Fixed RNA - GEM Recovery - Sequencing
Ksenija Sabic
Abstract
This protocol is a continuation of CG000527 (Step 3 and onward).
Before start
Prepare daily fresh 80% Ethanol for wash steps.
Steps
GEM Recovery and Pre-Amplification
Reagent Preparation (~15 mins):
Thaw Reducing Agent B at Room temperature, vortex, verify no precipitate, centrifuge briefly.
Thaw Pre-Amp Primers B at Room temperature, vortex, centrifuge briefly.
Keep Amp Mix On ice. Vortex and centrifuge briefly.
Prepare fresh 80% Ethanol (2.5mL for 4 GEM reactions).
Add 125µL Recovery Agent to each sample at room temperature.
DO NOT pipette mix or vortex the biphasic mixture.
Firmly secure the cap on the tube strip, ensuring that no liquid is trapped between the cap and the tube rim. Mix by inverting the capped tube strip 5x.
DO NOT invert without firmly securing the caps.
Wait 2 min.
The resulting biphasic mixture contains Recovery Agent/Partitioning Oil (pink) and aqueous phase (translucent/opaque). A smaller aqueous phase volume indicates a clog during GEM generation.
Centrifuge briefly.
Slowly remove and discard 125µL Recovery Agent/Partitioning Oil (pink) from the bottom of the tube. DO NOT aspirate any aqueous sample.
Proceed directly to Pre-Amplification PCR. No cleanup step is required.
Prepare Pre-Amplification Mix on ice. Vortex and centrifuge briefly.
| A | B | C | D |
|---|---|---|---|
| Pre-Amp Mix (add reagents in order listed) | PN | 1X (µl) | 4X + 10% (µl) |
| Amp Mix | 2000103 | 25.0 | 110.0 |
| Pre-Amp Primers B | 2000529 | 10.0 | 44.0 |
| Total | 35.0 | 154.00 |
Add 35µL Pre-Amplification Mix to aqueous sample from step 6.
Cap firmly and invert 8x to mix. Centrifuge briefly.
Incubate in a thermal cycler with the following protocol:
| A | B | C |
|---|---|---|
| Step | Temperature | Time (hh:mm:ss) |
| 1 | 98C | 00:03:00 |
| 2 | 98C | 00:00:15 |
| 3 | 63C | 00:00:20 |
| 4 | 72C | 00:01:00 |
| 5 | Go to Step 2, 7x (total 8 cycles) | |
| 6 | 72C | 00:01:00 |
| 7 | 4C | Hold |
Saved as 'pre-amp' under Fixed RNA folder.Lid Temperature: 105CReaction Volume: 100µlRun Time: ~30-45 min Store at 4°C for up to 72 h or −20°C for ≤1 week, or proceed to the next step.
Prepare Elution Solution. Vortex and centrifuge briefly.
| A | B | C |
|---|---|---|
| Elution Solution (add reagents in order listed) | PN | 1000µl |
| Buffer EB | 980 | |
| 10% Tween 20 | 10 | |
| Reducing Agent B | 2000087 | 10 |
| Total | 1000 |
Centrifuge the sample (PCR product) for 0h 0m 30s sec in a microcentrifuge and transfer 70µL of the upper layer to a new tube.
Presence of a cloudy precipitate at the interface between phases is normal. Avoid transferring the precipitate when transferring 70 μl at this step.
Vortex to resuspend the SPRIselect reagent. Add 126µL SPRIselect reagent (1.8X) to each sample and pipette mix 15x (pipette set to 180 μl).
Incubate 0h 5m 0s min at Room temperature.
Place on the magnet (high position) until the solution clears.
Remove the supernatant. DO NOT discard any beads.
With the tube still in the magnet, add 200µL 80% ethanol to the pellet. Wait 0h 0m 30s sec.
Remove the ethanol.
Repeat steps 18 and 18.1 for a total of 2 washes.
Centrifuge briefly and place on the magnet (low position).
Remove any remaining ethanol. DO NOT let the sample dry to ensure maximum elution efficiency.
Remove from the magnet. Add 101µL Elution Solution (from step 12). Wait 0h 1m 0s min before resuspending. Pipette mix 15x.
Incubate 0h 2m 0s min at Room temperature.
Place the tube strip on the magnet (high position) until the solution clears.
Transfer 100µL sample to a new tube strip.
Store at 4°C for ≤72h 0m 0s hours or at -20°C for ≤4 weeks, or proceed to the next step.
Library Construction
Reagent Preparation (~10 min):
Thaw Dual Index Plate TS Set A at Room temperature.
Keep Amp Mix On ice. Vortex and centrifuge briefly.
If planning to assess quality and concentration of library immediately after completion, thaw Agilent Bioanalyzer High Sensitivity Kit at Room temperature, keeping prepared gel and gel-dye away from light sources. Consult the following protocol for details:
Choose the appropriate sample index sets to ensure that no sample indices overlap in a multiplexed sequencing run. Record the 10x sample index name (PN-3000511 Dual Index Plate TS Set A well ID) used.
Prepare Sample Index PCR Mix on ice:
| A | B | C | D | E |
|---|---|---|---|---|
| Sample Index PCR (add reagents in the order listed) | PN | 1X (µl) | 1X + 10% (µl) | 4X + 10% (µl) |
| Amp Mix | 2000103 | 50 | 55 | 220 |
| Nuclease-free water | 10 | 11 | 44 | |
| Total | 60 | 66 | 264 |
Transfer ONLY 20µL of sample from the step 24 to a new tube strip.
Add 60µL Sample Index PCR Mix to 20 μl sample.
Add 20µLof an individual Dual Index TS Set A to each sample. Pipette mix 5x (pipette set to 90 μl). Centrifuge briefly.
Incubate in a thermal cycler with the following protocol:
| A | B | C |
|---|---|---|
| Step | Temperature | Time (hh:mm:ss) |
| 1 | 98C | 00:00:45 |
| 2 | 98C | 00:00:20 |
| 3 | 54C | 00:00:30 |
| 4 | 72C | 00:00:20 |
| 5 | Go to step 2, see table below for total # of cycles | |
| 6 | 72C | 00:01:00 |
| 7 | 4C | Hold |
Saved as 'Sample Index PCR' under Fixed RNA folder.Lid Temperature: 105CReaction Volume: 100µlRun Time: ~25 - 40 min
| A | B | C | D | E |
|---|---|---|---|---|
| Targeted Cell Recovery | Total Cycles* | |||
| for Cell Lines | for PBMCs & Nuclei | for Cells from Fixed & Dissociated Tissues** | for Cells from FFPE Tissue Sections | |
| 500 - 2,000 | 11 | 15 | 14 - 15 | 16 |
| 2,000 - 4,000 | 10 | 14 | 13 - 14 | 15 |
| 4,000 - 7,000 | 9 | 13 | 12 - 13 | 14 |
| 7,000 - 12,000 | 8 | 12 | 11 - 12 | 13 |
| 12,000 - 25,000 | 7 | 11 | 10 - 11 | 12 |
| 25,000 - 50,000 | 6 | 10 | 9 - 10 | 11 |
| 50,000 - 128,000 | 5 | 9 | 8 - 9 | 10 |
*Optimization of cycle number may be needed based on the total RNA content of the sample. Theideal target library concentration is 50 - 200 nM. However, if the concentration is between 10-50nM or between 200-500 nM and if the libraries do not contain low or high molecular weight peaks,sequencing can still be performed. If optimization is needed, additional Amp Mix can be obtainedusing the Fixed RNA Feature Barcode Kit (PN-1000419). For dissociated tumor cells, cyclenumbers for cell lines can be used as a starting point. For dissociated primary cells, cycle numbersfor PBMCs can be used as a starting point.**For cells derived from the fixed and dissociated tissue samples, the cycle number will depend onthe RNA expression level of the tissue and on overall quality of the tissue prior to fixation.Additional optimization may be required.
Store at 4°C for ≤72 h, or proceed to the next step.
Vortex to resuspend the SPRIselect reagent. Add 100µL SPRIselect Reagent (1.0X) to each sample. Pipette mix 15x (pipette set to 180 μl).
Incubate 0h 5m 0s min at Room temperature.
Place on the magnet (high position) until the solution clears.
Remove the supernatant. DO NOT discard any beads.
With the tube still in the magnet, add 200µL 80% ethanol to the pellet. Wait 0h 0m 30s sec.
Remove the ethanol.
Repeat steps 38 and 38.1 for a total of 2 washes.
Centrifuge briefly and place on the magnet (low position).
Remove any remaining ethanol. DO NOT let the sample dry to ensure maximum elution efficiency.
Remove from the magnet. Add 41µL Buffer EB . Pipette mix 15x.
Incubate 0h 2m 0s min at Room temperature.
Place on the magnet (low position) until the solution clears.
Transfer 40µL to a new labeled DNA LoBind tube.
Make a 1:80 dilution of the library for quantification.
Store at 4°C for up to 72 h, -20°C or at -80°C for long-term storage.
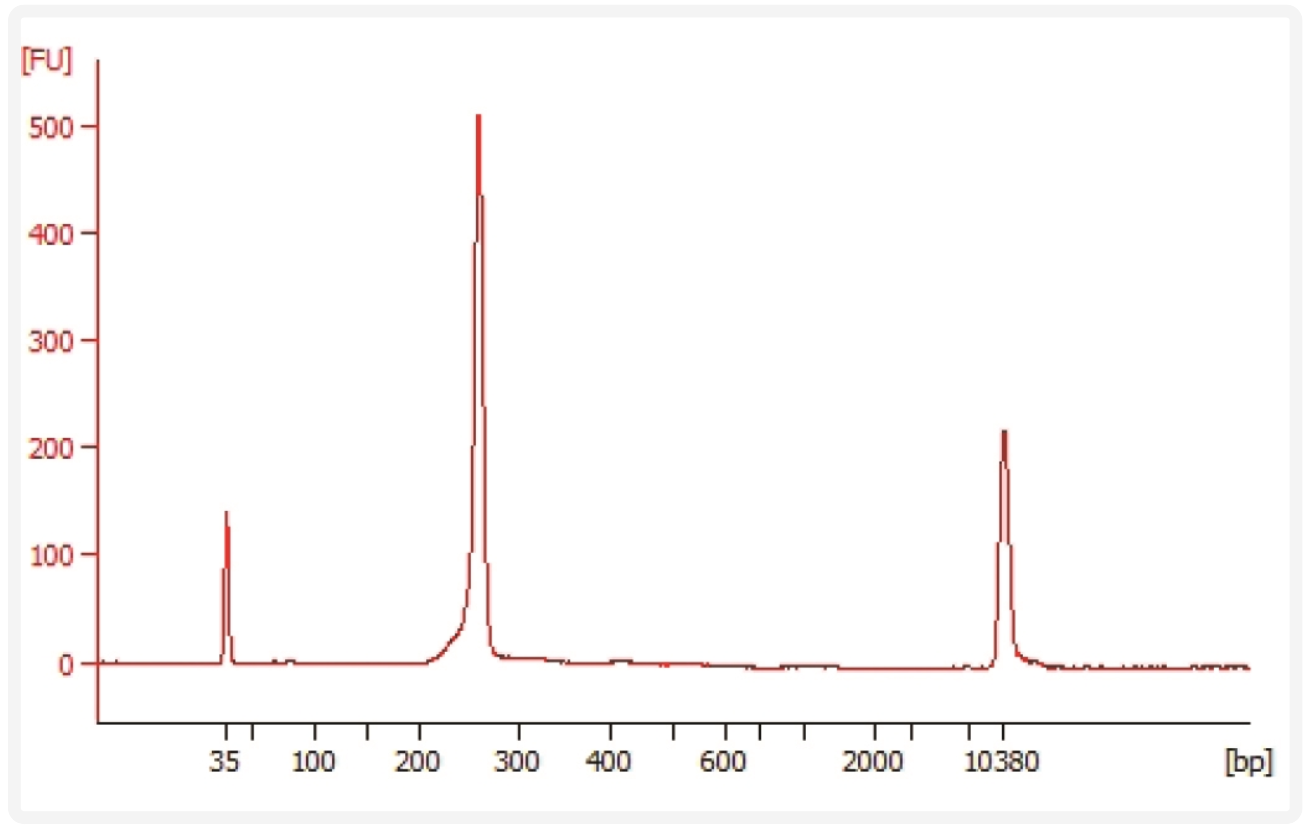
Run 1µL sample at 1:80 dilution on an Agilent Bioanalyzer High Sensitivity chip . Select the region between 150-300 bp to determine average size of the library. Consult the following protocol:
Sequencing
Consult the following protocol to send the libraries for sequencing: