High-Complexity One-Pot Golden Gate Assembly
Andrew P. Sikkema, Andrew P. Sikkema, S. Kasra Tabatabaei, S. Kasra Tabatabaei, Yan-Jiun Lee, Yan-Jiun Lee, Sean Lund, Sean Lund, Gregory J. S. Lohman, Gregory J. S. Lohman
data-optimized assembly design
Golden Gate Assembly
multigene constructs
synthetic biology
small genome assembly
Abstract
Golden Gate Assembly is a flexible method of DNA assembly and cloning that permits the joining of multiple fragments in a single reaction through predefined connections. The method depends on cutting DNA using a Type IIS restriction enzyme, which cuts outside its recognition site and therefore can generate overhangs of any sequence while separating the recognition site from the generated fragment. By choosing compatible fusion sites, Golden Gate permits the joining of multiple DNA fragments in a defined order in a single reaction. Conventionally, this method has been used to join five to eight fragments in a single assembly round, with yield and accuracy dropping off rapidly for more complex assemblies. Recently, we demonstrated the application of comprehensive measurements of ligation fidelity and bias data using data-optimized assembly design (DAD) to enable a high degree of assembly accuracy for very complex assemblies with the simultaneous joining of as many as 52 fragments in one reaction. Here, we describe methods for applying DAD principles and online tools to evaluate the fidelity of existing fusion site sets and assembly standards, selecting new optimal sets, and adding fusion sites to existing assemblies. We further describe the application of DAD to divide known sequences at optimal points, including designing one-pot assemblies of small genomes. Using the T7 bacteriophage genome as an example, we present a protocol that includes removal of native Type IIS sites (domestication) simultaneously with parts generation by PCR. Finally, we present recommended cycling protocols for assemblies of medium to high complexity (12-36 fragments), methods for producing high-quality parts, examples highlighting the importance of DNA purity and fragment stoichiometric balance for optimal assembly outcomes, and methods for assessing assembly success. © 2023 New England Biolabs, Inc. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Assessing the fidelity of an overhang set using the NEBridge Ligase Fidelity Viewer
Basic Protocol 2 : Generating a high-fidelity overhang set using the NEBridge GetSet Tool
Alternate Protocol 1 : Expanding an existing overhang set using the NEBridge GetSet Tool
Basic Protocol 3 : Dividing a genomic sequence with optimal fusion sites using the NEBridge SplitSet Tool
Basic Protocol 4 : One-pot Golden Gate Assembly of 12 fragments into a destination plasmid
Alternate Protocol 2 : One-pot Golden Gate Assembly of 24+ fragments into a destination plasmid
Basic Protocol 5 : One-pot Golden Gate Assembly of the T7 bacteriophage genome from 12+ parts
Support Protocol 1 : Generation of high-purity amplicons for assembly
Support Protocol 2 : Cloning assembly parts into a holding vector
Support Protocol 3 : Quantifying DNA concentration using a Qubit 4 fluorometer
Support Protocol 4 : Visualizing large assemblies via TapeStation
Support Protocol 5 : Validating phage genome assemblies via ONT long-read sequencing
INTRODUCTION
Modern molecular and synthetic biology depend on the rapid and accurate assembly of DNA from synthetic or genome-derived fragments into genes, expression systems, pathways, gene-editing arrays, and even large structures such as artificial chromosomes and small genomes (Fig. 1). In homology-directed assembly methods (e.g., Gibson assembly, polymerase cycling assembly (PCA), and yeast assembly), long homologous ends are assembled through some combination of exonuclease, polymerase, and ligase enzymes, or by taking advantage of native in vivo homologous recombination systems. Other assembly methods depend on the ligation of short complementary ends generated by an endonuclease, for example, restriction enzymes (BioBricks [Shetty et al., 2008] or Golden Gate Assembly [Engler et al., 2008]), AP lyases (USER cloning [Bitinaite et al., 2007]), and Argonautes (PlasmidMaker [Enghiad et al., 2022]). These methods rely on accurate generation of short, defined overhangs and specific ligation of only Watson-Crick-paired overhangs to generate the desired multifragment assembly. All methods can be used to generate large DNA constructs, but each has potential drawbacks and advantages. For example, homology-directed methods are difficult to use with highly repetitive DNA, as they can be prone to generating deletion products. The dependence on polymerase-driven template extension, though greatly improved by using high-fidelity PCR enzymes, can lead to errors such as indels and base substitutions near fusion sites. Ligation-directed methods are generally much less prone to introduction of mutations and more tolerant of repetitive DNA, but most require introduction of a target site for the endonuclease via PCR or in an earlier DNA synthesis step.

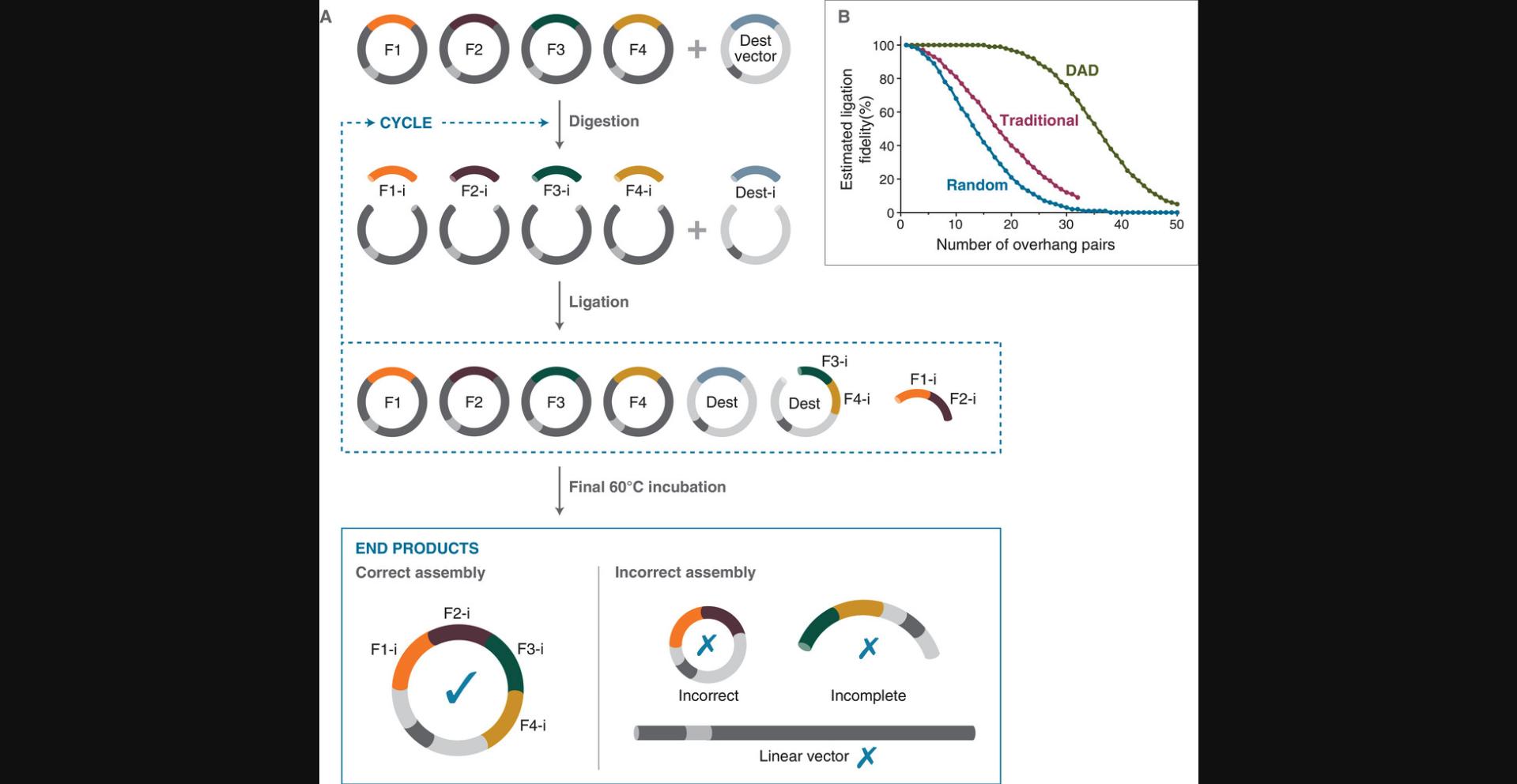
Golden Gate Assembly (GGA) is a widely used ligation-directed assembly that depends on the use of Type IIS restriction endonucleases to generate the short overhangs used as fusion sites in the assembly (Figs. 1 and 2). Type IIS enzymes cut outside of their recognition site, a feature that is key to the success of multifragment assembly (Szybalski et al., 1991). Fragments used in GGA are designed such that, upon restriction cleavage, the recognition site is separated from the insert, leaving the desired fusion site overhang behind. When a fragment is ligated to its desired assembly partner, the fused sequence no longer contains the recognition site and cannot be re-cut. If the fragment instead re-ligates to its precursor fragment, the cleavage site is regenerated and may be cut again. Repeated cycles of cutting and ligation thus lead to build-up of the assembled product and an ever-diminishing amount of fragments or holding vectors containing a recognition site (Fig. 2A). It is important to note that target sequences naturally containing one or more recognition sites for the selected Type IIS enzyme will be cut internally, leading to incomplete products. This often necessitates the removal of native recognition sites in target DNA, a process referred to as domestication.
Strategies exist to build large constructs through both homology-directed and ligation-directed assembly methods. In vivo recombination methods have been shown to successfully join dozens of fragments at once, but these methods are labor-intensive, requiring the introduction of precursor constructs into a target organism, growth of the organism, and re-isolation of the DNA before its final use (Gibson et al., 2008). In vitro methods, including GGA, are historically more limited in the number of fragments used in a single round, with higher-complexity assemblies resulting in inaccurate assembly or low yields. This limitation has been overcome using hierarchical assembly strategies, where fewer numbers of fragments (typically 2-8) are joined in a single reaction, and then multiple initial assemblies are joined via second- or third-round assemblies to achieve the final desired product (Bird et al., 2022; Current Protocols article Marillonnet & Grutzner, 2020). In addition to the time required for multiple rounds of assembly and passage through E. coli or another organism, these strategies often require a complex series of holding vectors and can be limited by issues with cloning, propagating, and assembling DNAs from certain sources. Bacteriophage DNA can be particularly challenging (Yeom et al., 2020), which has limited efforts to assemble bacteriophage genomes using Golden Gate Assembly (Liang et al., 2022).
Recent advances in assembly strategy have applied comprehensive DNA ligation fidelity (discrimination against mismatched sequences) and bias (preference for some sequences over others) measurements to inform the design of ligation-directed assembly (Potapov, Ong, Kucera, et al., 2018; Potapov, Ong, Langhorst, et al., 2018; Pryor et al., 2020). These data-optimized assembly design (DAD) strategies have been applied to GGA methods to successfully generate high-fidelity, high-efficiency assemblies of more than 35 parts in a single reaction (Pryor et al., 2022). DAD evaluates overhang sets by providing a maximizing fidelity score calculated based on observations in a highly multiplexed ligation fidelity experiment. Typically, many high-fidelity overhang sets can be identified at a given fragment complexity, allowing highly flexible assembly design of even native coding sequences. Specific overhangs can be required or excluded and fusion sites can be placed in desired regions to allow users to separate ORFs on their own fragments or subdivide potentially toxic sequences. In addition, domesticating mutations or other mutageneses can be conveniently designed into parts generated via PCR by simply requiring a fusion site be present near the site of introduction of any desired base changes. DAD has further enabled the assembly of small genomes (e.g., T7 bacteriophage) and the production of recombinant bacteriophages in one assembly step (Pryor et al., 2022).
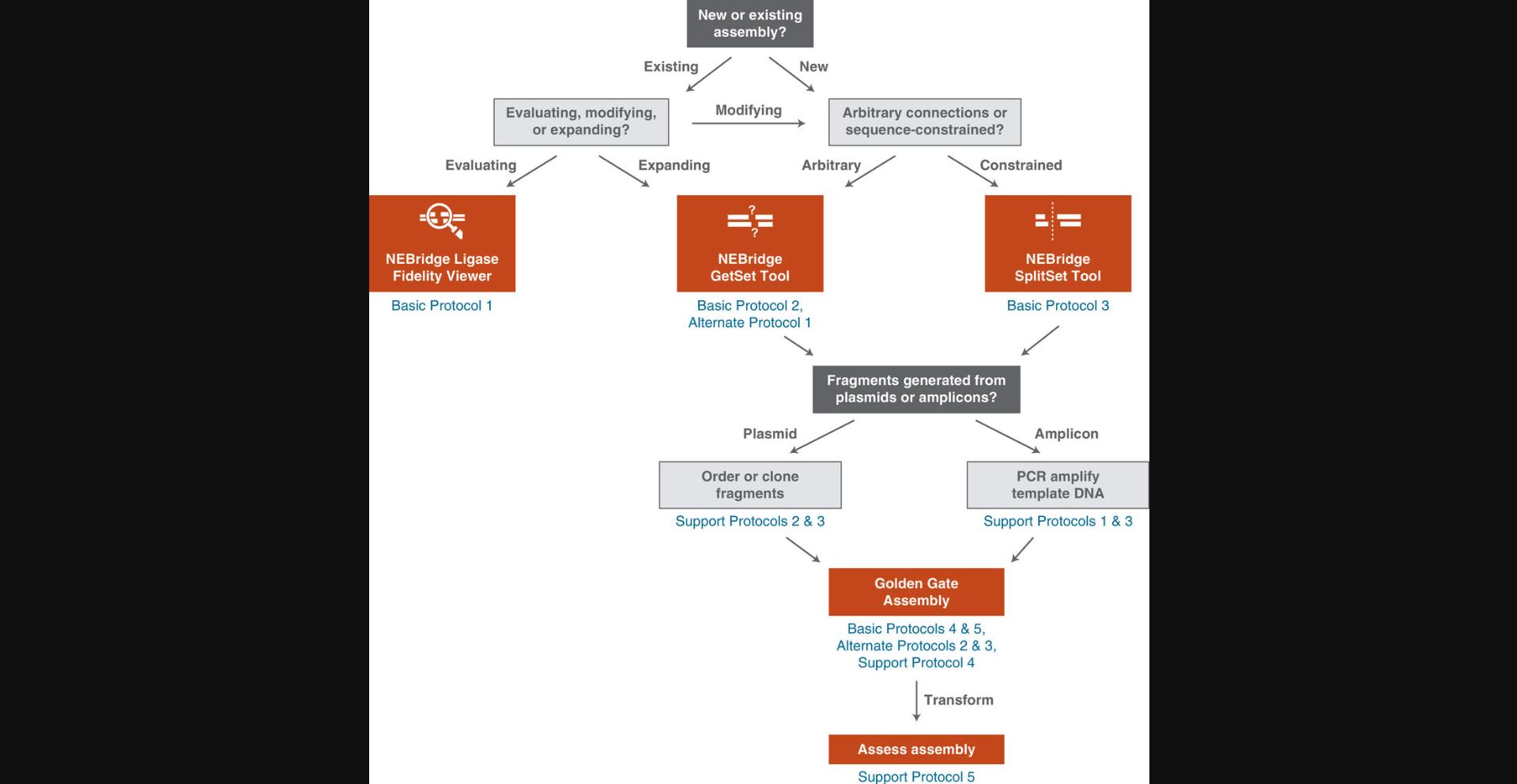
In this article, we present protocols for applying ligation fidelity data to assembly design using several online tools (https://ligasefidelity.neb.com; Pryor et al., 2020). Basic Protocol 1 describes the use of the Ligase Fidelity Viewer to check existing overhang sets for expected fidelity and identify potential mismatch ligation events. Basic Protocol 2 describes how to use the NEBridge GetSet Tool to generate sets of overhangs of desired size and to require or exclude certain connections. Alternate Protocol 1 describes the use of the NEBridge GetSet Tool to add additional compatible connections and use them to expand an existing assembly. Basic Protocol 3 describes the use of an updated version of the NEBridge SplitSet Tool to divide a known target sequence by selecting a set of high-fidelity fusion sites. Basic Protocol 4 describes the single-reaction assembly for 12 insert fragments, and Alternate Protocol 2 describes a modified protocol for 24 or more fragments. Basic Protocol 5 describes the assembly and rescue of T7 bacteriophage using a one-pot reaction created following the design rules of Basic Protocol 3. Support protocols describe the generation of high-quality parts via PCR (Support Protocol 1), cloning of parts into holding plasmids (Support Protocol 2), accurate fragment quantitation via Qubit (Support Protocol 3), visualization of large assemblies via TapeStation (Support Protocol 4), and validation of assemblies via Oxford Nanopore Technologies long-read sequencing (Support Protocol 5).
STRATEGIC PLANNING
Selection of Restriction Specificity
When planning a complex GGA reaction, a key consideration is which Type IIS restriction enzyme to use (Szybalski et al., 1991). The most common enzymes used in GGA have six-base recognition sites (BsaI: GGTCTCN, BsmBI/Esp3I: CGTCTCN, BbsI/BpiI: GAAGACNN), cut close to the recognition site, provide high cutting efficiency and accuracy, and produce four-base overhangs. On average, these enzymes are expected to have a recognition site every 2048 base pairs (roughly 20 in a 40-kb sequence). Less frequently used enzymes target seven-base recognitions sites, with SapI/BspQI (GCTCTTCN) producing three-base overhangs and AarI/PaqCI (CACCTGCNNNN) producing four-base overhangs (Kennedy et al., 2023). Enzymes with seven-base recognition sites are expected to have a naturally occurring site in an arbitrary DNA sequence every 8192 bp (roughly 4-5 in a 40-kb sequence). We have found that the above four-base-overhang-generating enzymes can support high-accuracy GGA of 36 or more fragments. SapI and BspQI are often overlooked for multifragment assembly because three-base overhangs give fewer possible fusion site pairs, but DAD ligation analysis has shown that at least 12 three-base overhang pairs can be joined in a single reaction with high accuracy and efficiency, which is sufficient for many applications (Pryor et al., 2020, 2022). Cycling protocols for most enzymes have been published elsewhere (Marillonnet & Grutzner, 2020; Pryor et al., 2022). Basic Protocol 4 and Alternate Protocol 2 present general protocols covering most Type IIS enzymes in high-complexity assemblies. A slightly modified protocol is provided in Basic Protocol 5 for AarI/PaqCI, which requires an activator oligonucleotide.
Previous work has shown that the identity of the restriction enzyme chosen has only a slight impact on fusion site overhang set fidelity (Pryor et al., 2020). Thus, enzyme selection will often be driven by the needs of the assembly system, such as the specificities present in the holding vectors used, e.g., BbsI/BpiI and BsaI for modular cloning (MoClo; Weber et al., 2011) or AarI/PaqCI and BsaI for Mobius assembly (Andreou & Nakayama, 2018). Additionally, any internal restriction sites for the selected enzymes that are present within the target assembly sequence will be cut during the reaction and lead to truncated assembly products. Therefore, any internal sites must be removed from fragments before assembly (i.e., domestication). This is most readily accomplished by making silent mutations in identified ORFs (HamediRad et al., 2019; Marillonnet & Grutzner, 2020; Zhang et al., 2015). Changes to intergenic regions should be avoided, especially in identifiable regulatory elements, and it is advisable to choose a different restriction specificity that would avoid the need for domestication mutagenesis within these regions. The best choice of enzyme is often the one that requires the fewest domesticating mutations (ideally zero), with any needed mutations placed within ORFs. Mutations may be introduced in silico if ordering fragments from a DNA synthesis vendor or via site-directed mutagenesis methods for fragments held in holding plasmids. It is also possible to incorporate mutations at recognition sites concurrent with part generation from genomic DNA (gDNA) if a break point is designed close enough to the recognition site that it can be encompassed in the PCR primer (see Basic Protocol 3).
Choice of DNA Ligase and Assembly Protocol
We have previously found that the fidelity of ligation has the largest effect on accurate assembly, with the ligation, buffer, cycling method, and ratio of restriction enzyme to ligase being important factors in ligation fidelity (Potapov, Ong, Kucera, et al., 2018; Pryor et al., 2020). In nearly all cases, T4 DNA ligase is the best ligase to use for high-complexity GGA. While T7 DNA ligase has been shown to have an overall higher fidelity than T4 DNA ligase, we have repeatedly found that this fidelity advantage is not easily realized due to the lower efficiency and higher bias of T7 DNA ligase (Bilotti et al., 2022). We consequently recommend using only T4 DNA ligase with validated high-fidelity overhang sets, as the application of DAD allows the high efficiency of T4 ligase to be utilized while minimizing assembly errors due to inaccurate ligation. In terms of protocol, we find that, while static 37°C incubation results in higher assembly fidelity, there is a significant trade-off of lower assembly efficiency that leads to a lower absolute number of correct assemblies. Static incubation can be used to minimize erroneous assembly of existing overhang sets with low predicted fidelity, but the use of DAD-selected overhangs combined with a cycling assembly protocol (see Basic Protocol 4) generally leads to higher overall fidelity and efficiency (Pryor et al., 2022).
Choice of Design Tools
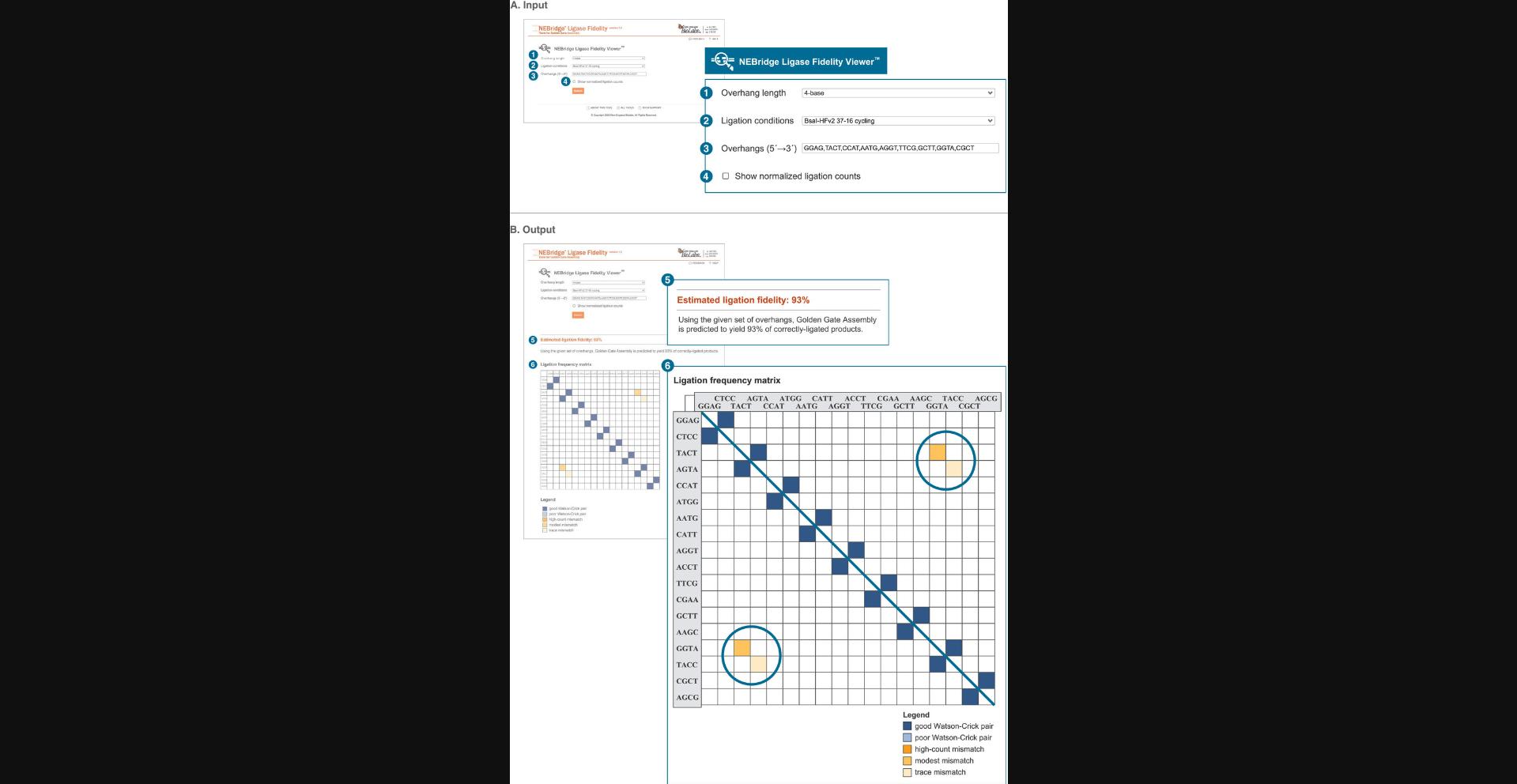
The Ligase Fidelity Viewer tool determines the estimated fidelity, a score that expresses overall mismatch potential, of an entered overhang set and can be used to identify ligase fidelity issues with an existing assembly (see Basic Protocol 1). The tool contains a database of comprehensive datasets, collected under variable ligase, restriction enzyme, and cycling protocols, that can be selected from to assess the entered overhang set. The NEBridge GetSet Tool (see Basic Protocol 2) is applied to generate sets of compatible fusion sites when there is no sequence restriction. This method can also be used to replace fusion sites in or add new sites to existing sets while optimizing overall fidelity. The NEBridge SplitSet Tool (see Basic Protocol 3) is designed for placing optimal fusion sites within a known sequence, including coding sequences, permitting fusion sites to be limited to specific regions or precise locations. The tool can also assist in selection of fusion sites appropriate for making mutations (including domestication) concomitant with parts generation. A flow chart to aid in the selection of appropriate tools and protocols is shown in Figure 3.
Determining an Appropriate Number of Fragments for Assembly
Figure 2B shows the predicted fidelity scores of different overhang selection methods as a function of the total number of fragments used (Pryor et al., 2020). High-complexity assemblies with many fragments have more opportunities for mismatch ligation, and thus lower predicted proportions of correct assembly. Even when using optimized overhang sets, increasing fragment number is generally associated with both decreased fidelity and efficiency. While there is no hard cutoff that will separate a successful assembly from an unsuccessful one, we typically find very high product yields (1,000s to 10,000s CFU/µl or PFU/µl) when using a DAD-optimized fusion site set up to 12 fragments; doubling to 24 fragments leads to approximately one order of magnitude fewer CFU/PFU, with further reduction at 36 fragments (see Supporting Information Fig. S1 and Understanding Results). Assemblies still result in many successful transformants even at this high fragment number, but the user must be increasingly mindful of factors such as DNA purity as complexity increases. We have previously shown it is possible to assemble up to 52 fragments in one reaction, but we feel this level of complexity represents the maximum when optimizing all factors, including having an extremely stringent selection mechanism whereby only complete assemblies can produce viable phage (Pryor et al., 2022). For best results, we recommend most users limit single assembly reactions to <40 fragments, with <25 considered routine when overhang selection is guided by DAD. A lower fidelity score for an assembly will lead to an increase in colonies that contain incorrect/incomplete assemblies, and thus may require screening more colonies post assembly to ensure isolation of the desired product. As most GGA errors will involve deletions or insertions of whole fragments, using colony PCR (see Current Protocols article Woodman et al., 2016) to measure insert size may be sufficient to identify correct assemblies, but full sequence verification of the final product is advisable (see Support Protocol 5).
In Silico Validation of Assembly
Before ordering parts or primers, it is recommended to visualize assembly designs to confirm that fragments are predicted to assemble in the correct order and with the final sequence desired. Many online tools are publicly available, including the NEBridge tool (https://goldengate.neb.com/#!/) and Geneious or SnapGene software. Note that some tools may evaluate fusion sites by older rules of thumb that frequently flag DAD-recommended pairings as incompatible; since the fusion sites chosen using DAD are based on empirical data and validated for use in Golden Gate Assemblies, these warnings may be safely ignored. Nevertheless, using an assembly tool allows for visualization of the order of assembly, ensures that parts will produce the intended overhangs, and provides a final assembly sequence, which should be checked carefully against the intended final construct. These tools can also flag internal restriction sites, which is important for confirming that all native sites are removed from the target sequence. If internal recognitions sites remain, either the assembly should be revised to eliminate these sites or a different Type IIS enzyme should be used. Careful validation at the design stage can save significant time and resource expenditure.
Generation of Fragments for Assembly
After the in silico design of parts is complete, a method or methods of parts generation must be selected. The general hierarchy of input DNA source quality is plasmid DNA > amplicon DNA (from gDNA or synthetic DNA template) > direct synthetic DNA (i.e., gBlocks). Assemblies can contain a mix of parts from multiple sources. An increasingly accessible solution is ordering the parts from a DNA vendor (see Understanding Results for example assemblies of the LacIZ cassette from synthetic fragments). Most vendors currently produce DNA starting from oligonucleotides synthesized through phosphoramidite synthesis, with larger fragments assembled via a method such as PCA (Fig. 1; Hoose et al., 2023). Fragments can be obtained as gBlocks (or similar linear dsDNA fragments) or as sequence-verified inserts in a holding vector. Synthesis of parts by a vendor can be convenient, as the needed Type IIS sites for GGA can be added simply in silico along with any domestication mutations or other desired modifications. Ordering synthetic DNA can also provide parts that cannot be obtained from natural sources due to the lack of suitable gDNA, including sequences from metagenomic and unculturable sources or from purely in silico designs. A major barrier to this route can be the cost of synthetic sequences, and sufficient lead time must be allowed for synthesis and shipping of DNA. Further, vendors have variable rates of success and may not be able to provide all ordered sequences. Sequence-verified fragments in plasmids are more expensive than linear dsDNA, but are characteristically higher purity. Purified plasmids are much less likely to contain contaminants, such as individual DNA molecules containing mutations, that lead to incorrect final products. However, not all sequences can be easily cloned due to issues like host toxicity. Linear dsDNA parts can be propagated using PCR, but this carries the risk of introducing mutations through successive rounds of amplification.
If available, using gDNA as template to generate parts by PCR is a practical alternative. When generating parts by PCR, the needed Type IIS sites are added via the primers (see Support Protocol 1). This method will very often allow production of fragments that cannot be obtained via DNA synthesis, and PCR amplicons may be used directly in GGA reactions (see Understanding Results for examples from the T7 bacteriophage genome). More care must be taken to ensure the purity of these fragments, as impurities derived from off-target amplification or primer dimers are substrates for the assembly reaction and can dramatically affect yield (Supporting Information Fig. S2). PCR fragments can be used directly with excellent results or can be propagated within a plasmid (though not all sequences can be successfully passaged in E. coli due to toxicity or genetic instability).
Basic Protocol 1: ASSESSING THE FIDELITY OF AN OVERHANG SET USING THE NEBridge LIGASE FIDELITY VIEWER
A critical component of an effective GGA is the fidelity of the overhang set (Potapov, Ong, Kucera, et al., 2018; Pryor et al., 2020). This is a predictive score calculated for a specific set of overhangs that expresses the overall mismatch potential between overhangs in the set. While not an absolute score, it can be used as a relative means of comparing sets. This protocol describes how to assess the fidelity of an existing overhang set using the NEBridge Ligase Fidelity Viewer, with application to assessing the fidelity of existing cloning toolsets and the compatibility of parts from different toolkits. Specific entries are provided for an example analysis described in Figure 4 and the Commentary (see Understanding Results).
Basic Protocol 2: GENERATING A HIGH-FIDELITY OVERHANG SET USING THE NEBridge GetSet Tool
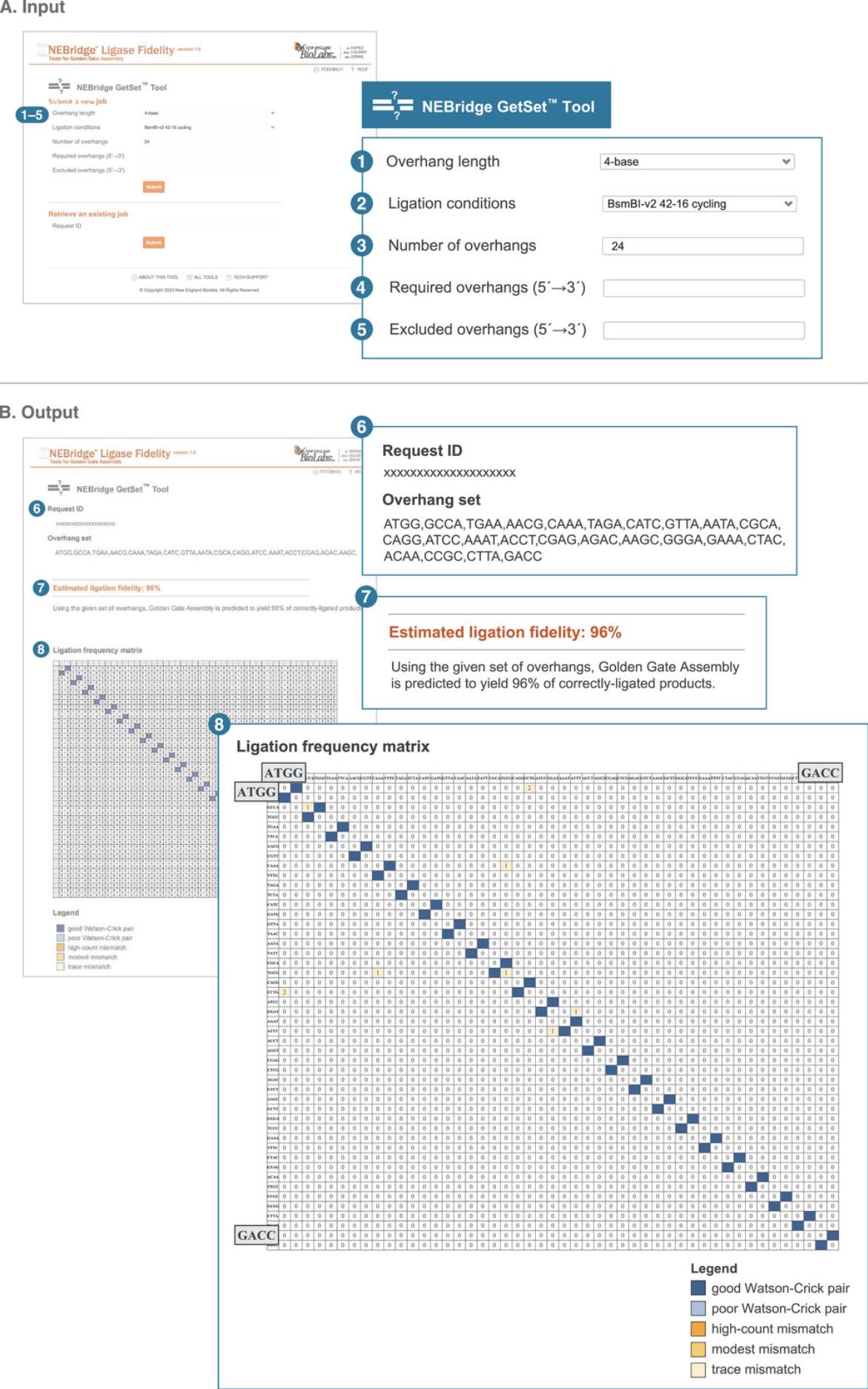
Being able to generate sets of overhangs with high fidelity enables the selection of arbitrary fusion sites between parts for modular assembly systems (Damalas et al., 2020; Malci et al., 2022; Stuttmann et al., 2021). To enable the easy creation of high-fidelity overhang sets, we have created a webtool called the NEBridge GetSet Tool based on our ligase fidelity data (Pryor et al., 2020). The tool allows the user to select the number of requested overhangs, Type IIS restriction enzyme, and ligation conditions to give optimal overhang sets for a specific use. Specific overhang sequences can also be required or excluded in the algorithmic search used by the program. These features allow for a large amount of control over the overhangs sets generated. Specific entries are provided for an example described in Figure 5 and the Commentary (see Understanding Results).
Necessary Resources
- Personal computer or other device with up-to-date web browser
1.Navigate to the NEBridge GetSet Tool (https://ligasefidelity.neb.com/getset/run.cgi).
2.Select the overhang length of your overhang set from the “Overhang length” drop-down menu. For our example, select 4-base.
3.Select the most representative ligation conditions from the “Ligation conditions” drop-down menu. For our example, select BsmBI-v2 42-16 cycling.
4.Enter the desired number of overhangs for the set in the “Number of overhangs” field. For our example, enter 24.
5.Input any required overhangs into the “Required overhangs (5′→3′)” field. For our example, leave this field blank.
6.Input any overhangs to be excluded into the “Excluded overhangs (5′→3′)” field. For our example, leave this field blank.
7.Click “Submit”.
Alternate Protocol 1: EXPANDING AN EXISTING OVERHANG SET USING THE NEBridge GetSet Tool
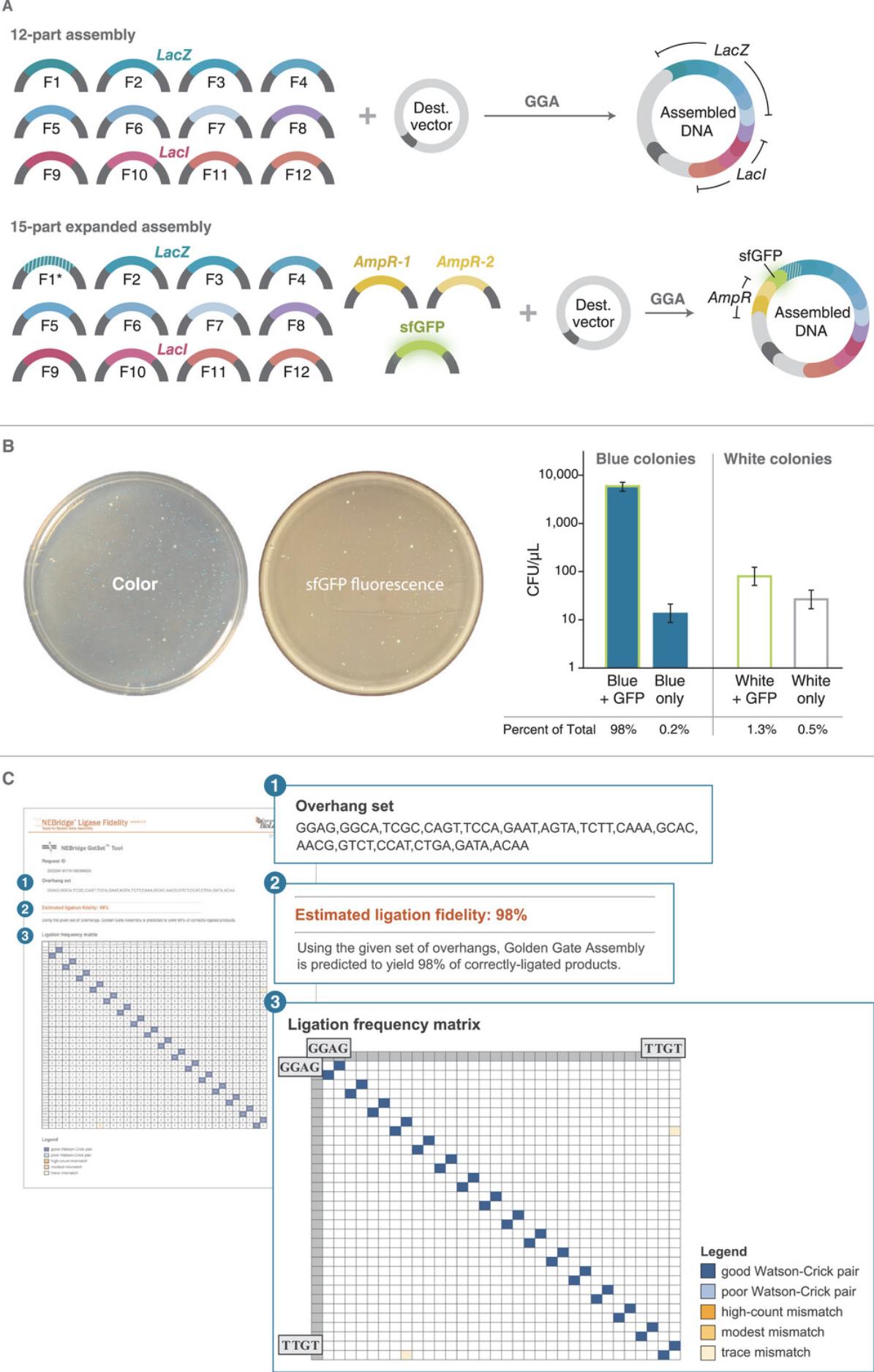
A particularly useful application of the NEBridge GetSet Tool is the expansion of an existing overhang set to add more compatible fusion sites while maintaining high set fidelity. This application of NEBridge GetSet supports adding new fragments to an existing assembly, finding optimal overhangs for subdividing existing fragments within an assembly, and expanding existing standard overhang sets (e.g., MoClo). This protocol covers how to use NEBridge GetSet to generate additional overhangs and how to redesign an existing assembly to incorporate the new overhangs. As an example, we describe how to expand the LacIZ assembly (see Basic Protocol 4, Supporting Information Table S1) to add three additional fragments (Fig. 6A) that encode two new selection markers, superfolder GFP (sfGFP, fluorescence) and an AmpR cassette (ampicillin resistance). NEBridge GetSet is used to select new fusion sites compatible with the existing assembly, and these new fusion sites are used to design new fragments and modify existing ones to generate the expanded assembly (Fig. 6A and Supporting Information Table S2).
Necessary Resources
-
Personal computer or other device with up-to-date web browser
-
Plain text, comma-delineated list of fusion site overhangs from the assembly to expand
-
Example set : GGAG, GGCA, TCGC, CAGT, TCCA, GAAT, AGTA, TCTT, CAAA, GCAC, AACG, GTCT, CCAT
-
Sequence files for known part sequences in plain text, FASTA, GenBank, or similar format
-
Example files : see Supporting Information GenBank files 12-part_LacIZ_Assembly, 15-part_Expanded_LacIZ_Assembly
1.Begin as in Basic Protocol 2, steps 1-3.For our example, select 4-base for overhang length and BsaI-HFv2 37-16 cycling for ligation conditions.
2.In the “Number of overhangs” field, enter the number of overhangs in the current set plus the desired number of new overhangs. For our example, enter 16.
3.Enter the current overhang set to be expanded into the “Required overhangs (5′→3′)” field. For our example, enter:
GGAG, GGCA, TCGC, CAGT, TCCA, GAAT, AGTA, TCTT, CAAA, GCAC, AACG, GTCT, CCAT
4.Input any overhangs to be excluded in the “Excluded overhangs (5′→3′)” field. For our example, leave this field blank.
5.Click “Submit”.
6.Identify new overhang sequences. The new overhang set is listed in the order of the required sequences as entered (step 3) followed by the newly generated overhangs (Fig. 6C). For our example, the new overhangs are CTGA, GATA, and ACAA.
7.Determine placement of the new fusion sites in the expanded assembly. For our example, two of the new overhang sequences are placed at new fusion sites and the third connects the new fragments to the existing assembly (Fig. 6A).
8.Assign new overhang sequences to new fusion sites. For our example, ACAA was placed at Fusion Site A, GATA was placed at Fusion Site B, and CTGA was placed at Fusion Site C.
9.Determine insert sequences for new fragments. After placement of the new overhangs in the assembly sequence, insert sequences can be extracted by taking the sequence between the two overhangs (inclusive of the overhangs). For our example, the insert sequences can be found in Supporting Information Table S2.
10.Convert insert sequences into Golden Gate fragments. The insert sequences extracted in the previous step lack Type IIS sites and associated spacers, which must be added. For our example, which uses BsmBI, CGTCTCA (spacer) was added to the 5′ ends of the insert sequences and TGAGACG was added to the 3′ end before synthesis and insertion into to the EcoRV site of pUC57-mini-BsaI-Free or pUC57-mini-Kana-BsmBI-Free (Supporting Information Table S3).
11.Modify an existing fragment to be compatible with the expanded assembly. The last step of expanding an assembly is resolving the conflict that results from inserting fragments into an existing fusion site. For our example, the new fragments are inserted at the GGAG fusion site of the existing assembly and the destination plasmid. This fusion site is part of the destination plasmid and is positionally locked; therefore, the fragment forming the other half of that fusion site must be changed. Here, that requires the GGAG overhang of fragment LacIZ-1 to be changed to the Fusion Site C overhang of CTGA. For final assembly sequence, see Supporting Information GenBank file 15-part_Expanded_LacIZ_Assembly.
12.Generate fragment DNA and perform assembly and transformation (see Basic Protocol 4). Plate transformants on LB plates containing ampicillin (100 µg/ml), chloramphenicol (25 µg/ml), IPTG (0.2 mM), and X-gal (80 µg/ml).
Basic Protocol 3: DIVIDING A GENOMIC SEQUENCE WITH OPTIMAL FUSION SITES USING THE NEBridge SplitSet Tool
One of the most powerful uses of DAD-driven Golden Gate Assembly is the division of existing sequences into fragments suitable for assembly using the NEBridge SplitSet Tool. NEBridge SplitSet can select high-fidelity overhang sets from a sequence using defined search windows and input parameters similar to NEBridge GetSet. As an example, we describe the division of the ∼40-kb T7 bacteriophage genome into 12 fragments. To perform the assembly from these 12 fragments, see Basic Protocol 5.
Necessary Resources
- Personal computer or other device with up-to-date web browser
- Geneious, SnapGene, or similar software for browsing and editing sequence data
- Sequence file for the target to be assembled as plain text, FASTA, GenBank or other format. The sequence must contain only canonical nucleotides with no ambiguous positions.
- Example : T7 genome sequence (https://www.ncbi.nlm.nih.gov/nuccore/V01146.1?report=fasta)
1.Using available sequence analysis software, find existing Type IIS enzyme recognition sites in the target sequence. For our example, PaqCI (AarI isochizomer) sites were identified in the T7 bacteriophage genome using Geneious. The wild-type T7 genome has five sites at positions 3942-3948, 8771-8777, 9387-9393, 9949-9955, and 17729-17735 (see Supporting Information Table S4 for gene contexts).
2.Navigate to the NEBridge SplitSet Tool at https://ligasefidelity.neb.com/splitset/run.cgi.
3.Enter assembly name into the “Assembly name” field. For our example, enter T7-PaqCI.
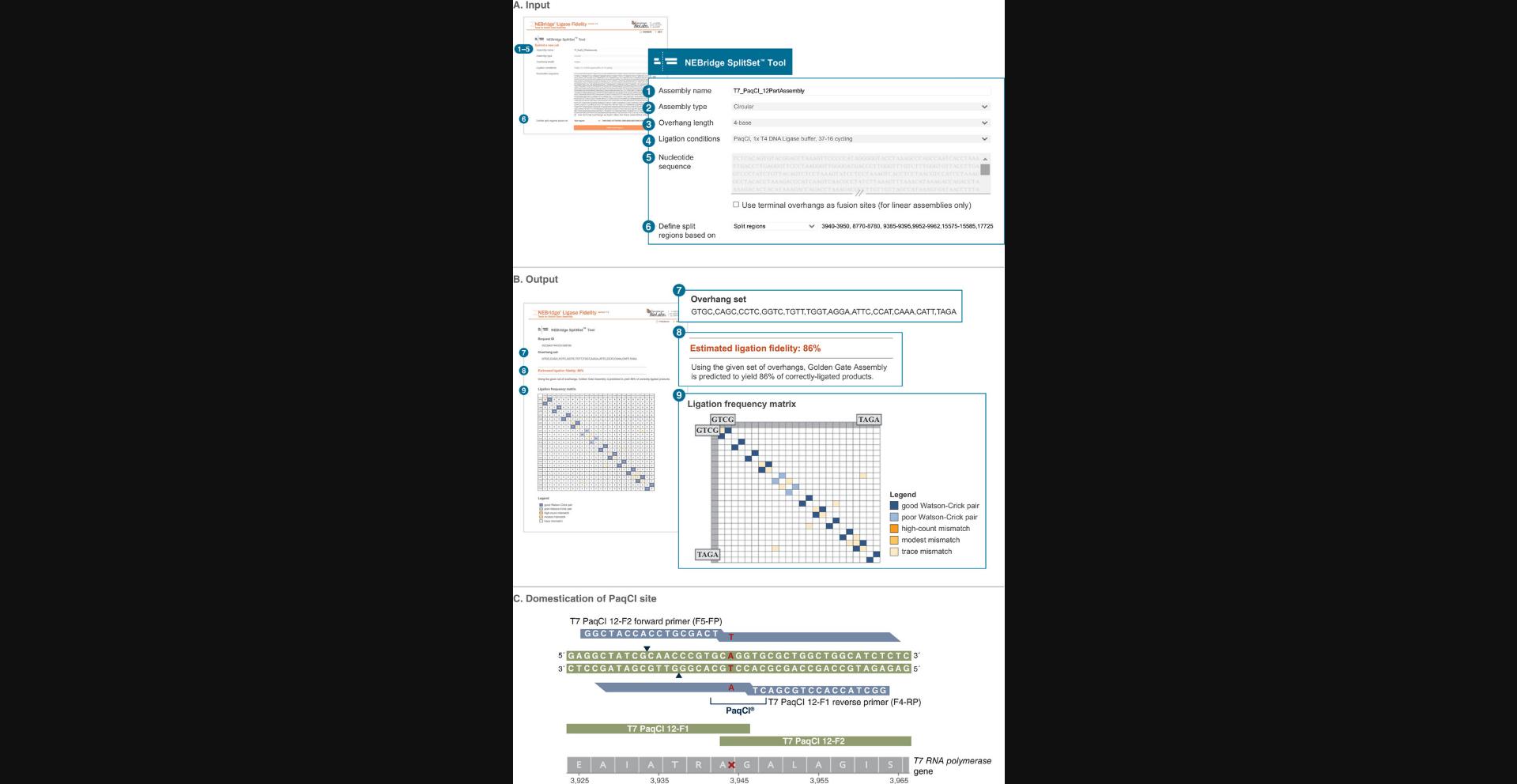
4.Select whether the assembly will be linear or circular. For our example, select circular.
5.Select the overhang length from the “Overhang length” drop-down menu. For our example, select 4-base.
6.Select the most representative ligation conditions from the “Ligation conditions” drop-down menu. For our example, select PaqCI, 1× T4 DNA Ligase buffer, 37-16 cycling.
7.Enter the full sequence to be assembled in the “Nucleotide Sequence” field. For our example, enter the full T7 bacteriophage genomic sequence.
8.Leave the box “Use terminal overhangs as fusion sites (for linear assemblies only)” unchecked.
9.For “Define split regions based on”, select the option appropriate for the assembly from the drop-down menu. For our example, select Split regions.
10.In the field following the drop-down menu, enter the predefined split regions for the assembly. For our example, enter:
3940-3950, 8770-8780, 9385-9395, 9952-9962, 15575-15585, 17725-17735, 23290-23300, 26630-26640, 29715-29725, 29945-29955, 33260-33270, 36584-36594
11.Click “Define split regions”.
12.Input any overhangs to be excluded in the “Excluded overhangs (5′→3′)” field. For our example, leave this field blank.
13.Click “Submit”.
14.Design primers to amplify the NEBridge SplitSet output fragments and append appropriate spacers and Type IIS sites. For this example, the sequence GGCTACCACCTGCGACT (spacers) was added to the 5′ end of all primers.
15.Introduce silent point mutations in appropriate primers to domesticate fragments. For our example, see Supporting Information Table S4 and S6 for mutation locations and a list of final primer sequences, respectively. For final assembly sequences, see Supporting Information GenBank files.
Basic Protocol 4: ONE-POT GOLDEN GATE ASSEMBLY OF 12 FRAGMENTS INTO A DESTINATION PLASMID
The most common application of GGA is the creation of plasmids assembled from multiple fragments. In this protocol, GGA is used for a one-pot assembly of 12 DNA fragments into a destination plasmid with a selectable antibiotic resistance marker. We recommend this protocol for assemblies of 12-23 fragments. In this example, illustrated in Figure 6A, the fragments assemble to form a cassette of LacI and LacZ , which, when plated on X-gal, produce blue colonies following correct assembly and white colonies following incorrect assembly (Potapov, Ong, Kucera, et al., 2018; Pryor et al., 2020). Fusion sites for the assembly were selected following Basic Protocol 3.The fragments are flanked on the 5′ and 3′ ends by Type IIS recognition sites (in this case BsmBI) and a spacer nucleotide. DNA fragments are cloned into holding plasmids, which are used directly in the assembly. See Supporting Information Table S1 for part sequences and overhangs used. See Strategic Planning and Critical Parameters for additional factors important in assembly design and parts preparation. Parts should be purified via miniprep (e.g., Current Protocols article Engebrecht et al., 1991) and quantified by Qubit (see Support Protocol 3), Nanodrop, or a similar method.
Materials
- Equimolar parts master mix of 12 LacIZ part vectors (LacIZ-12-F1 to LacIZ-12-F12; see Supporting Information Table S1 for part sequences)
- Destination plasmid: pGGAselect (New England Biolabs, cat. no. N0309A)
- 10× T4 DNA Ligase Reaction Buffer (New England Biolabs, cat. no. B0202)
- NEBridge Golden Gate Enzyme Mix (BsmBI-v2) (New England Biolabs, cat. no. M2617)
- Nuclease-free water (New England Biolabs, cat. no. B1500S)
- T7 Express Competent E. coli (New England Biolabs, cat. no. C2566)
- SOC Outgrowth Medium (New England Biolabs, cat. no. B9020S)
- LB plates (see Current Protocols article Elbing & Brent, 2019) containing 25 µg/ml chloramphenicol, 0.2 mM IPTG, and 80 µg/ml X-gal
-
- 0.2-ml PCR tubes (USA Scientific, cat. no. 1402-8108)
- Thermocycler (Bio-Rad T100 Thermal Cycler, cat. no. 1861096)
- 1.5-ml DNA LoBind tubes (Eppendorf, cat. no. 022431021)
- 42°C heat block
- Tube shaker
- Glass Plating Beads (Sigma-Aldrich, cat. no. 71013)
- 37°C incubator
Perform assembly
1.Combine the following components in a 0.2-ml PCR tube:
- Equimolar parts master mix at 3 nM final concentration
- 1.09 μl 55 nM (75 ng/μl) pGGAselect
- 2 μl 10× T4 DNA Ligase Reaction Buffer
- 2 μl NEBridge Golden Gate Enzyme Mix (BsmBI-v2)
- Nuclease-free water to 20 μl
2.Mix well by pipetting gently 5-10 times, then spin down in a microcentrifuge for 10 s.
3.Place in a thermocycler and run the following cycling protocol:
- (42°C, 5 min → 16°C, 5 min) × 30 cycles → 60°C, 5 min
4.Reduce temperature to 4°C until the assembly is used for transformation.
Transform product into E. coli
5.Thaw T7 Express cells on ice for 10 min.
6.Aliquot 50 μl T7 Express cells into a 1.5-ml DNA LoBind tube.
7.Add 5 μl assembly reaction, mix by flicking the tube gently four times, and incubate on ice for 30 min.
8.Heat-shock cells at 42°C for 10 s.
9.Return cells to ice for 5 min.
10.Add 950 μl SOC Outgrowth Medium and incubate at 37°C for 1 hr with shaking (225 rpm).
11.Using glass beads, plate 5 μl cells onto prewarmed LB plates containing chloramphenicol, IPTG, and X-gal. Incubate at 37°C overnight.
Assess assembly
12.Count blue colonies as positive for correct assembly and white colonies as negative.
13.Calculate the fidelity of the assembly as:
Alternate Protocol 2: ONE-POT GOLDEN GATE ASSEMBLY OF 24+ FRAGMENTS INTO A DESTINATION PLASMID
This protocol describes an alternative GGA protocol for one-pot assembly of 24 or more DNA fragments into a destination plasmid with a selectable antibiotic resistance marker. In this example, the insert fragments assemble the same LacI and LacZ cassette as in Basic Protocol 4, but using twice the number of insert fragments, each of which are roughly half the size. The insert fragments are split such that the overhangs of the 12-fragment assembly are reused in the 24-fragment assembly. The ability to split fragments in this manner is a good demonstration of the flexibility GGA, where fragments of the 12-fragment assembly can be replaced with 24-fragment counterparts, enabling easier part swapping and system engineering. The assembly used for this protocol also allows one to assess the effect of increasing fragment number on assembly efficiency using the same end product. Although fragment size in and of itself does not appear to be an important factor in assembly, very small (<25 bp) or very large (>>10 kb) fragments can experience normal issues with DNAs of those sizes. Namely, very small DNA duplexes are thermally unstable and can melt, while very large DNAs can experience shearing or other mechanical damage. Within these extremes, fragment size is arbitrary and can be dictated by a number of external factors, such as the length of synthetic DNA that can be acquired from a vendor. See Supporting Information Table S7 for fragment sequences and overhangs.
Additional Materials (also see Basic Protocol 4)
- Equimolar parts master mix of 24 LacIZ parts vectors (LacIZ-24-F1 to LacIZ-24-F24; see Supporting Information Table S7 for part sequences)
1.Set up the assembly reaction as described (see Basic Protocol 4, steps 1-2).
2.Place in a thermocycler and run the following cycling protocol:
- (42°C, 5 min → 16°C, 5 min) × 60 cycles → 60°C, 5 min
3.Perform transformation, plating, and assessment as described (see Basic Protocol 4, steps 4-13).
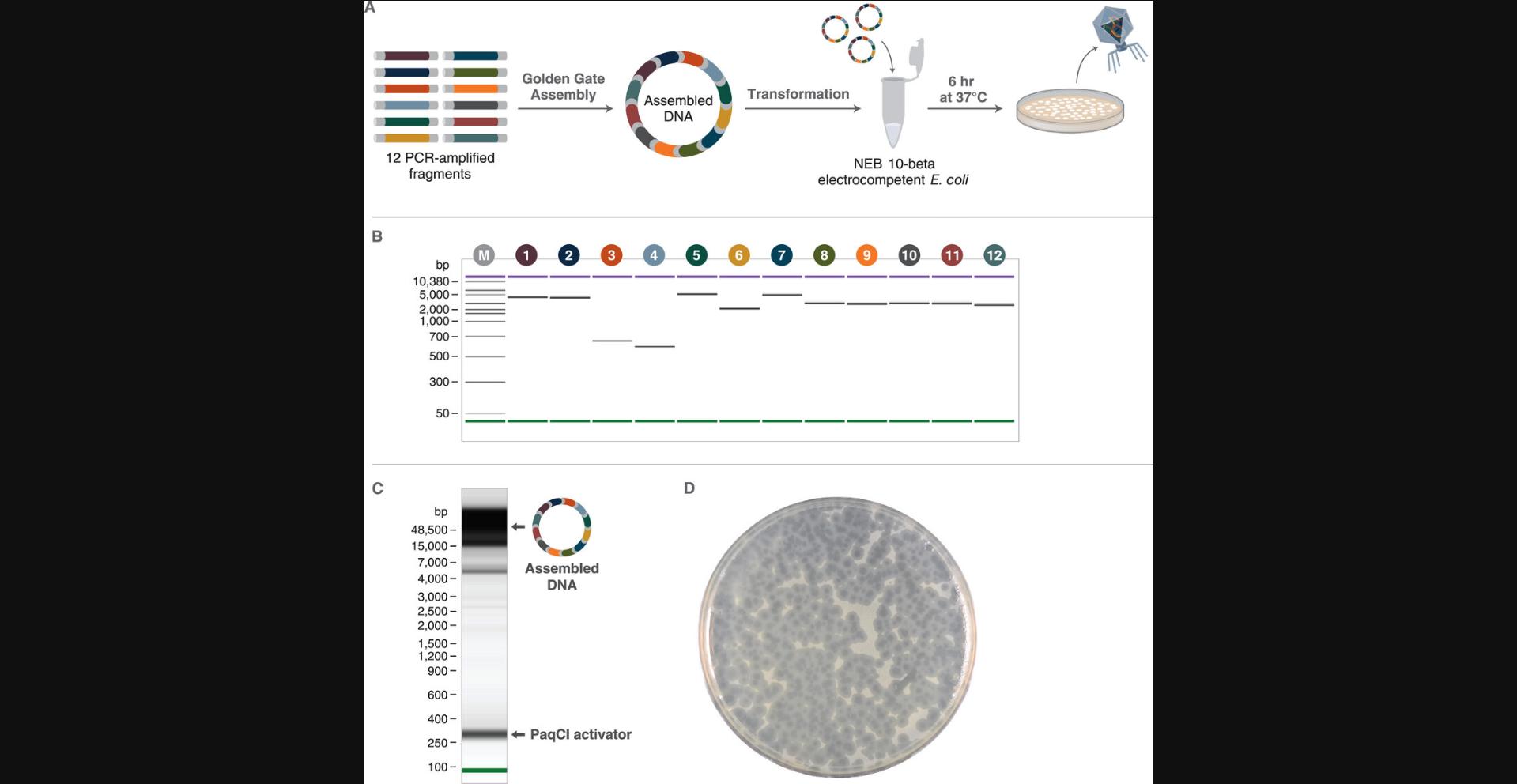
Basic Protocol 5: ONE-POT GOLDEN GATE ASSEMBLY OF THE T7 BACTERIOPHAGE GENOME FROM 12+ PARTS
Another compelling application of Golden Gate Assembly is the assembly of entire small genomes in a single reaction. This protocol describes the assembly and rescue of T7 bacteriophage using a one-pot assembly of 12, 24, or 36 PCR amplicon fragments (see Support Protocol 1) using PaqCI and T4 DNA ligase (Fig. 8A). The fragments were designed using as in Basic Protocol 3 (see Supporting Information Tables S6, S8-S10 and GenBank files for details on PCR primer and fragment sequences). This protocol assumes that parts have been prepared and purified following the guidelines of Support Protocol 1 and mixed in equimolar ratio. For more details on fragment design, see Figure 7, Strategic Planning, Critical Parameters, and Basic Protocol 3.For assembly design and execution, see Figure 8.
Additional Materials (also see Basic Protocol 4)
- Equimolar parts master mix of 12, 24, or 36 T7 parts: PaqCI-12-F1 to PaqCI-12-F12, PaqCI-24-F1 to PaqCI-24-F24, or PaqCI-36-F1 to PaqCI-36-F36 (see Support Protocol 1 for preparation; see Supporting Information Table S6 for primers; see Supporting Information Tables S8-10 for part sequences)
- 10 U/µl PaqCI (New England Biolabs, cat. no. R0745)
- 20 mM PaqCI activator (New England Biolabs, cat. no. S0532)
- 400 U/µl T4 DNA Ligase (New England Biolabs, cat. no. M0202S)
- NEB 10-beta Electrocompetent E. coli (New England Biolabs, cat. no. C3020K)
- NEB 10-beta/Stable Outgrowth Medium (New England Biolabs, cat. no. B9035S)
- Top agar (Elbing & Brent, 2019)
- LB agar plate(s) without antibiotics (Elbing & Brent, 2019)
-
- 1-mm-gap electroporation cuvettes (BTX, cat. no. 45-0134)
- Gene Pulser Xcel Electroporation System (Bio-Rad, cat. no. 1652660)
- 5-ml screw-cap tubes (MTC Bio, cat. no. C2540)
Perform assembly
1.Combine the following components in a 0.2-ml PCR tube:
- Equimolar parts master mix at 3 nM final concentration
- 2 μl 10× T4 DNA Ligase Buffer
- 2 μl PaqCI
- 0.5 μl PaqCI Activator
- 2 μl T4 DNA Ligase
- Nuclease-free water to 20 μl
2.Mix well by pipetting gently 5-10 times, then spin down in a microcentrifuge for 10 s.
3.Place in a thermocycler and run the following cycling protocol:
- (37°C, 5 min → 16°C, 5 min) × 90 cycles → 60°C, 5 min
4.Optional : Prior to transformation, validate the assembly by TapeStation (see Support Protocol 4; Fig. 8C) or other system suitable for visualizing large (∼40-kb) DNA constructs.
Perform phage boot-up
5.Pre-chill a 1-mm-gap electroporation cuvette on ice.
6.Thaw NEB 10-beta electrocompetent cells on ice for 10 min.
7.Aliquot 50 µl cells into a 1.5-ml DNA LoBind tube.
8.Add 1 µl assembly product and mix by pipetting gently 2-3 times.
9.Transfer DNA/cell mixture to the chilled cuvette and keep on ice.
10.Set up the Bio-Rad Gene Pulser Xcell electroporation system with the following settings:
- Voltage = 1800 V
- Capacitance = 25 µF
- Resistance = 200 Ω
- Cuvette = 1 mm
11.Wipe excess moisture from the outside of the cuvette and load the cuvette into the Xcell ShockPod cuvette chamber.
12.Press “Pulse” to electroporate the sample.
13.Immediately add 950 µl NEB 10-beta/Stable Outgrowth Medium and transfer cells to a new 1.5-ml DNA LoBind tube.
14.Allow cells to recover by outgrowth at 37°C for 1.5 hr with shaking (225 rpm).
15.Prewarm an LB agar plate (no antibiotics) to 37°C.
16.Add 100-500 µl outgrowth culture to a 5-ml screw-cap tube followed by enough molten (47°C) top agar to give a final volume of 3 ml.
17.Swirl gently for 1-2 s to homogenize.
18.Pour mixture onto the prewarmed LB agar plate, rock to spread the mixture evenly, and leave at room temperature for 10 min to allow the top agar to solidify.
19.Turn plate upside-down and incubate at 37°C until plaques form.
20.Remove plates from incubator and store at 4°C until assessed. See Figure 8D for an example plate containing plaques.
Assess efficiency
21.Count plaques and determine the viral assembly efficiency as the number of plaques formed per μl of assembly reaction:
where F is the fraction of outgrowth culture plated and V is the volume of assembly used for electroporation. For a plate with 100 plaques after 100 µl outgrowth culture was plated from an electroporation of 1 µl assembly:
Support Protocol 1: GENERATION OF HIGH-PURITY AMPLICONS FOR ASSEMBLY
When using PCR to generate fragments for high-complexity assemblies, high-purity amplicon DNA is required for robust results. Impurities such as primer dimers and off-target amplification products containing Type IIS sites can generate assembly-active overhangs that interfere with the production of the intended assembly product (see Troubleshooting and Supporting Information Figure S2). Rigorous purification reduces these impurities and allows more accurate quantification of assembly parts. This protocol describes the generation and purification of PCR DNA using Q5 DNA polymerase and the Monarch PCR & DNA Cleanup Kit.
Additional Materials (also see Basic Protocols 4-5)
- Q5 Hot Start High-Fidelity 2× Master Mix (New England Biolabs, cat. no. M0494L)
- 10 µM primers in nuclease-free water (see Supporting Information Table S6 for primers used to generate fragments in Basic Protocol 5)
- 1 ng/µl template DNA (to generate amplicons for Basic Protocol 5, use T7 gDNA, Boca Scientific, cat. no. 310005)
- Monarch PCR & DNA Cleanup Kit (5 μg) (New England Biolabs, cat. no. T1030L)
- HPLC-grade isopropanol (Sigma-Aldrich, cat. no. 34863)
- 200 proof ethanol (Sigma-Aldrich cat. no. E7023)
Perform PCR
1.Set up each PCR in a 0.2-ml PCR tube as follows:
- 25 μl Q5 Hot Start High-Fidelity 2× Master Mix
- 2.5 μl 10 µM forward primer
- 2.5 μl 10 µM reverse primer
- 1.0 µl 1 ng/μl template DNA
- 19 μl nuclease-free water
2.Microcentrifuge for 2-3 s.
3.Place in a thermocycler and run the following protocol with a lid temperature of 105°C:
98°C, 30 s → (98°C, 10 s → T a, 30 s → 72°C, 30 s/kb) × 35 cycles → 72°C, 2 min → hold at 10°C
Purify product
4.Prepare 1× working solutions of Binding Buffer and Wash Buffer according to the Monarch PCR & DNA Cleanup Kit instructions.
- Binding Buffer: add 63.6 ml isopropanol to concentrated buffer
- Wash Buffer: add 100 ml ethanol to concentrated buffer
5.Transfer PCR product to a 1.5-ml DNA LoBind tube.
6.For each 50 µl PCR product, add 250 µl Binding Buffer for products <2 kb or 100 µl for products >2 kb. Mix by pipetting up-and-down 10 times.
7.Place the DNA binding column into the collection tube provided.
8.Load diluted PCR product onto the column and centrifuge 1 min at 16,000 × g.
9.Discard the flowthrough and return column to the collection tube.
10.Add 200 µl Wash Buffer to the column and centrifuge 1 min at 16,000 × g.
11.Add another 200 µl Wash Buffer and centrifuge 1 min at 16,000 × g.
12.Discard flowthrough and return column to the collection tube.
13.Centrifuge a final time for 1 min at 16,000 × g.
14.Transfer column to a fresh 1.5-ml DNA LoBind tube.
15.Pipet 15 µl nuclease-free water directly to the membrane, being careful not to touch the membrane.
16.Incubate at room temperature for 3 min to elute bound DNA.
17.Centrifuge 1 min at 16,000 × g.
18.Remove and discard the column.
19.Assess fragment purity by bioanalysis, gel electrophoresis, or similar method. Measure sample concentration by Qubit (recommended, see Support Protocol 3) or Nanodrop.
Support Protocol 2: CLONING ASSEMBLY PARTS INTO A HOLDING VECTOR
For parts that are re-used frequently or require manipulation (e.g., site-directed mutagenesis), it is often convenient to clone PCR-generated or purchased synthesized fragments into a holding vector for ease of propagation, purification, and sequence verification. Here, we present a protocol for blunt cloning PCR fragments into the EcoRV site of the pUC57-mini-BsaI-Free vector (see Supporting Information Table S3 for sequence), a vector domesticated to remove recognition sites for common GGA enzymes with an ampicillin resistance marker. Note that it is not always possible to clone parts in E. coli ; this is not recommended when the parts may be toxic or include highly repetitive elements.
Additional Materials (see Basic Protocols 4-5 and Support Protocol 1)
- pUC57-mini-BsaI-Free (Supporting Information Table S3) or user provided holding vector at 100 ng/µl
- 10× CutSmart buffer (New England Biolabs, cat. no. B6004)
- 20 U/μl EcoRV-HF (New England Biolabs, cat. no. R3195S)
- 5 U/μl Quick CIP (New England Biolabs, cat. no. M0525S)
- Insert DNA: purified PCR amplicon (see Support Protocol 1) or commercial synthetic linear DNA
- Quick Blunting Kit (New England Biolabs, cat. no. E1201S) containing 10× Blunting Buffer, Blunting Enzyme Mix, and 1 mM Deoxynucleotide Solution Mix
- Quick Ligation Kit (New England Biolabs, cat. no. M2200S) containing Quick Ligase and 2× Quick Ligation Reaction Buffer
- NEB 5-alpha competent cell E coli (High Efficiency) (New England Biolabs, cat. no. C2987H)
- LB agar plate (Elbing & Brent, 2019) with 100 µg/ml ampicillin
Prepare vector
1.Linearize and dephosphorylate vector by setting up a 50-µl EcoRV-HF digest as follows:
- 10 µl 100 ng/µl pUC57-mini-BsaI-Free
- 5 µl 10× CutSmart buffer
- 2 µl 20 U/µl EcoRV-HF
- 2 µl 5 U/µl Quick CIP
- 31 µl nuclease-free water
2.Incubate at 37°C for 1 hr in a thermocycler.
3.Heat-inactivate EcoRV-HF and Quick CIP at 80°C for 10 min.
4.Purify digested DNA as described (see Support Protocol 1, steps 4-19). After determining the vector concentration, dilute to 10 nM using nuclease-free water.
Prepare insert
5.Dilute insert DNA to 60 nM using nuclease-free water.
6.Blunt and phosphorylate the insert DNA by setting up a 25-µl Quick Blunting reaction as follows:
- 5 µl 60 nM insert DNA
- 2.5 µl 10× Blunting Buffer
- 2.5 µl 1 mM Deoxynucleotide Solution Mix
- 1 µl Blunting Enzyme Mix
- 14 µl nuclease-free water
7.Incubate at 25°C for 20 min.
8.Incubation at 70°C for 10 min to inactivate the enzyme mix.
Ligate insert and vector
9.Set up a 20-µl ligation reaction as follows:
- 10 µl 2× Quick Ligation Reaction Buffer
- 5 µl 12 nM prepared insert
- 2 µl 10 mM linearized vector
- 2 µl nuclease-free water
- 1 µl Quick Ligase
10.Incubate 5 min at 25°C then hold at 4°C.
Transform into E. coli
11.Thaw one 50-µl tube of NEB 5-alpha competent E. coli cells on ice.
12.Add 2 µl ligation reaction, mix by flicking gently 4-5 times, and incubate on ice for 30 min.
13.Heat shock cells at 42°C for exactly 30 s, then return tube to ice for 5 min.
14.Add 950 µl room temperature SOC medium and incubate at 37°C for 1 hr with agitation.
15.Plate 100 µl outgrowth culture onto a prewarmed LB agar plate with 100 µg/ml ampicillin and incubate overnight at 37°C.
16.Identify colonies containing insert DNA using colony PCR (Woodman et al., 2016) and verify insert by sequencing.
17.To generate high-quality DNA for GGA from a cloned plasmid, follow a standard miniprep protocol (e.g., Engebrecht et al., 1991) or use a commercial plasmid purification kit.
Support Protocol 3: QUANTIFYING DNA CONCENTRATION USING A QUBIT 4 FLUOROMETER
Below we describe a protocol to measure the concentration of DNA for Golden Gate Assembly. This protocol allows the user to monitor the success of DNA amplification and plasmid purifications and confirm that the correct molar ratios are used in an assembly reaction.
Additional Materials (also see Basic Protocol 4)
- Qubit 1× dsDNA Broad Range (BR) Assay Kit (Thermo Fisher Scientific, cat. no. Q33265) including 1× dsDNA BR Working Solution and Qubit dsDNA BR Standards #1 and #2
- DNA samples to be assayed (generated using SP1, SP2, or purchased from a vendor as synthetic dsDNA or plasmid)
- Qubit Assay Tubes (Thermo Fisher Scientific, cat. no. Q32856)
- Qubit 4 Fluorometer (Thermo Fisher Scientific, cat. no. Q33238)
1.Add 190 µl of 1× dsDNA BR Working Solution to two Qubit Assay Tubes.
2.Add 10 µl standard #1 to one tube and 10 µl standard #2 to the other tube. Vortex for 2-3 s and microcentrifuge for 2-3 s.
3.Aliquot 199 µl of 1× dsDNA BR Working Solution into one Qubit Assay Tube for each sample to be measured.
4.Add 1 µl DNA sample to each tube. Vortex for 2-3 s and microcentrifuge for 2-3 s.
5.Leave all tubes at room temperature for 2 min.
6.On the Home screen of the fluorometer, select the 1× dsDNA Broad Range (BR) assay and follow the prompts to read the standards and the samples.
7.For samples with concentrations <100 ng/µl, stop at this step and use the concentration determined in step 6 to generate the parts master mix. For samples with concentrations >100 ng/µl, dilute a portion of the DNA sample to 20-50 ng/µl using nuclease-free water to a final volume ≥ 10 µl and measure again as in steps 8-11.
8.Aliquot 190 µl of 1× dsDNA BR Working Solution into one Qubit Assay Tube for each sample to be re-measured.
9.Add 10 µl diluted DNA sample to each tube. Vortex for 2-3 s and microcentrifuge for 2-3 s.
10.Repeat steps 5-6 to take measurements.
11.Determine the sample concentration by multiplying the measured concentration by the dilution factor in step 7.Use this concentration to generate the parts master mix.
Support Protocol 4: VISUALIZING LARGE ASSEMBLIES VIA TAPESTATION
Here, we describe a method to visualize final assembly products using the Agilent TapeStation system. This can be used as a semiquantitative tool to assess the success of the assembly reaction.
Additional Materials (also see Basic Protocol 4)
- Completed GGA reaction(s) (see Basic Protocols 4-5 and Alternate Protocol 2) or other DNA samples to analyze
- Genomic DNA Ladder and Sample Buffer (Agilent, cat. no. 5067-5366)
- Optical tube strips and caps (Agilent, cat. nos. 401428 and 401425)
- IKA MS3 vortex shaker with MS 3.5 PCR plate attachment (IKA, cat. nos. 0003319000 and 0003428000)
- 4200 TapeStation system (Agilent, cat. no. G2991BA)
- Loading Tips (Agilent, cat. no. 5067-5598)
- Genomic DNA ScreenTape (Agilent cat. no. 5067-5365)
1.Allow all reagents to equilibrate at room temperature for at least 30 min.
2.Spin down GGA assembly sample(s) for 10 s in a microcentrifuge.
3.Pipette 10 µl Genomic DNA Sample Buffer and 1 µl Genomic DNA Ladder into the first position of an optical tube strip.
4.For each sample, pipette 10 µl Genomic DNA Sample Buffer and 1 µl GGA reaction into subsequent wells of the tube strip.
5.Place caps on the tube strip and mix for 1 min on an IKA MS3 vortex shaker set to 2000 rpm.
6.Spin down samples for 1 min in a microcentrifuge.
7.Prepare the 4200 TapeStation per manufacturer's instructions. Ensure that sufficient loading tips are present in the instrument.
8.Insert the Genomic DNA ScreenTape into the 4200 TapeStation system.
9.In the TapeStation Controller software, select the appropriate sample positions and type in each sample name.
10.Load samples into the instrument, placing the ladder in position A1 on tube strip holder.
11.Click “Start” in the TapeStation Controller software.
Support Protocol 5: VALIDATING PHAGE GENOME ASSEMBLIES VIA ONT LONG-READ SEQUENCING
In many situations, it is important to verify the sequence of assemblies to check that all parts are present and in the correct order and that no unintended mutations were introduced during parts production and assembly. This protocol describes a workflow for sequence-verifying assembled T7 viral genomes isolated from cultured phages. The steps include isolation of viral genomic DNA, library preparation, data collection using Oxford Nanopore Technologies (ONT) long-read sequencing, and finally processing and sequence assembly of ONT sequencing data. Further information on handling and characterizing bacteriophages can be found in Current Protocols article Pelzek et al. (2013).
Materials
-
NEB 10-beta Electrocompetent E. coli (New England Biolabs, cat. no. C3020K) or other suitable T7 host strain
-
LB medium (Elbing & Brent, 2019)
-
LB plate containing T7 phage plaques (see Basic Protocol 5)
-
Phage dilution buffer (see recipe)
-
PEG 8000 (Sigma-Aldrich, cat. no 89510)
-
NaCl (Sigma-Aldrich, cat. no. 793566)
-
Monarch HMW DNA Extraction Kit for Tissue (New England Biolabs, cat. no. T3060S) containing:
-
Monarch HMW gDNA Tissue Lysis Buffer
-
Proteinase K (molecular biology grade)
-
Monarch Protein Separation Solution
-
Monarch 2 ml Tubes
-
Monarch DNA Capture Beads
-
Monarch gDNA Wash Buffer
-
Monarch Bead Retainers
-
Monarch Collection Tubes II
-
Monarch gDNA Elution Buffer II
-
HPLC-grade isopropanol (Sigma-Aldrich, cat. no. 34863)
-
Ligation Sequencing Kit (Oxford Nanopore Technologies, cat. no. SQL-LSK109)
-
Native Barcoding Expansion (Oxford Nanopore Technologies, cat. no. EXP-NBD104 or EXP-NBD114)
-
Adapter Mix II Expansion (Oxford Nanopore Technologies, cat. no. EXP-AMII001)
-
NEB Blunt/TA Ligase Master Mix (New England Biolabs, cat. no. M0367)
-
NEBNext Quick Ligation Reaction Buffer (New England Biolabs, cat. no. B6058)
-
NEBNext Companion Module for Oxford Nanopore Technologies Ligation Sequencing (New England Biolabs, cat. no. E7180S)
-
Agencourt AMPure XP beads (Beckman Coulter, cat. no. A63881)
-
MinION Flow Cell (R9.4.1) (Oxford Nanopores Technologies, cat. no. FLO-MIN106D)
-
Flow Cell Priming Kit (Oxford Nanopore Technologies, cat. no. EXP-FLP002)
-
- Culture tube (VWR, cat. no. 10545-946)
-
37°C shaking incubator
-
250-ml Erlenmeyer flask (VWR, cat. no. 10536-914)
-
1.5-ml DNA LoBind tubes (Eppendorf, cat. no. 022431021)
-
50-ml centrifuge tubes (VWR, cat. no. 21008-940 or similar)
-
Thermal mixer (e.g., Eppendorf, cat. no. 5382000023)
-
Tube rotator (e.g., VWR, cat. no. 10136-084)
-
Magnetic separator for 1.5-ml tubes (New England Biolabs, cat. no. S1509S)
-
MacOS with NCBI BLAST software (https://ftp.ncbi.nlm.nih.gov/blast/executables/blast/LATEST/)
-
NOTE : As of this writing, some of the ONT kits listed (SQL-LSK109, EXP-NBD104/114, and EXP-AMII001) are being replaced by a single kit (SQK-NBD, 114.24 Ligation Sequencing gDNA - Native Barcoding Kit 24 V14), which is used with NEB products M0367, M6630, E7546, and E6056.The protocol remains largely identical.
Propagate phage and isolate DNA
1.Inoculate 5 ml LB medium in a culture tube with NEB 10-beta cells and incubate overnight at 37°C with shaking (225 rpm) until saturated.
2.Inoculate 50 ml LB medium in a 250-ml Erlenmeyer flask with 0.5 ml overnight culture and incubate at 37°C (225 rpm) until the OD600 reaches ∼0.1.
3.Collect an agar plug of a single, isolated T7 phage assembly plaque by plunging a glass Pasteur pipet to the bottom of the plate through the agar. Wiggle the pipet slightly to loosen the plug from the plate and remove the pipet.
4.Eject the plug into 1 ml phage dilution buffer in a 1.5-ml DNA LoBind tube. Agitate the tube gently to diffuse the phage particles into the buffer.
5.Inoculate the bacterial culture with 100 µl phage suspension.
6.Incubate at 37°C (225 rpm) and monitor phage propagation by measuring the drop in OD600. Phage propagation is complete when the culture appears clear by eye and has an OD600 < 0.1.
7.Transfer the culture to a 50-ml centrifuge tube and pellet residual cells and cell debris by centrifuging 20 min at 12,000 × g , 4°C. Decant the supernatant into a new 50-ml tube.
8.Add 5 g PEG 8000 (final 10%) and 2.922 g NaCl (final 1 M) and mix by inversion until fully dissolved.
9.Incubate at 4°C for 4-8 hr or overnight.
10.Centrifuge 10 min at 12,000 × g , 4°C, and discard the supernatant.
11.Resuspend phage particles in 300 µl phage dilution buffer and transfer to a new DNA LoBind tube.
12.Microcentrifuge 5-10 s to pellet any insoluble particles and transfer supernatant to a new DNA LoBind tube.
13.Add 300 µl HMW gDNA Tissue Lysis Buffer and 20 µl proteinase K and incubate in a thermal mixer at 56°C for at least 45 min with agitation at 300 rpm.
14.Add 300 µl Protein Separation Solution, mix by inverting for 1 min, and centrifuge 10 min at 16,000 × g.
15.Transfer the upper DNA-containing phase to a Monarch 2-ml Tube and add two DNA Capture Beads.
16.Add 550 µl isopropanol and mix for 4 min on a tube rotator to bind DNA to the beads.
17.Discard liquid by pipetting, being careful not to remove any of the DNA wrapped around the beads.
18.Wash twice by adding 500 µl gDNA Wash Buffer, mixing by inversion 2-3 times, and removing the buffer by pipetting.
19.Place a Monarch Bead Retainer insert into a Monarch Collection Tube II and pour the beads into the Bead Retainer.
20.Remove residual alcohol by pulse spinning (≤ 1 s) in a microcentrifuge.
21.Transfer the Bead Retainer from the Collection Tube to a new DNA LoBind tube and set aside for later use.
22.Pour the beads from the collection tube to a new Monarch 2 ml Tube and immediately add 100 µl Elution Buffer II.
23.Incubate 5 min at 56°C in a thermal mixer with agitation at 300 rpm.
24.Pour the eluted DNA and beads into the Bead Retainer (step 21) and microcentrifuge 1 min at 12,000 × g.
25.Dissolve the eluted gDNA completely by incubating two to three times for 10 min each at 50°C with light mixing in between.
Prepare and sequence ONT Library
26.Prepare ONT library using the Ligation Sequencing Kit according to manufacturer's instructions along with the following reagents as indicated in the instructions:
- Native Barcoding Expansion (EXP-NBD104 or EXP-NBD114)
- Adapter Mix II Expansion
- NEB Blunt/TA Ligase Master Mix
- NEBNext Quick Ligation Reaction Buffer
- NEBNext Companion Module
- Agencourt AMPure XP beads
- Magnetic separator for 1.5-ml tubes
27.Prepare a MinION Flow Cell using the Flow Cell Priming Kit according to the instructions provided with the Ligation Sequencing Kit.
28.Load the sequencing library into the flow cell according to the instructions provided with the Ligation Sequencing Kit and initiate the sequencing run.
Assemble and analyze genome
29.Create a file containing the sequences of interest (i.e., the inserts) in FASTA format.
30.On your computer, open a Command Line Interface (CLI).
This protocol is written for MacOS, where the default CLI is called the “Terminal”. To open a Terminal window, simply press “Command + Option + T”.
For systems running Windows, the CLI is called the “Command Prompt” and can be accessed by pressing “Windows + R”, typing “cmd” in the dialog box, and pressing Enter. Note that the syntax in the Windows Command Prompt will be slightly different from the MacOS syntax used here.
31.Ensure that the BLAST executables are installed in your PATH. The latest version of BLAST can be downloaded at https://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/.
The PATH environment variable can be set using the command:
export PATH= ∼/home/user/ncbi-blast version+/bin:$PATH
32.Navigate to the directory folder where the fragment sequences are stored.
33.Create a local database using the command:
makeblastdb -in ../fragments.fasta -input_type fasta -dbtype nucl -title fragments -out fragments
where “fragments.fasta” is the file containing insert sequences created in step 29.
34.Prior to running the BLAST operation, define output parameters using the command: outfmt.
An example outfmt used in this study can be defined by the line:
outfmt="qseqid sseqid qstart qend sstart send evalue bitscore pident mismatch gaps"
Major outputs include:
| qseqid | query (i.e., genome) sequence id |
| sseqid | target (e.g., insert/fragment) sequence id |
| pident | percentage of identical matches |
| length | aligned length |
| mismatch | number of mismatches |
| gaps | number of gaps |
| qstart | start of alignment in query |
| qend | end of alignment in query |
| sstart | start of alignment in fragment |
| send | end of alignment in fragment |
| evalue | expectation value |
| bitscore | bit score |
35.Search for every fragment sequence present in your query by running the BLASTN command:
blastn -query query.fasta -db fragments.fasta -evalue X -out blastn1 -outfmt "n $outfmt"
| qseqid | sseqid | qstart | qend | sstart | send | evalue | bitscore | pident | mismatch | gaps |
|---|---|---|---|---|---|---|---|---|---|---|
| T7-Query_1 | F3 | 43190820 | 43191431 | 612 | 3 | 1e-3 | 1083 | 99.510 | 1 | 2 |
Reagents and Solutions
Phage dilution buffer
- 25 ml 1 M Tris·HCl, pH 7.5 (Thermo Fisher Scientific, cat. no. 15567027)
- 7.5 ml 5 M NaCl (Thermo Fisher Scientific, cat. no. AM9760G)
- 5 ml 1 M MgCl2 (Thermo Fisher Scientific, cat. no. AM9530G)
- Bring to 500 ml with distilled water
- Store up to 2 years at 25°C
Commentary
Background Information
Golden Gate Assembly has been used for over a decade as a method for multifragment, modular DNA assembly (Engler et al., 2008). It is especially attractive in synthetic biology applications where high-diversity protein libraries are needed, or where many variations of genes are to be assembled into functional operons or circuits, using highly reusable parts designed with standardized connections allowing easy swapping of variants. A common approach to date has been the adoption of “standards” that use defined sets of overhangs as fusion sites for assembling proteins or multigene constructs (many current standards are reviewed in Bird et al., 2022). A recent Current Protocols article (Marillonnet & Grutzner, 2020) addresses in detail the principles of this design and assembly strategy in the context of MoClo, one of the first and still most widely adopted modular hierarchical strategies. Each level of MoClo permits six fragments to be assembled using standardized, experimentally verified fusion sites, allowing large constructs to be assembled from many parts using multiple rounds of sub-assemblies (i.e., six initial transcription units, themselves assembled through GGA, assembled into one part, and six of these sub-assemblies united into a gene pathway made of up to 36 initial transcription units). Many similar standards exist, with defined plasmids and vetted sets of fusion sites used for assembly. Some of these standards are designed for systems targeting a particular organism, and the use of these standard sets by multiple labs studying the same system permits easy sharing of parts between research teams that can be used without modification.
A question not well addressed in GGA systems dependent on standardized overhangs is the initial selection of these sets, and specifically how to distinguish a set that results in high-fidelity, efficient assembly from one that leads to low efficiency or inaccurate assembly. The use of standardized connections is also typically limited to the assembly of transcription units, placing the connections between the coding regions or regulatory elements, because placing fusion sites within functional sequences requires identifying fusion sites from the pre-existing sequence without modification. In principle, the use of four-base overhangs allows up to 120 fragments if all non-palindromic, complementary pairs could be used in a single reaction. In practice, the vast number of possible cross-pairings results in slow assembly rates and leads to many products formed from the ligation of mismatched pairs. For a given assembly, a single erroneous fusion leads to an incomplete or incorrect construct, and the number of correct assemblies begins to drop precipitously for assemblies with more than six to eight parts. It is possible to select sets that are nearly perfect in terms of their fidelity by requiring that all chosen overhangs have at least two base pairs different from every other overhang in the set; in this case any undesired ligation event would require two mismatch pairs, which is very unlikely for cohesive-end ligation (Bilotti et al., 2022; Potapov, Ong, Kucera, et al., 2018). However, this requirement imposes a limit of eight fragments in a single reaction; going beyond this number requires more guesswork in selecting additional sites to determine overhang sets where erroneous ligation is limited.
The selection of fusion sites is even more of an issue when attempting to follow these rules of thumb while subdividing coding sequences (or other functional sequences), where the assembly is additionally limited to the exact sequence that needs to be assembled with no freedom to add additional nucleotides mid-sequence or change the existing sequence (aside from potentially including silent mutations in ORFs). These assemblies are often practically limited to five to eight fragments, with increasing fragment number requiring more screening to identify correct assemblies along with diminishing yields of full-length products. Consequently, transcriptional units are often assembled using Gibson assembly or another method that uses homology regions to assemble the elements to circumvent the need for fusion site selection, or by limiting the number of fragments assembled by GGA to as few as feasible.
Much greater flexibility in the design of Golden Gate Assemblies is possible by using DAD to select large sets of compatible overhangs that are predicted to have low or no mismatch ligation potential (Pryor et al., 2020). The ligation fidelity data confirmed the expectations that (1) sets where all fusion sites have at least two mismatches are expected to be assembled with high accuracy and (2) when adding fusion sites beyond this by guesswork, expected assembly accuracy drops off rapidly with middling fidelity expected around twelve fragments and almost no products expected to be accurately assembled around twenty fragments (Fig. 2B). DAD-guided overhang selection can generate sets of 20+ overhangs that have nearly perfect estimated fidelity, with the predicted fidelity not dropping to near zero until set sizes reach >50 overhangs. Absent sequence restraint, overhang set selection is accomplished by a simple data search within ligase fidelity sets, looking for sets with minimized predicted mismatch ligation potential. Using this method, it is possible to design new standards of compatible connections with high confidence that the reactions will assemble accurately. Further, although manually deducing one's way to a large high-fidelity set is unlikely, there are many high-fidelity sets possible up to 30 or more fusion sites. This means that sets in this complexity range can also be easily selected with additional constraints added, such as fusion sites that must be used or must be excluded. This feature can be used to expand existing overhang sets without sacrificing overall fidelity, or to analyze existing sets to identify and replace pairings that lead to high amounts of mismatch ligation (see Basic Protocol 2 and Alternate Protocol 1).
The flexibility of high-fidelity overhang set selection using DAD further simplifies the selection of fusion sites within coding sequences. By selecting approximate regions where fusion sites are desired, or simply the number of fragments one wishes to obtain, fusion site sets can be identified that are constrained to the existing sequence. This permits genes or small genomes to be divided into 36 (or more) parts predicted to assemble with high accuracy without the need to make any changes to the native sequence. This high complexity Golden Gate strategy has been applied to the assembly of full T7 phage genomes from 10 to >50 parts in a single assembly round, requiring only changes to remove native Type IIS restriction sites through silent mutations (Pryor et al., 2022). Indeed, the high flexibility of overhang selection allows placement of fusion sites near these native Type IIS sites, permitting domestication during parts generation via PCR (see Basic Protocol 3). This allows whole phage genomes to be assembled in a single step, bypassing the need to propagate sub-assemblies that are likely to be highly toxic to bacterial hosts. Other published applications of DAD to design high-complexity assemblies include the highly modular assembly of individual proteins for making high-complexity or combinatorial libraries (Oling et al., 2022), the simplified assembly of highly repetitive DNA elements such as CRISPR arrays (Volke et al., 2022; Yuan et al., 2022), and the simplification of construction for large (10 kb+) plasmid expression systems in yeast (Szent-Gyorgyi et al., 2022).
Importantly, as mentioned before, the key to high-complexity assembly is fusion site selection, whereas the actual assembly protocols and associated key considerations are the same as those for any Golden Gate Assembly (Marillonnet & Grutzner, 2020). Indeed, the cycling protocols recommended here closely match those previously reported, and the use of fidelity data collected under 37°C/16°C and 42°C/16°C cycling conditions allows selection of fusion sites under the conditions used in Golden Gate protocols. Although it has been shown previously that using static incubation at 37°C instead of a cycling protocol leads to even higher fidelities, most users will find excellent results using a vetted overhang set compatible with the lower-temperature cycling conditions (Potapov, Ong, Kucera, et al., 2018; Pryor et al., 2022). In practice, including the 16°C annealing/ligation step leads to faster and more complete assembly of full-length product with no appreciable increase in the amount of inaccurate assembly except in cases at the extreme of predicted ligase fidelity for cycled assemblies (Pryor et al., 2022). The increased ligation activity at 16°C can be crucial for driving the completion of many-fragment assemblies, as typically only full-length assemblies will be successfully transformed while accurate but incomplete constructs will not. Static elevated temperature incubation can overcome fidelity issues when one is limited to using pre-existing overhang sets predicted to have reduced fidelity, but in practice the reduced yield seen in many-fragment assemblies with 37°C static incubation makes it less useful for typical assemblies compared to selection of a high-fidelity overhang set.
Critical Parameters
During Golden Gate Assembly, any mismatch ligation leads to an irreversible joining of two fragments in an incorrect order. These mistakes lead to incorrect or incomplete products and limit the yield of full-length, error-free products (see below). These factors are less important for smaller numbers of fragments (less than six), as small sets are unlikely to contain two overhangs with high mismatch potential. It is straightforward to select high-fidelity overhang sets up to eight overhang pairs by ensuring that at least two base pairs differ among all overhangs in the set. Mismatch ligation between overhangs that meet this criterion is very unlikely. Conversely, when selecting sets larger than eight members, it is increasingly difficult to hit on a good set through guesswork or classic rules of thumb. A search algorithm utilizing ligation fidelity data greatly simplifies the identification of high-fidelity overhang sets and allows the addition of constraints based on sequence and the inclusion or exclusion of specific overhangs. The resultant estimated fidelity is a quality score noting how close to optimal the set is, rather than a true quantitative predictor of the percent of colonies/plaques that will contain full-length, correctly ordered assemblies. Often, many unique high-fidelity overhang sets with minimized mismatch ligation potential can be selected for any assembly design. However, there are many more suboptimal sets, and finding a high-fidelity set through guesswork is unlikely and becomes increasingly difficult as the size of the set increases (Fig. 2B).
Selection of an optimal overhang set maximizes the chances of success and the overall yield of assembled product, but other factors must be considered. Empty vector backgrounds, contaminating DNA (e.g., genomic DNA, aberrant amplicons, or primer dimers), and post-transformation elimination of toxic inserts can all contribute to increased numbers of colonies containing incorrect assemblies. It is also important to recognize that there are no “good” or “bad” overhangs, and ligation fidelity can only be estimated in the context of an overhang set. An overhang that is part of one high-fidelity set could have high mismatch potential in a different overhang set, which reinforces the need to check the fidelity of any new overhang set.
A related issue is the need to ensure that all native recognition sites for the restriction specificity used in the assembly have been removed from the target sequence, a process referred to as domestication. Internal sites present within the fragments will be recognized and cut by the Type IIS restriction enzyme used in the assembly. For example, the PaqCI assembly of T7 bacteriophage presented here (see Basic Protocol 3) removes the five native PaqCI sites during parts generation. Internal sites that are digested during assembly will generate additional overhangs that can interfere with assembly. In addition, many protocols include a 60°C end hold step, which inactivates the ligase but allows the restriction enzyme to remain active. Normally this ensures digestion of any empty vector to reduce empty vector background; however, the assembly will also be cut if internal sites are present, greatly reducing the yield of the desired product.
While selection of a high-fidelity overhang set and careful review of the in silico design will maximize chances of success, many other factors can influence the efficiency and accuracy of the reaction. The purity and accurate quantitation of DNA used in the assembly are key considerations. When fragments are produced by PCR, it is critical to ensure that the amplicons are free of contaminating primers and off-target amplification products. Such impurities do not simply complicate quantitation but can be catastrophic for assembly yields, as impurities contain Type IIS sites and consequently produce overhangs during digestion that are assembly active and can generate incorrect or incomplete assemblies. This effect compounds as more fragments containing impurities are present. For complex assemblies, the effect of even one impure PCR product can be drastic. In the case of a 12-fragment T7 phage genome assembly, fragments with notable impurities reduced the assembly yield as measured by PFU/µl by at least two orders of magnitude (Supporting Information Fig. S1). The molar balance of fragments is also important in complex assembly, although this typically has less impact than the purity of the DNA fragments. Fragments that are present in excess or are limiting will lead to a buildup of incomplete assemblies with incompatible ends. In this situation, assembly accuracy is not affected, but the yield of transformants/plaques can be reduced. For example, when quadrupling the concentration of an individual fragment within a 13- or 25-fragment assembly of the LacIZ cassette, the number of CFU was reduced by ∼50% (Supporting Information Fig. S3). While both purity and incorrect quantitation can reduce yield regardless of assembly complexity, these effects are compounded in high-complexity assemblies, which tend to have reduced efficiencies even when carefully designed and executed. When comparing 12-, 24-, and 36-fragment T7 phage assemblies using DAD-selected overhang sets, we see large drop-offs in PFU/µl across the series (Supporting Information Fig. S1A).
Finally, the transformation/rescue efficiency of the system can cause apparent failure, even with optimal design, due to insufficient concentration of the raw assembly. While this factor is unlikely to affect transformation of a standard plasmid system (outside attempted transformation of a gene with host toxicity issues), it may be an issue when attempting to rescue phage genomes or other large assemblies. For T7 phage, the genome is small enough that transformation is efficient and viable phage are readily generated from the naked genome once inside the cell. As each system will have different concerns and requirements for successful rescue, a robust transformation protocol specific to the system being studied is recommended, as well as an estimate of the amount of assembled DNA that will be required.
Troubleshooting
The most common factors leading to failed Golden Gate Assemblies are (1) the fidelity of the overhang set, (2) the efficiency of the assembly reaction, which is affected by the reaction buffer, enzyme amounts and ratios, part concentrations and purity, and cycling conditions, and (3) the viability and/or stability of the final product in the host organism. Following the protocols outlined here and the manufacturer recommendations when using commercial assembly kits will maximize the likelihood of successful assembly, but some factors (such as part purity and assembly toxicity) can be difficult to mitigate. Table 3 shows common failure modes that can occur when using the NEBridge Ligase Fidelity tools and performing Golden Gate Assemblies.
| Problem | Possible cause | Solution | |
|---|---|---|---|
| Basic Protocol 1 | Overhang sequence not accepted by NEBridge Ligase Fidelity Viewer | Presence of non-standard bases or characters, incorrect overhang length, duplicate overhang | Read error message returned by program and correct input |
| Basic Protocol 2, Alternate Protocol 1 | Overhang sequence not accepted by NEBridge GetSet | Presence of non-standard bases or characters, incorrect overhang length, duplicate overhang | Read error message returned by program and correct input |
| Basic Protocol 3 | Nucleotide sequence not accepted by NEBridge SplitSet | Presence of non-standard bases or characters | Check sequence; make sure only A, T, C, G bases are present |
| Fusion sites at incorrect locations | Entered split regions overlap | Double check start and end coordinates for each region | |
| Basic Protocol 4, Alternate Protocols 1-2 | Few to no colonies after assembly | Incorrect design | Verify primer design (if PCR), overhang sequences, and Type IIS sites and spacers |
| Basic Protocol 4 | Few to no colonies after assembly of vector from PCR products | Poor input DNA quality | Verify purity of input DNA, particularly for absence of primer dimers in PCR DNA. Verify and optimize PCR conditions and consider gel-purifying fragments. |
| Incorrect assemblies | Incorrect design | Verify primer design (if PCR), overhang sequences, and Type IIS sites and spacers | |
| Incorrect assemblies | Insert biologically interferes with E. coli | Consider attempting to clone lost part individually to verify ability to clone insert | |
| Failure to clone individual fragment | Insert biologically interferes with E. coli | First verify the design. The insert may have activity incompatible with propagation in E. coli. | |
| Basic Protocol 5 | Few/no plaques observed on plate | Assembly reaction failure | Check product of assembly reaction on TapeStation. If correct size is not observed, review assembly design and reaction conditions. |
| Low transformation efficiency | Make sure competent cells are viable. Transform a control plasmid (e.g., pUC19) and calculate transformation efficiency of competent cells. If it is too low, use fresh high-efficiency competent cells. Confirm correct antibiotics are present. | ||
| No assembly product or incorrect/multiple product sizes observed on TapeStation | Incorrect design | Verify primer design (if PCR), overhang sequences, and Type IIS sites and spacers | |
| Poor input DNA quality | Verify purity of input DNA, particularly for absence of primer dimers for PCR DNA. Verify and optimize PCR conditions and consider gel-purifying fragments. | ||
| Suboptimal GGA reaction | Confirm proper reaction conditions for design (e.g., cycle number and temperatures). Use fresh buffers/components. |
Understanding Results
Successful Golden Gate Assembly depends on all fragments being joined efficiently and in the correct order. Our previous work has demonstrated that ligation fidelity is the major determinant of accurate assembly, with application of comprehensive ligation fidelity datasets to selection of fusion site overhangs permitting dependable, flexible design of complex assemblies consisting of dozens of fragments in a single reaction. The protocols presented here demonstrate the application of these tools, along with best practices for preparation of parts and execution of assembly reactions.
Basic Protocol 1 covers the use of the NEBridge Ligase Fidelity Viewer webtool to evaluate the fidelity of an existing set of overhangs. As an example dataset, we used the standard Level 1 MoClo overhangs (Weber et al., 2011) as input (Fig. 4A and Supporting Information Table S11). The estimated ligation fidelity under the MoClo standard ligation conditions (BsaI-HFv2 37-16 cycling ligation conditions) is 93% (Fig. 4B). Below the estimated ligation fidelity is the ligation frequency matrix, which depicts the ligation potential of all possible overhang pairings in the given overhang set. The given overhang sequences and their reverse complements are listed along the x and y axes. Each box of the matrix represents the ligation frequency of the sequences indicated at the axes. Watson-Crick base pairing is indicated using blue (good) and light blue (poor, low efficiency) highlights, whereas mismatch parings are indicated using orange (high frequency mismatch event), light orange (modest frequency), and very light orange (trace mismatch ligation). Pairings without observations in the dataset are not highlighted. For our example MoClo overhang set, we note two pairings with predicted mismatch ligation potential. The pairing of GGTA with TACT has high mismatch potential and the pairing of TACC with AGTA has moderate mismatch potential.
Basic Protocol 2 demonstrates how to generate a high-fidelity set of overhangs de novo using the NEBridge GetSet Tool. Here, the number of required overhangs is entered and a list of high-fidelity fusion sites is returned. The graphical output of NEBridge GetSet is functionally identical to the output of the NEBridge Ligase Fidelity Viewer and can be interpreted in the same manner (Fig. 5B). Distinctions in the NEBridge GetSet output are the inclusion of the Request ID and overhang set outputs above the estimated ligation fidelity. As there are many overhang sets with similar fidelity for a given size, re-running the same set of input parameters will frequently return a different list of overhangs. Using the Request ID will recover the output from a previous run. The overhang set is the set selected by the program, with any required overhangs listed first followed by any new overhangs added by the tool. The number of overhangs requested is the largest factor in the final estimated ligation fidelity. Requests for sets of ≤20 overhangs have estimated fidelities near 100%, whereas requests for sets of up to ∼30 overhangs will have estimated fidelities near 90% (assuming no overhangs are required or excluded). Further increases in the number of requested overhangs result in significant decreases in estimated fidelity, with a minimum reached ∼50 overhangs for most cycling conditions (Fig. 2B). While there is no hard cutoff in estimated fidelity where an assembly will not work, we find rapidly diminishing returns below 50%.
An additional application of the NEBridge GetSet Tool is to suggest replacement overhangs for cloning standards found to have low predicted fidelity for the number of fusion sites. Often this can be accomplished by replacing only a few fusion sites. For example, returning to the MoClo example in Basic Protocol 1 (93% estimated fidelity), replacement of either the TACT/ACTA or GGTA/TACC overhang pair in the set would remove the predicted mismatches. When Basic Protocol 2 was carried out using the MoClo set minus TACT as the required overhangs and requesting nine overhangs (i.e., the eight required overhangs plus a replacement for TACT), the tool returned the new overhang GACT and an estimated ligation fidelity of 100% for the new set (Supporting Information Fig. S4). This demonstrates that a single base change (T to G) in one overhang can have significant effects on the estimated fidelity of even a relatively small overhang set.
Alternate Protocol 1 demonstrates the application of NEBridge GetSet to expand an existing assembly by adding compatible overhangs that minimally decrease the predicted fidelity. As an example, we took a 12-part BsmBI LacIZ assembly (based on the 12-part BsaI LacIZ assembly in Pryor et al., 2020; see Fig. 6A and Supporting Information Table S1 and GenBank files 12-part_LacIZ_Assembly) and expanded it to a 15-part assembly by adding new fragments containing an AmpR cassette and sfGFP (Pedelacq et al., 2006). These additional genes were chosen primarily because they would provide easy-to-distinguish phenotypes that can be assessed by antibiotic selection and fluorescence. For this demonstration we grouped the new sequences into a single block to minimize the number of existing parts that needed to be modified, but the ways to arrange DNA in silico are nearly limitless and many different arrangements of the genes are possible. Additionally, we inserted the new sequences between the existing assembly and the destination plasmid to minimize disruption to the LacIZ cassette (Fig. 6A). For this assembly, three new fragment overhangs were selected. This number of fusion sites ensures that no fragment is >1 kb and that no fragment contains more than one gene. A fragment size cap of 1 kb was chosen to reduce synthesis cost and to keep fragment size consistent with the existing fragments. Allowing only one gene per fragment allows for easy swapping of the antibiotic resistance and fluorophore genes. For example, having sfGFP wholly contained on its own fragment allows it to be replaced with a different fluorophore by manipulating only a single fragment.
For our AmpR and sfGFP expanded 15-part BsmBI LacIZ assembly, the GGCA, TCGC, CAGT, TCCA, GAAT, AGTA, TCTT, CAAA, GCAC, AACG, GTCT, and CCAT overhangs were used exactly as in the base 12-part assembly (i.e., flanking the same fragments). The GGAG overhang is the upstream overhang of the pGGAselect destination vector and must be used as the upstream overhang of the first fragment of the expanded assembly, and therefore must also be included as a required overhang. This means that all 13 overhangs from the base 12-part BsmBI LacIZ assembly are reused in the expanded assembly and must be entered as required overhangs (Supporting Information Fig. S5). Because we are adding three additional fragments (and therefore fusion sites) to the existing 13 overhangs, the required number of overhangs is 16. After the run is complete, the generated overhangs appear with the required overhangs first and in the order they were entered, followed by the newly generated overhangs. For this example, the program selected the overhangs CTGA, GATA, and ACAA and gave an estimated ligation fidelity of 98% for the entire set (Fig. 6C). It should be noted that the overhangs generated can vary between submissions, but any high-fidelity set of overhangs generated by NEBridge GetSet is expected to behave similarly.
With the new overhangs selected, we next needed to place those overhangs into the expanded assembly (Fig. 6A). For this assembly we wanted to divide the AmpR gene into two fragments without changing the native sequence, which required finding one of the selected new fusion sites within the existing sequence. We used an occurrence of ACAA present at the approximate midpoint of the AmpR gene as the fusion site between the AmpR-1 and AmpR-2 fragments. This demonstrates the ease with which an overhang can be located in a native sequence near a desired fusion site location. We next placed the overhang GATA within the spacer between AmpR and sfGFP, and placed the overhang CTGA between the end of LacZ and the sfGFP ribosome binding site (RBS). This latter change replaces the GGAG overhang from the end of the final fragment in the original LacIZ assembly, as that sequence pairs with the pGGAselect destination plasmid. The GGAG overhang was subsequently moved to the beginning of the AmpR-1 fragment to ensure that the assembly inserts properly into the destination plasmid, which completes the assembly (Fig. 6A). The four new fragment sequences were obtained from GenScript and replaced Fragment 1 of the 12-part BsmBI LacIZ in the parts master mix (see Supporting Information Table S2 and GenBank file, 15-part_Expanded_LacIZ_Assembly). The assembly was then carried out as in Basic Protocol 4 except that transformed cells were plated onto LB cam/amp IPTG X-gal plates. We observed that the expanded assembly retained similar fidelity to the base 12-part BsmBI LacIZ (Supporting Information Table S12). There was an ∼50% decrease in the overall number of colonies, but this is to be expected since transformation efficiency is typically reduced under double antibiotic selection. Of the colonies found on the double-antibiotic plates, 99.3% showed fluorescence, indicating that they contained the sfGFP gene (Fig. 6B and Supporting Information Table S12).
Basic Protocol 3 explains how to divide a genome into a certain number of fragments with an optimal set of overhangs using the NEBridge SplitSet Tool. The process is very similar to the selection of sites using NEBridge GetSet except that site selection is limited to the sequence within the indicated search windows. In addition to the selected overhang set, estimated fidelity, and ligation fidelity matrix (Fig. 7A), NEBridge SplitSet will also output the component fragments, including the 5′ and 3′ overhangs and the internal sequence (Supporting Information Fig. S6B). Importantly, the output sequences lack Type IIS sites, and appropriate recognition sites and spacers need to be added to the 5′ and 3′ ends of the sequences before assembly. For fragments generated by PCR, the recognition sites and spacers can be included as 5′ primer extensions. For fragments generated synthetically, the recognition sites and spacer can be appended to the sequence prior to ordering. Similar to NEBridge GetSet, there are many possible sets of high-fidelity overhangs and re-running the same input parameters can return multiple lists of equivalent overhang sets. In general, any such set will produce good assembly results and the user may select any high-fidelity set for designing parts.
As an example of Basic Protocol 3, we demonstrate how to divide the T7 bacteriophage genome into a 12-part circular assembly using PaqCI (Figs. 7 and 8, Supporting Information GenBank file PaqCI_12-part_T7_Assembly). Additionally, we used NEBridge SplitSet to divide the same sequence into 24- and 36- part versions (Supporting Information Fig. S1, Tables S7-S8, and GenBank Files PaqCI_24-part_T7_Assembly, PaqCI_36-part_T7_Assembly). We chose a circular design because we previously showed that a circular assembly of the T7 genome produced significantly more plaques than a linear assembly (Pryor et al., 2022) despite the T7 genome being normally injected as linear dsDNA during infection (Fujisawa & Morita, 1997). This can likely be attributed to the generally increased transformability of circular constructs. Assembly circularization is accomplished using a bridge fragment that spans from the 3′ end of the genome across the terminal repeat to the 5′ end of the genome generated by PCR of gDNA. We have not confirmed how this fragment is generated from linear gDNA ending in terminal repeats, but we believe the terminal repeats are functioning as homologous ends that can be extended through a PCA-like mechanism to generate the full bridge fragment, analogous to how terminal repeats function in vivo (Weigel & Seitz, 2006). We have confirmed through sequencing that the bridge fragment contains a single instance of the terminal repeat.
As the next step in assembly design, we identified the location of all native PaqCI/AarI recognition sites within the T7 bacteriophage sequence using Geneious (Supporting Information Table S4). We then restricted SplitSet to require fusion sites within 5 nucleotides of these sites, which would permit their removal during parts generation by PCR using primers containing the desired silent mutation (Supporting Information Tables S4 and S9). For example, the PaqCI site located at 9387 to 9393 of the T7 genome was removed by converting A9388 to T to create a silent mutation of Pro77 in gp2.5. Other fusion sites were spaced out arbitrarily with larger search windows permitted. A sample set of overhangs obtained using these parameters is GGCA, CTGG, CAGC, CCTC, GGTC, TGTT, TGGT, AGGA, ATTC, CAAA, CATT, TAGA and has a ligation fidelity score of 86%. Note that this score is relatively low for a 12-part assembly design; in this case, the restrictions to native sequence and tight windows near the required silent mutagenesis positions limit the possible overhang sets and are likely responsible for a lower fidelity than would be observed for an unrestricted set generated using NEBridge GetSet. For the 24- and 36-fragment versions, which share these initial five sites, both assemblies have predicted fidelities of 24% (Supporting Information Table S5).
Once the output fragments are obtained, one can design primers and perform PCR to amplify all parts using Support Protocol 1 or previously published protocols (Marillonnet & Grutzner, 2020). If domestication is needed, the forward and reverse primers covering the region must impose the desired mutation. Otherwise, regular primer design is performed. Once the annealed portion of the primers are designed, appropriate spacers and Type IIS recognition sites are added as 5′ extensions (see Alternate Protocol 1, step 10). Following the guidelines provided in Support Protocols 1 and 3 helps ensure that the amplicons are highly pure, free of undesired DNA pieces or primers, and of suitable concentration (Fig. 8B and Supporting Information Fig. S2). As an alternative, full fragments can be ordered from a DNA vendor, with the caveat that not all fragments may be synthesizable or cloneable, especially when dealing with bacteriophage genomes. After PCR, the amplicons are quantified and mixed in an equimolar ratio. We recommend preparing a master mix of all amplicons that can be aliquoted and used in multiple assembly reactions. For a typical reaction, we suggest a final concentration of 3 nM for each fragment.
Division of the same genome into 12, 24, and 36 parts permitted the direct evaluation of the effect of number of fragments on assembly. The assembly reactions for all T7 assemblies were performed according to Basic Protocol 5 and the results evaluated using PFU/µl (Fig. 8D and Supporting Information Fig. S1). The final products were also visualized on an Agilent TapeStation using Support Protocol 4 (Fig. 8C and Supporting Information Fig. S1). Doubling the number of fragments from 12 to 24 parts resulted in a one-order-of-magnitude decrease in PFU/µl; an additional smaller decrease was observed for 36 parts (Supporting Information Fig. S1B). These results show that higher complexity assemblies will result in fewer full-length assemblies even when optimizing overhang sets using DAD, consistent with previous work on LacIZ assemblies in a destination plasmid (Pryor et al., 2020, 2022). We also examined the effect of including an impure PCR fragment in an assembly. In this assembly, we posited that primer dimers and incorrect amplification products were assembly active and found a two-order-of-magnitude decrease in observed yield (Supporting Information Fig. S1B). This demonstrates that even one product showing significant amounts of off-target amplification can be catastrophic for assembly yields.
Time Considerations
Assessing a known set of overhangs using Basic Protocol 1 requires only a few minutes to enter the required information and <1 s to perform the search. Generating an overhang set in Basic Protocol 2 also requires a few minutes to enter information, but the search time depends on how many new overhangs are requested. A 50-overhang request requires ∼5 min.
Alternate Protocol 1 has similar time considerations, with additional time required to design the assembly and incorporate changes. Typically, this in silico work can be completed within a few hours depending on the complexity of the system and the user's familiarity with the system. The method chosen for generating and altering parts affects the time required to move from in silico design to assembly. If parts are generated by PCR, the time required to design and order primers is generally on the order of hours. Once primers arrive, the PCR, cleanup, and quantification can typically be completed in a day or two. If synthetic DNA is ordered from a vendor, the design and ordering process can be completed in a couple of hours, but delivery times range from weeks to months depending on the vendor. The assembly reactions described here range from 5 to 18 hr but low-complexity assemblies can be set up in ∼1 hr and completed in ∼1 hr more. Transformation, outgrowth, and plating take 2-3 hr followed by an overnight growth step before plates can be assessed.
Basic Protocol 3 requires an initial analysis of the genome to be assembled, which can take minutes to days depending on the complexity and needs of the assembly. Once parameters are established, NEBridge SplitSet requires careful entry and verification of information for the desired search windows for placement of breaks, which can take upwards of 15 min. The actual run time for NEBridge SplitSet is similar to NEBridge GetSet, scaling as the number of required fusion sites increases, but generally taking 5 min or less for even the largest searches.
Basic Protocols 4-5 and Alternate Protocol 2 all require ∼1 hr for preparation of the assembly reaction. The reaction time depends on cycling conditions and cycle number. The recommended 30 cycles for Basic Protocol 4 take 5 hr, the 60 cycles for Alternate Protocol 2 take 10 hr, and the 90 cycles for Basic Protocol 5 take 18 hr. Transformation, outgrowth, and plating take 2-3 hr followed by overnight growth before plates can be assessed.
Support Protocol 1 typically takes 2-3 hr for the PCR reaction depending on the size of the amplicons generated. Purification by spin column takes ∼1 hr and visualization by agarose gel electrophoresis or bioanalysis can take an additional 1-2 hr. It is often convenient to run multiple reactions in parallel, with modest increases in total time for each additional reaction.
Support Protocol 2 utilizes PCR fragments generated by Support Protocol 1, which are treated and ligated into a holding plasmid. Beyond generation of the PCR fragments, treatment and ligation take ∼2 hr, and transformation, outgrowth, and plating take 2-3 hr followed by overnight growth. Preparation of the holding plasmid takes ∼2 hr, followed by overnight growth and harvesting, and purification by miniprep, which takes 1 hr. It is often straightforward to clone, grow, and purify multiple fragments in parallel.
Support Protocol 3 takes up to 30 min to set up samples, depending on the number of samples, and <1 min per sample to run on the Qubit fluorometer. Support Protocol 4 requires 15-30 min to prepare samples and 2-3 min per sample to run on the TapeStation.
Support Protocol 5 requires 1 day to grow the phage, starting with an overnight culture of host bacteria. Harvesting phage particles and isolating DNA take ∼1 hr to remove cell debris and add precipitants, followed by an overnight incubation, and then 2-3 hr to isolate phage particles and purify the gDNA. Preparation of the ONT library takes 2-3 hr and ONT sequencing takes 12-72 hr depending on the number of reads collected. Data analysis takes 1-2 days including building the analysis pipeline, or 1-2 hr if a pipeline exists when using a standard reference sequence.
Acknowledgments
We are grateful to Tasha José for assistance with figure design and production, and thank Katharina Bilotti, Eric Cantor, Laurence Ettwiller, Rachel Keown, Vladimir Potapov, Lindsey Spiegelman, Nathan Tanner, and Michael Terns for critical feedback and careful reading of the manuscript. New England Biolabs funded the research described and paid the authors’ salaries. Funding for open access charge is from New England Biolabs.
Author Contributions
Andrew P. Sikkema : investigation; methodology; writing (original draft, review and editing); S. Kasra Tabatabaei : investigation; methodology; writing (original draft, review and editing); Yan-Jiun Lee : investigation, writing (original draft); Sean Lund : investigation, writing (original draft); Gregory J. S. Lohman : conceptualization; investigation; methodology; supervision; writing (original draft, review and editing).
Conflict of Interest
All authors are employees of New England Biolabs, a manufacturer and vendor of molecular biology reagents, including DNA ligases, Type IIS restriction enzymes, and DNA assembly kits. This affiliation does not affect the authors’ impartiality, adherence to journal standards and policies, or availability of data.
Open Research
Data Availability Statement
Data supporting the assemblies described in this study, along with annotated sequence files for the completed assemblies, can be found in the Supporting Information. Data tables used by the ligase fidelity tools have been published elsewhere (Pryor et al., 2020). The Overhang Optimizer Code used by the tools may be requested under a noncommercial use license here: https://www.neb.com/forms/overhang-optimizer-code.
Supporting Information
| Filename | Description |
|---|---|
| cpz1882-sup-0001-figureS1.eps5.1 MB | Figure S1: Comparison of 12-, 24-, and 36-part PaqCI T7 assemblies. |
| cpz1882-sup-0002-figureS2.eps2.3 MB | Figure S2: Examples of common issue with Golden Gate Assemblies. |
| cpz1882-sup-0003-figureS3.eps2 MB | Figure S3: Effect of single part imbalances on LacIZ assemblies. |
| cpz1882-sup-0004-figureS4.eps1.7 MB | Figure S4: NEBridge Ligase Fidelity Viewer output of the improved MoClo overhang set. |
| cpz1882-sup-0005-figureS5.eps1.1 MB | Figure S5: NEBridge GetSet Tool inputs for the 15-part expanded LacIZ assembly. |
| cpz1882-sup-0006-figureS6P1.eps2.4 MB | Figure S6: Complete input and output pages for NEBridge SplitSet for the 12-part T7 PaqCI assembly. |
| cpz1882-sup-0007-figureS6P2.eps3.5 MB | Figure S6: Complete input and output pages for NEBridge SplitSet for the 12-part T7 PaqCI assembly. |
| cpz1882-sup-0008-tableS1-S12.docx92.7 KB | Table S1: 12-part Lac assembly parts, sequences, and overhangs. Table S2: Expanded LacIZ assembly parts, sequences, and overhangs. Table S3: Carrier plasmid sequences. Table S4: T7 phage PaqCI sites and domesticating mutations. Table S5: T7 PaqCI assembly overhang sets. Table S6: T7 PaqCI assembly primers. Table S7: 24-part Lac assembly parts, sequences, and overhangs. Table S8: 12-part T7 PaqCI assembly parts, sequences, and overhangs. Table S9: 24-part T7 PaqCI assembly parts, sequences, and overhangs. Table S10: 36-part T7 PaqCI assembly parts, sequences, and overhangs. Table S11: Fidelity of MoClo overhang sets. Table S12: Comparison of the 12-part LacIZ and expanded LacIZ assemblies. |
| cpz1882-sup-0009-genbank.zip290.3 KB | GenBank file: 12-part_LacIZ_Assembly GenBank file: 15-part_Expanded_LacIZ_Assembly cpz1882-sup-0001-tableS1-S12.docx GenBank file: PaqCI_12-part_T7_Assembly GenBank file: PaqCI_24-part_T7_Assembly GenBank file: PaqCI_36-part_T7_Assembly |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Andreou, A. I., & Nakayama, N. (2018). Mobius assembly: A versatile Golden-Gate framework towards universal DNA assembly. PLoS ONE , 13(1), e0189892. https://doi.org/10.1371/journal.pone.0189892
- Bilotti, K., Potapov, V., Pryor, J. M., Duckworth, A. T., Keck, J. L., & Lohman, G. J. S. (2022). Mismatch discrimination and sequence bias during end-joining by DNA ligases. Nucleic Acids Research , 50(8), 4647–4658. https://doi.org/10.1093/nar/gkac241
- Bird, J. E., Marles-Wright, J., & Giachino, A. (2022). A user's guide to Golden Gate cloning methods and standards. ACS Synthetic Biology , 11(11), 3551–3563. https://doi.org/10.1021/acssynbio.2c00355
- Bitinaite, J., Rubino, M., Varma, K. H., Schildkraut, I., Vaisvila, R., & Vaiskunaite, R. (2007). USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Research , 35(6), 1992–2002. https://doi.org/10.1093/nar/gkm041
- Damalas, S. G., Batianis, C., Martin-Pascual, M., de Lorenzo, V., & Martins dos Santos, V. A. P. (2020). SEVA 3.1: Enabling interoperability of DNA assembly among the SEVA, BioBricks and Type IIS restriction enzyme standards. Microbial Biotechnology , 13(6), 1793–1806. https://doi.org/10.1111/1751-7915.13609
- Elbing, K. L., & Brent, R. (2019). Recipes and tools for culture of Escherichia coli. Current Protocols in Molecular Biology , 125(1), e83. https://doi.org/10.1002/cpmb.83
- Engebrecht, J., Brent, R., & Kaderbhai, M. A. (1991). Minipreps of plasmid DNA. Current Protocols in Molecular Biology , 15(1), 1.6.1–1.6.10. https://doi.org/10.1002/0471142727.mb0106s15
- Enghiad, B., Xue, P., Singh, N., Boob, A. G., Shi, C., Petrov, V. A., Liu, R., Peri, S. S., Lane, S. T., Gaither, E. D., & Zhao, H. (2022). PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction. Nature Communications , 13(1), 2697. https://doi.org/10.1038/s41467-022-30355-y
- Engler, C., Kandzia, R., & Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS ONE , 3(11), e3647. https://doi.org/10.1371/journal.pone.0003647
- Fujisawa, H., & Morita, M. (1997). Phage DNA packaging. Genes to Cells , 2(9), 537–545. https://doi.org/10.1046/j.1365-2443.1997.1450343.x
- Gibson, D. G., Benders, G. A., Axelrod, K. C., Zaveri, J., Algire, M. A., Moodie, M., Montague, M. G., Venter, J. C., Smith, H. O., & Hutchison, C. A., 3rd (2008). One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proceedings of the National Academy of Sciences of the United States of America , 105(51), 20404–20409. https://doi.org/10.1073/pnas.0811011106
- HamediRad, M., Weisberg, S., Chao, R., Lian, J., & Zhao, H. (2019). Highly efficient single-pot scarless Golden Gate assembly. ACS Synthetic Biology , 8(5), 1047–1054. https://doi.org/10.1021/acssynbio.8b00480
- Hoose, A., Vellacott, R., Storch, M., Freemont, P. S., & Ryadnov, M. G. (2023). DNA synthesis technologies to close the gene writing gap. Nature Reviews Chemistry , 7(3), 144–161. https://doi.org/10.1038/s41570-022-00456-9
- Kennedy, M. A., Hosford, C. J., Azumaya, C. M., Luyten, Y. A., Chen, M., Morgan, R. D., & Stoddard, B. L. (2023). Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI. Nucleic Acids Research , 51(9), 4467–4487. https://doi.org/10.1093/nar/gkad228
- Liang, J., Zhang, H., Tan, Y. L., Zhao, H., & Ang, E. L. (2022). Directed evolution of replication-competent double-stranded DNA bacteriophage toward new host specificity. ACS Synthetic Biology , 11(2), 634–643. https://doi.org/10.1021/acssynbio.1c00319
- Malci, K., Watts, E., Roberts, T. M., Auxillos, J. Y., Nowrouzi, B., Boll, H. O., Sousa do Nascimento, C. Z., Andreou, A., Vegh, P., Donovan, S., Fragkoudis, R., Panke, S., Wallace, E., Elfick, A., & Rios-Solis, L. (2022). Standardization of synthetic biology tools and assembly methods for Saccharomyces cerevisiae and emerging yeast species. ACS Synthetic Biology , 11(8), 2527–2547. https://doi.org/10.1021/acssynbio.1c00442
- Marillonnet, S., & Grutzner, R. (2020). Synthetic DNA assembly using Golden Gate cloning and the hierarchical modular cloning pipeline. Current Protocols in Molecular Biology , 130(1), e115. https://doi.org/10.1002/cpmb.115
- Oling, D., Lan-Chow-Wing, O., Martella, A., Gilberto, S., Chi, J., Cooper, E., Edström, T., Peng, B., Sumner, D., Karlsson, F., Volkov, P., Webster, C. I., & Roth, R. (2022). FRAGLER: A fragment recycler application enabling rapid and scalable modular DNA assembly. ACS Synthetic Biology , 11(7), 2229–2237. https://doi.org/10.1021/acssynbio.2c00106
- Pedelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C., & Waldo, G. S. (2006). Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology , 24(1), 79–88. https://doi.org/10.1038/nbt1172
- Pelzek, A. J., Schuch, R., Schmitz, J. E., & Fischetti, V. A. (2013). Isolation, culture, and characterization of bacteriophages. Current Protocols Essential Laboratory Techniques , 7(1), 4.4.1–4.4.33. https://doi.org/10.1002/9780470089941.et0404s07
- Potapov, V., Ong, J. L., Kucera, R. B., Langhorst, B. W., Bilotti, K., Pryor, J. M., Cantor, E. J., Canton, B., Knight, T. F., Evans, T. C., Jr., & Lohman, G. J. S. (2018). Comprehensive profiling of four base overhang ligation fidelity by T4 DNA ligase and application to DNA assembly. ACS Synthetic Biology , 7(11), 2665–2674. https://doi.org/10.1021/acssynbio.8b00333
- Potapov, V., Ong, J. L., Langhorst, B. W., Bilotti, K., Cahoon, D., Canton, B., Knight, T. F., Evans, T. C., Jr., & Lohman, G. J. S. (2018). A single-molecule sequencing assay for the comprehensive profiling of T4 DNA ligase fidelity and bias during DNA end-joining. Nucleic Acids Research , 46(13), e79–e79. https://doi.org/10.1093/nar/gky303
- Pryor, J. M., Potapov, V., Bilotti, K., Pokhrel, N., & Lohman, G. J. S. (2022). Rapid 40 kb genome construction from 52 parts through data-optimized assembly design. ACS Synthetic Biology , 11(6), 2036–2042. https://doi.org/10.1021/acssynbio.1c00525
- Pryor, J. M., Potapov, V., Kucera, R. B., Bilotti, K., Cantor, E. J., & Lohman, G. J. S. (2020). Enabling one-pot Golden Gate assemblies of unprecedented complexity using data-optimized assembly design. PLoS ONE , 15(9), e0238592. https://doi.org/10.1371/journal.pone.0238592
- Shetty, R. P., Endy, D., & Knight, T. F., Jr. (2008). Engineering BioBrick vectors from BioBrick parts. Journal of Biological Engineering , 2, 5. https://doi.org/10.1186/1754-1611-2-5
- Stuttmann, J., Barthel, K., Martin, P., Ordon, J., Erickson, J. L., Herr, R., Ferik, F., Kretschmer, C., Berner, T., Keilwagen, J., Marillonnet, S., & Bonas, U. (2021). Highly efficient multiplex editing: One-shot generation of 8× Nicotiana benthamiana and 12× Arabidopsis mutants. Plant Journal , 106(1), 8–22. https://doi.org/10.1111/tpj.15197
- Szent-Gyorgyi, C., Perkins, L. A., Schmidt, B. F., Liu, Z., Bruchez, M. P., & van de Weerd, R. (2022). Bottom-Up design: A modular Golden Gate assembly platform of yeast plasmids for simultaneous secretion and surface display of distinct FAP fusion proteins. ACS Synthetic Biology , 11(11), 3681–3698. https://doi.org/10.1021/acssynbio.2c00283
- Szybalski, W., Kim, S. C., Hasan, N., & Podhajska, A. J. (1991). Class-IIS restriction enzymes—a review. Gene , 100, 13–26. https://doi.org/10.1016/0378-1119(91)90345-c
- Volke, D. C., Martino, R. A., Kozaeva, E., Smania, A. M., & Nikel, P. I. (2022). Modular (de)construction of complex bacterial phenotypes by CRISPR/nCas9-assisted, multiplex cytidine base-editing. Nature Communications , 13(1), 3026. https://doi.org/10.1038/s41467-022-30780-z
- Weber, E., Engler, C., Gruetzner, R., Werner, S., & Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS ONE , 6(2), e16765. https://doi.org/10.1371/journal.pone.0016765
- Weigel, C., & Seitz, H. (2006). Bacteriophage replication modules. FEMS Microbiology Review , 30(3), 321–381. https://doi.org/10.1111/j.1574-6976.2006.00015.x
- Woodman, M. E., Savage, C. R., Arnold, W. K., & Stevenson, B. (2016). Direct PCR of intact bacteria (colony PCR). Current Protocols in Microbiology , 42, A.3D.1–A.3D.7. https://doi.org/10.1002/cpmc.14
- Yeom, H., Ryu, T., Lee, A. C., Noh, J., Lee, H., Choi, Y., Kim, N., & Kwon, S. (2020). Cell-free bacteriophage genome synthesis using low-cost sequence-verified array-synthesized oligonucleotides. ACS Synthetic Biology , 9(6), 1376–1384. https://doi.org/10.1021/acssynbio.0c00051
- Yuan, G., Martin, S., Hassan, M. M., Tuskan, G. A., & Yang, X. (2022). PARA: A new platform for the rapid assembly of gRNA arrays for multiplexed CRISPR technologies. Cells , 11(16), 2467. https://doi.org/10.3390/cells11162467
- Zhang, Z., Xu, K., Xin, Y., & Zhang, Z. (2015). An efficient method for multiple site-directed mutagenesis using type IIs restriction enzymes. Analytical Biochemistry , 476, 26–28. https://doi.org/10.1016/j.ab.2015.01.010
Key References
- Marillonnet & Grutzner (2020). See above.
Covers many important methods and considerations for GGA design and implementation, including preparation of parts and vectors and hierarchical assembly.
- Pryor et al. (2020). See above.
The original publication covering the development and use of the GGA ligase fidelity tools NEBridge Ligase Fidelity Viewer, NEBridge GetSet, and NEBridge SplitSet. It also contains underlying data that are used in the tools and describes the LacIZ cassette system in more detail.
- Pryor et al. (2022). See above.
Describes the initial use of the NEBridge SplitSet Tool to design T7 bacteriophage assemblies and examines the limits of GGA complexity possible with DAD.
Internet Resources
Houses the three main tools discussed here: NEBridge Ligase Fidelity Viewer, the NEBridge GetSet Tool, and the NEBridge SplitSet Tool.
Assists in visualizing GGAs and can assess overhangs, flag internal Type IIS sites, and assist with primer design.
Citing Literature
Number of times cited according to CrossRef: 1
- Shlomo Yakir Hoch, Ravit Netzer, Jonathan Yaacov Weinstein, Lucas Krauss, Karen Hakeny, Sarel Jacob Fleishman, GGAssembler: Precise and economical design and synthesis of combinatorial mutation libraries, Protein Science, 10.1002/pro.5169, 33 , 10, (2024).

