Pathogen-Oriented Low-cost Assembly & Re-sequencing (POLAR): A highly sensitive and high-throughput SARS-CoV-2 diagnostic based on whole genome sequencing
Per A. Adastra, Neva C. Durand, Namita Mitra, Saul Godinez, Ragini Mahajan, Alyssa Blackburn, Zane Colaric, Joshua W. M. Theisen, David Weisz, Olga Dudchenko, Andreas Gnirke, Suhas S.P. Rao, Parwinder Kaur, Erez Lieberman Aiden, Aviva Presser Aiden
Abstract
Here, we introduce a low-cost, high-throughput method for diagnosis of SARS-CoV-2 infection, dubbed Pathogen- Oriented Low-Cost Assembly & Re-Sequencing (POLAR), that enhances sensitivity by aiming to amplify the entire SARS-CoV-2 genome rather than targeting particular viral loci, as in typical RT- PCR assays. To achieve this goal, we combine a SARS-CoV-2 enrichment method developed by the ARTIC Network (https://artic.network/) with short-read DNA sequencing and de novo genome assembly. For details on our computational pipeline for automated data processing, including documentation and test set, please visit our Githuh at https://github.com/aidenlab/Polar.
The last step in this version contains a supplemental video with extra context and tips, as part of the protocols.io Spotlight series, featuring conversations with protocol authors.
Before start
Steps
RNA extraction
Collect 96 patient samples and record sample data. Add 200µL of the samples into each well of the Zymo-Spin™ I-96 plate. Keep plate On ice while preparing reagents in Step 2.
Prepare the following reagents before starting viral RNA extraction:1. Add beta-mercaptoethanol (user supplied) to the Viral RNA Buffer to a final dilution of 0.5% volume i.e., 500µL per 100 ml.2. Add 192mL of 100% ethanol (or 204mL of 95% ethanol) to the 48mL Viral Wash Buffer concentrate.
Add 400µL of Viral RNA Buffer to each sample well. Mix well by pipetting.
Mount the Zymo-Spin™ I-96 plate on a Collection Plate and centrifuge at 3000x g,0h 0m 0s-5000x g,0h 0m 0s for 0h 5m 0s. Discard the flow-through from the collection plate.
Add 500µLViral Wash Buffer to each well and centrifuge at 3000x g,0h 0m 0s-5000x g,0h 0m 0s for 0h 5m 0s. Discard the flowthrough from the collection plate. Repeat this step.
Add 500µL ethanol (95-100%) to each well and centrifuge at 3000x g,0h 0m 0s - 5000x g,0h 0m 0s for 0h 5m 0s.
To elute RNA, mount a new 96-well plate between the Zymo-Spin™ I-96 plate and an empty Collection Plate. Add 15µL DNase/RNase-Free Water directly to the column matrix of the Zymo-Spin™ I-96 plate wells and centrifuge at 3000x g,0h 0m 0s - 5000x g,0h 0m 0s for 0h 5m 0s. The eluted RNA extract will be collected in the middle 96-well plate.
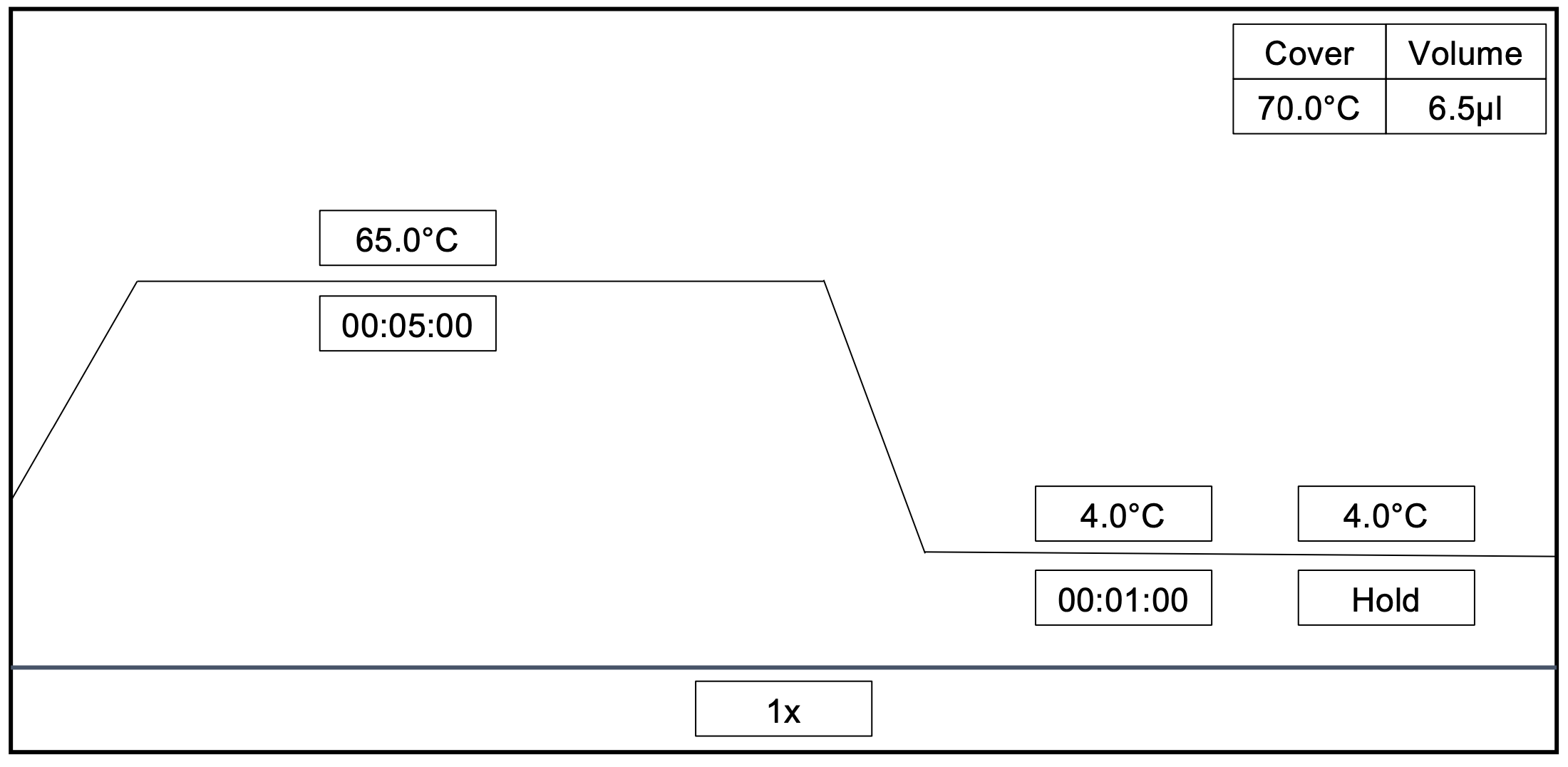
cDNA preparation
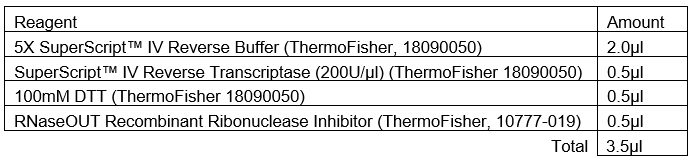
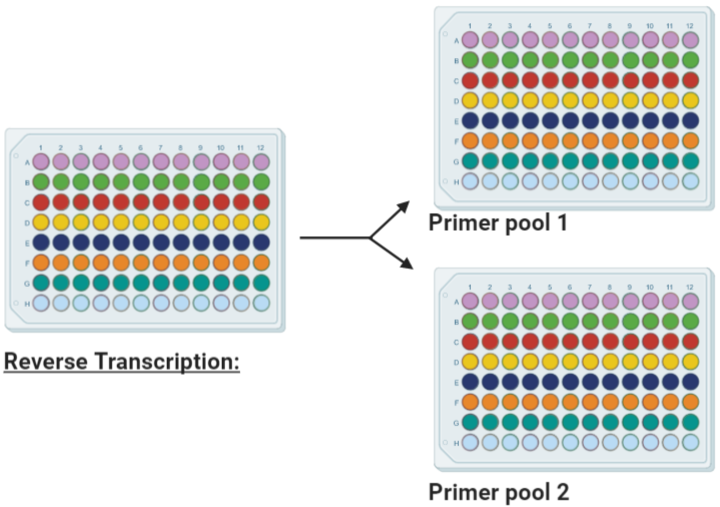
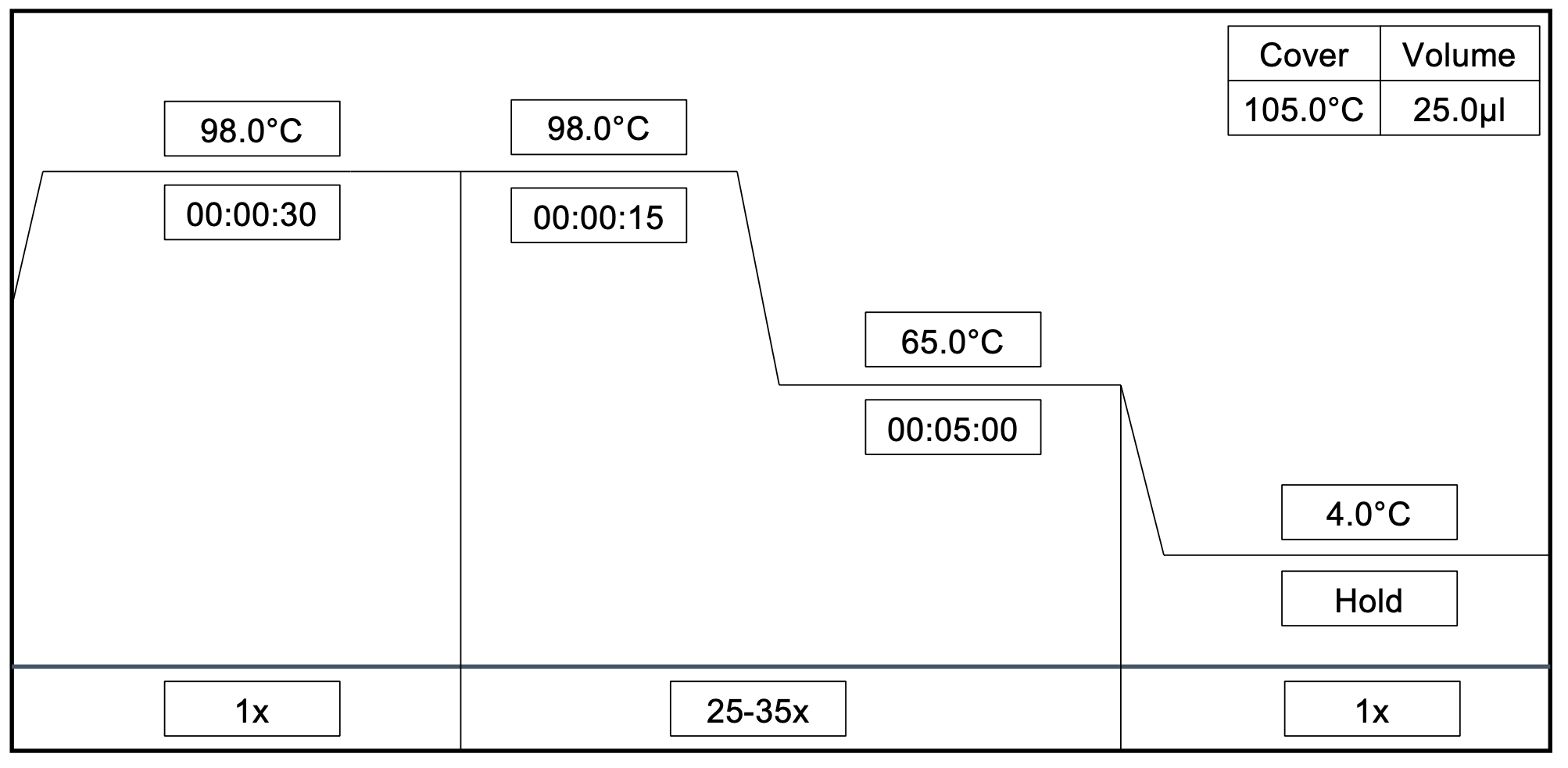
Multiplex PCR
Add 2.5µL of cDNA of each biological sample from from Step 11 to both Pool #1 and Pool #2 96-well plates.
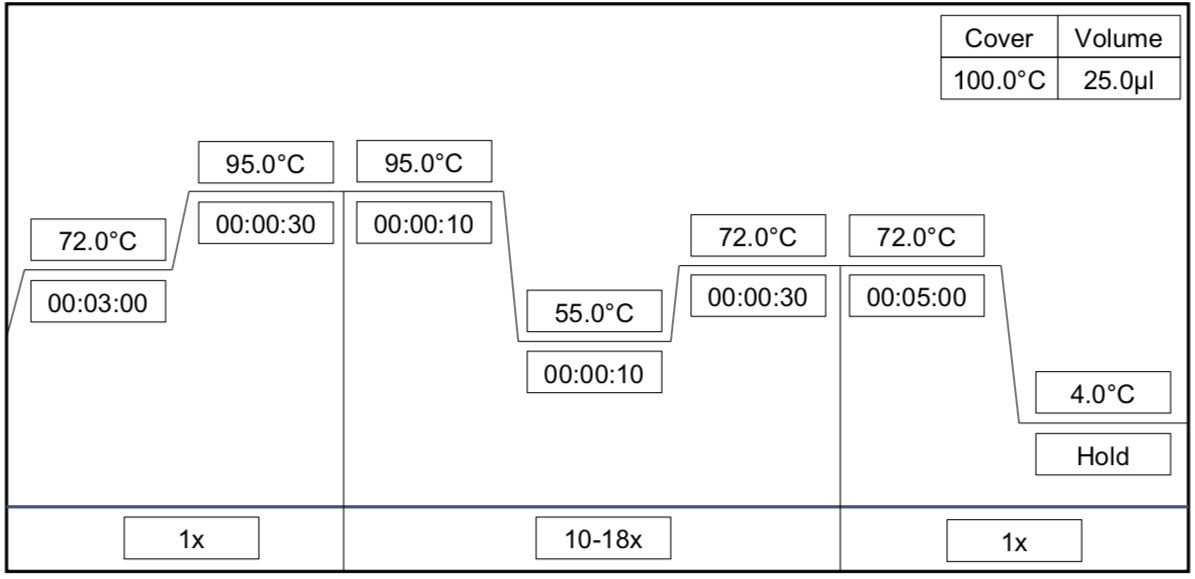
Post PCR Clean-up
Add an equal volume (1:1) of sparQ PureMag beads to each sample well in plates Pool #1 and Pool #2 and mix gently by either flicking or pipetting. For example, add 50µL sparQ PureMag beads to a 50µL reaction. Then pulse centrifuge to collect all liquid at the bottom of the tube.
Incubate for 0h 5m 0s at room temperature.
Place the plates on the Agencourt SPRIPlate Super Magnet Plate and incubate for 0h 2m 0s or until the beads have pelleted and the supernatant is completely clear. Then, while avoiding the bead pellet, carefully remove the clear supernatant.
Equipment
| Value | Label |
|---|---|
| SPRIPlate 96R Ring Super Magnet Plate | NAME |
| 96-well Magnet Plate | TYPE |
| Agencourt | BRAND |
| A32782 | SKU |
Keeping on magnetic plate, add 200µL of Room temperature 80% volume ethanol to the side of the well opposite to the pellet and let sit for 0h 0m 30s.
Avoid disturbing the bead pellet, carefully remove and discard ethanol. Wait for 0h 0m 10s then remove any remaining ethanol.
and repeat ethanol wash.
Add 20µLof 10millimolar (mM) Tris-HCl (Ph 8.0) and pipette to mix well. Incubate for 0h 2m 0s at 37°C .
Separate beads on the Agencourt SPRIPlate Super Magnet Plate for 0h 2m 0s or until the beads have pelleted.
Transfer supernatant from each biological sample in the two plates (Pool #1 and Pool #2) and combine into a single well per sample in a new 96-well plate.
Quantification of DNA concentration using a Qubit® High Sensitivity Kit:
Quantify the DNA concentration using the Qubit® High Sensitivity DNA kit from 1µL of each product using Steps 26-35.
Set up the required number of 0.5-mL tubes for standards and samples and label lids accordingly.
Prepare a working solution by diluting the Qubit® dsDNA HS Reagent 1:200 in Qubit® dsDNA HS Buffer. For example, add 199µL of buffer and 1µL of dye per sample to a tube and vortex.
Add 190µL of Qubit® working solution and 10µL of the standard to each of the standards tubes and vortex.
For each sample, add 199µL of Qubit® working solution and 1µL of the sample in each 0.5-mL sample tube and vortex.
Allow all tubes to incubate at room temperature for 0h 2m 0s
On the Home screen of the Qubit® 3.0 Fluorometer, press DNA, select dsDNA High Sensitivity, and press Read Standards to proceed.
Insert the tube containing Standard #1 into the sample chamber, close the lid, then press Read standard. When the reading is complete (~0h 0m 3s), remove Standard #1.
Repeat Step 32 with Standard #2.
To quantify samples, Press Run samples and select the sample volume (1µL ) and units.
Repeat Step 34 for all samples.
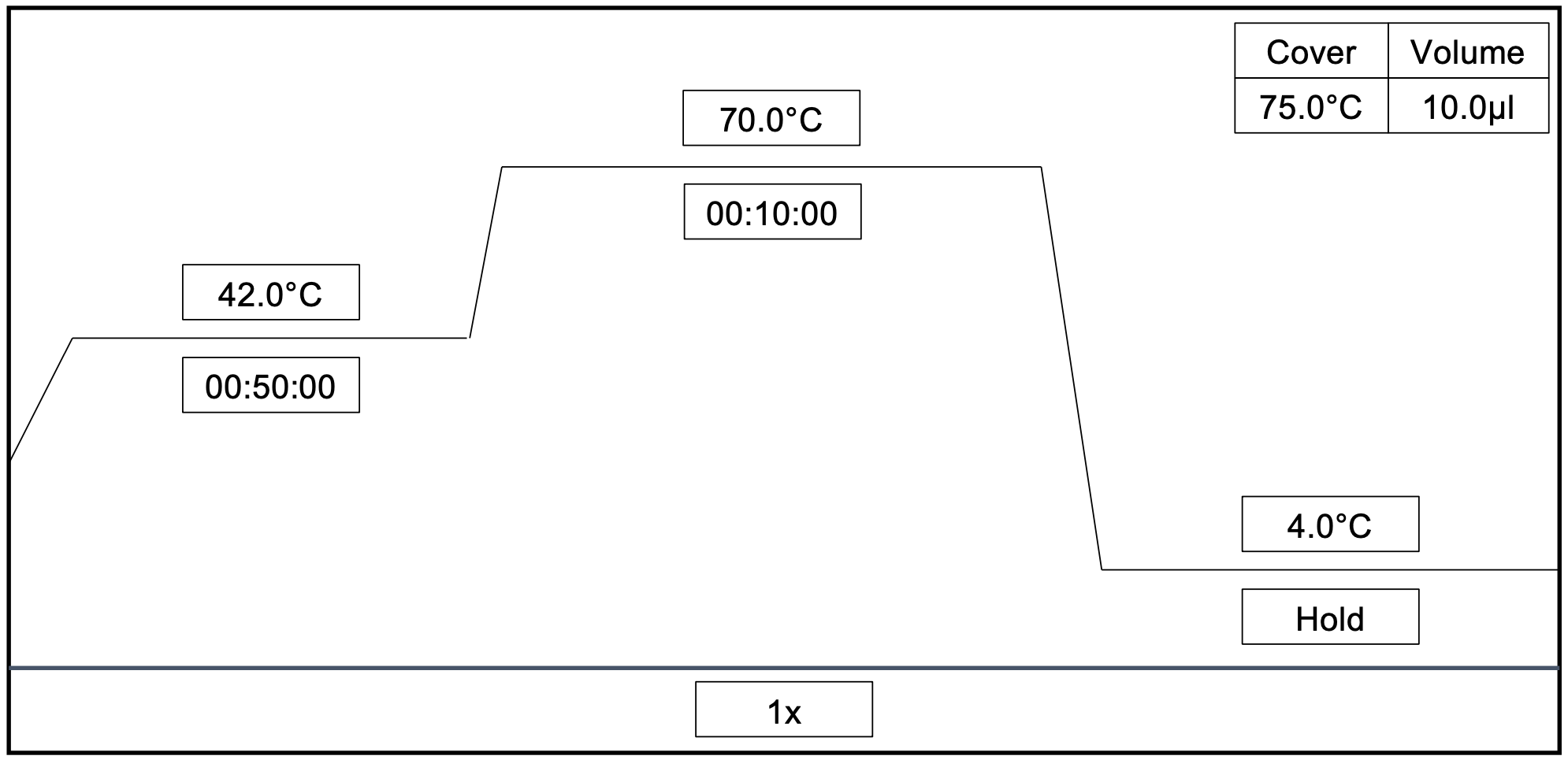
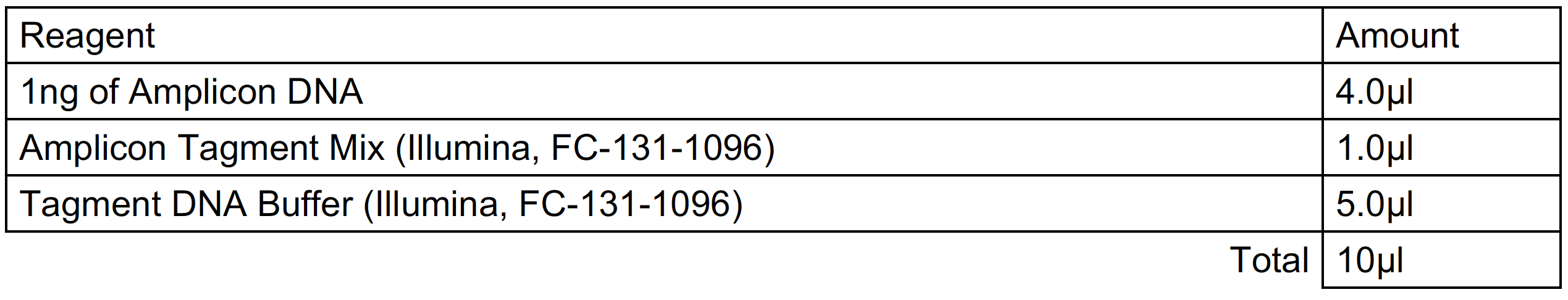
Nextera Library Preparation
Incubate at 55°C for0h 5m 0s, then lower to 10°C. Once at 10°C, immediately add 2.5µL of Neutralizing Tagment Buffer to stop the reaction. Mix by pipetting up and down.
Incubate at 10Room temperature for 0h 5m 0s. Centrifuge at 280x g,0h 0m 0s for 0h 1m 0s.
Final Library Cleanup
After PCR, bring up sample volume in each well to 50µL with nuclease free water.
Add a 1:1.8 ratio of sparQ PureMag beads to each well and mix gently pipetting. For example, add 90µL sparQ PureMag beads to a 50µL reaction.
Incubate for 0h 5m 0s at room temperature.
Place the plate on the Agencourt SPRIPlate Super Magnet Plate and incubate for 0h 2m 0s or until the beads have pelleted and the supernatant is completely clear. Then, while avoiding the bead pellet, carefully remove the clear supernatant.
While keeping on magnetic plate, add 200µL of Room temperature 80% volume ethanol to the side of the well opposite of the pellet and let sit for 0h 0m 30s.
Avoid disturbing the bead pellet, carefully remove and discard ethanol. Wait for 0h 0m 10s then remove any remaining ethanol.
Add 100µLof 10millimolar (mM) Tris-HCl (pH 8.0) , vortex, and pulse centrifuge to collect all liquid at the bottom of the tube. Incubate for 0h 2m 0s at 37°C.
Separate beads on the Agencourt SPRIPlate Super Magnet Plate and transfer 100µL of supernatant to a new plate for a second round of cleanup.
Add an equal volume (1:1) of sparQ PureMag beads to each well and mix gently by pipetting. For example add 100µL sparQ PureMag beads to a 100µL reaction.
Incubate for 0h 5m 0s at room temperature.
Separate beads on the Agencourt SPRIPlate Super Magnet Plate and incubate for 0h 2m 0s or until the beads have pelleted and the supernatant is completely clear. Then, while avoiding the bead pellet, carefully remove the clear supernatant.
While keeping on magnetic plate, add 200µL of Room temperature 80% volume ethanol to the side of the well opposite of the pellet and let sit for 0h 0m 30s.
Avoid disturbing the bead pellet, carefully remove and discard ethanol. Wait for 0h 0m 10s then remove any remaining ethanol.
and repeat ethanol wash.
Add 20µLof 10millimolar (mM) Tris-HCl (Ph 8.0) and mix well by pipetting. Incubate for 0h 2m 0s at 37°C
Place the plate on the Agencourt SPRIPlate Super Magnet Plate and incubate for 0h 2m 0s or until the beads have pelleted and the supernatant is completely clear. Then, while avoiding the bead pellet, transfer the 20µL final library to a new plate for subsequent steps and storage.
Library Normalization
Transfer 10µL supernatant from each well of the PCR plate to the corresponding well of a new midi plate.
Combine the following volumes in a 15mL conical tube to prepare the LN master mix. Multiply each volume by the number of samples being processed: 23µL of LNA1 and 4µL of LNA2.
Pipette 10 times to mix and pour the LN master mix into a trough.
Use a p200 multichannel pipette to transfer 22.5µL LN master mix to each sample well.
Seal the plate, and then use a plate shaker at 1800rpm,0h 0m 0s for 0h 30m 0s
Place on the magnetic stand and wait until the liquid is clear (~2 minutes). Without disturbing the beads, discard all the supernatant.
Wash two times as follows:
Add 22.5µL LNW1 to each well. Seal the plate, and then use a plate shaker at 1800rpm,0h 0m 0s for0h 5m 0s. Separate on a magnetic plate and wait until the liquid is clear (~2 minutes). Without disturbing the beads, remove and discard all supernatant.
Add 15µL 0.1 N NaOH to each well. Seal the plate, and then use a plate shaker at 1800rpm,0h 0m 0s for 0h 5m 0s
Add 15µL LNS1 to each well of a new 96-well PCR plate labeled SGP.
After the 0h 5m 0s elution completes, make sure that all samples in the midi plate are resuspended. If they are not, resuspend as follows:
Pipette 10 times to mix or lightly tap the sample plate on the bench. Seal the plate, and then use a plate shaker at 1800rpm,0h 0m 0s for 0h 5m 0s. Place on a magnet rack and transfer 15µL supernatant from each well of the midi plate to thecorresponding well of the SGP plate.
Seal the plate, and then centrifuge at 1000x g,0h 0m 0s for0h 1m 0s.
Place on a magnetic stand and wait until the liquid is clear ~ 0h 2m 0s.
Transfer 15µL supernatant from each well of the midi plate to the corresponding well of the SGP plate.
Seal the sample plate, and then centrifuge at1000x g,0h 0m 0s for 0h 1m 0s.
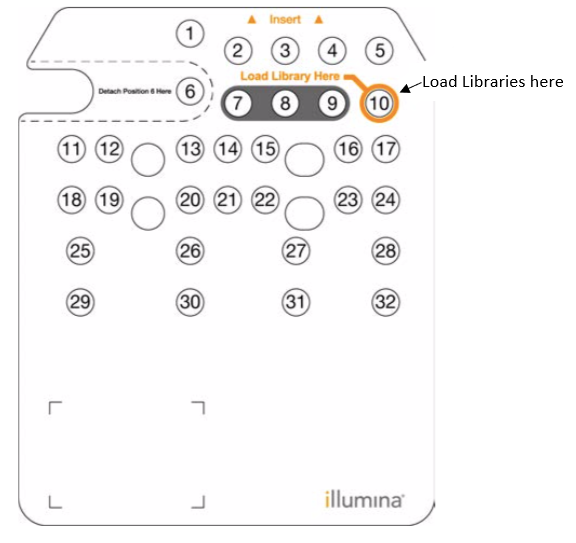
Loading the NextSeq550 sequencer
Remove a “Mid-Output Kit" reagent cartridge from -20°C freezer storage.
Place the cartridge right-side-up in a secondary container and fill the container with water at room temperature, but do not fully submerge the cartridge. Thaw cartridge in this water bath for about 1h 0m 0s .
When the reagent cartridge has been thawing for 0h 30m 0s, remove a "Mid-Output Kit" flow cell from the 4°C refrigerator but do not open the foil package. Place the flow cell on the bench for 0h 30m 0s to warm up to Room temperature
After the reagent cartridge is fully thawed, take it out of the water bath, tap on the bench to dislodge excess water, and dry with wipes.
Invert the cartridge to mix the reagents and make sure that the sequencing reagents in positions 29-32 are completely thawed. Then tap on the bench again to reduce air bubbles.
Remove the flow cell from its packaging and gently clean with a 70% ethanol wipe. Dry the glass with another wipe.
Inspect the flow cell to confirm: ports are free of obstructions, port gaskets are sealed and white plastic posts are visible, all 4 white retention clips are snapped over the edge of the black carrier plate and all 4 metal spring clips are laying flat against the black carrier plate.
Preheat an incubator to 98°C
Remove a tube of Hybridization Buffer (HT1) from the -20°C freezer and thaw at Room temperature . When thawed, place on ice.
In a fresh 1.5ml tube, combine an aliquot from each library pool such that the total volume is 5µL. For a Mid-Output kit, expect to obtain 150M reads in total.
Add 995µL of ice-cold Hybridization Buffer (HT1). Vortex briefly and centrifuge at 300x g,0h 0m 0s for 0h 1m 0s.
Transfer 750µLof diluted library to a new tube and add 750µL of ice-cold Hybridization Buffer (HT1).
Vortex briefly and centrifuge at 300x g,0h 0m 0s for 0h 1m 0s.
Place the tube on the 98°C heated incubator for 0h 2m 0s and immediately cool on ice. Leave on ice for 0h 5m 0s
In a fresh 1.5ml tube, combine 1203µL of ice-cold Hybridization Buffer (HT1) and 97µL diluted library for a final concentration of 1.5picomolar (pM). Invert to mix and pulse centrifuge. Place on ice until ready to load onto the reagent cartridge.
Add the entire 1.3ml of the 1.5picomolar (pM) library dilution into this reservoir.
The reagent cartridge, flow cell, and buffer pack are now ready to be loaded onto the NextSeq500 for sequencing.
Spotlight video
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
<iframe title="YouTube video player" src="https://www.youtube.com/embed/1vFvHcBK32k?si=chU20kQpZDQ3wfwG" height="315" width="560"></iframe>