Exploring Manually Curated Annotations of Intrinsically Disordered Proteins with DisProt
Federica Quaglia, Federica Quaglia, András Hatos, András Hatos, Edoardo Salladini, Edoardo Salladini, Damiano Piovesan, Damiano Piovesan, Silvio C. E. Tosatto, Silvio C. E. Tosatto
Abstract
DisProt is the major repository of manually curated data for intrinsically disordered proteins collected from the literature. Although lacking a stable three-dimensional structure under physiological conditions, intrinsically disordered proteins carry out a plethora of biological functions, some of them directly arising from their flexible nature. A growing number of scientific studies have been published during the last few decades to shed light on their unstructured state, their binding modes, and their functions. DisProt makes use of a team of expert biocurators to provide up-to-date annotations of intrinsically disordered proteins from the literature, making them available to the scientific community. Here we present a comprehensive description on how to use DisProt in different contexts and provide a detailed explanation of how to explore and interpret manually curated annotations of intrinsically disordered proteins. We describe how to search DisProt annotations, both using the web interface and the API for programmatic access. Finally, we explain how to visualize and interpret a DisProt entry, the SARS-CoV-2 Nucleoprotein, characterized by the presence of unstructured N-terminal and C-terminal regions and a flexible linker. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Performing a search in DisProt
Support Protocol 1 : Downloading options
Support Protocol 2 : Programmatic access with DisProt REST API
Basic Protocol 2 : Exploring the DisProt Ontology page
Basic Protocol 3 : Visualizing and interpreting DisProt entries–the SARS-CoV-2 Nucleoprotein use case
INTRODUCTION
Intrinsically disordered proteins (IDPs) are characterized by the presence of unstructured and highly flexible segments, termed “intrinsically disordered regions” (IDRs), that lack a stable three-dimensional structure. IDRs can be easily detected by several biophysical and biochemical methods, among which X-ray and NMR are the most commonly used (Tompa, 2010; van der Lee et al., 2014). Missing electron density regions that cannot be detected on X-ray crystal structures are due to unobserved atoms that fail to properly scatter X-rays, denoting their structural flexibility (Tompa, 2010, 201; Uversky & Dunker, 2010). NMR spectroscopy studies are also widely used to assess the presence of unstructured protein segments, being able to recognize disordered regions that in crystal structures are visible due to the formation of crystal contacts (Dyson & Wright, 2019). Several additional methods can assess the presence of intrinsic disorder in a protein, such as circular dichroism, sensitivity to proteolysis, and small-angle X-ray scattering (Kragelund and Skriver, 2020; Tompa, 2010).
Intrinsically disordered proteins can also exist as partially structured folding intermediates, pre-molten globules and molten globules, that exhibit a higher degree of secondary structure than random coils while being less compact than native structures (van der Lee et al., 2014). IDPs can play a crucial role in several biological processes, such as membrane localization and interaction with protein chaperones, to name a few (Uversky & Dunker, 2010). The lack of structure in IDR segments in their unbound state provides a multiplicity of advantages due to their largely extended conformation, such as: (1) the possibility for a single IDR to be involved in interactions with more structurally different partners; (2) several structured partners being able to bind to a single region; (3) the coupled folding and binding that give the ability for high specificity; and (4) a reduced binding strength that allows for transient interactions (Bugge et al., 2020; Dogan, Gianni, & Jemth, 2014). IDRs can undergo a disorder-to-order transition upon binding of a partner, enabling them to play a central role as protein hubs, as in the case of p53 (DisProt identifier: DP00086) and α-synuclein (DisProt identifier: DP00070), or as targets of a structured hub, e.g., TAZ and KIX (Cumberworth, Lamour, Babu, & Gsponer, 2013; Dosztányi, Chen, Dunker, Simon, & Tompa, 2006; Oldfield et al., 2008; Wright & Dyson, 2015). Finally, IDPs can also be involved in the regulation of several biological processes, interacting with different types of binding partners such as proteins, nucleic acids, lipids, and small molecules (Tompa, 2005; van der Lee et al., 2014). Strikingly, some of the most well characterized and crucial functions of IDPs arise from their flexible nature: they can be flexible linkers connecting structured domains of a protein, or they can act as entropic clocks, bristles, and springs due to their entropic features (Uversky & Dunker, 2010; van der Lee et al., 2014).
DisProt is a service of the Italian node of ELIXIR, the European infrastructure for biological data, and a key resource for the recently established ELIXIR IDP user community (Davey et al., 2019). It is also the largest repository of manually curated annotations of intrinsically disordered proteins (IDPs) collected from the literature (Hatos et al., 2020; Piovesan et al., 2017; Quaglia et al., 2022a). A team of expert DisProt curators looks for new data on IDPs/IDRs from relevant publications and annotates them through a dedicated curation interface by means of intrinsic disorder–related annotation terms. DisProt relies on three different ontologies to annotate intrinsically disordered regions: the Intrinsically Disordered Proteins Ontology (IDPO), the Gene Ontology (GO), and the Evidence and Conclusion Ontology (ECO). IDPO is used to describe structural aspects of an IDP/IDR, self-functions and functions directly associated with their disordered state. Gene Ontology (Ashburner et al., 2000; Gene Ontology Consortium, 2021) is used to describe functional aspects of an IDP/IDR. The Evidence and Conclusion Ontology (Nadendla et al., 2022) describes the technique associated with an annotation. A DisProt entry corresponds to a protein isoform and unambiguously maps to a UniProt entry. DisProt annotations describe local properties of the protein sequence (e.g., intrinsically disordered regions), which are always supported by experimental evidence taken from the literature. Each DisProt annotation is uniquely identified by the DisProt entry accession number followed by a suffix starting with a lowercase letter r (example DP00086r003).
In this article, we provide detailed protocols explaining how to perform a search in DisProt (Basic Protocol 1), explore the ontologies used in DisProt (Basic Protocol 2), and visualize and interpret annotations of a DisProt entry (Basic Protocol 3). We also describe the downloading options in DisProt (Support Protocol 1) and programmatic access with the DisProt REST API (Support Protocol 2).
Basic Protocol 1: PERFORMING A SEARCH IN DisProt
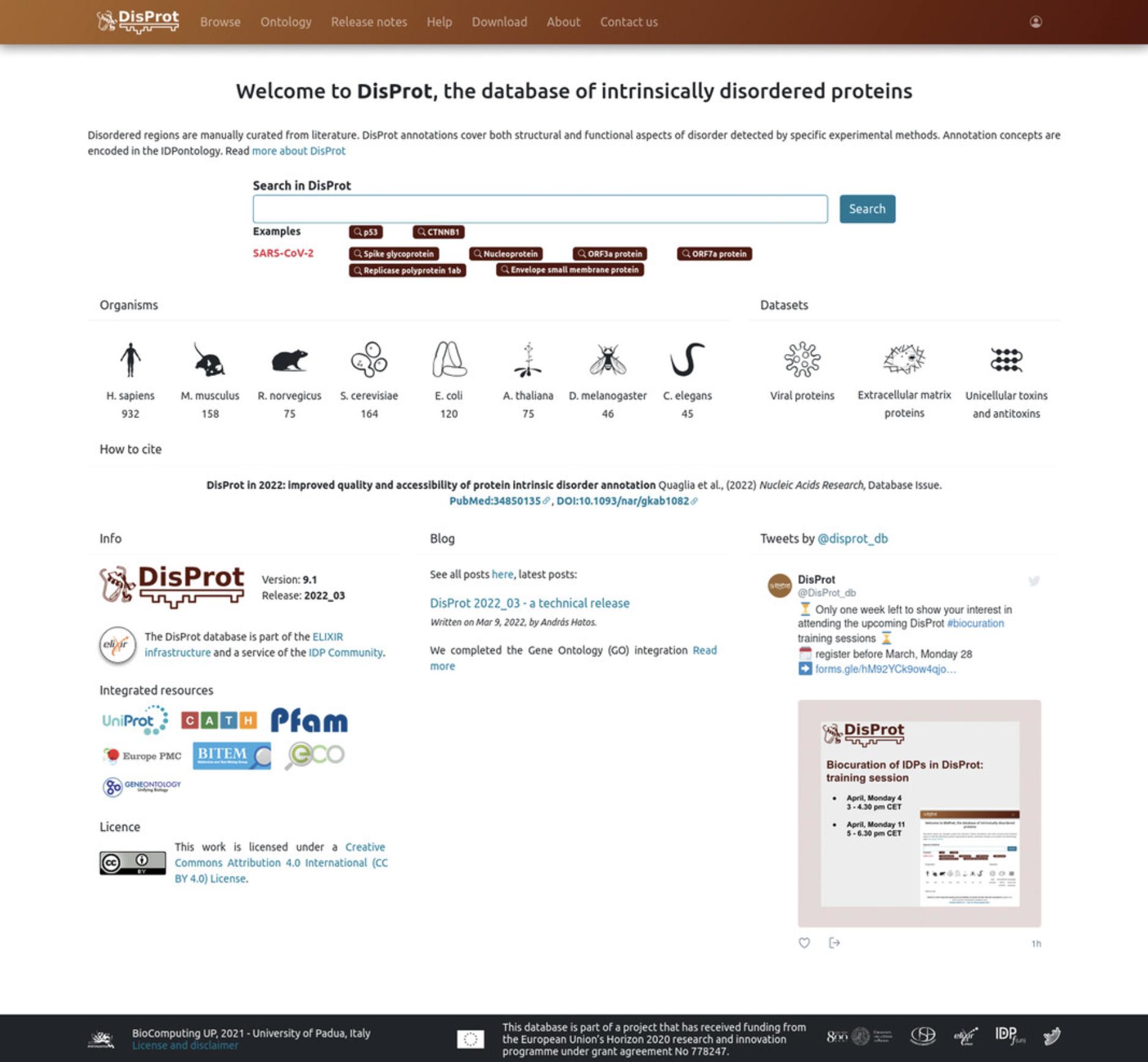
DisProt is freely accessible at https://disprot.org/. This protocol describes how to search entries and to retrieve information in DisProt. From the home page, users can also navigate the DisProt blog (https://disprot.org/blog) to read posts describing our updates or explore the DisProt Twitter account (https://twitter.com/disprot_db) (Fig. 1).
Necessary Resources
Hardware
- While DisProt works best on laptop or desktop computers, it is also easily accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser, e.g., Firefox (http://www.mozilla.org/firefox), Google Chrome (http://www.google.com/chrome), or Safari (http://www.apple.com/safari)
Input data
Free text search against the database
Performing a text search
1.Open a web browser and connect to DisProt at https://disprot.org/.
2.Searches in DisProt can be performed either using the “Search” box on the top-middle of the DisProt home page, or by clicking on the “Browse” button available on the top-left of the home page.
- Users can perform a search using the “Search” box on the top-middle of the DisProt home page to look for protein entries or entries referencing a specific publication.
Users can look for specific proteins, e.g., nucleoproteins, by typing the protein nameNucleoprotein. Users will be redirected to a list of all the nucleoprotein entries available in DisProt, e.g., Nucleoprotein from Measles virus (DisProt entry: DP00640).
Users might also be interested in looking for a specific publication. In this case, enter the corresponding PubMed identifier (PMID) of the publication in the search box. All entries that have at least one evidence referencing that publication will be displayed.
-
Alternatively, it is possible to perform an advanced search by clicking on the “Browse” button available on the top-left of the home page. Users will be redirected to an advanced search page, where they can refine their search and look for a specific query or a combination of them (Fig.2), e.g., a protein name and an organism.
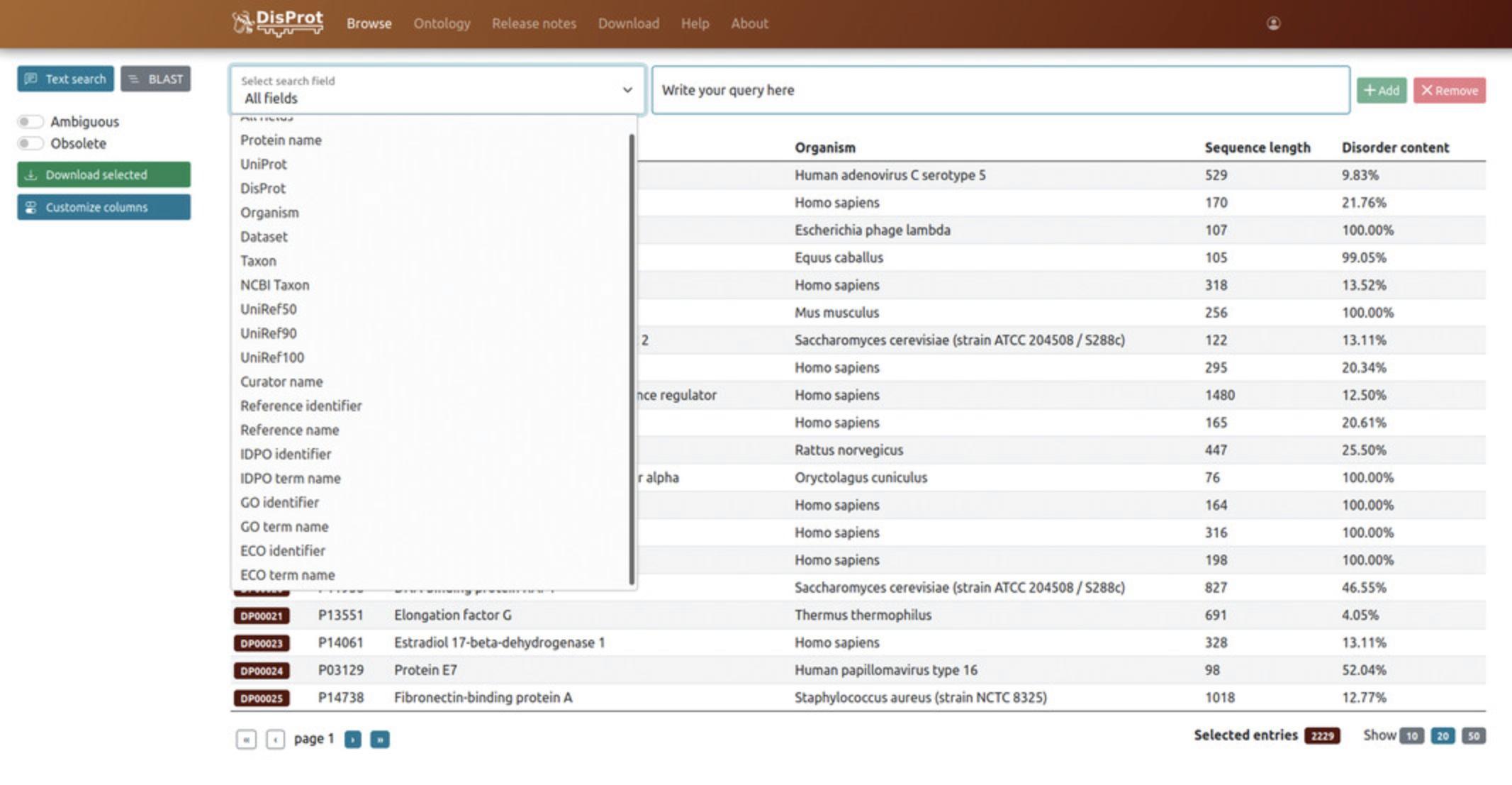
3.Select “Text search” on the top-left side of the Browse page, then select a term from the drop-down menu.
Users can look for the following aspects:
-
A specific protein: select a “Protein name”, e.g., Nucleoprotein, or “UniProt”, e.g., P0DTC9.
-
A specific DisProt entry: select “DisProt”, e.g., DP03212.
-
A set of proteins from a specific organism: choose an “Organism”, e.g., “Gallus”, the “Taxon”, or “NCBI Taxon”.
-
UniProt Reference Clusters (UniRef). UniRef databases cluster UniProtKB sequences by gathering together proteins based on their sequence similarity (Suzek, Wang, Huang, McGarvey, & Wu,2015). Terms available are “UniRef50”, “UniRef90”, and “UniRef100” (clustering the sequences at 50%, 90% and 100% identity, respectively).
-
Entries from a specific curator: select the “Curator name” term and start typing the name you are looking for.
-
A specific reference: users can look for a specific PMID, e.g., 8632448, by selecting the “Reference identifier” term or for the title of the corresponding publication, e.g., “Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein”, by selecting the “Reference name” term.
-
A specific term from the ontologies adopted in DisProt:An IDPO term: select a “IDPO identifier”, e.g., “flexible linker/spacer”, and “IDPO term name”, e.g., IDPO:00502.A Gene Ontology (GO) term: select a “GO identifier”, e.g., “modulation by virus of host cell cycle”, or the “GO term name”, e.g., GO:0060153. An Evidence and Conclusion Ontology (ECO) term: select a “ECO identifier”, e.g., “modulation by virus of host cell cycle”, or the “ECO term name”, e.g., ECO:0006163.
Users that wish to have a better insight on the terms of our ontology and read their descriptions can refer to the Ontology page available athttps://disprot.org/ontology.
-
Entries from a specific dataset: select “Dataset”, e.g., “Viral proteins”.
-
It is also possible to perform a free text search by selecting the “all fields” term in the drop-down menu.
4.It is possible to customize the table columns to visualize more details of an entry in the displayed results. Default columns include “ DisProt ID ”, “ UniProt Accession ”, “ Protein Name ”, “ Organism ”, “Sequence length”, and “ Disorder content ”. We suggest adding at least the “ annotated terms ” column to have an insight on the disorder aspects available for each entry.
5.Download the search results using the “Download selected” button at the top-left of the Browse page. Users can also choose to include ambiguous and/or obsolete entries by selecting the corresponding buttons above “Download selected”.
-
Select the type of pieces of evidence you want to download among: structural state (IDPO), structural transition (IDPO), disorder function (IDPO), molecular function (GO), biological process (GO), or cellular component (GO)
-
Select the type of desired data, i.e., “regions” or “consensus”.
-
Select the file format. Available options for download are JSON, TSV, FASTA, and GAF.
Performing a sequence similarity search
6.Open a web browser and connect to DisProt at https://disprot.org/.
7.Click on the “Browse” button on the top-left side of the home page (Fig. 3) to be redirected to the advanced search page.
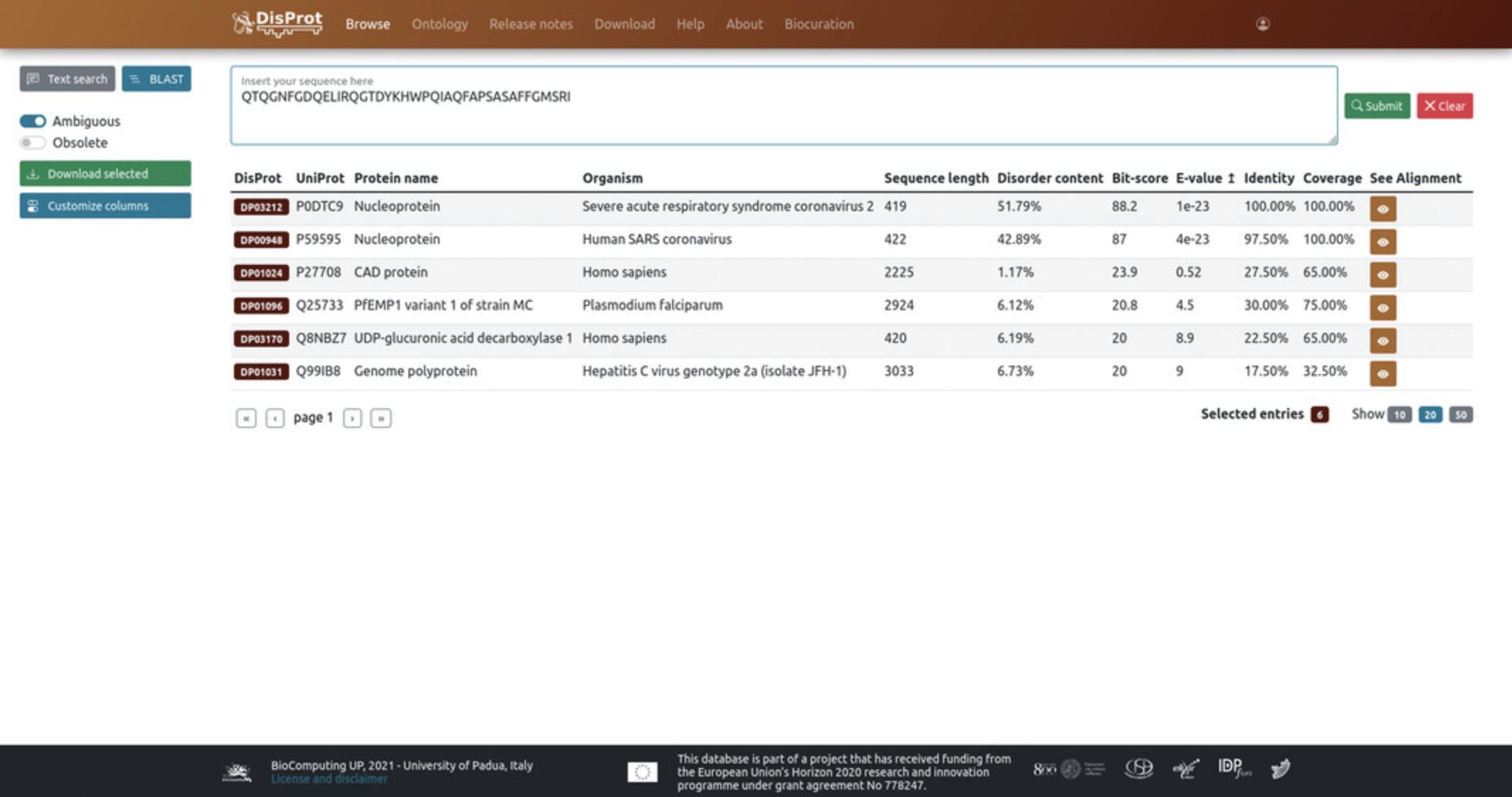
8.Select “BLAST” on the top-left side of the Browse page to perform a BLAST (Altschul, Gish, Miller, Myers, & Lipman, 1990) sequence similarity search against DisProt entries.
9.Insert a protein sequence in the corresponding box and click on “Submit”.
10.It is possible to customize the table columns to visualize more details of an entry in the displayed results. Default columns include “ DisProt ”, “ UniProt ”, “ Protein name ”, “ Organism ”, “ Sequence length ”, and “ Disorder content ” along with “ Bit-score ”, “ E-value ”, “ Identity ”, and “ Coverage ”.
11.Click on “ See alignment ” to visualize where the query and the subject sequences align.
12.Download the search results using the “Download selected” button at the top-left of the Browse page. Users can also choose to include ambiguous and/or obsolete entries by selecting the corresponding buttons above “Download selected”.
-
Select the type of pieces of evidence you want to download among: structural state (IDPO), structural transition (IDPO), disorder function (IDPO), molecular function (GO), biological process (GO), and cellular component (GO).
-
Select the type of desired data, i.e., “regions” or “consensus”.
-
Select the file format. Available options for download are JSON, TSV, FASTA, and GAF.
Support Protocol 1: DOWNLOADING OPTIONS
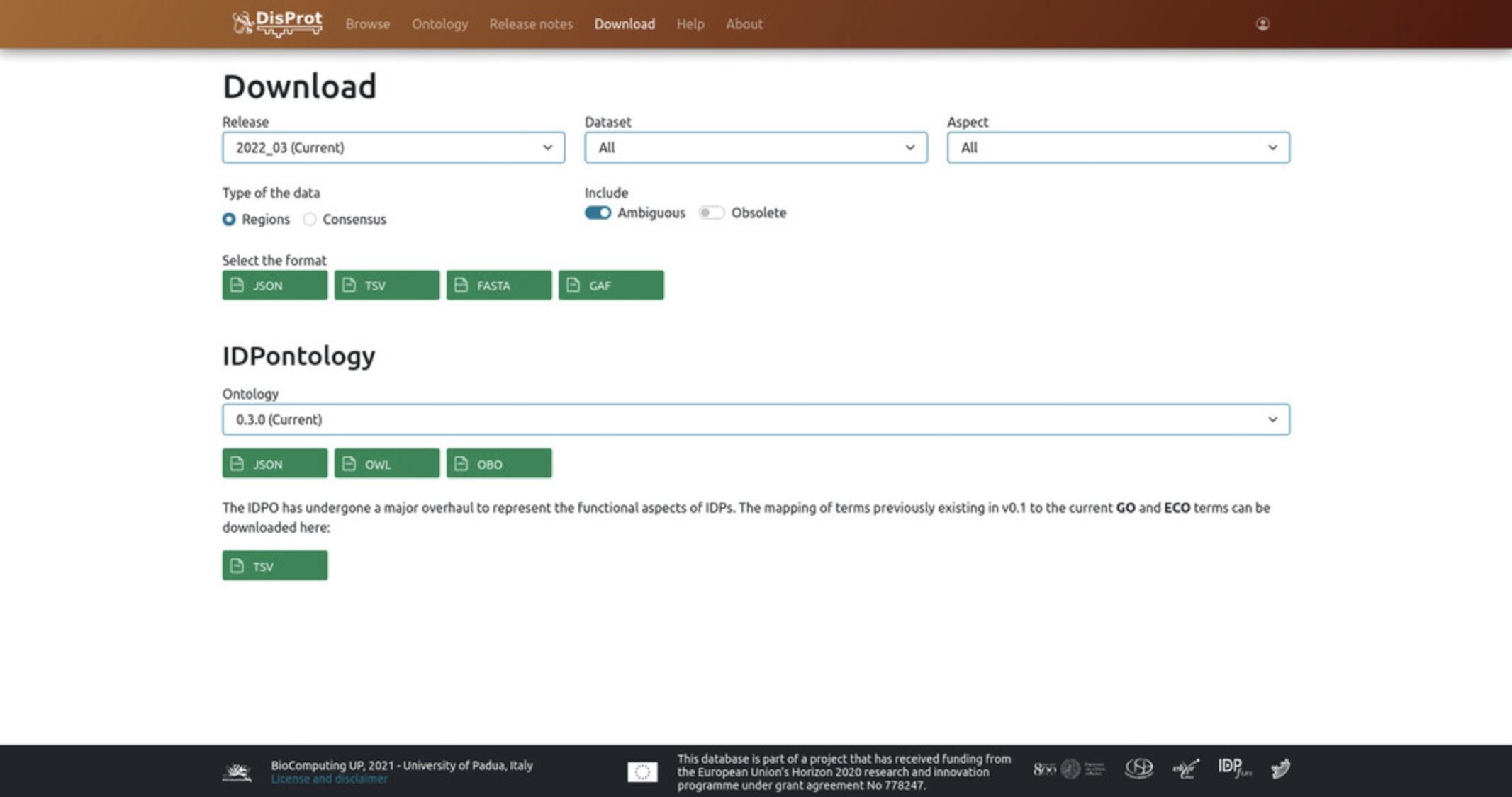
From the DisProt “Download” page (https://disprot.org/download), users can download a specific release of the database, datasets and annotated aspects, or a specific version of the IDP ontology (Fig. 4).
Necessary Resources
Hardware
- While DisProt works best on laptop or desktop computers, it is also easily accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser, e.g., Firefox (http://www.mozilla.org/firefox), Google Chrome (http://www.google.com/chrome), or Safari (http://www.apple.com/safari)
Input data
No input data are required
Downloading a release, a dataset, or a specific ontology aspect of DisProt
1.Open a web browser and connect to DisProt at https://disprot.org/.
2.Click on the “Download” button on the top bar of DisProt (https://disprot.org/download).
3.Users can select the DisProt data they want to download, by using the dedicated drop-down menus, i.e.:
-
Release: a specific release of the database, e.g., “2021_12”.
-
Dataset: a specific thematic dataset, e.g., “viral proteins”.
-
Aspect: a specific ontology aspect annotated in DisProt, e.g., “Structural state (IDPO)” or “Molecular Function (GO)”.
4.Users can choose the type of data they want to download, i.e., “regions” or “consensus”. It is possible to include ambiguous and/or obsoleted regions by selecting them from the “Include” options; otherwise leave the corresponding boxes unchecked.
5.Select the format of the output file. Available options for download are JSON, TSV, FASTA, and GAF formats.
Downloading the IDP ontology
6.Open a web browser and connect to DisProt at https://disprot.org/.
7.Click on the “Download” button on the top bar of DisProt (https://disprot.org/download).
8.Users can select a version of the ontology they are interested in, e.g., 0.3.0 (Current) , from the “Ontology” drop-down menu.
9.Select the format of the output file. Available options for download are JSON, OWL, and OBO formats. OBO and OWL formats correspond to the Biomedical Ontology and Web Ontology Language, respectively.
Support Protocol 2: PROGRAMMATIC ACCESS WITH DisProt REST API
DisProt can be accessed programmatically via REST API to retrieve a single entry (or region) and to perform large-scale database searches. DisProt API documentation (https://disprot.org/api) is available as a Swagger representation that follows OpenAPI specifications.
All API endpoints are available from https://disprot.org/api/{endpoint_name}. In this support protocol we introduce three different endpoints—the first one can be used to retrieve a single entry, the other two to search entries in the database.
Necessary Resources
Hardware
Laptop or desktop computer. An active and stable internet connection is required.
Software
Python 3.5+, Requests Python library 2.23+
Input data
No input data are required
1.Get a single entity.
Users can retrieve a single entity, i.e., a protein entry or one of its manually curated regions, by using its corresponding identifier. The following syntax must be used to retrieve a single entity from DisProt disprot.org/api/{identifier}, where the “identifier” must be a valid DisProt ID, DisProt region ID, or UniProt accession. The query is customizable with various parameters, e.g., file format and release. Here we provide two pieces of code to retrieve a single entry in JSON format written on the standard output (Sample code 1) and write a file in FASTA format (Sample code 2). In Sample code 2 the API version of DisProt is also specified.
Sample code 1
-
#!/usr/bin/env python3
-
import requests
-
disprot_id = "DP00086"
-
url = "https://disprot.org/api/" + disprot_id
-
resp_json = requests.get(url).json()
-
print(resp_json)
Sample code 2
-
#!/usr/bin/env python3
-
import requests
-
disprot_id = "DP00086"
-
params = {
- 'format':'fasta',
-
}
-
url = "https://disprot.org/api/" + disprot_id
-
resp_fasta = requests.get(url, params=params).text
-
f = open("DP00086.fasta", "w")
-
f.write(resp_fasta)
-
f.close()
2.Results.
DisProt currently provides three output formats: JSON (default), FASTA, and TSV. Due to the inherent limitations of the FASTA and TSV file formats, the JSON format renders the most comprehensive description of intrinsic disorder. The TSV and FASTA files provide details about regions or different types of consensus.
Searching entries in DisProt database
3.Performing a text search.
DisProt provides an extensively customizable search engine. It is possible to perform a free text search or formulate complex queries against combined fields, e.g., organism and UniRef50. The search query is sent to https://disprot.org/api/search with URL parameters. Note that whitespace and other special characters must be converted into a valid ASCII format; the space is usually replaced with %20. Multiple search fields can be combined in the same query by joining them with an AND operator (& symbol), e.g., disprot.org/api/search?organism=homo%20sapiens&name=kinase returns all the human proteins with “kinase” in the protein name. Given that some fields are interpreted as regular expressions, it is also possible to use the OR operator (| symbol). This is the case with the following query, e.g., https://disprot.org/api/search?organism=homo%20sapiens|mus%20musculus, which returns both human and mouse entries. The user can choose to customize the output format. Currently available output formats are JSON, FASTA, and TSV. By default, the endpoint returns the results in JSON; however, users can select another format using the “format” field in the parameters or headers. It is possible to use an older version of the API for legacy reasons by specifying accept-version in the URL header of a request. By default, the server responds with the latest version of the API.
Sample code
-
#!/usr/bin/env python3
-
import requests
-
parameters= {
- 'ncbi_taxon_id':'9606',
- 'format':'tsv'
-
}
-
url = "https://disprot.org/api/search"
-
resp_tsv = requests.get(url, params=parameters).text
-
print(resp_tsv)
4.Results.
DisProt returns an object with “data” and “size” fields. “Data” contains a list of entries, and these entry objects are the same described in the previous section. “Size” corresponds to the number of matched entries. Note that when the pagination parameters are provided, only the data field is affected, whereas the size field always refers to the full query result.
Sequence Similarity Searches in DisProt Database
5.Performing a sequence similarity search.
The users can also perform a BLAST sequence similarity search against the database with a POST request to https://disprot.org/api/blast.
Sample code
-
#!/usr/bin/env python3
-
import requests
-
data = {
- 'seq':'KKPLDGEYFTLQIRGRERFEMFRELNEALELKDAQAGKEPGGSRAHSSHLKSKKGQSTSR',
-
}
-
url = "https://disprot.org/api/blast"
-
resp_json = requests.post(url, data=data).json()
-
print(resp_json)
6.Results.
The output provided is the same as that available for the text search described above, i.e., JSON (by default), TSV, or FASTA. In addition, DisProt returns the corresponding “Bit-score”, “E-value”, “Identity”, and “Coverage”, as provided by BLAST.
Basic Protocol 2: EXPLORING THE DisProt ONTOLOGY PAGE
The ontologies adopted in DisProt are available at https://disprot.org/ontology. DisProt relies on three ontologies to provide structured annotations of IDRs: (i) the IDPontology (IDPO) to describe structural states, transitions, and disorder-associated functions, (ii) the Gene Ontology (GO) to describe functional aspects of an IDP/IDR, (iii) the Evidence and Conclusion Ontology (ECO) to describe the methods used to assess the presence of disorder or one of its associated aspects. From the Ontology page (Fig. 5), users can explore the available ontology terms used in DisProt.

Necessary Resources
Hardware
- While DisProt works best on laptop or desktop computers, it is also easily accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser, e.g., Firefox (http://www.mozilla.org/firefox), Google Chrome (http://www.google.com/chrome), or Safari (http://www.apple.com/safari)
Input data
No input data are required
Exploring the IDPontology terms
1.Open a web browser and connect to DisProt at https://disprot.org/.
2.Click on the “Ontology” button on the top bar of DisProt (https://disprot.org/ontology).
3.The Intrinsically Disordered Proteins Ontology (IDPO) features three disorder-related branches, i.e., structural state, structural transitions, and disorder function.
4.Users can explore the IDPO terms by using the filter option or, alternatively, by opening the ontology branch of interest and looking for the available child terms.
5.By typing in the “Filter” box an IDPO term, such as flexible linker/spacer and hitting the “Search” button, users will visualize the definition of the term and its parent term entropic chain.
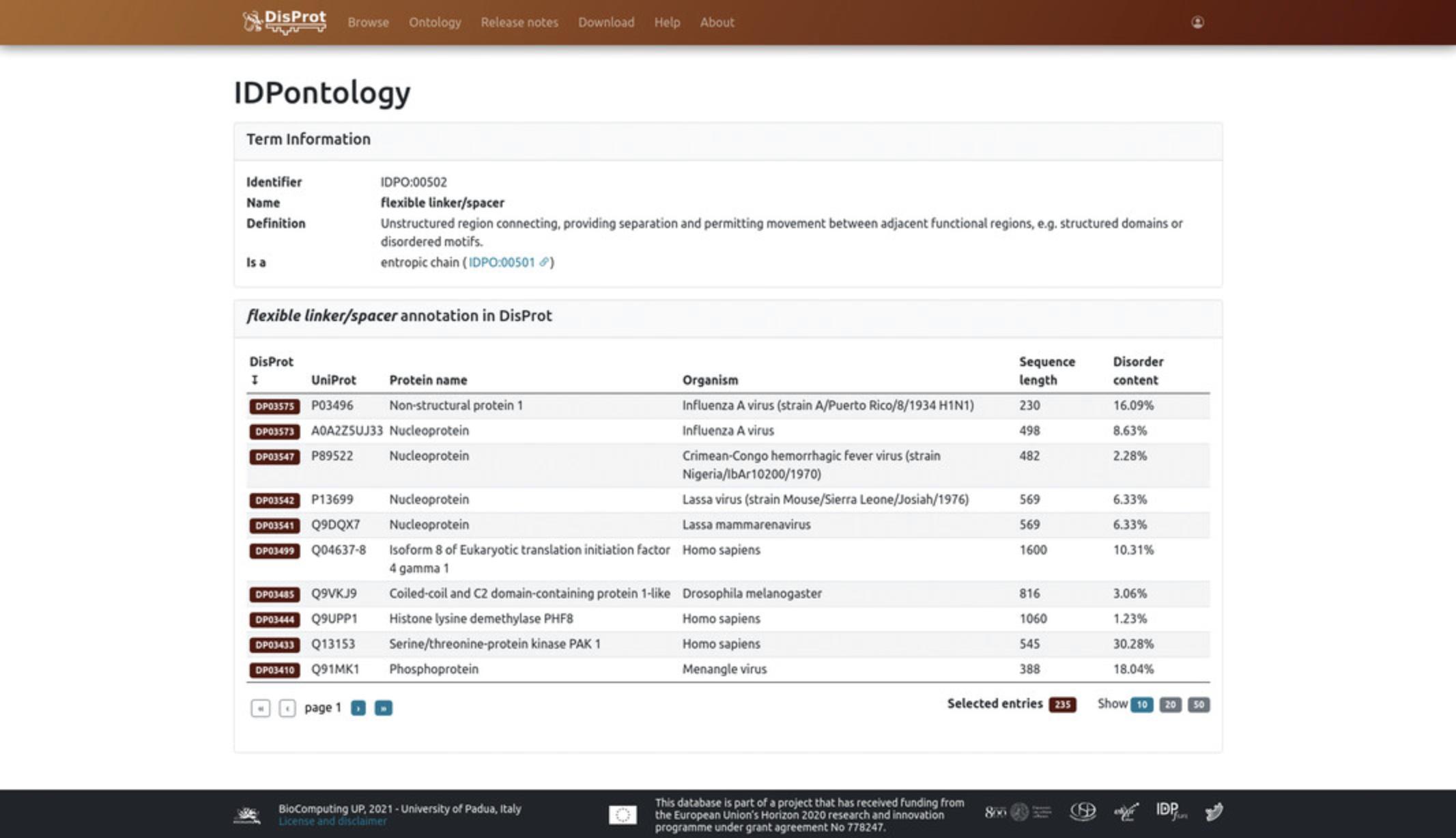
6.Clicking on the IDPO identifier in brackets, users will be redirected to the dedicated IDPontology page of that term (https://disprot.org/idpo/IDPO:00502). Each IDPontology page is divided in two sections (Fig. 6):
-
The first section describes the information about that specific term, i.e.,Identifier(IDPO:00502),Name(flexible linker/spacer),Definition(unstructured region connecting, providing separation, and permitting movement between adjacent functional regions, e.g., structured domains or disordered motifs), and its parent term,Is a(entropic chain, IDPO:00501).
-
The second section lists all the available DisProt entries with at least one piece of evidence annotated using that term, e.g., DP00018.
Exploring the Gene Ontology (GO) terms
7.Open a web browser and connect to DisProt at https://disprot.org/.
8.Click on the “Ontology” button on the top bar of DisProt (https://disprot.org/ontology).
9.Users can explore all the GO terms, classified into one of the three aspects—i.e., Molecular Function, Biological Process, and Cellular Component—by clicking on the AmiGO 2 (http://amigo.geneontology.org/amigo; Carbon et al., 2009) or on the QuickGO (https://www.ebi.ac.uk/QuickGO/; Binns et al., 2009) tools linked.
Exploring the Evidence and Conclusion Ontology (ECO) terms
10.Open a web browser and connect to DisProt at https://disprot.org/.
11.Click on the “Ontology” button on the top bar of DisProt (https://disprot.org/ontology).
12.Users can explore all the ECO terms available for annotation in DisProt, along with their child terms, i.e.:
-
Author inference used in manual assertion (ECO:0006216): A type of author inference that is used in a manual assertion.
-
Author statement used in manual assertion (ECO:0000302): A type of author statement that is used in a manual assertion.
-
Combinatorial evidence used in manual assertion (ECO:0000244): A type of combinatorial analysis that is used in a manual assertion.
-
Combinatorial experimental and curator inference evidence used in manual assertion (ECO:0007014): A type of combinatorial evidence from curator knowledge and experimental evidence that is used in a manual assertion.
-
Curator inference used in manual assertion (ECO:0000305): A type of curator inference that is used in a manual assertion.
-
Experimental evidence used in manual assertion (ECO:0000269): A type of experimental evidence that is used in a manual assertion.
13.By typing in the “Filter” box the technique of interest, such as circular dichroism and hitting the “Search” button, users will visualize all the available terms along with the parent term, e.g.:
- a.Circular dichroism evidence used in manual assertion (ECO:0006200): A type of circular dichroism evidence that is used in a manual assertion.
- i.Far-UV circular dichroism evidence used in manual assertion (ECO:0006204): A type of far-UV circular dichroism evidence that is used in a manual assertion.
- ii.Near-UV circular dichroism evidence used in manual assertion (ECO:0006206): A type of near-UV circular dichroism evidence that is used in a manual assertion.
- iii.Synchrotron radiation circular dichroism evidence used in manual assertion (ECO:0006202): A type of synchrotron radiation circular dichroism evidence that is used in a manual assertion
Basic Protocol 3: VISUALIZING AND INTERPRETING DisProt ENTRIES–THE SARS-CoV-2 NUCLEOPROTEIN USE CASE
Here, we present a use case, the SARS-CoV-2 Nucleoprotein (DisProt entry: DP03212), to explain how to visualize and interpret a DisProt entry page and its annotations. The SARS-CoV-2 Nucleoprotein entry, also shown among the SARS-CoV-2 home page examples, has been recently released (DisProt release 2021_12) and currently includes more than 30 pieces of evidence annotated from nine scientific articles. The SARS-CoV-2 Nucleoprotein is characterized by the presence of three intrinsically disordered regions, i.e., the N- and C-termini (Cubuk et al., 2021; Schiavina, Pontoriero, Uversky, Felli, & Pierattelli, 2021) and a flexible linker that connects the RNA-binding domain (RBD) with the dimerization domain (Cubuk et al., 2021; Schiavina et al., 2021). The N-terminal IDR plays a role in phase separation (Perdikari et al., 2020), and a deletion of the flexible linker has been associated with a reduction of turbidity and of LLPS-associated droplet formation (Perdikari et al., 2020), while the C-terminus appears to be involved in droplet formation and in contributing to the protein RNA-binding activity (Wu et al., 2021). Overall, up to 50% of the SARS-CoV-2 Nucleoprotein consists of disordered regions. Interestingly, Nucleoprotein mutation hotspots cluster in disordered regions: 89% of mutations occurring in the 12 major variants of SARS-CoV-2 map to these IDRs, while in the Omicron variant and its lineages (BA.1 and BA.2), all the mutated positions localize in unstructured regions (Quaglia et al., 2022b).
DisProt entries are annotated by biocurators that collect all experimental evidence related to disorder available from a publication. In DisProt, an entry corresponds to a protein isoform, and each IDR annotation is an evidence about its flexible nature or function. The minimal information required to annotate a region in DisProt include reference to the publication (PMID or a DOI); the boundaries of the region (start and end position on the amino acid sequence); the Evidence and Conclusion Ontology (ECO) term that defines the experimental technique and the type of information, i.e., an IDPontology term (structural state , structural transition , disorder-derived functions); or a Gene Ontology term defining a molecular function, biological process, or cellular component associated with the annotated IDRs. To support annotations, curators report authors’ statements as snippets of text from the corresponding publication. Finally, a selected team of reviewers carefully check all annotations, to ensure a high-quality standard. Each entry page consists of two main sections. The first provides information about the protein and includes a feature viewer to visualize DisProt region annotations on the sequence. The second section lists all annotations in a tabular format.
Necessary Resources
Hardware
- While DisProt works best on laptop or desktop computers, it is also easily accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser, e.g., Firefox (http://www.mozilla.org/firefox), Google Chrome (http://www.google.com/chrome), Safari (http://www.apple.com/safari)
Input data
No input data are required
1.Select the “Nucleoprotein” example, DP03212, from the DisProt home page (https://disprot.org/).
2.On the top of each entry page, the following details are available: the DisProt identifier (DP03212) and protein name (Nucleoprotein); organism (Severe acute respiratory syndrome coronavirus 2); sequence length (419); disorder content (51.8%); and cross references with other databases, MobiDB (Piovesan et al., 2018) and UniProt (UniProt Consortium, 2019; UniProt accession code: P0DTC9).
3.Users can select the release they want to visualize from the history of the entry by clicking the “Entry history” button on the top-right of the entry page.
4.The feature viewer, which can be expanded and collapsed, allows users to visualize regions’ annotations on the sequence (Fig. 7). By default, two tracks are shown, the first showing DisProt annotations and the other including domain data as defined by Pfam (El-Gebali et al., 2019), which provides conserved domain families, and Gene3D (Lewis et al., 2018), which provides globular domains. It is possible to expand the feature viewer to visualize the sub-tracks and each disorder evidence annotated for a specific structural or functional aspect. By hovering each region on the sequence viewer, a tooltip provides additional information such as annotated terms, identifiers, cross-references, the name of the curator who annotated the region, the experimental method, and the reference supporting that annotation.
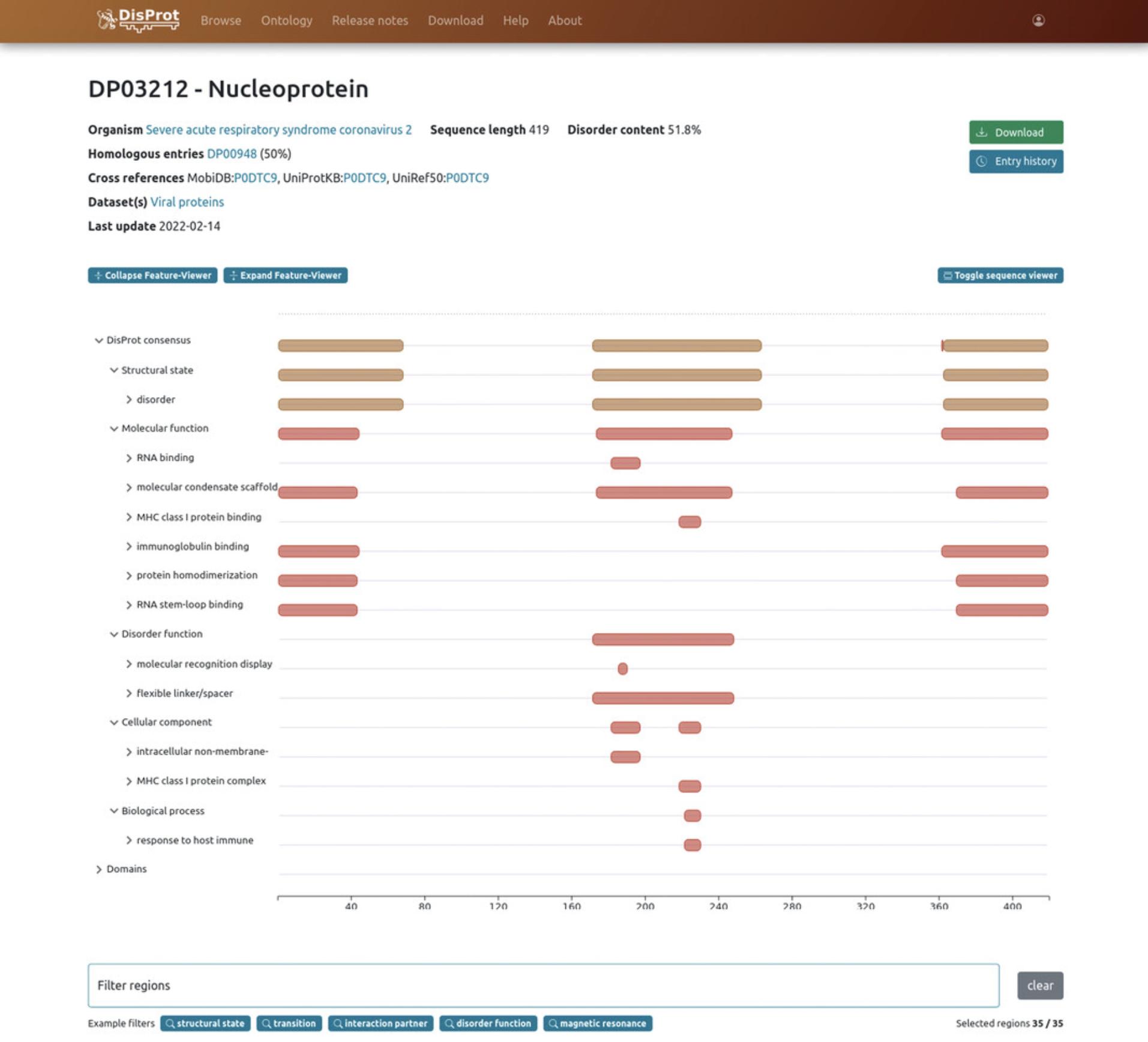
5.Users can open (“toggle”) the sequence viewer, which dynamically highlights amino acids of the selected IDR directly on the protein sequence.
6.It is also possible to select a subset of annotations using the “Filter” box under the sequence viewer.

GUIDELINES FOR UNDERSTANDING RESULTS
In DisProt, a team of expert professional and community biocurators manually annotates experimental intrinsic disorder data from peer-reviewed publications. Each DisProt entry corresponds to a UniProt entry, i.e., the canonical sequence or one of its isoforms. An entry consists of a set of manually curated intrinsically disordered regions—each one of them is an evidence , together with all the information about its flexible nature and other associated aspects. The minimal information included in the evidence is the reference (PMID or DOI) to a scientific publication, the (ECO) term that defines the experimental technique used to detect the annotated aspect, the start and end positions of the region, and a disorder aspect associated with the IDR. The aspects annotated in DisProt cover the main features of an IDR: the structural state and the structural transition, along with disorder-related functions–defined by IDPontology terms–and Gene Ontology–derived functions. Curators also add statements , i.e., sentences from the publication that support the disordered nature of the region or one of its aspects, to provide the users with an exhaustive description of each protein region. Additional information useful to unambiguously characterize a disorder-related experiment can be annotated using the MIADE (Minimum Information About Disorder Experiments) standard. A standardized curation effort is one of the main goals of DisProt. In line with this, DisProt curators benefit from a regularly updated curation manual describing in detail the DisProt curation process, along with dedicated training sessions. Researchers interested in contributing to DisProt, whether by volunteering in curation or by sharing articles from their research groups, can find detailed information in the DisProt Biocuration page (https://disprot.org/biocuration).
COMMENTARY
Background Information
The DisProt database is the main resource for manually curated annotations of Intrinsically Disordered Proteins (IDPs) and regions (IDRs) from literature. The database features more than 2300 annotated entries, each one of them corresponding to a UniProt accession (Suzek et al., 2015).
The database includes not only data about disordered regions, but also information on their state transitions, functions, and interactions with other proteins, nucleic acids, and small molecules. In addition, DisProt holds specific information on the experimental setup and conditions by implementing the Minimum Information about a Disorder Experiment (MIADE) guidelines. To improve interoperability, DisProt now relies on two ontologies, the Evidence and Conclusion Ontology (Nadendla et al., 2022) and the Gene Ontology (Ashburner et al., 2000; Gene Ontology Consortium, 2021).
Critical Parameters
Each DisProt identifier is mapped to a specific UniProt accession number. Please keep in mind that the DisProt identifier can refer to an isoform of the protein—e.g., the DisProt entry DP02025 is mapped to the canonical isoform of Cell death protein 4 (UniProt accession: P30429), while DP03045 is associated with the second isoform of Cell death protein 4 (UniProt accession: P30429-2). DisProt also provides the corresponding protein amino acid sequence. However, given the fact that the updating of DisProt is not synchronized with the UniProt releases, the user may experience differences between the sequences in both resources. For any application, we recommend that users compare the sequences (or the checksums) to verify the synchronicity and the boundaries of IDR regions.
Moreover, polyproteins in UniProt are provided with a single UniProt accession. Similarly, each polyprotein corresponds to a specific DisProt entry. This may cause issues when interpreting data such as the “disorder content”, as users should consider that the disorder content refers to the whole polyprotein sequence and not to the single smaller proteins that compose the polyprotein.
Troubleshooting
Potential errors that may arise with the basic and support protocols described in this article can be addressed at the email address available on the “About” page (https://disprot.org/about) under the “Contact Us” section. Finally, database documentation is provided on the DisProt “Help” page (https://disprot.org/help).
Acknowledgments
DisProt is a service of the Italian node of ELIXIR, the research infrastructure for life-science data. This paper is part of a project that has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No 778247. Funding from the Italian Ministry of University and Research (MIUR), PRIN No. 2017483NH8.
Open Access Funding provided by Universita degli Studi di Padova within the CRUI-CARE Agreement.
Author Contribution
Federica Quaglia : conceptualization, data curation, formal analysis, writing original draft; András Hatos : formal analysis, software, writing original draft; Edoardo Salladini : data curation, formal analysis, writing original draft; Damiano Piovesan : supervision, writing original draft; Silvio Tosatto : supervision, writing original draft, funding acquisition, project administration.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are openly available in DisProt at https://disprot.org/.
Literature Cited
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology , 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
- Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., … Sherlock, G. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics , 25, 25–29. doi: 10.1038/75556
- Binns, D., Dimmer, E., Huntley, R., Barrell, D., O'Donovan, C., & Apweiler, R. (2009). QuickGO: A web-based tool for gene ontology searching. Bioinformatics , 25, 3045–3046. doi: 10.1093/bioinformatics/btp536
- Bugge, K., Brakti, I., Fernandes, C. B., Dreier, J. E., Lundsgaard, J. E., Olsen, J. G., … Kragelund, B. B. (2020). Interactions by disorder–a matter of context. Frontiers in Molecular Biosciences , 7, 110. doi: 10.3389/fmolb.2020.00110
- Carbon, S., Ireland, A., Mungall, C. J., Shu, S., Marshall, B., Lewis, S., & AmiGO Hub, and Web Presence Working Group. (2009). AmiGO: Online access to ontology and annotation data. Bioinformatics , 25, 288–289. doi: 10.1093/bioinformatics/btn615
- Cubuk, J., Alston, J. J., Incicco, J. J., Singh, S., Stuchell-Brereton, M. D., Ward, M. D., … Holehouse, A. S. (2021). The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nature Communications , 12, 1936. doi: 10.1038/s41467-021-21953-3
- Cumberworth, A., Lamour, G., Babu, M. M., & Gsponer, J. (2013). Promiscuity as a functional trait: Intrinsically disordered regions as central players of interactomes. The Biochemical Journal , 454, 361–369. doi: 10.1042/BJ20130545
- Davey, N. E., Babu, M. M., Blackledge, M., Bridge, A., Capella-Gutierrez, S., Dosztanyi, Z., … Tosatto, S. C. E. (2019). An intrinsically disordered proteins community for ELIXIR. F1000Research , 8, ELIXIR–1753. doi: 10.12688/f1000research.20136.1
- Dogan, J., Gianni, S., & Jemth, P. (2014). The binding mechanisms of intrinsically disordered proteins. Physical Chemistry Chemical Physics: PCCP , 16, 6323–6331. doi: 10.1039/C3CP54226B
- Dosztányi, Z., Chen, J., Dunker, A. K., Simon, I., & Tompa, P. (2006). Disorder and sequence repeats in hub proteins and their implications for network evolution. Journal of Proteome Research , 5, 2985–2995. doi: 10.1021/pr060171o
- Dyson, H. J., & Wright, P. E. (2019). Perspective: The essential role of NMR in the discovery and characterization of intrinsically disordered proteins. Journal of Biomolecular NMR , 73, 651–659. doi: 10.1007/s10858-019-00280-2
- El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., … Fin, R. D. (2019). The Pfam protein families database in 2019. Nucleic Acids Research , 47, D427–D432. doi: 10.1093/nar/gky995
- Gene Ontology Consortium. (2021). The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Research , 49, D325–D334. doi: 10.1093/nar/gkaa1113
- Hatos, A., Hajdu-Soltész, B., Monzon, A. M., Palopoli, N., Álvarez, L., Aykac-Fas, B., … Piovesan, D. (2020). DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Research , 48, D269–D276.
- B. B. Kragelund, & K Skriver. (Eds.). (2020). Intrinsically disordered proteins: Methods and protocols. New York: Humana Press.
- Lewis, T. E., Sillitoe, I., Dawson, N., Lam, S. D., Clarke, T., Lee, D., … Lees, J. (2018). Gene3D: Extensive prediction of globular domains in proteins. Nucleic Acids Research , 46, D1282. doi: 10.1093/nar/gkx1187
- Nadendla, S., Jackson, R., Munro, J., Quaglia, F., Mészáros, B., Olley, D., … Giglio, M. G. (2022). ECO: The Evidence and Conclusion Ontology, an update for 2022. Nucleic Acids Research , 50, D1515–D1521. doi: 10.1093/nar/gkab1025
- Oldfield, C. J., Meng, J., Yang, J. Y., Yang, M. Q., Uversky, V. N., & Dunker, A. K. (2008). Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics , 9, (Suppl 1), S1. doi: 10.1186/1471-2164-9-S1-S1
- Perdikari, T. M., Murthy, A. C., Ryan, V. H., Watters, S., Naik, M. T., & Fawzi, N. L. (2020). SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. The EMBO Journal , 39, e106478. doi: 10.15252/embj.2020106478
- Piovesan, D., Tabaro, F., Mičetić, I., Necci, M., Quaglia, F., Oldfield, C. J., … Tosatto, S. C. E. (2017). DisProt 7.0: A major update of the database of disordered proteins. Nucleic Acids Research , 45, D219–D227. doi: 10.1093/nar/gkw1056
- Piovesan, D., Tabaro, F., Paladin, L., Necci, M., Micetic, I., Camilloni, C., … Tosatto, S. C. E. (2018). MobiDB 3.0: More annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Research , 46, D471–D476. doi: 10.1093/nar/gkx1071
- Quaglia, F., Mészáros, B., Salladini, E., Hatos, A., Pancsa, R., Chemes, L. B., … Piovesan, D. (2022a). DisProt in 2022: Improved quality and accessibility of protein intrinsic disorder annotation. Nucleic Acids Research , 50, D480–D487. doi: 10.1093/nar/gkab1082
- Quaglia, F., Salladini, E., Carraro, M., Minervini, G., Tosatto, S. C. E., & Le Mercier, P. (2022b). SARS-CoV-2 variants preferentially emerge at intrinsically disordered protein sites helping immune evasion. The FEBS Journal , 50(D1), D480–D487. doi: 10.1111/febs.16379
- Schiavina, M., Pontoriero, L., Uversky, V. N., Felli, I. C., & Pierattelli, R. (2021). The highly flexible disordered regions of the SARS-CoV-2 nucleocapsid N protein within the 1-248 residue construct: Sequence-specific resonance assignments through NMR. Biomolecular NMR Assignments , 15, 219–227. doi: 10.1007/s12104-021-10009-8
- Suzek, B. E., Wang, Y., Huang, H., McGarvey, P. B., Wu, C. H., & UniProt Consortium. (2015). UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics , 31, 926–932. doi: 10.1093/bioinformatics/btu739
- Tompa, P. (2010). Structure and function of intrinsically disordered proteins. Boca Raton: Chapman & Hall/CRC Press.
- Tompa, P. (2005). The interplay between structure and function in intrinsically unstructured proteins. FEBS Letters , 579, 3346–3354. doi: 10.1016/j.febslet.2005.03.072
- UniProt Consortium. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research , 47, D506–D515. doi: 10.1093/nar/gky1049
- Uversky, V. N., & Dunker, A. K. (2010). Understanding protein non-folding. Biochimica Et Biophysica Acta , 1804, 1231–1264. doi: 10.1016/j.bbapap.2010.01.017
- van der Lee, R., Buljan, M., Lang, B., Weatheritt, R. J., Daughdrill, G. W., Dunker, A. K., … Babu, M. M. (2014). Classification of intrinsically disordered regions and proteins. Chemical Reviews , 114, 6589–6631. doi: 10.1021/cr400525m
- Wright, P. E., & Dyson, H. J. (2015). Intrinsically disordered proteins in cellular signalling and regulation. Nature Reviews Molecular Cell Biology , 16, 18–29. doi: 10.1038/nrm3920
- Wu, C., Qavi, A. J., Hachim, A., Kavian, N., Cole, A. R., Moyle, A. B., … Leung, D. W. (2021). Characterization of SARS-CoV-2 nucleocapsid protein reveals multiple functional consequences of the C-terminal domain. iScience , 24, 102681. doi: 10.1016/j.isci.2021.102681
Citing Literature
Number of times cited according to CrossRef: 2
- Federica Quaglia, Anastasia Chasapi, Maria Victoria Nugnes, Maria Cristina Aspromonte, Emanuela Leonardi, Damiano Piovesan, Silvio C E Tosatto, Best practices for the manual curation of intrinsically disordered proteins in DisProt, Database, 10.1093/database/baae009, 2024 , (2024).
- Maria Cristina Aspromonte, Maria Victoria Nugnes, Federica Quaglia, Adel Bouharoua, Vasileios Sagris, Vasilis J Promponas, Anastasia Chasapi, Erzsébet Fichó, Galo E Balatti, Gustavo Parisi, Martín González Buitrón, Gabor Erdos, Matyas Pajkos, Zsuzsanna Dosztányi, Laszlo Dobson, Alessio Del Conte, Damiano Clementel, Edoardo Salladini, Emanuela Leonardi, Fatemeh Kordevani, Hamidreza Ghafouri, Luiggi G Tenorio Ku, Alexander Miguel Monzon, Carlo Ferrari, Zsófia Kálmán, Juliet F Nilsson, Jaime Santos, Carlos Pintado-Grima, Salvador Ventura, Veronika Ács, Rita Pancsa, Mariane Goncalves Kulik, Miguel A Andrade-Navarro, Pedro José Barbosa Pereira, Sonia Longhi, Philippe Le Mercier, Julian Bergier, Peter Tompa, Tamas Lazar, Silvio C E Tosatto, Damiano Piovesan, DisProt in 2024: improving function annotation of intrinsically disordered proteins, Nucleic Acids Research, 10.1093/nar/gkad928, 52 , D1, (D434-D441), (2023).

