Immunocapture of virion from body fluids
Jeffrey A. Johnson, Sarah Sabour, Jin-fen Li, Jonathan Lipscomb
Disclaimer
The performance of this protocol is claimed by the authors and does not necessarily represent the official view of the CDC.
Abstract
Procedure for immunocapturing HIV virions from blood and seminal plasma, cerebral spinal fluid, and cell culture supernatant by monoclonal antibody-targeting source cell markers in virion envelopes.
Attachments
Steps
Concentrate Antibody
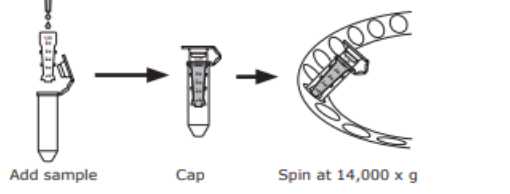
Use Amicon Ultra-0.5 Centrifugal Filter Devices.
Insert the Amicon device into the microcentrifuge tube.
Add up to 500µL Ab (0.1-0.2) to the filter device and cap it.
Insert the capped Amicon Ultra device into a centrifuge tube and place in the centrifuge rotor.
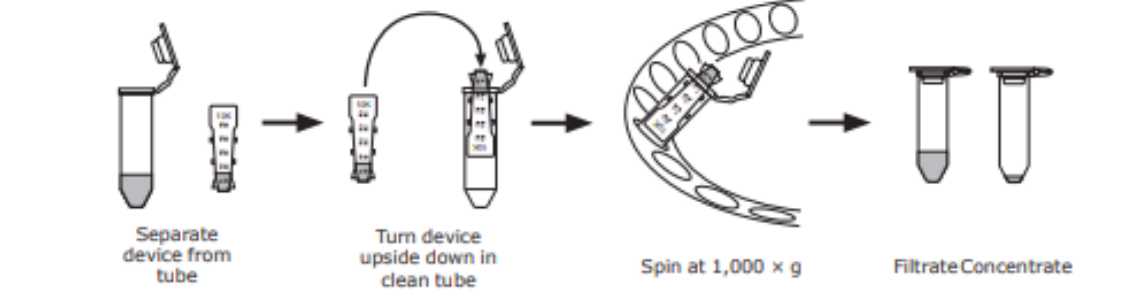
Remove the device and place it upside down in a clean tube, place in centrifuge, aligning the open cap strap, toward the center of the rotor.
Add SPB (0.1Molarity (M) sodium phosphate buffer, 7.2) to achieve a final volume 100µL mAb at a concentration of 1-2.
Antibody Biotinylation (Sigma BTAG)
Add 30µL DMSO to the vial of Biotinylation Reagent (BAC-SulfoNHS), and then add 970µL 0.1Molarity (M) sodium phosphate buffer.
Immediately add 2µL of Biotinylation Reagent to the antibody solution with gentle stirring.
Incubate with gentle stirring for 0h 30m 0s at Room temperature or 2h 0m 0s at 2°C-8°C.
Isolation of Labeled Antibody (Sigma BTAG)
Place the column G-50 in a 1.5 ml Eppendorf tube, pre-spin the column for 0h 1m 0s at 700x g,0h 0m 0s (3000rpm,0h 0m 0s).
Add 200µL PBS (7.4) to the column, spin the column for 0h 1m 0s at 700x g,0h 0m 0s (3000rpm,0h 0m 0s).
Repeat two times.
Label two of 1.5 ml Eppendorf tube.
Place column in tube 1 and apple the biotinylation reaction mix to the column.
Centrifuge the column for 0h 2m 0s at 700x g,0h 0m 0s and collect flow-through (fraction 1).
Place column in tube 2 and add 200 up to the column, spin the column for 0h 2m 0s at 700x g,0h 0m 0s. collect flow-through (fraction 2).
Determine Ab Concentration
Use Bicinchoninic Acid Kit, 96 well Immulon II plate assay.
Prepare standard curve dilutions:
| A | B | C | D |
|---|---|---|---|
| Protein Ci (µg/mL) | Protein Input Volume (uL) | PBS (µL) | Protein Cf (ug/mL) |
| 1000 | - | - | 1000 |
| 1000 | 400 | 100 | 800 |
| 800 | 375 | 125 | 600 |
| 600 | 333 | 166 | 400 |
| 400 | 250 | 250 | 200 |
| 200 | 250 | 250 | 100 |
Prepare BCA Working Reagent: Mix Reagent A(50) and Reagent B(1).
Add 25µL protein standard solution, PBS, and Ab samples into well of 96 well plate. Duplicate.
Add 200µL of BCA working to each well (1:8 protein/BCA ratio).
Cover the plate with film and incubate 37°C for 0h 30m 0s.
Read the absorbance at 562 (540-590).
Calculate mAb concentration against the standard curve.
ELISA to Check Biotinylated Antibody
Using Immulon II 96 well plate.
Coat three wells with a dilution series of mAb beginning with 1µL mAb in 99µL PBS (1:100) continuing with two more 10-fold dilutions. Incubate 0h 30m 0s at 4°C.
Wash plate 4 times with PBS+0.05% Tween.
Add 100µL blocking buffer to each well, incubate at 37°C for 1h 0m 0s.
Wash plate 4 times with PBS+0.05% Tween.
Add 100µL of 1:5000 ExtrAvidin_Peroxidase diluted with blocking buffer to each well. Cover plate and incubate at 37°C for 1h 0m 0s.
Wash plate 4 times with PBS+0.05% Tween.
Add 100µL TMB substrate to each well.
Develop plate at Room temperature in the dark for 0h 15m 0s.
Add 100µL of stop solution to each well.
Read the absorbance of each well at 450 and 550. OD values of the 1:100 dilution (first well) of ≥0.6 indicates adequate biotin labelling of antibody.
Streptavidin coated beads_ Biotinylated Ab + HIV → bead-Ab_HIV complex
Dilute Biotinylated Ab to 0.4 with 0.1Molarity (M) sodium phosphate buffer.
Incubate 100µL of Streptavidin coated beads with 5µL PBS (negative Ab control) or 2µg (5µL of 0.4) biotinylated Ab for 0h 10m 0s at Room temperature on a roller platform.
Centrifuge bead-Ab complex at 8000rpm.
Remove supernatant and wash pellet with 100µL PBS+1% BSA +1% Tween 20, centrifuge beads Ab complex at 8000rpm. Wash 3 times.
Add 100µL Blocking buffer (PBS+1% BSA +1% Tween 20) to the tube and incubate at 4°C 0h 10m 0s.
Centrifuge bead-Ab complex at 8000rpm and then remove supernatant.
If working with tissue culture supernatants first DNase treat and inactivate.
Add 200µL HIV-positive material (plasma, CSF, Semen, Culture or flow-through) to the designated bead-Ab complex and incubate for 0h 30m 0s at Room temperature. Mixing gently on a roller-mixer.
Prepare µMACs column
Attach µMACs column to the magnetic multistand.
Add 100µL equilibration buffer for nucleic acid applications to the column.
Rinse column with 100µL wash buffer (PBS+1% BSA +1% Tween 20), twice.
Binding HIV-bead-Ab complex to the column and collecting the flow-through
Apply HIV-bead-Ab complex onto the top of column, collecting the flowthrough in a clean microfuge tube or eluting directly into the next tube of biotinylated mAb-bead complex. Let reaction pass through the column completely, captured virus will be retained on the column and flow-through will contain non-target virus (see figure below).
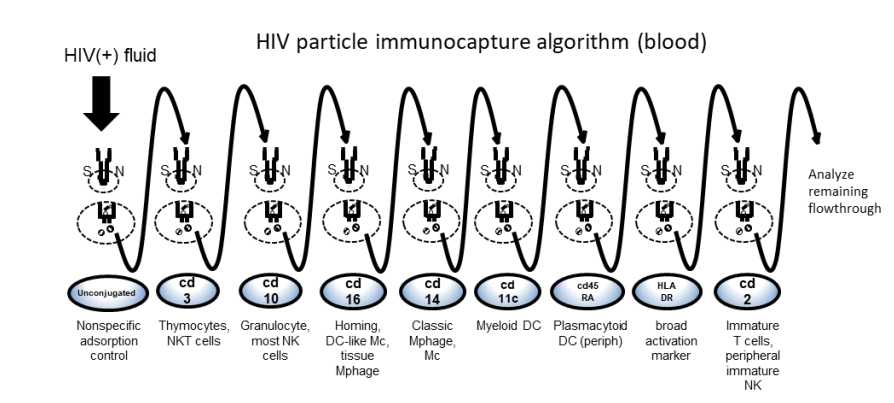
Add 30µL wash buffer to the column and collect the flow.
Incubate the flowthrough with next mAb-bead complex for 0h 30m 0s on the roller-mixer at
Room temperature.
To the just-eluted column, rinse the column 3 times with 400µL of wash buffer to remove nonspecifically bound material, allowing the column drain completely. Discard the wash.
Repeat this process until all mAb-bead columns in the series are completed.
Elute target virion RNA from the column (using the QIAamp Viral RNA Mini kit)
After washing the column, place the column of bound virion in a new 1.5 mL Eppendorf tube.
Add 50µL AVL lysis buffer to the column and pass through the column completely.
Add another 150µL AVL lysis buffer to the column and pass through the column completely.
Add 360µL AVL lysis buffer to the tube of eluted lysate and incubate tube at Room temperature for 0h 10m 0s. Continue with the extraction kit instructions as follows.
RNA extraction: QIAamp Viral RNA Mini Kit
Add 560µL ethanol (96–100%) to the sample and mix by pulse-vortexing for 0h 0m 15s. After mixing, briefly centrifuge the tube to remove drops from inside the lid.
Carefully apply 630µL of the sample solution to the QIAamp Mini column (in a 2 ml collection tube) without wetting the rim. Close the cap, and centrifuge at 6000x g,0h 0m 0s (8000rpm,0h 0m 0s) for 0h 1m 0s. Place the QIAamp Mini column into a clean 2 ml collection tube and discard the tube containing the filtrate.
Repeat this step until all of the lysate has been loaded onto the spin column.
Add 500µL Buffer AW1. Close the cap, and centrifuge at 6000x g,0h 0m 0s (8000rpm,0h 0m 0s) for 0h 1m 0s. Place the QIAamp Mini column in a clean 2 ml collection tube.
Add 500µL Buffer AW2. Close the cap and centrifuge at full speed (20000x g,0h 0m 0s; 14000rpm,0h 0m 0s) for 0h 3m 0s.
Place the QIAamp Mini column in a new 2 mL collection tube and discard the old collection tube with the filtrate. Centrifuge at 20000x g,0h 0m 0s (full speed) for 0h 1m 0s.
Place the QIAamp Mini column in a clean 1.5 ml microcentrifuge tube. Discard the old collection tube containing the filtrate. Carefully open the QIAamp Mini column and add 60µL Buffer AVE equilibrated to Room temperature.
Close the cap and incubate at Room temperature for 0h 1m 0s. Then centrifuge at 6000x g,0h 0m 0s (8000rpm,0h 0m 0s) for 0h 1m 0s.
RT PCR: SuperScript™ III One-Step RT-PCR System with Platinum™ Taq High Fidelity DNA Polymerase
Thaw, vortex briefly to mix and centrifuge each component before use.
Prepare 45µL reaction mast mix in a PCR workstation.
| A | B |
|---|---|
| Component | Volume (uL) |
| 2x Reaction Mix | 25 |
| F primer (10 μM) | 1 |
| R primer (10 μM) | 1 |
| SuperScript III RT/Platinum Taq High Fidelity Enzyme Mix | 2 |
| RNA Inhibitor (40 U/µL) | 1 |
| Water | 15 |
| Total | 45 |
Add 5µL of template RNA. Final reaction volume is 50µL.
Gently mix and make sure that all the components are at the bottom of the amplification tube.
Place the reaction in the preheated thermal cycler programmed as described above. Collect the data and analyze the results.
Nested PCR (nPCR): Platinum™ SuperFi II PCR Master Mix
Thaw, vortex briefly to mix and centrifuge each component before use.
For each sample, prepare 48µL reaction master mix in a PCR workstation as follows:
| A | B |
|---|---|
| Component | Volume (uL) |
| Platinum SuperFi II PCR Master Mix | 25 |
| F primer (10 μM) | 1 |
| R primer (10 μM) | 1 |
| Water | 21 |
| Total | 48 |
Transfer new reaction microfuge tubes and RT-PCR samples to Nested PCR room. Add 2µL of each RT-PCR sample per tube.
DNA is quantified and PCR amplicon size is verified via the Agilent 2200 Tapestation after nested PCR is performed for sequencing. Alternatively, bands can be checked by agarose gel.
Identify samples with clean amplicon bands for further analysis.
Perform Sanger sequencing with available platform.
Sequence analysis
Compare relatedness of HIV sequences in alignment software (e.g., Geneious) and MEGA to generate neighbor-joining trees and perform genetic distance analysis. Perform best model fit (typically Tamura 92 is the best fit)