Compare_Genomes: A Comparative Genomics Workflow to Streamline the Analysis of Evolutionary Divergence Across Eukaryotic Genomes
Jefferson Paril, Jefferson Paril, Tannaz Zare, Tannaz Zare, Alexandre Fournier-Level, Alexandre Fournier-Level
Abstract
The dawn of cost-effective genome assembly is enabling deep comparative genomics to address fundamental evolutionary questions by comparing the genomes of multiple species. However, comparative genomics analyses frequently deploy multiple, often purpose-built frameworks, limiting their transferability and replicability. Here, we present compare_genomes, a transferable and extensible comparative genomics workflow package we developed that streamlines the identification of orthologous families within and across eukaryotic genomes and tests for the presence of several mechanisms of evolution (gene family expansion or contraction and substitution rates within protein-coding sequences). The workflow is available for Linux, written as a Nextflow workflow that calls established genomics and phylogenetics tools to streamline the analysis and visualization of eukaryotic genome divergence. This workflow is freely available at https://github.com/jeffersonfparil/compare_genomes, distributed under the GNU General Public License version 3 (GPLv3). © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Comparative genomics with Nextflow and Conda
INTRODUCTION
The genomes of eukaryotic organisms represent records of evolutionary histories along the tree of life. We can infer the drivers of their evolution by analyzing the signatures of eukaryotic genome evolution during polyploidization events, gene duplication or loss, or selection of adaptive mutations between species. This comparative genomics framework scrutinizes alternative evolutionary histories across different species within and between clades, which will offer insights into how the spatiotemporal dynamics and interactions between the biosphere and the environment prefer one group of biological solutions to fitness over others.
The increasing availability of affordable high-throughput DNA sequencing has allowed the assembly of the genomes of multiple species, going beyond the early set of model species toward species of specific biological relevance. This enabled the use of deep comparative genomics to answer fundamental evolutionary questions using multiple species within and across clades. However, the pipelines for these analyses are often study specific and are rarely described with enough details to be fully reproducible. This is a major impediment to the generalization or meta-analysis of the results and hinders the transfer of these workflows. Hence, the bioinformatics community would benefit from a unified but open-source and portable comparative genomics workflow.
Web-based comparative genomics workflows exist (for example, PLAZA; Van Bel et al., 2022). However, the centralized design limits potential extension, and usage is inherently limited to the computational resources provided, making it unsuitable for high-throughput or high-bandwidth usage. Various portable frameworks have been developed to run comparative genomics pipelines in a reproducible way (e.g., Snakemake by Mölder et al., 2021, and Nextflow by Di Tommaso et al., 2017). These tools work synergistically with package and environment management systems such as Docker (https://www.docker.com/) or Conda (https://docs.conda.io/en/latest/). They have been used to generate genome assemblies and annotations, sequence alignments, variant callings, and transcriptomic data analyses. However, there is a noticeable lack of fully transparent and easily transferable comparative genomics analysis workflows. In this article, we address this gap with compare_genomes, a comparative genomics workflow built under the Nextflow framework with packages managed by Conda.
Basic Protocol: COMPARATIVE GENOMICS WITH NEXTFLOW AND CONDA
We developed compare_genomes, a transferable and extendible comparative genomics workflow for eukaryotic species built using the Nextflow framework and Conda package management system. It provides a wieldy pipeline to test for non-random evolutionary patterns that can be mapped to evolutionary processes to help identify the molecular basis of specific biological properties of the species analyzed. Additionally, it provides a template that other comparative genomics pipelines can use for improved reproducibility. It is also worth mentioning that other comparative genomics analysis methods exist that are not included in this workflow. These include syntenic block detection, as well as population genetics tools such as demographic history estimation to infer divergence times between populations or species.
A detailed user manual describing how to install, set up, and run the workflow is presented in the README page of the compare_genomes project repository (https://github.com/jeffersonfparil/compare_genomes). We included a tutorial analyzing four Arabidopsis species. Here, we detail the steps needed to run this example.
Necessary Resources
- To run this workflow using the provided example dataset: A minimum of one CPU core, one gigabyte (GB) of random-access memory (RAM), and 100 GB of free storage (our tests using the example dataset revealed that leveraging 32 CPU cores operating at base clock speeds of 2 GHz and using 50 GB of RAM enabled the completion of the analysis within 6 hr)
Download and run the example
1.Download the compare_genomes repository:
2.Install Conda. For more information, please follow the official Conda installation guide (https://conda.io/projects/conda/en/latest/user-guide/install/index.html). Installation on Linux can be achieved via:
- wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
- sh./Miniconda3-latest-Linux-x86_64.sh
3.Import and activate the compare_genomes Conda environment:
- conda env create -n compare_genomes --file compare_genomes/compare_genomes.yml
- conda activate compare_genomes
4.Edit the configuration file, i.e., compare_genomes/config/params.config, by replacing line number 2, dir = ‘/data/TEST’, with the absolute path on the user's computer that will be used as the output directory for the workflow:
- cd compare_genomes
- nano config/params.config
- Replace “dir='/data/TEST'” with your path.
5.Run the example:
- chmod +x run.sh
- time ./run.sh
Edit configuration files
6.To set up the workflow for a new analysis, edit the seven configuration files located in compare_genomes/config/.
- a.
‘urls.txt’: list of web links or absolute paths to the genome sequences, genome annotations, coding DNA sequences, and amino acid sequences for at least three species to be included in the analyses. It is formatted as a headerless, two-columned, comma-separated file. Column 1 contains the filenames of the genome sequence, genome annotation, coding DNA sequence, and amino acid sequences (species names and extension names should be the consistent across these files, e.g., ‘.fna’ for the genomes, ‘.gff’ for the annotations, ‘.cds’ for the coding DNA sequences, and ‘.faa’ for the amino acid sequences). Column 2 contains the URL (uniform resource locator) of the zipped (‘.gz’ or ‘.zip’) or unzipped files for download. Alternatively, this can be the absolute path to the pre-downloaded zipped or unzipped files on a local computer.
- b.
‘dates.txt’: list of pairwise divergence times between species. This information can be found at http://timetree.org. Divergence times between all pairs of species are not required. Pick at least two pairs of species divergence times, ideally including the outgroup species. This file is formatted as a headerless, two-columned, tab-delimited file. Column 1 contains the pair of species separated by a comma with the same names used in the ‘urls.txt’. Column 2 contains the time in million years, e.g., ‘-160’ for 160 million years ago.
- c.
‘comparisons_4DTv.txt’: list of species and pairs of species to be included in the estimation of transversion rates among four-fold-degenerate sites (4DTv). This statistic is used to set the molecular clock, with more mutations at the third codon position meaning more divergence time between a pair of sequences. By default, 4DTv is estimated using genes present with two copies. Edit line 34 of ‘compare_genomes/modules/assess_WGD.nf’ to include genes with more than two copies. This is formatted as a headerless, one-columned file. Column 1 contains the species and/or pairs of species names, which should match the names in ‘urls.txt’, and species pairs should be written as, for example, “Zea_mays X Oryza_sativa”.
- d.
‘venn_species_max_5.txt’: list of at most five species to be plotted in the Venn diagram comparing the differences and commonalities of gene families between species. It is currently not possible to fit more than five species in the Venn diagram because of the limitations of the plotting package used. This is formatted as a headerless, one-columned file. Column 1 contains the species names matching those in ‘urls.txt’.
- e.
‘genes.txt’: links to the gene sequences to be tested for significant expansion/contraction and for nonsynonymous/synonymous mutation (Ka/Ks) rates between pairs of sequences within and among species. It is formatted as a headerless, three-columned, comma-separated file. Column 1 contains phenotype names or some identifier (noncritical information). Column 2 contains the species names from which the gene sequence was derived and can be a species not included in ‘urls.txt’ (noncritical information). Column 3 contains the URL of the genes to be downloaded and analyzed.
- f.
‘params.config’: configuration file listing the parameter values for the specific analyses to be conducted.
1.‘dir’: output directory.
2.‘species_of_interest’: a single species of interest, which should match one of the species listed in ‘urls.txt’.
3.‘species_of_interest_panther_HMM_for_gene_names_url’: URL to the Panther HMM database to extract gene names from, preferably from the species used for the gene ontology (GO) term enrichment analysis. See the current release list at <http://data.pantherdb.org/ftp/sequence_classifications/current_release/PANTHER_Sequence_Classification_files/>.
4.‘urls’: location of ‘urls.txt’.
5.‘dates’: location of ‘dates.txt’.
6.‘comparisons_4DTv’: location of ‘comparisons_4DTv.txt’.
7.‘venn_species_max_5’: location of ‘venn_species_max_5.txt’.
8. ‘genes’: location of ‘genes.txt’.
9. ‘cafe5_n_gamma_cats’: number of the Gamma values (parameter of the substitution model) to use for the assessment of significant gene family expansion and contraction using CAFE5. If this is equal to 1, then we use the substitution model without the gamma function.
10. ‘cafe5_pvalue’: significance threshold for the gene family expansion and contraction test.
11. ‘go_term_enrich_genome_id’: genome ID for the species specified in ‘species_of_interest_panther_HMM_for_gene_names_url’ or some closely related species. Find the appropriate taxon ID at <https://pantherdb.org/services/oai/pantherdb/supportedgenomes>.
12. ‘go_term_enrich_annotation_id’: code for the gene ontology level to be used, e.g., “GO:0008150” for "Biological Process". See the list of GO codes at <https://pantherdb.org/services/oai/pantherdb/supportedannotdatasets>.
13. ‘go_term_enrich_test’: GO term enrichment test, which can be set to either “FISHER” (Fisher's exact test) or “BINOMIAL” (binomial distribution test).
14. ‘go_term_enrich_correction’: multiple testing correction which can be set to “NONE”, “FDR” (false-discovery rate), or “BONFERRONI” (Bonferroni correction).
15. ‘go_term_enrich_ngenes_per_test’: number of randomly sampled genes to include in each GO term enrichment analysis.
16. ‘go_term_enrich_ntests’: number of GO term enrichment test replications to perform.
- g.
‘process.config’: configuration file setting the computing resource allocation. Assign the number of ‘cpus’ and ‘memory’ capacity to use for low- and high-resource tasks with ‘LOW_MEM_LOW_CPU’ and ‘HIGH_MEM_HIGH_CPU’, respectively.
Workflow of compare_genomes
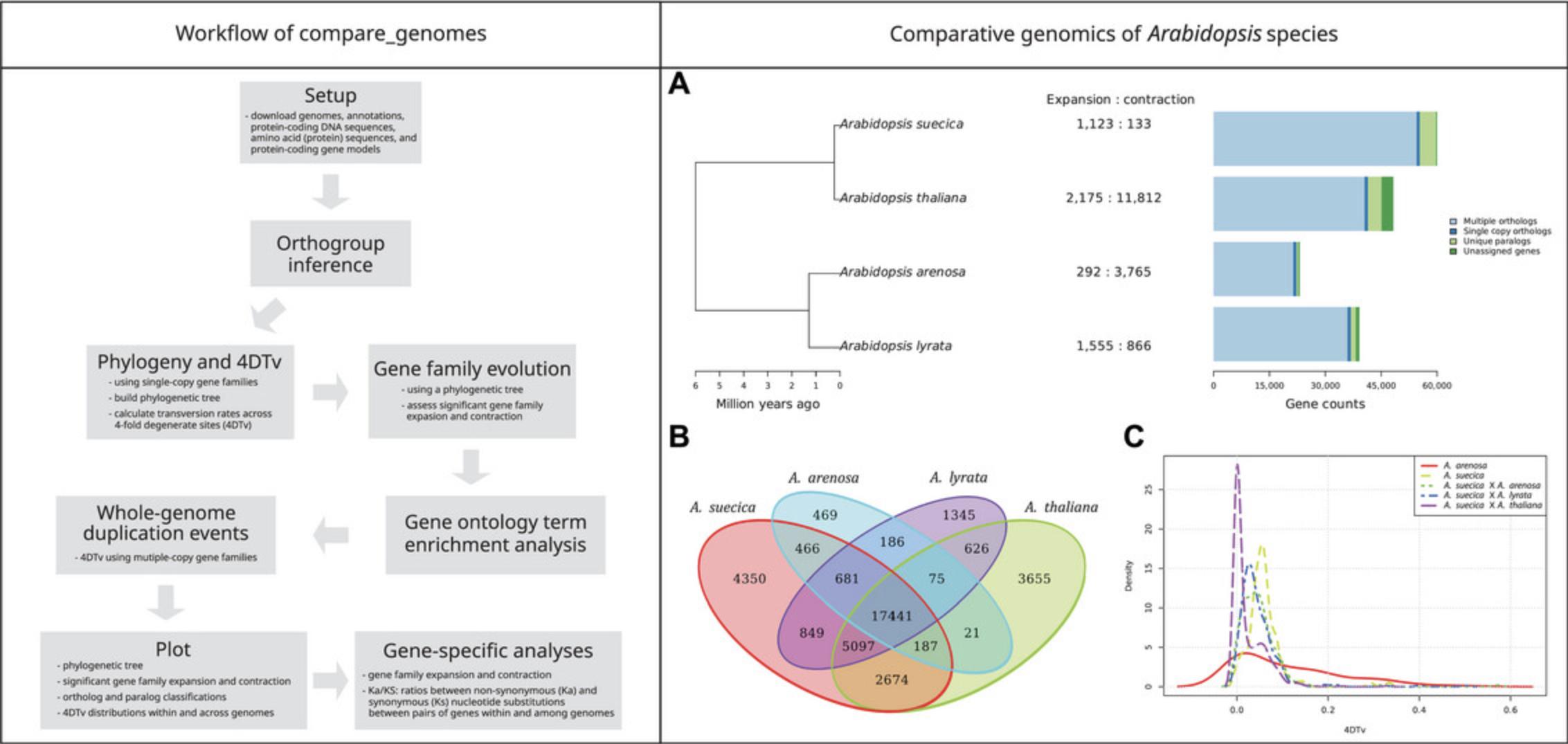
7.The compare_genomes workflow consists of nine analysis steps under the default setup (Fig. 1, left).
- a.
Download the user-defined genome datasets: genome sequences (fasta, .fna), annotations (general feature format, .gff), coding DNA sequences or CDS (.cds), protein sequences (fasta, .faa), protein-coding gene models (probabilistic protein model format, .hmm), corresponding gene ontology terms (.txt), and protein sequences of specific genes of interest (.faa).
- b.
Identify orthogroups using OrthoFinder (Emms & Kelly, 2019) and gene families for each orthogroup using HMMER3 (Mistry et al., 2013) and Panther HMMs (protein-coding gene family models; Thomas et al., 2022). An orthogroup is a set of genes descended from a single gene from the last common ancestor of all the species included in the analysis.
- c.
Infer phylogenetic trees for each orthogroup using IQ-TREE 2 (Minh et al., 2020) based on CDS alignments generated by MACSE (Ranwez et al., 2011) and the most likely nucleotide substitution model inferred by ModelFinder (Kalyaanamoorthy et al., 2017).
- d.
Infer the rate of sequence divergence based on transversion rates among four-fold-degenerate sites (4DTv) in single-copy genes between pairs of species using the custom Julia script for this purpose that ships with compare_genomes. 4DTv is a proxy for time, where the accumulation of transversion mutations at the degenerate (neutral) site is proportional to the amount of time passed.
- e.
Identify whole-genome duplication events using 4DTv computed from multi-copy gene families. If the paralogs within a genome show an accumulation of 4DTv that is greater than 0, it is likely that a genome duplication or polyploidization occurred from which the multiple paralogs are derived.
- f.
Test for significant gene family expansion or contraction across genomes using CAFE (version 5; De Bie et al., 2006). Gene family expansion and contraction are calculated relative to the gene family count in the ancestral species given the phylogenetic tree from IQ-TREE 2. Expanded and contracted gene families can be indicative of adaptation.
- g.
Analyze gene ontology (GO) term enrichment for significantly expanded gene families using the Panther GO API (Mi et al., 2019). To test the significantly contracted gene families, replace all instances of the term ‘expanded’ with ‘contracted’ in lines 47-59 of ‘compare_genomes/modules/GO_enrichment.nf’.
- h.
Visualize a summary of the results (i.e., see Fig. 1, right, for a sample output). This generates the summary output of the whole-genome-level analysis.
- i.
This optional step is available for testing hypotheses involving specific genes: analyze user-defined genes of interest, i.e., gene family expansion/contraction analyses with CAFE, and estimate non-synonymous to synonymous nucleotide substitution rates (Ka/Ks) using KaKs_Calculator 2.0 (Wang et al., 2009) and custom R script.
COMMENTARY
Background Information
The compare_genomes workflow was initially designed to compare the newly released, high-quality reference genome of annual ryegrass (Lolium rigidum ; an important weed in winter cropping) to related grass species, including the pasture crop perennial ryegrass (Lolium perenne). This analysis showed significant expansion of herbicide-resistance-related gene families, including detoxification genes, in the noxious weed annual ryegrass (Paril et al., 2022).
Critical Parameters
The configuration files (i.e., ‘compare_genomes/config/*.txt’) need to be checked for typos to make sure that the links to files and databases exist and are publicly accessible without the need for authentication. At least three species should be included, making sure species names are used consistently across files. Avoid spaces, non-alphanumeric characters, and quotes in species and gene names.
Troubleshooting
In cases where IQ-TREE 2 fails to build a time tree (‘#{dir}/ORTHOGROUPS_SINGLE_GENE.NT.timetree.nex’), review the estimated divergence times between species in ‘compare_genomes/config/dates.txt’ and remove divergence time estimates with low confidence based on the number of literature reports and dating studies. Empty files generated by the workflow are indicative of steps that did not completely finish successfully. It is encouraged for users to submit issues through the GitHub repository to track the development of fixes: https://github.com/jeffersonfparil/compare_genomes/issues.
Understanding Results
The main output of the workflow is a figure in scalable vector graphics format (.svg) summarizing the species phylogeny, number of genes within genomes, gene family expansion/contraction, and whole-genome duplication events (Fig. 1, right). Other important output files include the:
-
1.OrthoFinder results in ‘#{dir}/ORTHOGROUPS/OrthoFinder/’,
-
2.GO annotations for each orthogroup in ‘#{dir}/ORTHOGROUPS/orthogroups_gene_counts_families_go.out’,
-
3.gene family expansion/contraction results in ‘#{dir}/CAFE_results/’,
-
4.4DTv results in ‘#{dir}/*.4DTv’,
-
5.GO term analysis results in ‘#{dir}/*.goout’, and
-
6.results of specific gene analyses:
-
a. gene family expansion/contraction (‘#{dir}/SPECIFIC_GENES/*.conex’),
-
b. Ka/Ks plots across 15-bp windows: (‘#{dir}/SPECIFIC_GENES/*kaks.svg’), and
-
c. significant Ka/Ks peak Ka/Ks plots (‘#{dir}/SPECIFIC_GENES/*PEAKS.csv’).
The workflow generates other intermediate files, the purposes of which can be understood by reading the Nextflow modules (‘compare_genomes/modules/*.nf’).
To illustrate the performance and transferability of our pipeline, we compared the genomes of four well-characterized Arabidopsis species to test its ability to recapture the evolutionary patterns expected in these species. The summary of the output for this analysis is shown in Figure 1, right. It shows that the phylogenetic relationship between species (Fig. 1A, left subpanel) was accurately recapitulated (Novikova et al., 2016) and also shows the patterns of gene family contraction and expansion (Fig. 1A, right subpanel). Arabidopsis suecica , an allopolyploid hybrid of A. thaliana and A. arenosa (Novikova et al., 2017; Burns et al., 2021), experienced gene family expansion. A similar expansion was observed in A. lyrata , an outbreeder diverged from A. thaliana around 5 million years ago (Schmickl et al., 2010). Similarities in gene family composition between species are presented as a Venn diagram (Fig. 1B). Whole-genome duplication events were identified using the 4DTv density plot (Fig. 1C). This analysis recaptured the recent polyploidization event of the A. arenosa sub-genome within the A. suecica allopolyploid genome detected by Novikova and colleagues in 2017. The Ka/Ks ratio analyses presented in the Supporting Information show evidence of selection across several 15-bp windows in the GSTU13 (glutathione transferase–tau class; size of the window can be modified).
Time Considerations
The main drivers of computational complexity are the number of species and genes. Computation time is approximately exponential as a function of the number of species included, the number of genes per genome, and the number of user-defined genes of interest. It is advisable to start with the minimum number of species, i.e., three, and one gene of interest to gauge the computational resources and time required for specific analyses.
Acknowledgments
We thank the Department of Agriculture, Fisheries and Forestry of Australia together with the Grains Research and Development Corporation (grant ID: 4-FY9JQPE), Australian Research Data Commons (grant ID: DP727), and the Commonwealth Scientific and Industrial Research Organisation for funding, and the University of Melbourne for hosting this study.
Open access publishing facilitated by The University of Melbourne, as part of the Wiley - The University of Melbourne agreement via the Council of Australian University Librarians.
Author Contributions
Jefferson Paril : Conceptualization, software, validation, visualization, writing—original draft, writing—review and editing. Tannaz Zare : Validation, writing – review and editing. Alexandre Fournier-Level : Supervision, writing—original draft, writing—review and editing.
Conflict of Interest
The authors declare no conflicting interests.
Open Research
Data Availability Statement
The data used in this study are all publicly available. The list of links for the Arabidopsis genomes, annotations, coding DNA sequences, and proteomes from the US National Center for Biotechnology Information (NCBI) can be found in the file https://github.com/jeffersonfparil/compare_genomes/blob/main/config/urls.txt.
Supporting Information
| Filename | Description |
|---|---|
| cpz1876-sup-0001-FigureS1.eps548.8 KB | Ka/Ks ratio analyses of |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Burns, R., Mandáková, T., Gunis, J., Soto-Jiménez, L. M., Liu, C., Lysak, M. A., Yu Novikova, P., & Nordborg, M. (2021). Gradual evolution of allopolyploidy in Arabidopsis suecica. Nature Ecology & Evolution, 5(10), 1367–1381. https://doi.org/10.1038/s41559-021-01525-w
- de Bie, T., Cristianini, N., Demuth, J. P., & Hahn, M W. (2006). CAFE: A computational tool for the study of gene family evolution. Bioinformatics , 22(10), 1269–1271. https://doi.org/10.1093/bioinformatics/btl097
- di Tommaso, P., Chatzou, M., Floden, E. W., Barja, P. P., Palumbo, E., & Notredame, C. (2017). Nextflow enables reproducible computational workflows. Nature Biotechnology , 35(4), 316–319. https://doi.org/10.1038/nbt.3820
- Emms, D. M., & Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biology , 20(1), 238. https://doi.org/10.1186/s13059-019-1832-y
- Kalyaanamoorthy, S., Minh, B. Q., Wong, T K. F., von Haeseler, A., & Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods , 14(6), 587–589. https://doi.org/10.1038/nmeth.4285
- Mi, H., Muruganujan, A., Ebert, D., Huang, X., & Thomas, P. D. (2019). PANTHER version 14: More genomes, a new PANTHER go-slim and improvements in enrichment analysis tools. Nucleic Acids Research , 47(D1), D419–D426. https://doi.org/10.1093/nar/gky1038
- Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., & Lanfear, R. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution , 37(5), 1530–1534. https://doi.org/10.1093/molbev/msaa015
- Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., & Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Research , 41(12), e121. https://doi.org/10.1093/nar/gkt263
- Mölder, F., Jablonski, K. P., Letcher, B., Hall, M. B., Tomkins-Tinch, C. H., Sochat, V., Forster, J., Lee, S., Twardziok, S. O., Kanitz, A., Wilm, A., Holtgrewe, M., Rahmann, S., Nahnsen, S., & Köster, J. (2021). Sustainable data analysis with Snakemake. F1000Research , 10, 33. https://doi.org/10.12688/f1000research.29032.1
- Novikova, P. Y., Hohmann, N., Nizhynska, V., Tsuchimatsu, T., Ali, J., Muir, G., Guggisberg, A., Paape, T., Schmid, K., Fedorenko, O. M., Holm, S., Säll, T., Schlötterer, C., Marhold, K., Widmer, A., Sese, J., Shimizu, K. K., Weigel, D., Krämer, U., … Nordborg, M. (2016). Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nature Genetics , 48(9), 1077–1082. https://doi.org/10.1038/ng.3617
- Novikova, P. Y., Tsuchimatsu, T., Simon, S., Nizhynska, V., Voronin, V., Burns, R., Fedorenko, O. M., Holm, S., Säll, T., Prat, E., Marande, W., Castric, V., & Nordborg, M. (2017). Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Molecular Biology and Evolution , 34(4), 957–968. https://doi.org/10.1093/molbev/msw299
- Paril, J., Pandey, G., Barnett, E. M., Rane, R. V., Court, L., Walsh, T., & Fournier-Level, A. (2022). Rounding up the annual ryegrass genome: High-quality reference genome of Lolium rigidum. Frontiers in Genetics , 13, 1012694. https://doi.org/10.3389/fgene.2022.1012694
- Ranwez, V., Harispe, S., Delsuc, F., & Douzery, E. J. P. (2011). MACSE: Multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS ONE , 6(9), e22594. https://doi.org/10.1371/journal.pone.0022594
- Schmickl, R., Jørgensen, M. H., Brysting, A. K., & Koch, M. A. (2010). The evolutionary history of the Arabidopsis lyrata complex: A hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier. BMC Evolutionary Biology , 10(1), 98. https://doi.org/10.1186/1471-2148-10-98
- Thomas, P D., Ebert, D., Muruganujan, A., Mushayahama, T., Albou, L.-P., & Mi, H. (2022). PANTHER: Making genome-scale phylogenetics accessible to all. Protein Science , 31(1), 8–22. https://doi.org/10.1002/pro.4218
- Van Bel, M., Silvestri, F., Weitz, E. M., Kreft, L., Botzki, A., Coppens, F., & Vandepoele, K. (2022). PLAZA 5.0: Extending the scope and power of comparative and functional genomics in plants. Nucleic Acids Research , 50(D1), D1468–D1474. https://doi.org/10.1093/nar/gkab1024
- Wang, D.-P., Wan, H.-L., Zhang, S., & Yu, J. (2009). γ-MYN: A new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biology Direct , 4(1), 20. https://doi.org/10.1186/1745-6150-4-20
- Wang, D., Zhang, Y., Zhang, Z., Zhu, J., & Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics, Proteomics & Bioinformatics, 8(1), 77–80. https://doi.org/10.1016/S1672-0229(10)60008-3
Internet Resources
“Conda—Conda Documentation” (accessed January 5, 2023).
“Docker: Accelerated, Containerized Application Development” (accessed January 5, 2023).
Citing Literature
Number of times cited according to CrossRef: 2
- Tannaz Zare, Jeff F. Paril, Emma M. Barnett, Parwinder Kaur, Rudi Appels, Berit Ebert, Ute Roessner, Alexandre Fournier‐Level, Comparative genomics points to tandem duplications of SAD gene clusters as drivers of increased α‐linolenic (ω‐3) content in S. hispanica seeds, The Plant Genome, 10.1002/tpg2.20430, 17 , 1, (2024).
- Kai Liu, Nan Xie, Yuxi Wang, Xinyi Liu, The Utilization of Reference-Guided Assembly and In Silico Libraries Improves the Draft Genome of Clarias batrachus and Culter alburnus, Marine Biotechnology, 10.1007/s10126-023-10248-x, 25 , 6, (907-917), (2023).

