Efficient Electrotransformation of Bacillus thuringiensis for Gene Manipulation and Expression
Ephrem Debebe Zegeye, Ephrem Debebe Zegeye, Marina Aspholm, Marina Aspholm
Abstract
Genetic manipulation of Bacillus spp., such as B. thuringiensis and B. cereus , is laborious and time consuming due to challenges in transformation of the plasmid DNA construct. Larger shuttle plasmids, such as pMAD, that are commonly used in markerless gene replacement are particularly difficult to transform into Bacillus spp. Here, we present robust protocols that work efficiently for the transformation of both small and large plasmid constructs into B. thuringiensis. Our protocols involve preparation of efficient electrocompetent Bacillus cells by cultivating the cells in the presence of a cell wall–weakening agent, followed by washing the cells with optimized solutions. The protocols further highlight the importance of using unmethylated plasmid DNA for the efficient transformation of B. thuringiensis. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Preparation of electrocompetent B. thuringiensis
Basic Protocol 2 : Transformation of B. thuringiensis
INTRODUCTION
Efficient transformation of DNA is a prerequisite for successful genetic manipulation of bacterial strains. Whereas some Bacillus spp./strains are naturally competent (i.e., able to take up DNA naturally), the vast majority of them require special treatments to make them take up exogenous DNA. There are two major barriers that need to be overcome for successful transformation of DNA. Firstly, the plasmid DNA construct should be able to readily pass through the bacterial cell wall to reach the cell's interior. This implies that the composition of the cell walls of bacteria, as evident in Gram-positive and Gram-negative species, could potentially influence their permeability to DNA (Trevors, Chassy, Dower, & Blaschek, 1992). Some studies also suggest that the transformation efficiency is affected by the size of the plasmid (Chan, Dreolini, Flintoff, Lloyd, & Mattenley, 2002; Ohse, Takahashi, Kadowaki, & Kusaoke, 1995). Secondly, the methylation (modification) state of the plasmid DNA to be used for transformation has to match the restriction-modification (RM) systems of the host cell to avoid degradation. The lack of optimal protocols that address the major barriers to transformation has made genetic manipulation of Bacillus spp. laborious and time consuming.
Here, we describe protocols for efficient electrotransformation of B. thuringiensis. Basic Protocol 1 is based on the glycine-mediated cell wall–weakening method, which has previously been demonstrated to improve the electrocompetence of B. mycoides , B. cereus , and B. thuringiensis (Peng et al., 2009; Turgeon, Laflamme, Ho, & Duchaine, 2006; Yi & Kuipers, 2017). This protocol involves growing the cells in the presence of glycine, which significantly improves the permeability of the wall for DNA, combined with optimized washing in an electroporation solution. Furthermore, in Basic Protocol 2, we employ a demethylation step for the plasmid DNA to be used for transformation, which is critical for successful transformation of some strains, particularly B. thuringiensis (subsp. kurstaki).
CAUTION : Use protective gloves, lab coats, and safety eyeglasses when handling Bacillus spp. Because Bacillus spp. form endospores, contaminated surfaces should be disinfected using sporicidal agents such as sodium hypochlorite. Glassware/waste should be autoclaved before washing or disposal.
Basic Protocol 1: PREPARATION OF ELECTROCOMPETENT B. thuringiensis
In this protocol, we describe preparation of super-competent B. thuringiensis cells for transformation by electroporation (Basic Protocol 2). Briefly, cells are grown to approximately mid-exponential phase, followed by addition of a glycine solution to a final concentration of 2%. After 1 hr of incubation with glycine, the cells are harvested, thoroughly washed, resuspended in an electroporation solution, and flash-frozen. Aseptic techniques are strictly followed throughout this procedure.
Materials
-
B. thuringiensis (subsp. kurstaki) strain (GEVI1716, acquired from Prof. Henri De Greve's lab, VUB, Belgium)
-
LB agar (or blood agar) plates (see recipe)
-
Brain-heart infusion (BHI) broth (see recipe), 30°C
-
20% (w/v) glycine solution (see recipe), 30°C
-
Washing/electroporation solution (see recipe; make fresh)
-
95% to 100% ethanol (Antibac, #600068)
-
Dry ice
-
1-µl inoculating loops (VWR, #89126-808)
-
30°C incubator (e.g., FRIOCELL 222-Eco line)
-
Sterile 50-ml conical tubes (Falcon, #352070)
-
Vortex (e.g., Stuart SA8)
-
30°C shaking incubator (e.g., Innova 42 incubator shaker series)
-
500-ml Erlenmeyer flasks (VWR, #1214-1133)
-
Spectrophotometer (e.g., Ultraspec 10 cell density meter, Biochrom)
-
Cuvettes (e.g., Brand UV cuvette micro, Merck, #BR759220)
-
Standard tabletop centrifuge (e.g., Heraeus Megafuge 16, Thermo Scientific)
-
Styrofoam box or another suitable container
-
Low-temperature thermometer (e.g., FLUKE 51 II digital thermometer, RS, #353-4449)
-
Sterile 1.5-ml microcentrifuge tubes (Eppendorf AG, #0020125.150)
-
1.5-ml tube rack
CAUTION : Work in a well-ventilated area and avoid inhaling fumes generated from the dry ice during preparation of the dry ice/ethanol bath (see steps 8 and 11). Avoid frostbite by using insulated gloves when handling dry ice or supercooled ethanol and wear safety glasses.
NOTE : Note that steps 1 to 5 require strict sterile conditions, and therefore, inoculation and pipetting are preferably carried out inside a Class II biosafety cabinet (e.g., NinoSAFE class II).
Cultivate Bacillus strain to exponential phase
1.To prepare a starter culture, streak out B. thuringiensis (subsp. kurstaki) strain from a −80°C glycerol stock onto an LB agar (or blood agar) plate using a 1-µl inoculating loop and incubate at 30°C.
2.The next day, make an overnight culture by transferring a single colony into 10 ml pre-warmed (30°C) BHI broth in a 50-ml conical tube and mix well by vortexing. Incubate overnight at 30°C with shaking (200 rpm) in a shaking incubator.
3.Prepare a larger culture by transferring 1 ml overnight culture into 100 ml pre-warmed (30°C) BHI broth in a 500-ml Erlenmeyer flask and incubate at 30°C with shaking (200 rpm).
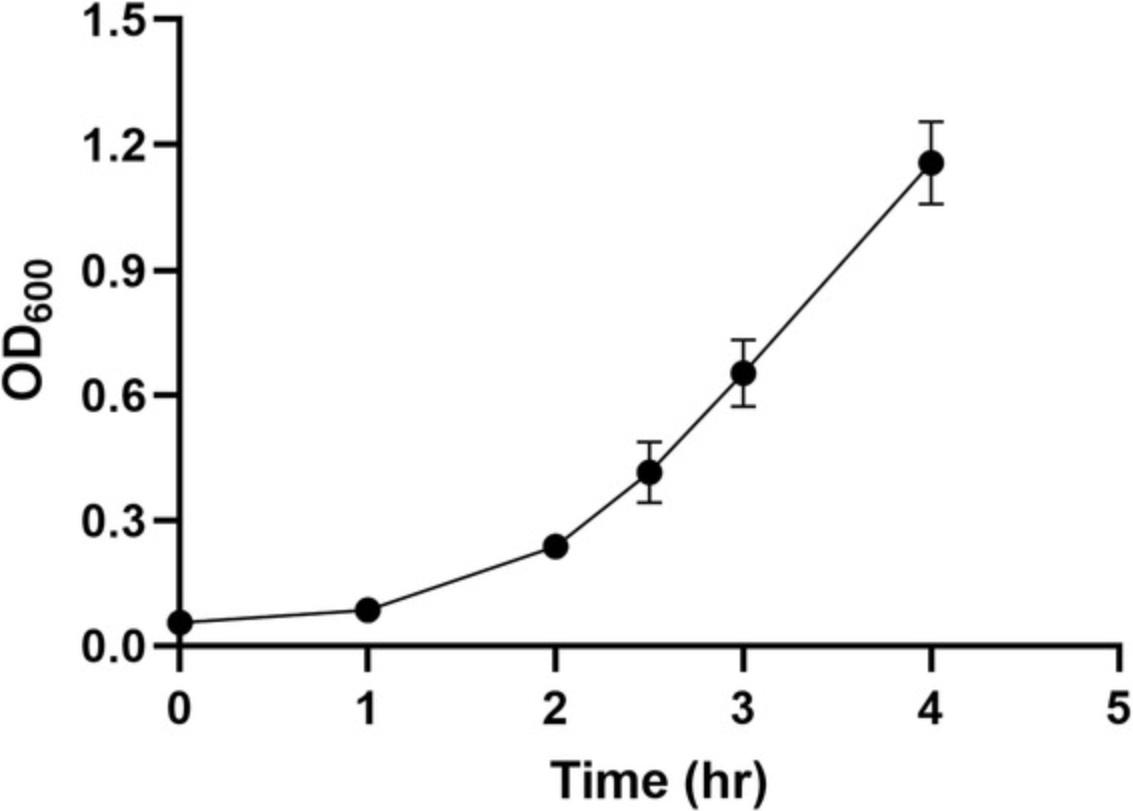
4.Monitor bacterial growth by measuring the optical density at 600 nm (OD600) regularly by removing 0.5 to 1 ml culture aseptically every 0.5 to 1 hr and measuring the absorbance using a spectrophotometer and cuvettes (Fig. 1). Use sterile BHI broth as a blank.
Add glycine solution
5.When the OD600 reaches about 0.5 to 0.7, add pre-warmed (30°C) 20% glycine solution to a final concentration of 2% to the culture and continue incubating at 30°C and 200 rpm for one more hour.
Harvest and wash cells
6.To harvest cells, divide the approximately 100- to 106-ml culture into multiple sterile 50-ml conical tubes and centrifuge 10 min at 1690 × g at room temperature. Discard supernatant.
7.Wash cell pellets three times using washing/electroporation solution:
-
Resuspend cells in 10 ml solution per tube by pipetting up and down or by gentle vortexing.
-
Once resuspended, combine samples into one 50-ml conical tube and adjust volume to 40 ml with the same solution.
-
Centrifuge 5 min at 3005 ×gat room temperature and discard supernatant.
-
Repeat resuspension (in 40 ml) and centrifuge as in step 7c two more times.
-
Remove any traces of supernatant by pipetting.
Be aware that the cell pellet could be loosely attached to the tube because of the glycerol.
8.During one of the centrifugations in step 7, prepare a dry ice/ethanol bath in a styrofoam box or another suitable container by mixing 95% to 100% ethanol and pieces of dry ice. Continue adding dry ice until temperature is below −70°C, monitoring the temperature with a low-temperature thermometer.
Resuspend cells in washing/electroporation medium, snap-freeze, and store
9.Resuspend cell pellet in ∼1 ml washing/electroporation solution by pipetting up and down.
10.Aliquot 100 μl cells into sterile 1.5-ml microcentrifuge tubes.
11.Snap-freeze cells by submerging the cell aliquot tubes in a 1.5-ml tube rack in the dry ice/ethanol bath (see step 8) for ∼5 min. Store at −80°C until use.
Basic Protocol 2: TRANSFORMATION OF B. thuringiensis
In this protocol, a plasmid DNA construct is electroporated into the B. thuringiensis competent cells prepared in Basic Protocol 1.The shuttle vector pMAD has been used for genetic manipulation of Gram-positive species, including B. cereus , Staphylococcus aureus , and Listeria monocytogenes (Arnaud, Chastanet, & Débarbouillé, 2004). A slightly modified version of pMAD, pMAD-I-SceI, in which a restriction site for I-SceI has been inserted to promote double crossover (Janes & Stibitz, 2006; Lindbäck et al., 2012), has been widely used for genetic manipulation of B. thuringiensis, B. cereus , and B. licheniformis (Borch-Pedersen et al., 2016; Lindbäck et al., 2012; Pradhan et al., 2021). In this protocol, we electroporate the pMAD-I-SceI-Δ ena2B construct, which contains the start and stop codons of the ena2B (ATGTAA) gene (Pradhan et al., 2021) flanked by 577-bp upstream and 634-bp downstream homology sequences (Fig. 2A), into B. thuringiensis.
The plasmid pHT315 is a low-copy-number shuttle vector derived from B. thuringiensis (Fig. 2B). It is stably maintained in B. thuringiensis and B. cereus (Arantes & Lereclus, 1991) and has hence been widely used for gene expression from plasmids in various Bacillus spp. (Borch-Pedersen et al., 2016; Pradhan et al., 2021; Wang et al., 2020). In this protocol, we also transform B. thuringiensis with pHT315 to demonstrate the enhancement of electrocompetence resulting from supplementation of the growth medium with 2% glycine. The plasmid maps used for the transformation and the corresponding DNA sequences of the DNA fragments are shown in Figure 2 and the Supporting Information (Files 1 and 2), respectively.
Materials
-
Shuttle plasmid construct of interest [e.g., pMAD-I-SceI-B. thuringiensis Δ ena2B (10,894 bp; available from Prof. Marina Aspholm's lab on request); pHT315 (6531 bp; Center of Analytical and Genetic Engineering Research, Institute of Microbiology, the National Academy of Sciences of Belarus, #038.A.I3-4)]
-
One ShotTM INV110 chemically competent Escherichia coli (Thermo Fisher Scientific, #C717103)
-
LB-ampicillin (100 µg/ml) agar plates (see recipe)
-
One ShotTM Top10 chemically competent E. coli (Thermo Fisher Scientific, #C404010)
-
QIAprep Spin Miniprep Kit (Qiagen, #27115)
-
Nuclease-free water (Qiagen, #129117)
-
B. thuringiensis (subsp. kurstaki) competent cells (see Basic Protocol 1)
-
BHI broth (see recipe)
-
LB-erythromycin (5 µg/ml) agar plates (see recipe)
-
LB-erythromycin/X-gal agar plates (see recipe; make fresh)
-
2× Dream Taq PCR master mix (Thermo Scientific, #K1071)
-
pMAD primers (5'-3') (10 µM):
- Forward: CCATCAGACGGTTCGATCTT
- Reverse: GTCCCTGATGGTCGTCATCT
-
pHT315 primers (5’-3’) (10 µM):
- Forward: GGAGAAAATACCGCATCAGG
- Reverse: TTTATGCTTCCGGCTCGTAT
-
30°C and 37°C incubators (e.g., FRIOCELL 222-Eco line)
-
1-µl inoculating loops (VWR, #89126-808)
-
Micro-volume spectrophotometer (e.g., mySPEC, VWR, #732-2533)
-
Horizontal electrophoresis system (e.g., Mini-Sub Cell GT, Bio-Rad, #1704406)
-
Gel imaging instrument (e.g., C300, Azure Biosystems)
-
Electroporation cuvettes, 0.2-cm gap (Gene Pulser, Bio-Rad, #1652086)
-
Electroporator (e.g., Eppendorf Eporator)
-
Sterile 5-ml centrifuge tubes (VWR, #525-0666)
-
30°C and 37°C shaking incubators (e.g., Innova 42 incubator shaker series)
-
Sterile 1.5-ml microcentrifuge tubes (Eppendorf AG, #0020125.150)
-
Vortex (e.g., Stuart sA8)
-
Microwave oven (e.g., Electrolux)
-
PCR tubes (SARSTEDT, #72.991.002)
-
Thermocycler (Veriti Thermal Cycler, Thermo Fisher Scientific)
-
Additional reagents and equipment for 0.7% and 1% agarose gel electrophoresis (see Current Protocols article: Voytas, 2000), such as agarose (Lonza, #50004), gel stain (SYBR® Safe DNA gel stain, Invitrogen, #S33102), gel-loading dye (NEB, #B7024S), and a DNA ladder (GeneRuler 1 kb, Thermo Scientific, #SM0313)
NOTE : Experiments involving PCR require careful technique to prevent contamination. Use DNase-free pipet tips.
Prepare unmethylated DNA
1.Transform shuttle plasmid construct of interest (e.g., the pMAD-I-SceI-Δ ena2B or pHT315 construct) into One-ShotTM INV110 chemically competent E. coli cells according to the manufacturer's recommendations. At the last step, plate on an LB-ampicillin (100 µg/ml) agar plate. As a control, prepare methylated DNA by transforming plasmid of interest into One ShotTM Top10 chemically competent E. coli.
2.Pick one transformant colony and streak it onto an LB-ampicillin (100 µg/ml) agar plate. Incubate overnight at 37°C.
3.Take approximately two loopfuls of cells (using a 1-μl inoculating loop) from agar surface and purify plasmid with a QIAprep Spin Miniprep Kit according to the manufacturer's recommendations. At the last step, use about 30 to 35 µl nuclease-free water to elute the plasmid DNA.
4.Measure concentration of the plasmid DNA using a micro-volume spectrophotometer.
5.Check quality of the purified DNA by performing 0.7% agarose gel electrophoresis (Fig. 3) using a horizontal electrophoresis system and gel imaging instrument.
Perform electroporation
6.Chill electroporation cuvettes on ice prior to electroporation.
7.Thaw an aliquot of B. thuringiensis (subsp. kurstaki) competent cells from −80°C freezer on ice for ∼5 min.
8.Add 1 to 2 µg plasmid DNA (2 to 5 µl) from step 3 into tube containing the competent cells and gently flick side of the tube a few times to mix. For a negative-control tube, add an equivalent amount of nuclease-free water.
9.Incubate DNA/cell mixture for 1 to 2 min on ice.
10.Transfer DNA/cell mixture to the pre-chilled electroporation cuvette from step 6.Avoid air bubbles during pipetting.
11.Wipe surface of the electroporation cuvette with a paper towel.
12.Pulse at ∼11 kV/cm (∼5 ms) with an electroporator.
13.Immediately add 500 µl room-temperature BHI broth and transfer into a sterile 5-ml centrifuge tube.
14.Allow cells to recover by incubating at 30°C (for pMAD constructs) or 37°C (for pHT315) for 3 to 3.5 hr with shaking (200 rpm) in a shaking incubator.
15.Make serial dilutions (1:10) using BHI broth for pHT315 transformation and plate cells on an LB-erythromycin (5 µg/ml) agar plate.
Verify transformation by colony PCR
16.For pMAD constructs, select a few isolated blue colonies (Fig. 4), transfer them to new LB-erythromycin/X-gal agar plates, and incubate overnight at 30°C. For pHT315 transformants, select a few isolated colonies, transfer them to new LB-erythromycin agar plates, and incubate overnight at 37°C.
17.Prepare cell lysates by transferring a tiny amount of cells, using a 1-µl inoculating loop, into sterile 1.5-ml microcentrifuge tubes containing 100 µl nuclease-free water. Vortex well and boil in a microwave oven for 3 min at 900 W.
18.To run colony PCR on the transformants, prepare the following PCR mixture in a PCR tube, with pMAD or pHT315 primers:
| Component | Volume (µl) |
| 2× Dream Taq PCR master mix | 12.5 |
| Forward primer (10 µM) | 0.5 |
| Reverse primer (10 µM) | 0.5 |
| Cell lysate (step 17) | 2 |
| Nuclease-free water | 9.5 |
| Total | 25 |
19.Run PCR using a thermocycler according to the manufacturer's recommendations. For the pMAD and pHT315 primers, follow the thermal cycling conditions below:
| Initial step: | 2 min | 95°C |
| 30 cycles: | 30 s | 95°C |
| 30 s | 52°C | |
| 1.5 min | 72°C | |
| 1 cycle: | 5 min | 72°C |
| Final step: | indefinitely | 4°C. |
20.Resolve PCR products by 1% agarose gel electrophoresis for 1 hr at 100 V.
REAGENTS AND SOLUTIONS
Ampicillin solution, 100 mg/ml
Dissolve 1 g ampicillin (GBiosciences, #A051-B) in ∼8 ml distilled water. Adjust volume to 10 ml. Filter-sterilize, aliquot, and store ≤6 months at −20°C.
Brain-heart infusion (BHI) broth
Suspend 37 g BD Bacto™ BHI powder (Difco Laboratories, #237500) in 1 L distilled water. Mix thoroughly, heat with frequent agitation, boil for 1 min, and sterilize by autoclaving at 121°C for 15 min. Store ≤2 months at 4°C.
Erythromycin solution, 5 mg/ml
Dissolve 50 mg erythromycin (Sigma, #E5389) in ∼8 ml 95% to 100% ethanol (Antibac, #600068). Adjust volume to 10 ml with ethanol. Filter-sterilize (0.22 µm), aliquot, and store ≤1 year at −20°C.
Glycine solution, 20%
Weigh out 10 g glycine (Calbiochem, #357002) and add distilled water to a total volume of ∼45 ml in 50-ml conical tube. After complete dissolution, adjust volume to 50 ml with distilled water. Sterilize by autoclaving at 121°C for 15 min. Store ≤1 year at room temperature.
LB agar
Dissolve 37 g LB agar (Merck, #110283) in 1 L distilled water, heat to boiling while stirring, and autoclave at 121°C for 15 min. Pour ∼20 ml per plate (9-cm Petri dishes, HEGER AS, #1065). Store ≤2 months at 4°C.
LB-ampicillin agar plates
Dissolve 37 g LB agar (Merck, #110283) in 1 L distilled water, heat to boiling while stirring, and autoclave at 121°C for 15 min. After the medium has cooled to ∼50°C, add 1 ml 100 mg/ml ampicillin solution (see recipe; to 100 µg/ml). Use a magnetic stirrer to mix antibiotic into the medium uniformly. Pour ∼20 ml per plate (9-cm Petri dishes, HEGER AS, #1065). Store ≤2 weeks at 2-8°C.
LB-erythromycin agar plates
Dissolve 37 g LB agar (Merck, #110283) in 1 L distilled water, heat to boiling while stirring, and autoclave at 121°C for 15 min. After the medium has cooled to about 50° to 55°C, add 1 ml 5 mg/ml erythromycin solution (see recipe; to 5 µg/ml). Use a magnetic stirrer to mix antibiotic into the medium uniformly. Pour ∼20 ml per plate (9-cm Petri dishes, HEGER AS, #1065). Store ≤5 weeks at 4°C.
LB-erythromycin/X-gal agar plates
Prepare LB-erythromycin agar plates (see recipe). Spread 40 µl 40 mg/ml X-gal (see recipe) on surface of plates using an L-shaped spreader (Microspec, #E3) prior to plating cells. Prepare fresh immediately before use.
Washing/electroporation solution
Dissolve 45.4 g D-sorbitol (Sigma-Aldrich, #W302902), 94.6 g D (+) trehalose (VWR, #28719.290), and 118 ml 85% glycerol (VWR, #24384.290) in 800 ml distilled water. Adjust volume to 1 L and sterilize by autoclaving at 121°C for 15 min. Prepare fresh immediately before use.
X-gal solution, 40 mg/ml
Dissolve 0.4 g X-gal (5-bromo-4-chloro-3-indolyl β-D-galactopyranoside, Calbiochem, #7240-90-6) in 8 ml N, N-dimethylformamide (Sigma, #D4551). Adjust volume to 10 ml. Store ≤4 weeks at 4°C in the dark.
COMMENTARY
Background Information
High transformation efficiency is needed for conducting successful mutagenesis studies. Electroporation is a simple, rapid, and efficient procedure for transformation of bacterial cells. During electroporation, the electric pulse generates transient pores in cell membranes (Tokman et al., 2013), thus facilitating the influx of exogenous nucleic acids into cells. Generally, the electrotransformation efficiency of Gram-positive bacteria is lower than that of their Gram-negative counterparts, probably due to the presence of a peptidoglycan-rich cell wall in the former group of bacteria (Trevors et al., 1992). In the Gram-positive bacterium Corynebacterium glutamicum , the electrotransformation efficiency was enhanced by nearly 20-fold by mutating a gene involved in peptidoglycan synthesis (Liu et al., 2018). Indeed, conditions that are less optimal for cell wall synthesis have positive effects on transformation efficiency (Trevors et al., 1992). In particular, supplementation of the bacterial growth medium with glycine, DL-threonine, or penicillin G improved the transformation efficiency of different bacterial species (see review by Aune & Aachmann, 2010). When bacteria are grown in the presence of glycine, glycine can be incorporated in place of L-alanine and D-alanine in the peptidoglycan cell wall (Hammes, Schleifer, & Kandler, 1973). This will reduce the density of crosslinking of the muropeptides (Hammes et al., 1973) and thereby promote the passage of DNA during electroporation. Using this principle, the transformation efficiencies of several Bacillus spp., including B. cereus (Turgeon et al., 2006), B. mycoides (Yi & Kuipers, 2017), and B. thuringiensis (Macaluso & Mettus, 1991; Peng et al., 2009), have been improved.
Studies suggest that the methylation state of the plasmid DNA influences transformation efficiency in Bacillus spp. The RM systems present in cells could degrade the imported DNA (Sitaraman, 2016), thus preventing its retention or integration into the genome. Indeed, transformation efficiency is enhanced when using unmethylated DNA compared to methylated DNA in some Bacillus spp. (Macaluso & Mettus, 1991; Yi & Kuipers, 2017).
Using the shuttle plasmid pMAD-I-SceI (Arnaud et al., 2004; Lindbäck et al., 2012) as a tool for the genetic manipulation of Bacillus spp., only one or a few blue colonies (potential single crossover candidates) are required to proceed to the next step according to the markerless gene replacement method (Janes & Stibitz, 2006). In the second step, competent cells prepared from the blue colonies are transformed with a plasmid (pBKJ223) that expresses the nuclease enzyme I-SceI. The enzyme I-SceI inserts a double-strand break at the 18-bp recognition site present in the backbone of pMAD-I-SceI, which promotes a second crossover leading to the desired gene replacement (Janes & Stibitz, 2006).
Critical Parameters
Basic Protocol 1
During the preparation of competent cells, time is very critical. It is highly recommended to avoid any delays during harvesting and washing. It is recommended to not prepare more than two strains at a time. Failing to wash the cells well may lead to a carryover of salts, which causes arcing during electroporation.
Basic Protocol 2
Using unmethylated DNA is important for efficient transformation of B. thuringiensis (subsp. kurstaki), as shown in Figures 6A and B.
Troubleshooting
When a problem is encountered in getting transformants, it is recommended to review every step involved in preparation of the competent cells and electroporation, as there are many factors that could influence the transformation efficiency of the cells. A few common problems and solutions are listed in Table 1.
| Problem | Possible cause | Potential solution |
|---|---|---|
| Arcing | Salts in cells | Resuspend cells well during the washing steps |
| OD600 drops to a low level after addition of glycine | Lysis | Reduce glycine concentration to 1% |
| No transformants | Restriction-modification incompatibility | Ensure that you are using unmethylated DNA. Alternative E. coli strains include Dam-/Dcm- competent E. coli (NEB, #C2925). |
| Few transformants | Poor DNA quality | Check quality of the DNA by running agarose gel electrophoresis |
| Unexpected lawn of cells | Incorrect antibiotic type/concentration | Plate untransformed cells and check for growth |
Understanding Results
Due to the large size of the gene-replacement pMAD construct, many rounds of transformation attempts (Basic Protocol 2) are required to obtain a single or a few transformants from competent cells prepared using existing protocols (Mahillon et al., 1989). However, as shown in Figure 6, by using 2% glycine in the growth medium and an optimal washing/electroporation solution, we were able to obtain hundreds of blue colonies with a single transformation experiment. B. thuringiensis grows to an OD600 of ∼0.6 within 3 to 4 hr. After addition of 2% glycine to the medium and incubating for an additional hour, the OD600 will double to reach ∼1.3 or, sometimes, ∼1.5 (Fig. 1). Unlike other Bacillus spp. such as B. subtilis , B. pumilis , B. polymyxa , and B. atrophaeus , that appear to lyse in the presence of 2% glycine, the B. thuringiensis , B. cereus , and B. weihenstephanensis strains that we have tested so far grow without problem in the presence of 2% glycine (Zegeye & Aspholm, unpub. observ.). Hence, it is important to monitor the OD600 before harvesting. In the case of cell lysis, the OD600 drops, and hence, it is important to pay attention to lysis for the strain of interest. Harvesting and subsequent washing of the cells are done at room temperature. According to the literature (Morales-Ruiz, López-Ceballos, & Maldonado-Mendoza, 2019) and our experience, keeping the cells on ice has a negative effect on electrocompetence.
The shuttle plasmid pMAD (Fig. 2A) contains a beta-galactosidase (bgaB) gene, driven by a constitutive pclpB promoter, giving the transformants a blue color (Fig. 4) when the medium contains X-gal substrate (Arnaud et al., 2004). As mentioned above, the blue colonies may contain a pMAD plasmid construct integrated into the genome by a single crossover. It is therefore important to proceed with the second stage of transformation for double crossover following the markerless allelic exchange method developed by Janes & Stibitz (2006). The transformants are verified to retain the gene replacement construct by PCR using pMAD-specific primers (Fig. 5A).
The pHT315 plasmid (Fig. 2B) is a low-copy-number plasmid (Arantes & Lereclus, 1991) widely used for the expression of genes in Bacillus spp. The gene of interest, along with its promoter region, is cloned into the multiple cloning sites of the pHT315 plasmid for complementation of gene deletion mutants (Borch-Pedersen et al., 2016; Pradhan et al., 2021). Here, we transformed our competent cells grown in the presence or absence of 2% glycine with empty pHT315 vector. As shown in Figure 6, the competent cells grown in the absence of glycine had a very low transformation efficiency compared to those grown in the presence of glycine. Additionally, using unmethylated DNA appears to be very critical for the transformability of the B. thuringiensis strain we tested (Fig. 6).
Time Considerations
See Table 2 for the approximate time needed for competent cell preparation (Basic Protocol 1) and transformation (Basic Protocol 2).
| Protocol | Estimated time needed | Details |
|---|---|---|
| Preparation of electrocompetent B. thuringiensis | 3 days |
Day 1: 15 min Day 2: 15 min Day 3: 5-6 hr |
| Preparation of unmethylated DNA | 3 days |
Day 1: 90 min Day 2: 15 min Day 3: 45 min |
| Transformation of pMAD constructs | 1.5-2 days |
Day 1: 5 hr Day 2: 30 min |
| Verification of transformation by PCR | 2 days |
Day 1: 30 min Day 2: 3 hr |
| Transformation of pHT315 constructs | 1 day | Day 1: 5 hr |
Acknowledgments
We thank Kristin O'Sullivan for preparing the pMAD-Δ ena2B construct, Yohannes B. Mekonnen for MALDI-TOF analysis of Bacillus spp., and Toril Lindbäck for fruitful discussions about pMAD. This research was supported by a Norwegian University of Life Sciences research grant.
Author Contributions
Ephrem Debebe Zegeye : Conceptualization, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing; Marina Aspholm : Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Raw data files are available from the authors on request.
Supporting Information
| Filename | Description |
|---|---|
| cpz1588-sup-0001-SuppMat.dna41.8 KB | Supplementary Information |
| cpz1588-sup-0002-SuppMat.gb17.9 KB | Supplementary Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Arantes, O., & Lereclus, D. (1991). Construction of cloning vectors for Bacillus thuringiensis. Gene , 108, 115–119. doi: 10.1016/0378-1119(91)90495-W
- Arnaud, M., Chastanet, A., & Débarbouillé, M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Applied and Environmental Microbiology , 70, 6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004
- Aune, T. E. V., & Aachmann, F. L. (2010). Methodologies to increase the transformation efficiencies and the range of bacteria that can be transformed. Applied Microbiology and Biotechnology , 85, 1301–1313. doi: 10.1007/s00253-009-2349-1
- Borch-Pedersen, K., Lindbäck, T., Madslien, E. H., Kidd, S. W., O'Sullivan, K., Granum, P. E., & Aspholm, M. (2016). The cooperative and interdependent roles of GerA, GerK, and Ynd in germination of Bacillus licheniformis spores. Applied and Environmental Microbiology , 82, 4279–4287. doi: 10.1128/AEM.00594-16
- Chan, V., Dreolini, L. F., Flintoff, K. A., Lloyd, S. J., & Mattenley, A. A. (2002). The effect of increasing plasmid size on transformation efficiency in Escherichia coli. Journal of Experimental Microbiology and Immunology (JEMI) , 2, 207–223.
- Hammes, W., Schleifer, K. H., & Kandler, O. (1973). Mode of action of glycine on the biosynthesis of peptidoglycan. Journal of Bacteriology , 116, 1029–1053. doi: 10.1128/jb.116.2.1029-1053.1973
- Janes, B. K., & Stibitz, S. (2006). Routine markerless gene replacement in Bacillus anthracis. Infection and Immunity , 74, 1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006
- Lindbäck, T., Mols, M., Basset, C., Granum, P. E., Kuipers, O. P., & Kovács, Á. T. (2012). CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environmental Microbiology , 14, 2233–2246. doi: 10.1111/j.1462-2920.2012.02766.x
- Liu, J., Wang, Y., Lu, Y., Ni, X., Guo, X., Zhao, J., … Ma, Y. (2018). Mutations in peptidoglycan synthesis gene ponA improve electrotransformation efficiency of Corynebacterium glutamicum ATCC 13869. Applied and Environmental Microbiology , 84, e02225–18. doi: 10.1128/AEM.02225-18
- Macaluso, A., & Mettus, A. M. (1991). Efficient transformation of Bacillus thuringiensis requires nonmethylated plasmid DNA. Journal of Bacteriology , 173, 1353–1356. doi: 10.1128/jb.173.3.1353-1356.1991
- Mahillon, J., Chungjatupornchai, W., Decock, J., Dierickx, S., Michiels, F., Peferoen, M., & Joos, H. (1989). Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiology Letters , 60, 205–210. doi: 10.1111/j.1574-6968.1989.tb03447.x
- Morales-Ruiz, E., López-Ceballos, A., & Maldonado-Mendoza, I. E. (2019). Transformation of the rhizospheric Bacillus cereus sensu lato B25 strain using a room-temperature electrocompetent cells preparation protocol. Plasmid , 105, 102435. doi: 10.1016/j.plasmid.2019.102435
- Ohse, M., Takahashi, K., Kadowaki, Y., & Kusaoke, H. (1995). Effects of plasmid DNA sizes and several other factors on transformation of Bacillus subtilis ISW1214 with plasmid DNA by electroporation. Bioscience, Biotechnology, and Biochemistry , 59, 1433–1437. doi: 10.1271/bbb.59.1433
- Peng, D., Luo, Y., Guo, S., Zeng, H., Ju, S., Yu, Z., & Sun, M. (2009). Elaboration of an electroporation protocol for large plasmids and wild-type strains of Bacillus thuringiensis. Journal of Applied Microbiology , 106, 1849–1858. doi: 10.1111/j.1365-2672.2009.04151.x
- Pradhan, B., Liedtke, J., Sleutel, M., Lindbäck, T., Zegeye, E. D., O´Sullivan, K., … Remaut, H. (2021). Endospore Appendages: A novel pilus superfamily from the endospores of pathogenic Bacilli. The EMBO Journal , 40(17), e106887. doi: 10.15252/embj.2020106887
- Sitaraman, R. (2016). The Role of DNA restriction-modification systems in the biology of Bacillus anthracis. Frontiers in Microbiology , 7, 11. doi: 10.3389/fmicb.2016.00011
- Tokman, M., Lee, J. H., Levine, Z. A., Ho, M.-C., Colvin, M. E., & Vernier, P. T. (2013). Electric field-driven water dipoles: Nanoscale architecture of electroporation. PLOS ONE , 8, e61111. doi: 10.1371/journal.pone.0061111
- Trevors, J. T., Chassy, B. M., Dower, W. J., & Blaschek, H. P. (1992). Electrotransformation of bacteria by plasmid DNA. In D. C. Chang, B. M. Chassy, J. A. Saunders, & A. E. Sowers (Eds.), Guide to electroporation and electrofusion (pp. 265–290). Academic Press. doi: 10.1016/C2009-0-21564-9
- Turgeon, N., Laflamme, C., Ho, J., & Duchaine, C. (2006). Elaboration of an electroporation protocol for Bacillus cereus ATCC 14579. Journal of Microbiological Methods , 67, 543–548. doi: 10.1016/j.mimet.2006.05.005
- Voytas, D. (2000). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51(1), 2.5A.1–2.5A.9. doi: 10.1002/0471142727.mb0205as51
- Wang, Y., de Boer, R., Vischer, N., van Haastrecht, P., Setlow, P., & Brul, S. (2020). Visualization of germination proteins in putative Bacillus cereus germinosomes. International Journal of Molecular Sciences , 21(15), 5198. doi: 10.3390/ijms21155198
- Yi, Y., & Kuipers, O. P. (2017). Development of an efficient electroporation method for rhizobacterial Bacillus mycoides strains. Journal of Microbiological Methods , 133, 82–86. doi: 10.1016/j.mimet.2016.12.022
Citing Literature
Number of times cited according to CrossRef: 1
- Unni Lise Jonsmoen, Dmitry Malyshev, Mike Sleutel, Elise Egeli Kristensen, Ephrem Debebe Zegeye, Han Remaut, Magnus Andersson, Marina Elisabeth Aspholm, The role of endospore appendages in spore–spore interactions in the pathogenic Bacillus cereus group, Environmental Microbiology, 10.1111/1462-2920.16678, 26 , 9, (2024).

