High Throughput RNA Extraction and PCR Inhibitor Removal of Settled Solids for Wastewater Surveillance of SARS-CoV-2 RNA
Aaron Topol, Krista Wigginton, Alexandria B Boehm, marlene.wolfe, Bradley J White
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Please note that while this protocol is for TNA extraction using the Perkin Elmer Chemagic 360, RNA extraction with resuspended solids from this protocol has been verified to perform well using the Kingfisher MagMax kit as another high throughput, automated option and two manual Qiagen kits - the All Prep Powerviral DNA/RNA Kit and the Qiamp Viral RNA Mini Kit. this protocol has been verified to perform well using the Kingfisher MagMax kit as another high throughput, automated option and two manual Qiagen kits - the All Prep Powerviral DNA/RNA Kit and the Qiamp Viral RNA Mini Kit.
This process instruction describes the steps for purification of nucleic acids from wastewater solids and preparation for downstream quantitative analysis with Reverse Transcriptase droplet digital Polymerase Chain Reaction (RT-ddPCR). Due to the large quantities of substances that have inhibitory effects on PCR in wastewater samples, a subsequent PCR inhibitor removal step is required after nucleic acid purification. Both steps of the process are carried out in a 96-well plate format.
This method uses the resuspended solids generated using this protocol: High Throughput pre-analytical processing of wastewater settled solids for SARS-CoV-2 RNA analyses.
RNA purification is carried out using a kit optimized for the purification of viral on for the Perkin Elmer Chemagic 360. Although only RNA is used in the downstream applications from this protocol, DNA is also eluted in this process. A crucial component of the purification kit are the magnetic particles coated with poly vinyl alcohol (M-PVA Magnetic Beads) which have a hydrophilic surface giving them an affinity for nucleic acids but not many other biological molecules. The workflow involves binding nucleic acids in a sample to the beads which are then transferred through a series of wash buffers to remove debris with a robotic head with magnetic rods.
The OneStep PCR Inhibitor Removal Kits are PCR inhibitor clean up kits that contain all the components needed for efficient removal of contaminants that can inhibit downstream enzymatic reactions (e.g. PCR and RT) from DNA and RNA preparations. The column matrices in these PCR inhibitor clean up kits have been specifically designed for the efficient removal of polyphenolic compounds, humic/fulvic acids, tannins, melanin, etc. from the most impure DNA and RNA preparations.
This process instruction applies to extraction of RNA from wastewater samples using the Chemagic™ Viral DNA/RNA 300 Kit H96 for the Perkin Elmer Chemagic 360 followed by PCR Inhibitor Removal with the Zymo OneStep-96 PCR Inhibitor Removal Kit.
Steps
Preparation
If necessary, dissolve lyophilized Proteinase K in 11mLof nuclease free water and resuspend Poly A following manufacturer directions.
Label one 50 mL conical tube as Lysis buffer.
Label a deep-well-plate as “Sample Plate”.
If the Chemagic 360 has not been used within 24 hours, run the protocol “Check Manifolds All.che” to prime the buffer manifolds.
Equipment
| Value | Label |
|---|---|
| Chemagic 360 | NAME |
| Nucleic Acid Extractor | TYPE |
| Perkin Elmer | BRAND |
| 2024-0020 | SKU |
| http://www.perkinelmer.com/ | LINK |
Elution Plates and Magbead Plates are generated in batches as needed with 60µL Elution Buffer and 150µL Magbeads per well. This can be done using a liquid handler such as the Agilent Bravo.
If there is condensation on the seal of the prepared Elution Plate, briefly spin the plate to collect the buffer in the bottom of the well.
With the plate sealed, resuspend the magnetic beads in the prepared Magbead Plate by shaking in the BioShake XP at 1350rpm for 0h 1m 0s
Check that the Viral RNA Buffers are connected to the Chemagic tubing. If not, perform a normal cleaning procedure, switch to Viral RNA Buffers and prime the system.
RNA Extraction with the Chemagic 360 Process Steps
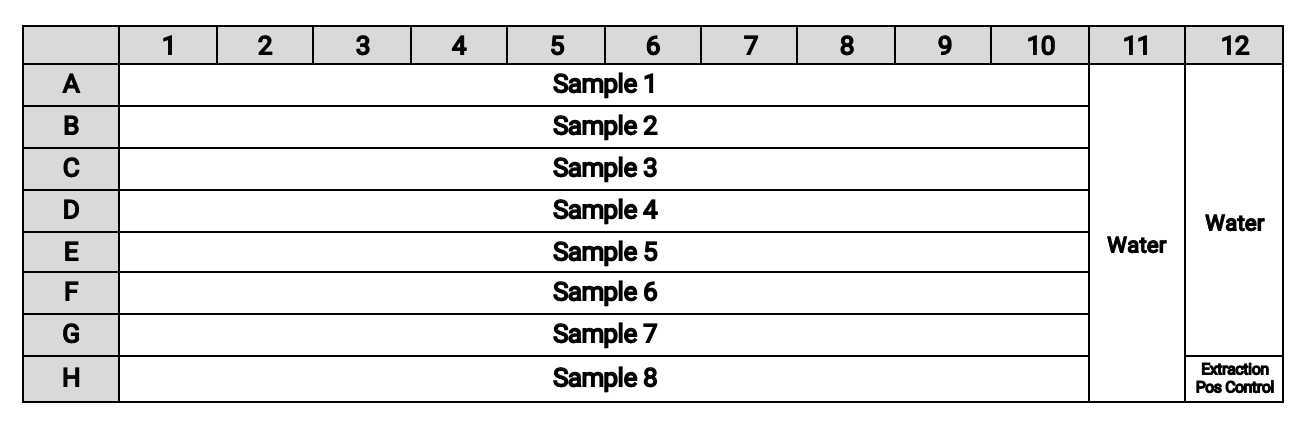
Using a wide orifice 1000 µL pipette tip, transfer 300µL per well of homogenized sample (prepared as specified in High Throughput pre-analytical processing of wastewater settled solids for SARS-CoV-2 RNA analyses) to each well of the Sample Plate as follows:
Add 300µL of DNA/RNA shield stock solution with BCoV spike-in from pre-analytical processing to one well of the Sample Plate to serve as an extraction positive control.
Using a wide orifice tip, add the samples one at a time to ensure that a full 300µL is pulled up and no clumps prevent the aspiration of the sample.
Using normal pipette tips:
- Add
300µLof water to the indicated wells in the figure below to create extraction negative controls. - Add
10µLof gRNA SARS-CoV-2 control (at a concentration of 50 cp/µl) to the indicated wells to represent the positive extraction control. Add4µLPoly A in wells with gRNA only.
Mix the Lysis buffer according to the following volumes per sample/plate:
| A | B | C |
|---|---|---|
| Reagents | Volume per Sample | Volume for Full Plate (with dead volume) |
| Proteinase K | 10 µL | 1 mL |
| Lysis Buffer 1 | 300 µL | 30 mL |
Add 310µLof Lysis buffer solution mixed in step 11 to each well of the Sample Plate.
Set up the plate deck in the Chemagic 360 as follows:
Position 1: Rack with Disposable Tips
Position 2: Magbead plate, low-well-plate prefilled with 150 µL Magnetic Beads
Position 3: Sample Plate
Position 4: Empty Deep Well Plate
Position 5: Empty Deep Well Plate
Position 6: Empty Deep Well Plate
Position 7: Elution Plate, Deep Well Plate with 60 µL Elution Buffer
Position 8: Empty
Equipment
| Value | Label |
|---|---|
| Chemagic 360 | NAME |
| Nucleic Acid Extractor | TYPE |
| Perkin Elmer | BRAND |
| 2024-0020 | SKU |
| http://www.perkinelmer.com/ | LINK |
Select the protocol: “chemagic Viral300 360 H96 drying prefilling VD141210.che”
Make sure that all wells are selected and start the protocol.
When the protocol is complete, immediately proceed to PCR Inhibitor Removal or seal the Elution Plate and store on ice.
Chemagic Clean Up
Dispose of empty Tip Rack (Position 1), empty Magbead plate (position 2) and Deep Well Plate with tips in it (Position 6) in a red biohazard waste bin.
Wrap Deep Well Plates in Positions 3, 4 and 5 in a blue absorbent pad, seal with lab tape, and dispose of as chemical waste.
PCR Inhibitor Removal with OneStep-96 PCR Inhibitor Removal Kit
Prepare Silicon-A™-HRC Plate
Mount the Silicon-A™-HRC Plate onto an Elution Plate
Add 150µL Prep Solution to the wells on the Silicon-A™-HRC Plate by piercing the cover foil in the middle with the pipette tip.
Incubate for 0h 5m 0s
Centrifuge the plate at exactly 3500x g,0h 0m 0s for 0h 5m 0s.
Discard Elution Plate with prep solution.
Add RNA extracts to prepped Silicon-A™-HRC Plate
While preparing the Silicon-A™-HRC Plate, allow the Chemagic Elution Plate (from step 16) to sit on the ALPAQUA Magnum EX Universal Magnet Plate for 2 minutes to pull down residual magnetic beads.
Equipment
| Value | Label |
|---|---|
| Magnum EX Universal Magnet Plate | NAME |
| Magnet Plate | TYPE |
| ALPAQUA | BRAND |
| A000380 | SKU |
| https://www.alpaqua.com/ | LINK |
Mount the prepped Silicon-A™-HRC Plate with the RNA extracts onto a new Elution Plate.
Transfer the full volume of extracted RNA in the Chemagic Elution Plate to the Silicon-A™-HRC Plate.
Centrifuge the plate at exactly 3500x g,0h 0m 0s for 0h 3m 0s.
Immediately seal the Elution Plate and store on ice until ddPCR analysis.
Discard the Silicon-A™-HRC Plate.

