thermo Fisher, cat. no. 12587010)
- 100 U/ml penicillin-streptomycin (10,000 U/ml stock; Thermo Fisher, cat. no. 15140122)
- 1% (v/v) GlutaMAX (Thermo Fisher, cat. no. 35050061)
- Filter sterilize using filter bottle (e.g., Millipore, cat. no. SCGVU02RE)
- Store ≤2 weeks at 4°C
NPC medium
- Complete neurobasal medium (see recipe)
- 20 ng/ml recombinant human FGFb (PeproTech, cat. no. 100-18C)
- 20 ng/ml recombinant human EGF (Thermo Fisher, cat. no. PHG0311)
- Store small volume ≤1 week at 4°C
Serum-free medium
- KnockOut DMEM/F-12 (Thermo Fisher, cat. no. 12660012)
- 15% (v/v) KnockOut Serum Replacement (Thermo Fisher, cat. no. 10828028)
- 1% (v/v) GlutaMAX (Thermo Fisher, cat. no. 35050061)
- 1% (v/v) MEM Non-Essential Amino Acids (NEAA; Thermo Fisher, cat. no. 11140050)
- 100 U/ml penicillin-streptomycin (10,000 U/ml stock; Thermo Fisher, cat. no. 15140122)
- 0.001% (v/v) beta-mercaptoethanol (Thermo Fisher, cat. no. 21985023)
- Filter sterilize using filter bottle (e.g., Millipore, cat. no. SCGVU02RE)
- Store ≤2 weeks at 4°C
COMMENTARY
Background Information
Over the past decade, iPSC-based technologies have demonstrated many advantages to model a wide range of human diseases (Liu, Oikonomopoulos, Sayed, & Wu, 2018). Their unique versatility and ability to retain the genetic background of the patient donor offer to yield new perspectives in disease biology and spark the promise of more translational drug discovery efforts. Mono-cultures, co-cultures, and 3D cultures provide many opportunities to study cell-autonomous and non-cell-autonomous mechanisms of disease onset and progression (Gonzalez, Gregory, & Brennand, 2017; Passier, Orlova, & Mummery, 2016). Differentiation of iPSCs into cell types of interest such as dopaminergic neurons and astrocytes is critical to the success of iPSC-based models. Whereas dual SMAD inhibition has been established as a neural-induction standard to produce midbrain-patterned NPCs and dopaminergic neurons (Kriks et al., 2011), differentiation of astrocytes can be achieved through multiple approaches (TCW et al., 2017). The methods described here are highly relevant to the study of Parkinson's disease because they take advantage of the well-established dual SMAD inhibition strategy to generate neurons and astrocytes from the same regionally patterned NPCs. These neurons and astrocytes can be studied separately, as mono-cultures, to describe cell-autonomous effects of Parkinson's disease, or they can be co-cultured to elucidate non-cell-autonomous mechanisms. Importantly, the co-culture system allows culture of wildtype and mutant cells to characterize the cell type-specific effects of mutations.
Critical Parameters
Good cell culture practices
It is critical to prevent cross-contamination of the cells. The risk of accidentally cross-contaminating plates increases with the number of cell types maintained in an incubator. It is highly recommended to organize the cell culture routine in a systematic manner. For example, process each cell type independently and clean the hood and aspirator between two different cell types. Avoid working with plates of different cell types in the biosafety cabinet simultaneously.
Sterility
It is critical to ensure that all steps of the protocols are performed under sterile conditions. Media and buffers need to be filtered through a 0.22-µm filter. It is highly recommended to prepare aliquots of media and buffers for daily use; this technique prolongs the shelf-life of large bottles of media and buffers and reduces the risk of contamination.
Quality of the iPSCs
The health and quality of the iPSCs (Basic Protocol 1) are critical parameters for the success of all the cell types produced in Basic Protocols 1 to 3. iPSCs must be monitored and the medium replaced daily. To reduce variability and maintain iPSC viability, we suggest culturing iPSCs in a feeder-free system, on Geltrex-coated plates, and in mTeSR1 medium. Our laboratory has had success using mTeSR1 Plus (Stemcell Technologies, cat. no. 05825) to reduce the frequency of medium changes and maintain tight, undifferentiated colonies. iPSCs need to be karyotyped regularly (every 2 months) to ensure their suitability for experiments and their karyotype stability. When necessary, differentiated cells in the iPSC culture must be removed manually and under sterile conditions.
Quality control of all cell types produced
It is critical to perform quality-control experiments on all the cell types produced. Depending on the line, the quality control can be done by qPCR or immunofluorescence and should also include functional assays. The Support Protocol, Basic Protocol 2 (step 10e), and the Understanding Results section contain suggestions of the most critical quality-control experiments to perform.
Troubleshooting
Please see Table 2 for a troubleshooting guide.
| Problem | Possible cause | Solution |
|---|---|---|
| Significant cell death |
|
|
| Cell death around Day 5 of iPSC-to-NPC differentiation |
|
|
| iPSCs do not form a confluent monolayer at Day 0 |
|
|
| Astrocytes grow too slowly or too fast | Growth of different lines of iPSCs/NPCs at different paces | Adjust the starting number of wells for differentiation accordingly. For example, astrocyte differentiation of a slow-growing line may need to be started in three wells of a 6-well plate instead of the usual one individual well of a 6-well plate. |
| Astrocytes do not express genes of interest at the same level | Poor or no quality control of NPCs | Confirm that the NPCs pass quality control (see Support Protocol) and make new astrocytes from the same batch of NPCs. If the NPCs do not pass quality control, make new astrocytes from a different batch of NPCs. |
| NPC culture fails before producing dopaminergic neurons (due to extensive cell death or lack of differentiation)a |
|
|
| Low number of dopaminergic neurons by immunofluorescence |
|
|
- a It is normal for a significant number of NPCs to die during the first days of differentiation. The extent of cell death varies between lines. If unsure, maintain the culture for up to 15 days, as it may be a “slow producer” of neurons.
Understanding Results
The protocols provide detailed instructions to prepare iPSC-derived cells (Basic Protocols 1 to 3) and co-cultures (Basic Protocol 4) relevant to the study of neurobiology and neurodegenerative diseases. Basic Protocol 1 describes differentiation of iPSCs into midbrain-patterned NPCs. Validation of midbrain patterning is critical to ensure the NPCs’ ability to efficiently produce dopaminergic neurons. NPCs self-renew and can be maintained for several weeks, as long as they are fed daily and passaged weekly. Basic Protocol 2 describes differentiation of NPCs into astrocytes. It is important to be aware that the midbrain patterning of the NPCs may influence the gene expression profile and biological functions of the NPC-derived astrocytes. Basic Protocol 3 describes differentiation of midbrain-patterned NPCs into dopaminergic neurons. The differentiation results in a mixture of neurons and undifferentiated NPCs. It is important to remove these NPCs, as they may influence experimental results. The quality of the neuronal culture needs to be assessed by estimating the number of neurons positive for the dopaminergic-neuron marker tyrosine hydroxylase (TH). A successful culture contains ∼50% TH+ neurons a few days after differentiation and ∼70% after several weeks in culture. Early after differentiation, young neurons are immature, and maturation strategies include extended time in culture and treatment with exogenous factors such as progerin (Miller et al., 2013; Sandoe & Eggan, 2013). Neurons co-cultured with astrocytes develop features of mature neurons, such as increased formation of synapses (Tang et al., 2013). Basic Protocol 4 describes how to set up and maintain a co-culture of neurons and astrocytes. The most critical parameter in this culture system is to identify culture conditions suitable for both neurons and astrocytes. Most of the factors used to differentiate dopaminergic neurons negatively affect astrocyte viability; therefore, they are omitted from the co-culture medium. Furthermore, iPSC-derived astrocytes require supplementation with 0.5% FBS to maintain a healthy culture. Our laboratory routinely maintains co-cultures for 2 weeks, but they could be maintained for longer periods of time. In 2 weeks, the neurons develop extended networks of neurites.
Time Considerations
Preparation and maintenance of NPCs (Basic Protocol 1) require 20 min (visual inspection, medium change) to 1 hr (passaging) every day. The protocol from initial plating of the iPSCs to freezing the first passage of NPCs is accomplished in 18 days. The cultures can be maintained for at least seven passages, until they are not able to produce neurons and astrocytes. Cultures should be discarded if the cells display unusual morphology and growth patterns as well as abnormal cell death.
Quality control of the NPCs by qPCR (Support Protocol) requires 1 day, from RNA extraction to data analysis.
Differentiation of NPCs into astrocytes (Basic Protocol 2) is accomplished in 1 month. It requires 20 min (visual inspection, medium change) to 1 hr (passaging) every other day. At the final stage of differentiation (Day 28), freezing astrocytes for cryostorage requires ∼1.5 hr. Depending on the line, the astrocytes can be maintained until passage 5 or later.
Differentiation of NPCs into dopaminergic neurons (Basic Protocol 3) is accomplished in 7 to 10 days. It requires 20 min (visual inspection, medium change) to 2 hr (MACS depletion and plating) per day. The dopaminergic neurons can be maintained for at least 60 days when plated on Geltrex-coated plates.
NPCs and astrocytes can be cryopreserved and thawed as necessary. These cell types can therefore be prepared in large batches and thawed as needed. Preparation of astrocyte/neuron co-culture requires ≥3 days: 1 day to plate the astrocytes (40 min of active time), 1 day to treat the astrocytes with AraC (15 min), and 1 day to harvest and plate the dopaminergic neurons (2 hr). In our laboratory, co-cultures are maintained for 14 days, but they could be maintained longer.
Acknowledgments
I would like to thank Erin K. O'Shea, W. Matt Reilly, and Deepika Walpita (Janelia Research Campus) as well as Jessica Young (University of Washington) for helpful discussions. This work was funded by the Howard Hughes Medical Institute.
Literature Cited
- Chai, H., Diaz-Castro, B., Shigetomi, E., Monte, E., Octeau, J. C., Yu, X., … Khakh, B. S. (2017). Neural circuit-specialized astrocytes: Transcriptomic, proteomic, morphological, and functional evidence. Neuron , 95(3), 531–549 e539. doi: 10.1016/j.neuron.2017.06.029.
- Chandrasekaran, A., Avci, H. X., Leist, M., Kobolak, J., & Dinnyes, A. (2016). Astrocyte differentiation of human pluripotent stem cells: New tools for neurological disorder research. Frontiers in Cellular Neuroscience , 10, 215. doi: 10.3389/fncel.2016.00215.
- Cheng, L., Hu, W., Qiu, B., Zhao, J., Yu, Y., Guan, W., … Pei, G. (2015). Cell Research , 25(5), 645–646. doi: 10.1038/cr.2015.55.
- Clarke, L. E., Liddelow, S. A., Chakraborty, C., Munch, A. E., Heiman, M., & Barres, B. A. (2018). Normal aging induces A1-like astrocyte reactivity. Proceedings of the National Academy of Sciences of the United States of America , 115(8), E1896–E1905. doi: 10.1073/pnas.1800165115.
- di Domenico, A., Carola, G., Calatayud, C., Pons-Espinal, M., Munoz, J. P., Richaud-Patin, Y., … Consiglio, A. (2019). Patient-specific iPSC-derived astrocytes contribute to non-cell-autonomous neurodegeneration in Parkinson's disease. Stem Cell Reports , 12(2), 213–229. doi: 10.1016/j.stemcr.2018.12.011.
- Di Giorgio, F. P., Boulting, G. L., Bobrowicz, S., & Eggan, K. C. (2008). Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell , 3(6), 637–648. doi: 10.1016/j.stem.2008.09.017.
- Emdad, L., D'Souza, S. L., Kothari, H. P., Qadeer, Z. A., & Germano, I. M. (2012). Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells and Development , 21(3), 404–410. doi: 10.1089/scd.2010.0560.
- Gonzalez, D. M., Gregory, J., & Brennand, K. J. (2017). The importance of non-neuronal cell types in hiPSC-based disease modeling and drug screening. Frontiers in Cell and Developmental Biology , 5, 117. doi: 10.3389/fcell.2017.00117.
- Hasel, P., Dando, O., Jiwaji, Z., Baxter, P., Todd, A. C., Heron, S., … Hardingham, G. E. (2017). Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nature Communications , 8, 15132. doi: 10.1038/ncomms15132.
- Ioannou, M. S., Jackson, J., Sheu, S. H., Chang, C. L., Weigel, A. V., Liu, H., … Liu, Z. (2019). Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell , 177(6), 1522–1535 e1514. doi: 10.1016/j.cell.2019.04.001.
- Kirkeby, A., Grealish, S., Wolf, D. A., Nelander, J., Wood, J., Lundblad, M., … Parmar, M. (2012). Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Reports , 1(6), 703–714. doi: 10.1016/j.celrep.2012.04.009.
- Kostuk, E. W., Cai, J., & Iacovitti, L. (2019). Subregional differences in astrocytes underlie selective neurodegeneration or protection in Parkinson's disease models in culture. Glia , 67(8), 1542–1557. doi: 10.1002/glia.23627.
- Kriks, S., Shim, J. W., Piao, J., Ganat, Y. M., Wakeman, D. R., Xie, Z., … Studer, L. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature , 480(7378), 547–551. doi: 10.1038/nature10648.
- Liu, C., Oikonomopoulos, A., Sayed, N., & Wu, J. C. (2018). Modeling human diseases with induced pluripotent stem cells: From 2D to 3D and beyond. Development , 145(5), pii: dev156166. doi: 10.1242/dev.156166.
- Miller, J. D., Ganat, Y. M., Kishinevsky, S., Bowman, R. L., Liu, B., Tu, E. Y., … Studer, L. (2013). Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell , 13(6), 691–705. doi: 10.1016/j.stem.2013.11.006.
- Passier, R., Orlova, V., & Mummery, C. (2016). Complex tissue and disease modeling using hiPSCs. Cell Stem Cell , 18(3), 309–321. doi: 10.1016/j.stem.2016.02.011.
- Sandoe, J., & Eggan, K. (2013). Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nature Neuroscience , 16(7), 780–789. doi: 10.1038/nn.3425.
- Tang, X., Zhou, L., Wagner, A. M., Marchetto, M. C., Muotri, A. R., Gage, F. H., & Chen, G. (2013). Astroglial cells regulate the developmental timeline of human neurons differentiated from induced pluripotent stem cells. Stem Cell Research , 11(2), 743–757. doi: 10.1016/j.scr.2013.05.002.
- TCW, J., Wang, M., Pimenova, A. A., Bowles, K. R., Hartley, B. J., Lacin, E., … Brennand, K. J. (2017). An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports , 9(2), 600–614. doi: 10.1016/j.stemcr.2017.06.018.
- Tyzack, G. E., Hall, C. E., Sibley, C. R., Cymes, T., Forostyak, S., Carlino, G., … Lakatos, A. (2017). A neuroprotective astrocyte state is induced by neuronal signal EphB1 but fails in ALS models. Nature Communications , 8(1), 1164. doi: 10.1038/s41467-017-01283-z.
- Wakeman, D. R., Hiller, B. M., Marmion, D. J., McMahon, C. W., Corbett, G. T., Mangan, K. P., … Kordower, J. H. (2017). Cryopreservation maintains functionality of human iPSC dopamine neurons and rescues parkinsonian phenotypes in vivo. Stem Cell Reports , 9(1), 149–161. doi: 10.1016/j.stemcr.2017.04.033.
- Yeh, T. H., Lee, D. Y., Gianino, S. M., & Gutmann, D. H. (2009). Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia , 57(11), 1239–1249. doi: 10.1002/glia.20845.
Citing Literature
Number of times cited according to CrossRef: 19
- Elena S. Yarkova, Elena V. Grigor’eva, Sergey P. Medvedev, Denis A. Tarasevich, Sophia V. Pavlova, Kamila R. Valetdinova, Julia M. Minina, Suren M. Zakian, Anastasia A. Malakhova, Detection of ER Stress in iPSC-Derived Neurons Carrying the p.N370S Mutation in the GBA1 Gene, Biomedicines, 10.3390/biomedicines12040744, 12 , 4, (744), (2024).
- Elena S. Yarkova, Elena V. Grigor’eva, Sergey P. Medvedev, Sophia V. Pavlova, Suren M. Zakian, Anastasia A. Malakhova, IPSC-Derived Astrocytes Contribute to In Vitro Modeling of Parkinson’s Disease Caused by the GBA1 N370S Mutation, International Journal of Molecular Sciences, 10.3390/ijms25010327, 25 , 1, (327), (2023).
- Alena E. Kopytova, George N. Rychkov, Alexander A. Cheblokov, Elena V. Grigor’eva, Mikhail A. Nikolaev, Elena S. Yarkova, Diana A. Sorogina, Farid M. Ibatullin, Galina V. Baydakova, Artem D. Izyumchenko, Daria A. Bogdanova, Vitali M. Boitsov, Akim V. Rybakov, Irina V. Miliukhina, Vadim A. Bezrukikh, Galina N. Salogub, Ekaterina Y. Zakharova, Sofya N. Pchelina, Anton K. Emelyanov, Potential Binding Sites of Pharmacological Chaperone NCGC00241607 on Mutant β-Glucocerebrosidase and Its Efficacy on Patient-Derived Cell Cultures in Gaucher and Parkinson’s Disease, International Journal of Molecular Sciences, 10.3390/ijms24109105, 24 , 10, (9105), (2023).
- Elena V. Grigor’eva, Alena E. Kopytova, Elena S. Yarkova, Sophia V. Pavlova, Diana A. Sorogina, Anastasia A. Malakhova, Tuyana B. Malankhanova, Galina V. Baydakova, Ekaterina Y. Zakharova, Sergey P. Medvedev, Sofia N. Pchelina, Suren M. Zakian, Biochemical Characteristics of iPSC-Derived Dopaminergic Neurons from N370S GBA Variant Carriers with and without Parkinson’s Disease, International Journal of Molecular Sciences, 10.3390/ijms24054437, 24 , 5, (4437), (2023).
- Yee Jie Yeap, Tng J. W. Teddy, Mok Jung Lee, Micaela Goh, Kah Leong Lim, From 2D to 3D: Development of Monolayer Dopaminergic Neuronal and Midbrain Organoid Cultures for Parkinson’s Disease Modeling and Regenerative Therapy, International Journal of Molecular Sciences, 10.3390/ijms24032523, 24 , 3, (2523), (2023).
- Mathias Busch, Hugo Brouwer, Germaine Aalderink, Gerrit Bredeck, Angela A. M. Kämpfer, Roel P. F. Schins, Hans Bouwmeester, Investigating nanoplastics toxicity using advanced stem cell-based intestinal and lung in vitro models, Frontiers in Toxicology, 10.3389/ftox.2023.1112212, 5 , (2023).
- Martyna M. Grochowska, Federico Ferraro, Ana Carreras Mascaro, Domenico Natale, Amber Winkelaar, Valerie Boumeester, Guido J. Breedveld, Vincenzo Bonifati, Wim Mandemakers, deCLUTTER2+ – a pipeline to analyze calcium traces in a stem cell model for ventral midbrain patterned astrocytes, Disease Models & Mechanisms, 10.1242/dmm.049980, 16 , 6, (2023).
- A. de Rus Jacquet, M. Alpaugh, H. L. Denis, J. L. Tancredi, M. Boutin, J. Decaestecker, C. Beauparlant, L. Herrmann, M. Saint-Pierre, M. Parent, A. Droit, S. Breton, F. Cicchetti, The contribution of inflammatory astrocytes to BBB impairments in a brain-chip model of Parkinson’s disease, Nature Communications, 10.1038/s41467-023-39038-8, 14 , 1, (2023).
- Anna Kreutz, Guang Hu, Erik Tokar, Pluripotent Stem Cell-derived Dopaminergic Neurons for Studying Developmental Neurotoxicity, Stem Cell Reviews and Reports, 10.1007/s12015-023-10555-9, 19 , 7, (2120-2130), (2023).
- Parivash Nouri, Anja Zimmer, Stefanie Brüggemann, Robin Friedrich, Ralf Kühn, Nilima Prakash, Generation of a NES-mScarlet Red Fluorescent Reporter Human iPSC Line for Live Cell Imaging and Flow Cytometric Analysis and Sorting Using CRISPR-Cas9-Mediated Gene Editing, Cells, 10.3390/cells11020268, 11 , 2, (268), (2022).
- Reed Berlet, Dorothy Anne Galang Cabantan, Daniel Gonzales-Portillo, Cesar V. Borlongan, Enriched Environment and Exercise Enhance Stem Cell Therapy for Stroke, Parkinson’s Disease, and Huntington’s Disease, Frontiers in Cell and Developmental Biology, 10.3389/fcell.2022.798826, 10 , (2022).
- Emilie M. Legault, Julie Bouquety, Janelle Drouin-Ouellet, Disease Modeling of Neurodegenerative Disorders Using Direct Neural Reprogramming, Cellular Reprogramming, 10.1089/cell.2021.0172, 24 , 5, (228-251), (2022).
- Razan Sheta, Maxime Teixeira, Walid Idi, Marion Pierre, Aurelie de Rus Jacquet, Vincent Emond, Cornelia E. Zorca, Benoît Vanderperre, Thomas M. Durcan, Edward A. Fon, Frédéric Calon, Mohamed Chahine, Abid Oueslati, Combining NGN2 programming and dopaminergic patterning for a rapid and efficient generation of hiPSC-derived midbrain neurons, Scientific Reports, 10.1038/s41598-022-22158-4, 12 , 1, (2022).
- Heather Branscome, Pooja Khatkar, Sarah Al Sharif, Dezhong Yin, Sheela Jacob, Maria Cowen, Yuriy Kim, James Erickson, Christine A. Brantner, Nazira El-Hage, Lance A. Liotta, Fatah Kashanchi, Retroviral infection of human neurospheres and use of stem Cell EVs to repair cellular damage, Scientific Reports, 10.1038/s41598-022-05848-x, 12 , 1, (2022).
- Reilly L. Allison, Emily Welby, Guzal Khayrullina, Barrington G. Burnett, Allison D. Ebert, Viral mediated knockdown of GATA6 in SMA iPSC‐derived astrocytes prevents motor neuron loss and microglial activation, Glia, 10.1002/glia.24153, 70 , 5, (989-1004), (2022).
- Aurelie de Rus Jacquet, Jenna L Tancredi, Andrew L Lemire, Michael C DeSantis, Wei-Ping Li, Erin K O'Shea, The LRRK2 G2019S mutation alters astrocyte-to-neuron communication via extracellular vesicles and induces neuron atrophy in a human iPSC-derived model of Parkinson’s disease, eLife, 10.7554/eLife.73062, 10 , (2021).
- Alla B. Salmina, Marina R. Kapkaeva, Anna S. Vetchinova, Sergey N. Illarioshkin, Novel Approaches Used to Examine and Control Neurogenesis in Parkinson′s Disease, International Journal of Molecular Sciences, 10.3390/ijms22179608, 22 , 17, (9608), (2021).
- Aurélie de Rus Jacquet, Hélèna L. Denis, Francesca Cicchetti, Melanie Alpaugh, Current and future applications of induced pluripotent stem cell-based models to study pathological proteins in neurodegenerative disorders, Molecular Psychiatry, 10.1038/s41380-020-00999-7, 26 , 7, (2685-2706), (2021).
- Yong H. Park, Joshua D. Snook, Iris Zhuang, Guofu Shen, Benjamin J. Frankfort, Optimized culture of retinal ganglion cells and amacrine cells from adult mice, PLOS ONE, 10.1371/journal.pone.0242426, 15 , 12, (e0242426), (2020).

