Adenine Nucleotide and Nicotinamide Adenine Dinucleotide Measurements in Plants
Alisdair R. Fernie, Alisdair R. Fernie, Youjun Zhang, Youjun Zhang, Ina Krahnert, Ina Krahnert, Antje Bolze, Antje Bolze, Yves Gibon, Yves Gibon
Abstract
As the principal co-factors of many metabolic pathways, the measurement of both adenine nucleotides and nicotinamide adenine dinucleotide provides important information about cellular energy metabolism. However, given their rapid and reversible conversion as well as their relatively low concentration ranges, it is difficult to measure these compounds. Here, we describe a highly sensitive and selective ion-pairing HPLC method with fluorescence detection to quantify adenine nucleotides in plants. In addition, nicotinamide adenine dinucleotide is a crucially important redox-active substrate for multiple catabolic and anabolic reactions with the ratios of NAD+/NADH and NADP+/NADPH being suggested as indicators of the general intracellular redox potential and hence metabolic state. Here, we describe highly sensitive enzyme cycling−based colorimetric assays (with a detection limit in the pmol range) performed subsequent to a simple extraction procedure involving acid or base extraction to allow the measurement of the cellular levels of these metabolites. © 2020 The Authors.
Basic Protocol 1 : Preparation of plant material for the measurement
Basic Protocol 2 : Measurement of ATP, ADP, and AMP via HPLC
Basic Protocol 3 : NAD+/NADP+ measurements
Basic Protocol 4 : NADH/NADPH measurements
Basic Protocol 5 : Data analysis and quality control approaches
INTRODUCTION
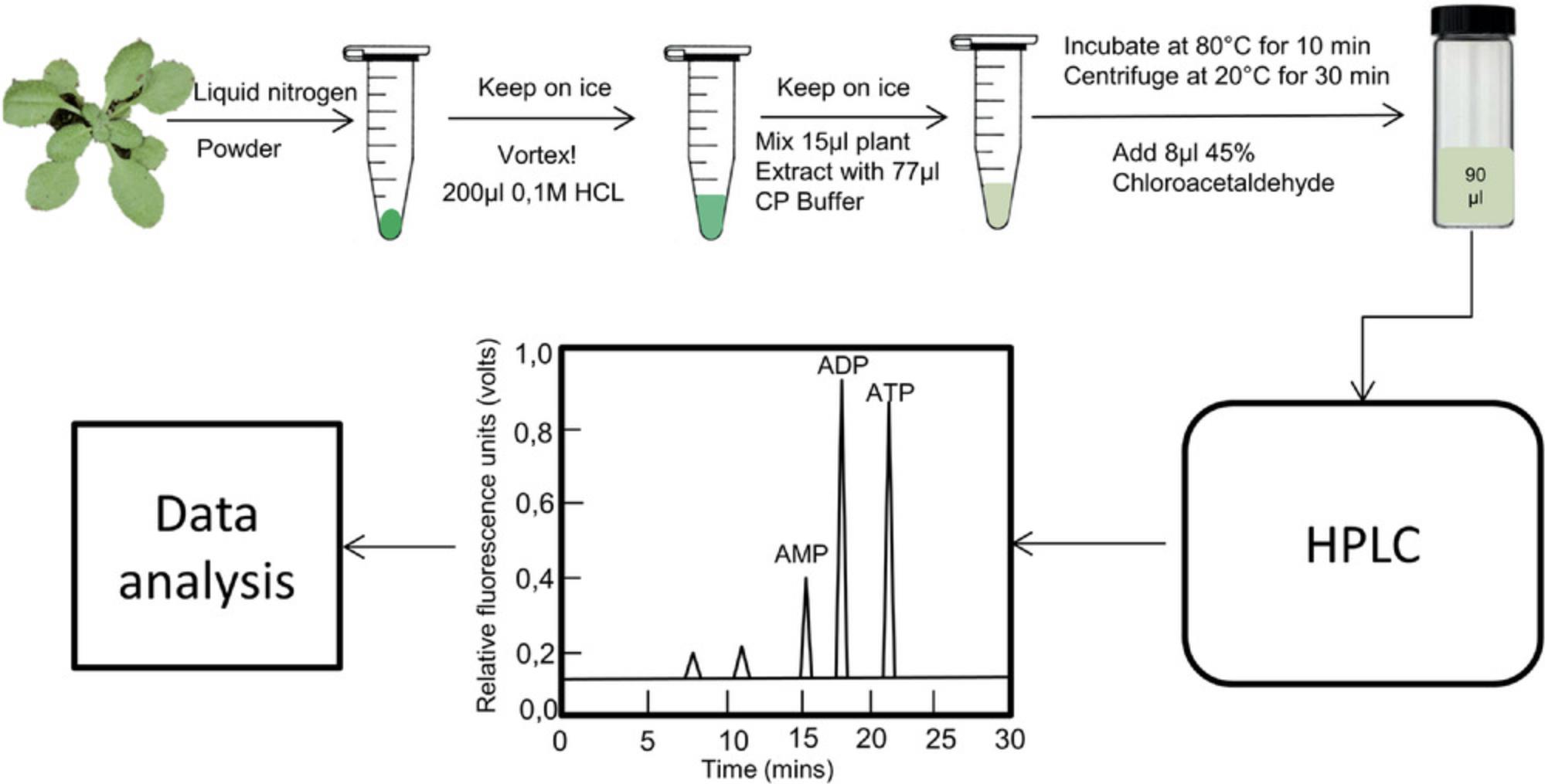
Both adenine nucleotides and nicotinamide adenine dinucleotide are essential for energy transfer as wells as enzyme regulation, functioning as co-factors in various signal transduction pathways in living plant cells (Atkinson, 1971; Srivastava & Bernhard, 1985). Adenine nucleotides, including adenosine monophosphate (AMP), adenosine diphosphate (ADP), and adenosine triphosphate (ATP), are a group of organic molecules that can also act as building blocks for both DNA and RNA (Bhatt, Chen, Geiger, & Rosenberger, 2012). AMP has the typical structure of a nucleotide, including a purine base, adenine, attached to a five-carbon sugar and one phosphate group. AMP can be reversibly converted into ADP or ATP by the addition of one and two phosphate groups, respectively. These reversible reactions store and release energy by the establishment or breaking of phosphate bonds, and are thereby essential for energy transfer and supply in multiple cellular reactions, especially cellular respiration. Given the important role of adenine nucleotides in various plant growth conditions, the adenylate energy charge (AEC, [ATP + (0.5ADP)]/[AMP + ADP + ATP]) has often been used to describe the energetic status of cells or tissues (Pradet & Raymond, 1983). However, the measurement of adenine nucleotides from living plant cells is a challenging task, often requiring high amounts of sample in order to obtain detectable concentrations, or the use of indirect modes of measurement that employ radiotracers or enzyme-linked assays (Bhatt et al., 2012). Here, we describe a highly sensitive and selective method to quantify adenine nucleotides and calculate the AEC in plant cells (Fig. 1).
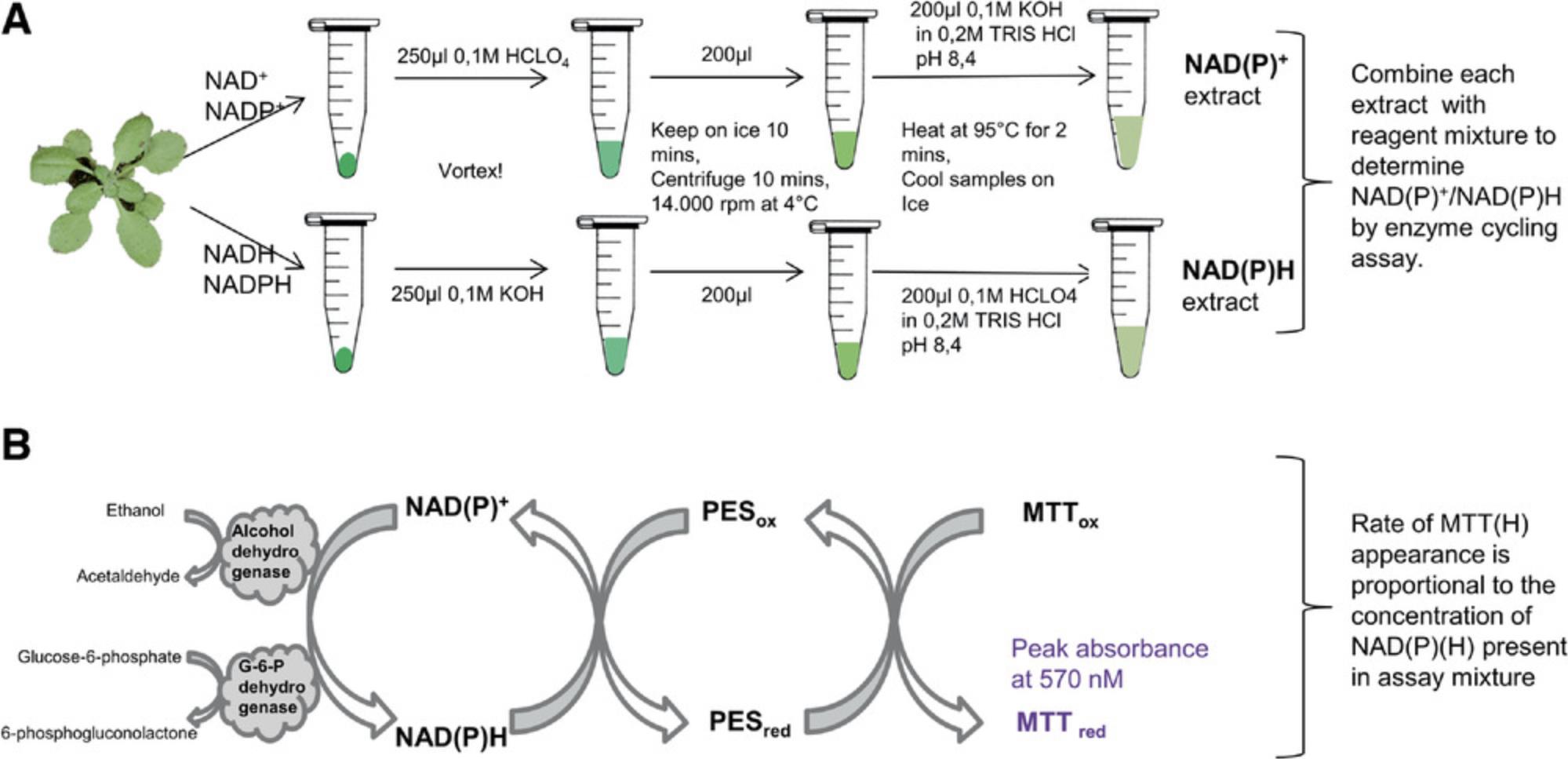
As enzyme cofactors, both oxidized nicotinamide adenine dinucleotide (NAD+) and oxidized nicotinamide adenine dinucleotide phosphate (NADP+) can be reversibly reduced by the additional of two hydrogen ions to form NADH and NADPH (Palmer & Møller, 1982; Riebel, Gibbs, Wellborn, & Bommarius, 2003). In this process, NADH can donate electrons to the respiratory chain through NADH dehydrogenase, regenerating NAD+, which participates in redox transformations involved in the oxidative conversion of substrates to metabolic end products, such as CO2 and H2O in the case of sugar metabolism (Rasmusson & Wallström, 2010). The reversible conversion of NADP+ and NADPH is commonly involved in anabolic reactions, including lipid and nucleic acid biosynthesis and the Calvin cycle (Kern, Price-Whelan, & Newman, 2014; Tamoi, Miyazaki, Fukamizo, & Shigeoka, 2005). NAD(H) and NADP(H) can be converted by coupling NADP+ reduction to NADH oxidation at the expense of moving one proton from the periplasm to the cytoplasm by the membrane-bound transhydrogenase enzyme. This conversion is reversible under native physiological conditions and dynamic equilibrium of appropriate cellular NAD(H) and NADP(H) redox levels (Kern et al., 2014). Thus, the measurement of reduced and oxidized nicotinamide cofactors will help us to further understand the regulation of plant metabolic pathways (Liang et al., 2016). Here, we describe an approach to nicotinamide adenine dinucleotide detection using an optimized in vitro colorimetric enzyme-mediated assay of cell extracts (Fig. 2A).
The basic protocols for adenine nucleotide and nicotinamide adenine dinucleotide measurements can be subdivided into five steps: (i) preparation of plant material (Basic Protocol 1), (ii) extraction and measurement of ATP, ADP, and AMP via HPLC (Basic Protocol 2), (iii) extraction and measurement of NAD+ and/or NADP+ (Basic Protocol 3), (iv) extraction and measurement of NADH and/or NADPH (Basic Protocol 4), and (v) data analysis and quality control (Basic Protocol 5).
STRATEGIC PLANNING
The programs for both the HPLC with the Hyperclone C18 (ODS) column and plate reader need to be prepared for the measurement before starting.
1.HPLC program (Estavillo et al., 2011)
- Column for HPLC: Hyperclone C18 (ODS) column (Phenomenex)
- Gradient flowrate 0.8 ml/min (50 min/sample)
- ○ 0-0.2 min equilibration: 95% Buffer A: 5% Buffer B
- ○ 0.2-53 min (linear gradient): up to 50% Buffer B
- ○ Recalibration: 7 min with 5% Buffer B
2.Microplate reader (e.g., Biotek EL 808 with 570-nm filter)
Basic Protocol 1: PREPARATION OF PLANT MATERIAL FOR THE MEASUREMENT
Both adenylate and pyrimidine nucleotide act as “currency” within living cells and play an important role in regulating plant growth and development (Gakière, Fernie, & Pétriacq, 2018; Geigenberger, Riewe, & Fernie, 2010). Although several publications use different approaches for measurement of adenylate and pyrimidine nucleotide, there is no detailed protocol for the measurement of these compounds in plants. Here, we present detailed protocols for sampling and extraction of whole seedlings, leaves, or roots. Because of the extremely high turnover rates of adenylate and pyrimidine nucleotides (down to milliseconds), plant material needs to be harvested as rapidly as possible, then instant-frozen, ideally by using the freeze-clamping technique (Ap Rees, Fuller, & Wright, 1977). When working with leaves, it is important to quench the samples directly in light, and not to alter the incident irradiance via shading. Given the instability of these metabolites, the plant materials should be frozen at –80°C for no more than 1 year. Here, we use Arabidopsis leaves as an example.
Materials
-
Plant material
-
Liquid nitrogen
-
2-ml microcentrifuge tubes
-
1.5 ml Safe-Lock tubes
-
Ball mill (Retsch)
-
Balance
1.Harvest around 100 mg of Arabidopsis or other plant leaf material in a 2.0-ml tube and snap-freeze it in liquid nitrogen.
2.Homogenize the plant material in a ball mill by adding a clean and pre-cooled metal ball to the leaf material, and grind for 30-45 s at medium speed.
3.Transfer aliquots of approximately 25 mg (±2 mg) to new 1.5-ml tubes and record the exact weight.
4.If assays cannot be performed immediately, store samples at –80°C until measurement.
Basic Protocol 2: MEASUREMENT OF ATP, ADP, AND AMP VIA HPLC
As the “molecular unit of currency” of intracellular energy transfer, ATP is a metabolite that provides energy to multiple processes in the living cell, including protein synthesis, chemical synthesis, metabolism, cell division, and transport processes. It can be converted to either ADP or AMP when consumed in living cells. Measurement of ATP, ADP, and AMP have been described in several articles (Liang et al., 2016; Pham et al., 2015; Zhang et al., 2018; Zhu et al., 2019), but detailed protocols are unavailable. Here, we describe in detail the process of ATP, ADP, and AMP measurement in plant samples (Fig. 1).
Materials
-
ATP disodium salt trihydrate (Sigma-Aldrich 10519987001)
-
ADP (Sigma-Aldrich 01905)
-
AMP disodium salt (Sigma-Aldrich 01930)
-
Sample: Arabidopsis leaf material prepared as in Basic Protocol 1
-
0.1 M HCl
-
CP buffer (see Table 1)
-
Chloroacetaldehyde (Sigma-Aldrich 317276-5 ml; HIGHLY TOXIC!)
-
Adenosine (Sigma-Aldrich A9251)
-
Running buffer A for HPLC (see Table 2)
-
Running buffer B for HPLC (see Table 3)
-
Refrigerated centrifuge
-
Heated shaker
-
Glass vials for HPLC
-
Hyperclone C18 (ODS) column (Phenomenex)
-
HPLC system (Dionex)
| Reagent | Source | Final conc. | For 10 ml | For 25 ml |
|---|---|---|---|---|
| Citric acid monohydrate, 1 M | Commonly available | 62 mM | 0.62 ml | 1.55 ml |
| (Na)2HPO4·2H2O, 1 M | Commonly available | 76 mM | 0.76 ml | 1.90 ml |
| Water | 8.62 ml | 21.55 ml |
- a
Adjust the pH to 4 with NaOH.
| Reagent | Source | Final conc. | For 2 L | For 5 L |
|---|---|---|---|---|
| Tetrabutylammonium hydrogen sulfate (TBAS) | Sigma 155837 | 5.7 mM | 3.87 g | 9.68 g |
| KH2PO4 | Roth 3904.2 | 30.5 mM | 8.30 g | 20.76 g |
- a
Adjust the pH to 5.8 with KOH.
| Reagent | Source | Final conc. | For 2 L | For 5 L |
|---|---|---|---|---|
| Acetonitrile, HPLC-grade | Commonly available | 67% | 1340 ml | 3350 ml |
| Running buffer A | See Table 2 | 33% | 660 ml | 1650 ml |
Preparation of adenylate standards for the measurement (always prepare fresh immediately before measurement)
1.Separately weigh defined amounts of AMP, ADP, and ATP powder to make 100 mM solutions. Dilute these solutions to 1 mM and then to 100 µM using 0.1 M HCl.
2.Take an equal amount of each of the 100 µM solutions of the three compounds to obtain 25, 12.5, 6.25, and 3.125 µM mixed solutions of the three compounds together.
3.Also prepare separate 25, 12.5, 6.25, and 3.125 µM solutions of each of the three individual compounds.
4.Take 15 µl of each of the different concentrations of mixed solution and each individual standard at different concentrations in a new 1.5-ml tube and keep on ice for the HPLC measurement.
ATP, ADP, and AMP measurement via HPLC
5.Add 200 µl of 0.1 M HCl to a 25-mg aliquot of the Arabidopsis leaf material in a 1.5-ml tube, prepared as in Basic Protocol 1, step 3, and vortex for 15 s prior to putting on ice for 30 s. Repeat vortexing for 15s, then centrifuge 10 min at 12,000 × g , 4°C.
6.Take 15 µl of the supernatant for HPLC measurement.
7.Dilute the 15 µl of plant extract from step 6 (sample) and each of the 15-μl aliquots of standard mixtures/single standards at different concentrations (step 4) with 77 µl CP buffer and keep on ice.
8.Add 8 µl 45% chloroacetaldehyde (HIGHLY TOXIC!) to each sample (to a volume of 100 µl).
9.Incubate at 80°C for 10 min.
10.Centrifuge 30 min at 12,000 × g , 20°C.
11.Transfer 90 µl of the supernatant to glass HPLC vials.
12.Run all the samples and standards on HPLC.
HPLC program (also see Strategic Planning and Basic Protocol 5)
The analysis of adenosines is performed by reversed-phase HPLC on a Hyperclone C18 (ODS) column connected to an HPLC system (Dionex). The HPLC analysis is carried out as described previously (Estavillo et al., 2011). The gradient for separation of adenosine derivatives is optimized as follows: equilibration of column for 0.2 min with 95% (v/v) of running buffer A (Table 2) and 5% (v/v) running buffer B (Table 3), linear gradient for 53 min up to 50% (v/v) of running buffer B, and re-equilibration for 7 min with 5% (v/v) running buffer B. The flowrate is set to 0.8 ml/min.
See Basic Protocol 5 for data interpretation.
Basic Protocol 3: NAD+/NADP+ MEASUREMENT
As the cofactor of the central metabolism, nicotinamide adenine dinucleotide (NAD) carries electrons from one reaction to another and is deeply involved in the regulation of redox. Oxidized nicotinamide adenine dinucleotide (NAD+) accepts electrons from other molecules and converts to the reduced form (NADH), especially in glycolysis and the mitochondrial TCA cycle (Zhang & Fernie, 2018; Zhang et al., 2017; Youjun Zhang et al., 2018). Nicotinamide adenine dinucleotide phosphate (NADP) is a cofactor in anabolic reactions, such as lipid and nucleic acid biosynthesis and the Calvin cycle, which use NADPH as a reducing agent (Kern et al., 2014). NAD+ can be converted to NADP+ by NAD+ kinase via the addition of a further phosphate group at the 2′ position of the ribose ring that carries the adenine moiety.
In this protocol, the reduced forms, i.e., NADH or NADPH, are first selectively destroyed at low pH and high temperature; then, NAD+ and NADP+ are measured using highly sensitive cycling assays. Alcohol dehydrogenase catalyzes the oxidation of ethanol, which involves the reduction of NAD+ (Fig. 2), and glucose-6-phosphate dehydrogenase catalyzes the oxidation of glucose 6-phosphate, which involves the reduction of NADP+ (Fig. 2B). Phenazine ethosulfate then catalyzes the reduction of MTT into deep-purple formazan, the absorbance of which can be measured at 570 nm. Concomitantly, the re-oxidation of NADH or NADPH makes it possible to re-commence the oxidation of ethanol or glucose-6-phosphate. This cycle, which obeys a zero-order kinetic rate law, is maintained at a constant rate as long as its substrates, i.e., ethanol or glucose-6-phosphate, and MTT, are saturating.
Materials
-
β-nicotinamide adenine dinucleotide hydrate (NAD+, Roche 10127990001)
-
β-nicotinamide adenine dinucleotide phosphate disodium salt (NADP+, Roche 10128031001)
-
0.1 M HCl
-
Liquid nitrogen
-
0.1 M perchloric acid (HClO4)
-
Sample: Arabidopsis leaf material prepared as in Basic Protocol 1
-
0.1 M KOH in 0.2 M Tris⋅HCl, pH 8.4
-
Double-distilled water
-
Detection mix for NAD+/NADH (Table 4)
-
Detection mix for NADP+/NADPH (Table 5)
-
Refrigerated centrifuge with rotor for microcentrifuge tubes, capable of speed of 20,000 × g
-
1.5-ml microcentrifuge tubes
-
Heat block
-
96-well microplates (Sarstedt; maximum well volume, 300 µl; transparent flat bottom)
-
Microplate reader (e.g., Biotek EL 808 with 570-nm filter)
| Reagent with final conc. | Stock conc. | Source | Amt. stock for 1 reaction | Amt. stock for 100 reactions |
|---|---|---|---|---|
| 0.3 M Tricine/KOH, pH 9.0 | 1 M | Carl Roth 6977.3 | 15 µl | 1500 µl |
| 12 mM disodium EDTA, pH 8.0 | 0.5 M | Commonly available | 1.2 µl | 120 µl |
| 0.3 mM PESa | 30 mM (water) | Sigma-Aldrich P4544 | 0.5 µl | 50 µl |
| 1.8 mM MTTb | 9 mM (water) | Sigma-Aldrich M2128 | 10 µl | 1000 µl |
| 1.5 M ethanol | 100% (17 M) | Commonly available | 4.4 µl | 440 µl |
| 18 U/ml alcohol dehydrogenase | 2000 U/ml | Sigma-Aldrich A3263 | 0.45 µl | 45 µl |
| Water | 18.45 µl | 1845 µl |
- a
Ethyldibenzopyrazine ethyl sulfate salt.
- b
Thiazolyl Blue tetrazolium bromide.
| Reagent with final conc. | Stock conc. | Source | Amt. stock for 1 reaction | Amt. stock for 100 reactions |
|---|---|---|---|---|
| 0.3 M Tricine/KOH, pH 9.0 | 1 M | Carl Roth 6977.3 | 15 µl | 1500 µl |
| 12 mM disodium EDTA, pH 8.0 | 100 mM | Commonly available | 1.2 µl | 120 µl |
| 0.3 mM PMSa | 30 mM | Sigma-Aldrich P9625 | 0.5 µl | 50 µl |
| 1.8 mM MTTb | 9 mM | Sigma-Aldrich M2128 | 10 µl | 1000 µl |
| 9 mM Glucose-6-phosphate | 180 mM | Sigma-Aldrich G7879 | 2.5 µl | 250 µl |
| 9 U/ml Glucose-6-phosphate dehydrogenase, grade I | 700 U/ml | Roche 10127990001 | 0.645 µl | 64.5 µl |
| Water | 20.155 µl | 2015.5 µl |
- a
Methylphenazinium methyl sulfate.
- b
Thiazolyl Blue tetrazolium bromide.
Preparation of NAD+/NADP+ standards for the measurement
1.Prepare 1 ml each of 20 mM NAD+ and NADP+ in 0.1 M HCl.
2.Divide the NAD**+** and NADP**+** solutions into 50-µl aliquots, freeze in liquid nitrogen, and store in a –80°C freezer.
Method for NAD+/NADP+ measurement
3.Add 200 µl of 0.1 M HClO4 to a 25-mg aliquot of Arabidopsis leaf material in a 1.5-ml tube, prepared as in Basic Protocol 1.
4.Immediately mix tube by tapping, vortex the tube for 15 s (keeping on ice), and repeat vortexing until the leaf powder has become homogenized.
5.Incubate tube 10 min on ice, then centrifuge 10 min at 12,000 × g , 4°C.
6.Transfer 200 µl of supernatant into a new 1.5-ml tube.
7.Heat tube to 95°C for 2 min, then cool down on ice.
8.Add 200 µl of 0.1 M KOH in 0.2 M Tris⋅HCl, pH 8.4, to neutralize the extract. Vortex.
9.Incubate tubes on ice for a minimum of 15 min.
10.Heat up the 20 mM NAD+ and NADP+ standards (each 50-µl aliquot prepared in step 2) to 95°C for 10 min, then cool down on ice.
11.Dilute the 20 mM standards to 1 µM with double-distilled water. Do this shortly before you start pipetting, because the 1 µM standard solution is very unstable and can only be used for maximum 4 hr.
12.Pipette samples (from step 9) and standards in a 96-well microplate with a final volume of 100 µl per well. Standard final concentration should be in the 0-25 pmol range (see Table 6). Do not put the plate on ice. Normally, we use the first two rows for the standards and the third row for the leaf samples.
| Final conc. in pmol per well | 0 | 5 | 10 | 15 | 20 | 25 |
| X µl 1 µM standard | 0 | 5 | 10 | 15 | 20 | 25 |
| X µl water | 100 | 95 | 90 | 85 | 80 | 75 |
13.First, make a test plate with two samples and the standards to check if the amount of NAD+ and NADP+ being produced is in the range of the standards.
14.For the test plate, pipette 2, 5, 10, and 25 µl of the two samples and fill the well to 100 µl with double-distilled water.
15.Prepare detection mix as in Table 4 for NAD+ and as in Table 5 for NADP+, adding PES for NAD+ and PMS for NADP+ shortly before adding the mix to the samples and standards. Add 50 µl of detection mix per well of the test plate under dim light.
Acquire data and calculate results
16.Basic Protocol 5 describes data analysis.
17.Read absorbance at 570 nm with the microplate reader for 45 min at 30°C.
18.Use V mean to calculate the amount of NAD(P)+.
19.Check the amount of NAD+ and NADP+ in the samples and whether the values are in the range of the standards—if not, dilute or concentrate the samples accordingly. Repeat the measurement with all samples. Here, we suggest processing three technical replicates for standards and samples. If you have six to eight biological replicates, you can decrease the number of technical replicates to two.
Basic Protocol 4: NADH/NADPH MEASUREMENT
Oxidized forms, i.e., NAD+ or NADP+, are first selectively destroyed at high pH and high temperature; then NADH and NADPH are measured using the same assays as in Basic Protocol 3 (Fig. 2B).
Materials
-
β-Nicotinamide adenine dinucleotide, reduced disodium salt hydrate (NADH, Roche 10107735001)
-
β-Nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt hydrate (NADPH, Roche 10621692001)
-
0.1 M KOH
-
Liquid nitrogen
-
Sample: Arabidopsis leaf material prepared as in Basic Protocol 1
-
0.1 M HClO4 in 0.2 M Tris pH 8.4 (mix 10 µl of 1 M HClO4 and 200 µl of 1 M Tris base, add 60 µl of ddH2O, adjust pH to 8.4, and make up volume to 100 µl with ddH2O)
-
Double-distilled water
-
Detection mix for NAD+/NADH (Table 4)
-
Detection mix for NADP+/NADPH (Table 5)
-
Refrigerated centrifuge with rotor for microcentrifuge tubes, capable of speed of 20,000 × g
-
1.5-ml microcentrifuge tubes
-
Heat block
-
96-well microplates (Sarstedt; maximum well volume, 300 µl; transparent flat bottom)
-
Microplate reader
Preparation of NADH/NADPH standards for the measurement
1.Prepare 1 ml each of 20 mM NADH and NADPH in 0.1 M KOH.
2.Divide the NADH and NADPH solutions into 50-µl aliquots, freeze them in liquid nitrogen, and store in a –80°C freezer.
Method for NADH/NADPH measurement
3.Add 0.1 M KOH to a 25-mg aliquot of Arabidopsis leaf powder material in a 1.5-ml tube, prepared as in Basic Protocol 1.
4.Immediately mix tube by tapping, vortex the tube for 15 s (keeping on ice), and repeat vortexing until the leaf powder has become homogenized.
5.Incubate tube 10 min on ice, then centrifuge 10 min at 12,000 × g , 4°C.
6.Transfer 200 µl of the supernatant into a new 1.5-ml tube.
7.Heat tube to 95°C for 2 min, then cool down on ice.
8.Add 200 µl of 0.1 M HClO4 in 0.2 M Tris·HCl, pH 8.4, to neutralize the extract. Vortex.
9.Incubate tubes on ice for a minimum of 15 min.
10.Heat up the 20 mM NADH and NADPH standards (each 50-µl aliquot prepared in step 2) to 95°C for 10 min, then cool down on ice.
11.Dilute the 20 mM standards to 1 µM with double-distilled water. Do this shortly before you start pipetting, because the 1 µM standard solution is very unstable and can only be used for a maximum of 4 hr.
12.Pipette samples and standards in a 96-well microplate for a final volume of 100 µl per well. Standard final concentration should be in the 0-25 pmol range (see Table 6). Do not put the plate on ice. Normally, we use the first two rows for the standards and the third row for the leaf samples.
13.First make a test plate with two samples and the standards to check if the amount of NADH and NADPH being produced is in the range of the standards.
14.For the test plate, pipette 2, 5, 10, 25 µl of two samples and fill up to 100 µl with double-distilled water
15.Prepare detection mix as in Table 4 for NADH and as in Table 5 for NADPH, adding PES for NADH and PMS for NADPH shortly before adding the mix to the samples and standards. Add 50 µl of detection mix per well of the test plate under dim light.
Acquire data and calculate results
16.Basic Protocol 5 describes data analysis.
17.Read absorbance at 570 nm at 30°C with the microplate reader until the rate stabilizes.
18.Use V mean (calculated by the software of the machine) to calculate the amount of NAD(P)H.
19.Check the amount of NADH and NADPH in the samples and whether the values are in the range of the standards—if not, dilute or concentrate the samples accordingly. Repeat the measurement with all samples. For standards and samples you should have three technical replicates. If you have three to four biological replicates, you can decrease the number of technical replicates to two.
Basic Protocol 5: DATA ANALYSIS AND QUALITY CONTROL
After completing the HPLC and plate reading, the raw data can be used for the analysis of the final levels of the metabolites. The normalized values are processed with at least six biological replicates in our laboratory and used to calculate the concentration of the adenine nucleotides and nicotinamide adenine dinucleotide. It is very important to note that the values of samples should not be above or below the range of standards used for the standard curves. Here, we use the wild-type Arabidopsis (Col-0 accession) seedling as example to analyze the concentration of adenine nucleotides and nicotinamide adenine dinucleotide.
Data analysis
1.In the ATP/ADP/AMP measurement (Basic Protocol 2), we use 200 µl extraction buffer and 15 µl reaction buffer. 10-µl samples are injected into the column. Here we use ATP as the example and the analysis ADP or AMP methods are same as for ADP (Fig. 3A).
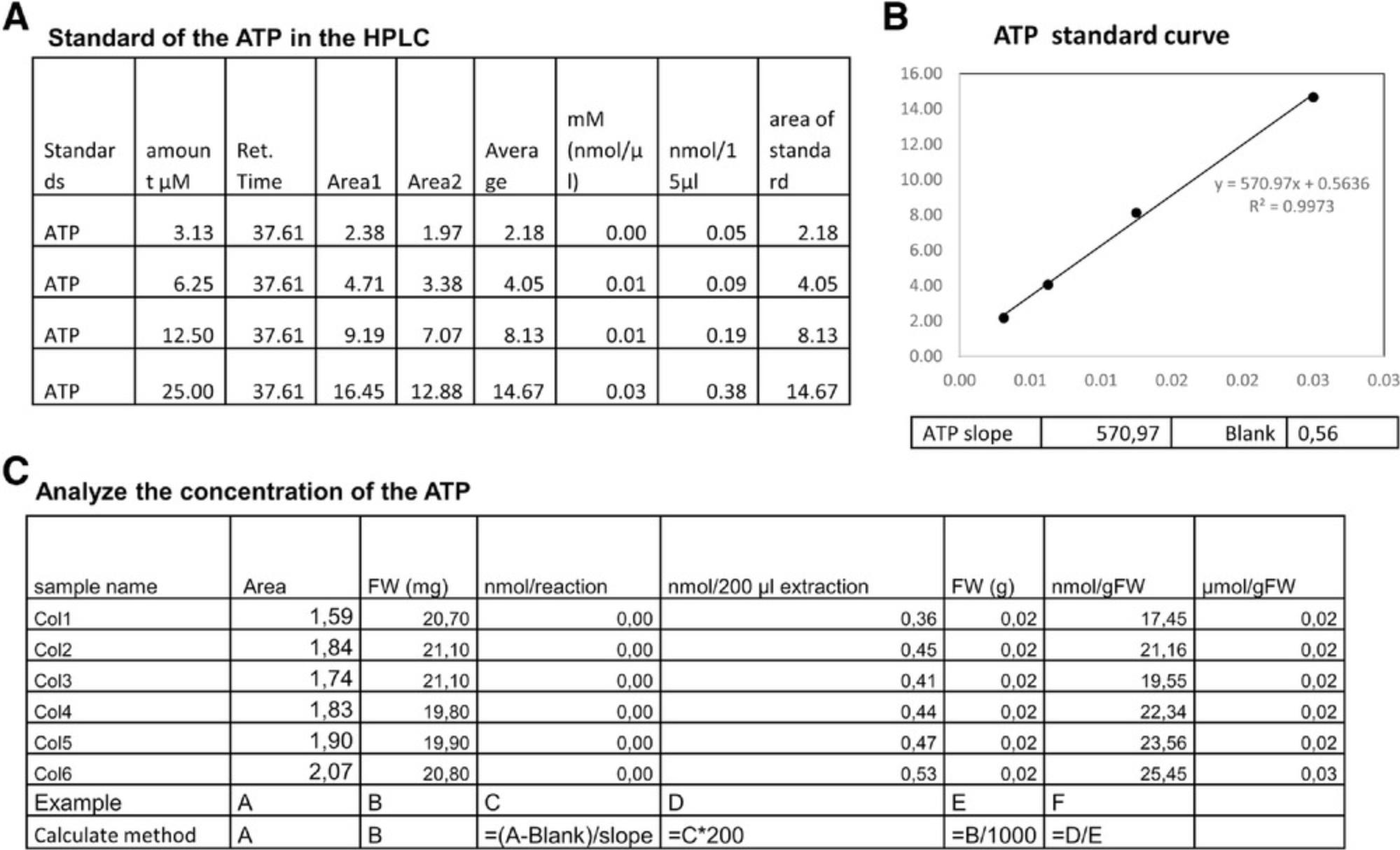
2.First, we analyze the ATP standards from the HPLC and get the values of slope and blank (Fig. 3B).
3.After we get the slope, we can calculate concentrations within samples as shown in Fig. 3C. Briefly, the reaction concentration is calculated as (value − blank)/slope and normalized by dilution factor and fresh weight (FW). Normally, the final level is normalized to provide a value with the units µmol/gFW (Fig. 3C), where gFW refers to the fresh weight of the plant material in grams.
4.Similarly to the ATP analysis, the concentrations of pyrimidine nucleotide are also normalized from the standard. Here, we use NAD+ as an example (Fig. 4A).
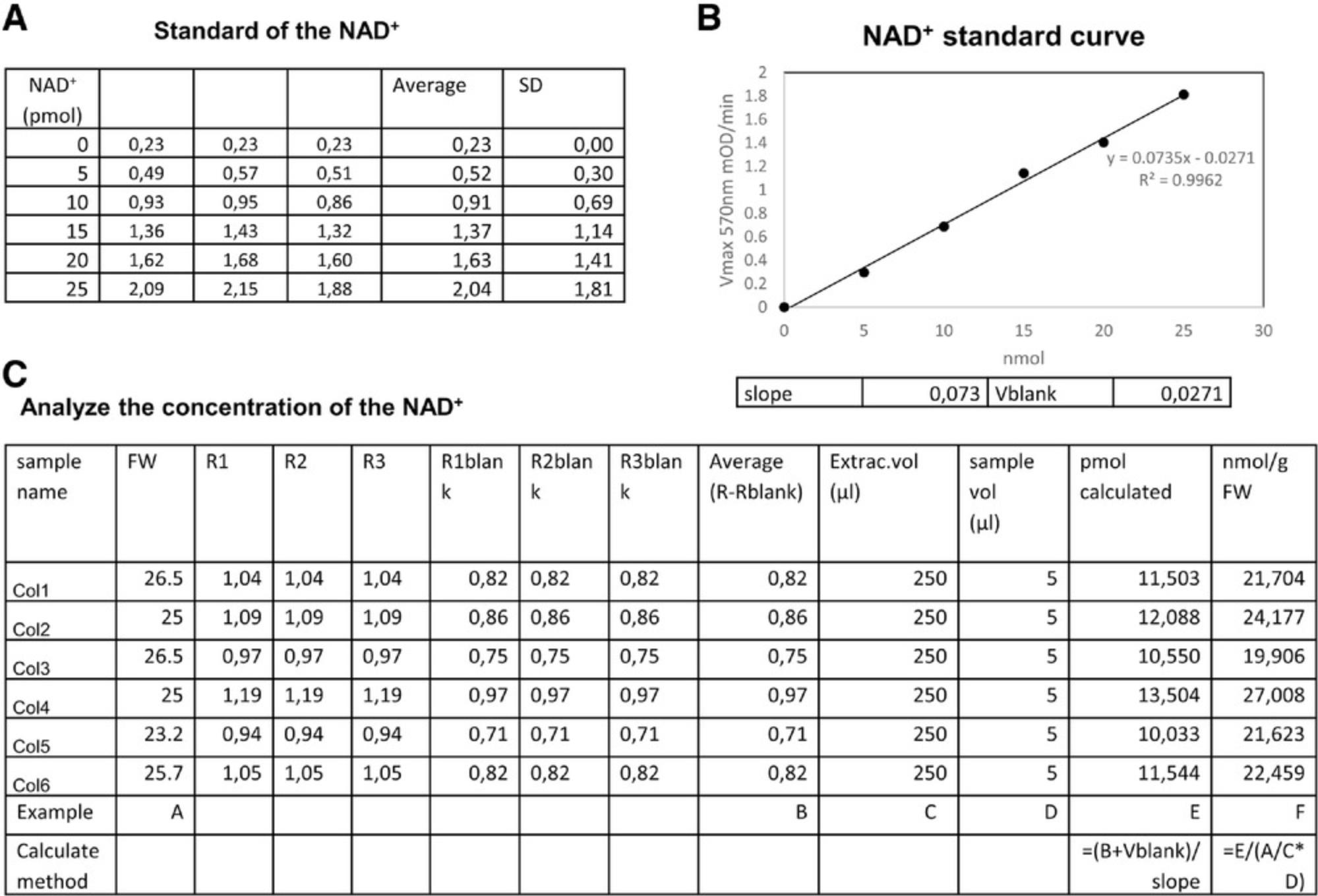
5.We first use the raw data of the standard to get the slope and V blank (Fig. 4B).
6.After we get the slope, we can go back to the concentration of samples as shown in Figure 4C. Briefly, the concentration is calculated as (value + blank)/slope and normalized by dilution factor, sample volume, and fresh weight. Usually, the final level is normalized to provide a value with the units nmol/gFW, where gFW refers to the fresh weight of the plant material in grams.
COMMENTARY
Background Information
As the universal energy donor molecules of living cells, ATP and ADP are representative of cellular energy and health. There are several methods that have been used to measure the concentration of ATP, ADP, and AMP, including the direct measurement of adenine nucleotide fluorescence from cell extract by HPLC (Bhatt et al., 2012), monitoring of single-wavelength genetically encoded fluorescent sensors for imaging of extracellular and cytosolic ATP (Lobas et al., 2019), and enzyme-mediated assays of cell extracts (Lee et al., 2012). The direct measurement of adenine nucleotide fluorescence includes several analytical procedures utilizing either isocratic or gradient reversed-phase, ion-exchange, or ion-pairing HPLC in combination with UV or fluorescence detection (Bhatt et al., 2012). Due to its ability to provide stable and reproducible results, we have used ion-pairing reversed-phase HPLC combined with UV or fluorescence detection to directly measure adenine nucleotide fluorescence. The detection mode varies; UV detection can be used for the micromole to nanomole range and fluorescence detection for the nanomole to picomole range, depending upon the amount of sample available for analysis. Therefore, to study energy metabolism in plant materials, we developed an ion-pairing HPLC method combined with fluorescence detection to accurately detect adenine nucleotides and calculate adenylate energy charge. This approach was optimized to limit the loss of native ATP/ADP/AMP due to hydrolysis. It allows baseline separation to quantify picomolar levels of nucleotides and is suitable for detecting small variations in levels of both adenine nucleotides and adenosine. Due to the importance of ATP/ADP/AMP metabolism and our interest in understanding the role of plant energy metabolism under different physiological condition, the method described here provides a clear advantage in quantifying the concentration of ATP/ADP/AMP in vitro and calculating the adenylate energy charge.
As the coenzymes of many dehydrogenase enzymes, NAD+ and NADP+ are linked to many biochemical reactions via the transfer of hydrogen groups between molecules to regulate their redox status (Geigenberger & Fernie, 2014). Accurate analysis of these dinucleotide redox cofactors from biological samples is important to understanding cellular redox homeostasis. In addition, these compounds can be detected by several methods, including direct measurement of NADH fluorescence by light absorbance at 340 nm or fluorescent emission of light at 445 nm (Chen, Xia, & Ju, 2003; Hung, Albeck, Tantama, & Yellen, 2011), measurement of NAD(H)-binding GFP fluorescence (Hung et al., 2011), colorimetric enzyme-mediated assays of cell extracts in vitro (Price-Whelan, Dietrich, & Newman, 2007), and time-resolved fluorescence profiling of cells in situ using multiphoton microscopy (Sullivan, Tzeranis, Wang, So, & Newman, 2011). It is important to note that direct detection of the concentration of NAD(P)(H) in the plant cell using only spectral properties may also be affected by other endogenous chemicals that have similar spectral properties, such as the iron chelator pyoverdine and 1-hydroxyphenazine (Kern et al., 2014; Sullivan et al., 2011). Due to the simplicity, sensitivity, and specificity of such assays, a time-resolved enzymatic colorimetric assay is used for the plant materials based on well-developed procedures for other species (Bernofsky & Swan, 1973; Kern et al., 2014; San et al., 2002). This reversible experimental procedure is specific to either NADP(H) or NAD(H), which could be detected from a single set of mixtures with an exchange of substrate and enzymes (Gibon et al., 2004) (Fig. 2). With high sensitivity, NADP(H) and NAD(H) concentrations as low as 0.05 μM (<1 pmol in the assay mixture, equivalent to ∼5 nmol/109 cells) can be detected.
Critical Parameters and Troubleshooting
The following troubleshooting guide does not include common issues that may arise when using reagents other than those recommended in the protocol.
Plant harvest
It is important to harvest the plant material as quickly as possible, without shading, and snap freeze the harvested material in liquid nitrogen. Given the rapidly reversible changes of ATP/ADP, NAD+/NADH, and NADP+/NADPH, sample collection should be as simple as possible; otherwise, time delays will significantly affect these ratios. In our experiment, we also found that light significantly affects these metabolites. It is therefore important to avoid the shadowing of plant leaves during harvest either by the harvesters themselves or any laboratory equipment.
HPLC detection
Regarding the different detection modes for HPLC, UV detection can measure in the micromolar to nanomolar range while fluorescence detection can be used to measure in the nanomolar to picomolar range. We strongly suggest using fluorescence detection.
Sensitivity to light
In the nicotinamide adenine dinucleotide measurement, PES is very sensitive to light and degrades to form a compound that is dark in color, which interferes with the A 570 readings of reduced MTT. When pipetting PES or the reagent mix, turn off bright lights and perform these steps in dim lighting, away from windows. Cover tubes of PES aliquots (which may be stored at −20°C) and the reagent mix in foil. Protect the final mix of samples and reagents from light by covering the 96-well plate with foil until it is moved inside the plate reader. In order to minimize measurement error due to pipetting, include technical replicates of the samples and standards on the 96-well plate. Use of multichannel pipets is strongly recommended.
Avoid measuring out of the range of the standard curve
It is very important to keep the value of the measurement in the range of the standards. Thus, the pre-test is essential for the experiment. If the value of the sample is too high, it can be diluted. If the value is two low, a larger amount of starting plant material can be sampled.
Understanding Results
The adenine nucleotide and nicotinamide adenine dinucleotide measurement protocols presented in this article have been used by us and our collaborators to characterize the levels of these metabolites. These protocols should enable both novices and skilled scientists alike to obtain valuable and meaningful information about these metabolites and help in understanding the regulation of plant metabolism.
In the following paragraphs, we will detail some of the studies in which we have used these protocols. Most recently we have studied the two diphosphoinositol pentakisphosphate kinases VIH1/2 that affect plant growth and lead to constitutive Pi starvation responses. Mutations in the kinase active site lead to increased ATP levels that change significantly under different Pi growth conditions (Zhu et al., 2019; Zhu et al., 2019). In other research, RAPTOR1B can alter the hormone and metabolite compositions of Arabidopsis seeds. Here, the NAD+/NADH ratio was found to be changed in mutant lines while the NADP+/NADPH ratio was not found to change (Zhang et al., 2018). In addition, in research into whether the mitochondrial-localized thioredoxin o1 regulates photorespiratory metabolism, there were no significant alterations in NAD+ or NADH levels, and consequently also no changes in the deduced NADH/NAD+ ratio (Reinholdt et al., 2019). Moreover, the Arabidopsis thaliana purple acid phosphatase 2 (AtPAP2) can improve the plant growth and biomass of several species (Sun et al., 2012; Zhang et al., 2012; Zhang et al., 2014). Measurements of NADP+/NADPH content in the AtPAP2 and wild-type (WT) plant at all three time points (t = 0, 1, and 8 hr) indicate that the ATP levels and ATP/NADPH ratios were significantly increased in the OE line at all time points (Liang et al., 2015; Liang et al., 2016). An ATP/ADP translocator (NTT) and a plastidial soluble inorganic pyrophosphatase (psPPase) for starch composition and tuber formation in potato (Solanum tuberosum) were investigated by individual and simultaneous down-regulation of the corresponding endogenous genes (Andersson et al., 2018). Measurements of NAD(P)(H) made it possible to show that a novel calmodulin/calcium−dependent NAD kinase contributes to an increase in the cellular NADP+ concentration and to the amplification of the oxidative burst induced by the bacterial elicitor flagelin22 (Dell'Aglio et al., 2019). The evaluation of changes in NAD(P)(H) content was useful in complementing a survey of the expression of genes involved in pyridine nucleotide metabolism during tomato fruit development (Decros et al., 2019).
Time Considerations
If the plant materials are already prepared, the measurement of adenine nucleotides and nicotinamide adenine dinucleotide comprises several separate steps that could be accomplished within 1 week for 50 samples. The project is easily divided to adenine nucleotide measurement and nicotinamide adenine dinucleotide detection.
The time needed for determining adenine nucleotide and nicotinamide adenine dinucleotide concentration depends on (i) harvest of the plant, (ii) setup of the HPLC measurement system and the plate reader program, and (iii) measurement of the standards to establish the conditions of the individual experiment. In our experiment, the full measurement can be finished within 1 week.
Acknowledgments
This work was supported by funding from the Max-Planck Society (Y.Z. and A.R.F.), and A.R.F and Y.Z. would like to thank the European Union's Horizon 2020 research and innovation program and project PlantaSYST (SGA-CSA no 664621 and no 739582 under FPA no. 664620) for supporting their research. Y.G. acknowledges funding from PHENOME (ANR-11-INBS-0012). Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Youjun Zhang : Conceptualization; data curation; investigation; methodology; project administration; writing-original draft; writing-review & editing. Ina Krahnert : Methodology. Antje Bolze : Methodology. Yves Gibon : Writing-review & editing. Alisdair R. Fernie : Writing-review & editing.
Literature Cited
- Andersson, M., Turesson, H., Arrivault, S., Zhang, Y., Fält, A.-S., Fernie, A. R., & Hofvander, P. (2018). Inhibition of plastid PPase and NTT leads to major changes in starch and tuber formation in potato. Journal of Experimental Botany , 69(8), 1913–1924. doi: 10.1093/jxb/ery051.
- Ap Rees, T., Fuller, W., & Wright, B. (1977). Measurements of glycolytic intermediates during the onset of thermogenesis in the spadix of Arum maculatum. Biochimica et Biophysica Acta (BBA)-Bioenergetics , 461(2), 274–282. doi: 10.1016/0005-2728(77)90177-3.
- Atkinson, D. E. (1971). Adenine nucleotides as stoichiometric coupling agents in metabolism and as regulatory modifiers: The adenylate energy charge. In Metabolic regulation (pp. 1–21). New York: Academic Press.
- Bernofsky, C., & Swan, M. (1973). An improved cycling assay for nicotinamide adenine dinucleotide. Analytical Biochemistry , 53(2), 452–458. doi: 10.1016/0003-2697(73)90094-8.
- Bhatt, D. P., Chen, X., Geiger, J. D., & Rosenberger, T. A. (2012). A sensitive HPLC-based method to quantify adenine nucleotides in primary astrocyte cell cultures. Journal of Chromatography B , 889, 110–115. doi: 10.1016/j.jchromb.2012.02.005.
- Chen, F., Xia, Q., & Ju, L.-K. (2003). Aerobic denitrification of Pseudomonas aeruginosa monitored by online NAD(P)H fluorescence. Applied and Environmental Microbiology , 69(11), 6715–6722. doi: 10.1128/AEM.69.11.6715-6722.2003.
- Decros, G., Beauvoit, B., Colombié, S., Cabasson, C., Bernillon, S., Arrivault, S., … Baldet, P. (2019). Regulation of pyridine nucleotide metabolism during tomato fruit development through transcript and protein profiling. Frontiers in Plant Science , 10, 1201. doi: 10.3389/fpls.2019.01201.
- Dell'Aglio, E., Giustini, C., Kraut, A., Couté, Y., Costa, A., Decros, G., … Finazzi, G. (2019). Identification of the Arabidopsis calmodulin-dependent NAD+ kinase that sustains the elicitor-induced oxidative burst. Plant Physiology , 181(4), 1449–1458. doi: 10.1104/pp.19.00912.
- Estavillo, G. M., Crisp, P. A., Pornsiriwong, W., Wirtz, M., Collinge, D., Carrie, C., … Javot, H. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. The Plant Cell , 23(11), 3992–4012. doi: 10.1105/tpc.111.091033.
- Gakière, B., Fernie, A. R., & Pétriacq, P. (2018). More to NAD+ than meets the eye: A regulator of metabolic pools and gene expression in Arabidopsis. Free Radical Biology and Medicine , 122, 86–95. doi: 10.1016/j.freeradbiomed.2018.01.003.
- Geigenberger, P., & Fernie, A. R. (2014). Metabolic control of redox and redox control of metabolism in plants. Antioxidants & Redox Signaling, 21(9), 1389–1421.
- Geigenberger, P., Riewe, D., & Fernie, A. R. (2010). The central regulation of plant physiology by adenylates. Trends in Plant Science , 15(2), 98–105. doi: 10.1016/j.tplants.2009.11.004.
- Gibon, Y., Blaesing, O. E., Hannemann, J., Carillo, P., Höhne, M., Hendriks, J. H., … Stitt, M. (2004). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. The Plant Cell , 16(12), 3304–3325. doi: 10.1105/tpc.104.025973.
- Hung, Y. P., Albeck, J. G., Tantama, M., & Yellen, G. (2011). Imaging cytosolic NADH-NAD+ redox state with a genetically encoded fluorescent biosensor. Cell Metabolism , 14(4), 545–554. doi: 10.1016/j.cmet.2011.08.012.
- Kern, S. E., Price-Whelan, A., & Newman, D. K. (2014). Extraction and measurement of NAD(P)+ and NAD(P)H. Pseudomonas methods and protocols (pp. 311–323). New York: Springer.
- Lee, M.-S., Park, W.-S., Kim, Y. H., Ahn, W. G., Kwon, S.-H., & Her, S. (2012). Intracellular ATP assay of live cells using PTD-conjugated luciferase. Sensors , 12(11), 15628–15637. doi: 10.3390/s121115628.
- Liang, C., Cheng, S., Zhang, Y., Sun, Y., Fernie, A. R., Kang, K., … Lim, B. L. (2016). Transcriptomic, proteomic and metabolic changes in Arabidopsis thaliana leaves after the onset of illumination. BMC Plant Biology , 16(1), 43. doi: 10.1186/s12870-016-0726-3.
- Liang, C., Zhang, Y., Cheng, S., Osorio, S., Sun, Y., Fernie, A. R., … Lim, B. L. (2015). Impacts of high ATP supply from chloroplasts and mitochondria on the leaf metabolism of Arabidopsis thaliana. Frontiers in Plant Science , 6, 922. doi: 10.3389/fpls.2015.00922.
- Lobas, M. A., Tao, R., Nagai, J., Kronschläger, M. T., Borden, P. M., Marvin, J. S., … Khakh, B. S. (2019). A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nature Communications , 10(1), 1–13. doi: 10.1038/s41467-019-08441-5.
- Palmer, J. M., & Møller, I. M. (1982). Regulation of NAD(P)H dehydrogenases in plant mitochondria. Trends in Biochemical Sciences , 7(7), 258–261. doi: 10.1016/0968-0004(82)90039-1.
- Pham, P. A., Wahl, V., Tohge, T., de Souza, L. R., Zhang, Y., Do, P. T., … Fernie, A. R. (2015). Analysis of knockout mutants reveals non-redundant functions of poly(ADP-ribose) polymerase isoforms in Arabidopsis. Plant Molecular Biology , 89(4–5), 319–338. doi: 10.1007/s11103-015-0363-5.
- Pradet, A., & Raymond, P. (1983). Adenine nucleotide ratios and adenylate energy charge in energy metabolism. Annual Review of Plant Physiology , 34(1), 199–224. doi: 10.1146/annurev.pp.34.060183.001215.
- Price-Whelan, A., Dietrich, L. E., & Newman, D. K. (2007). Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. Journal of Bacteriology , 189(17), 6372–6381. doi: 10.1128/JB.00505-07.
- Rasmusson, A. G., & Wallström, S. V. (2010). Involvement of mitochondria in the control of plant cell NAD(P)H reduction levels. London: Portland Press Ltd.
- Reinholdt, O., Schwab, S., Zhang, Y., Reichheld, J.-P., Fernie, A. R., Hagemann, M., & Timm, S. (2019). Redox-regulation of photorespiration through mitochondrial thioredoxin o1. Plant Physiology , 181(2), 442–457. doi: 10.1104/pp.19.00559.
- Riebel, B. R., Gibbs, P. R., Wellborn, W. B., & Bommarius, A. S. (2003). Cofactor regeneration of both NAD+ from NADH and NADP+ from NADPH: NADH oxidase from Lactobacillus sanfranciscensis. Advanced Synthesis & Catalysis, 345(6–7), 707–712.
- San, K.-Y., Bennett, G. N., Berríos-Rivera, S. J., Vadali, R. V., Yang, Y.-T., Horton, E., … Blackwood, K. (2002). Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metabolic Engineering , 4(2), 182–192. doi: 10.1006/mben.2001.0220.
- Srivastava, D., & Bernhard, S. (1985). Mechanism of transfer of reduced nicotinamide adenine dinucleotide among dehydrogenases. Biochemistry , 24(3), 623–628. doi: 10.1021/bi00324a013.
- Sullivan, N. L., Tzeranis, D. S., Wang, Y., So, P. T., & Newman, D. (2011). Quantifying the dynamics of bacterial secondary metabolites by spectral multiphoton microscopy. ACS Chemical Biology , 6(9), 893–899. doi: 10.1021/cb200094w.
- Sun, F., Suen, P. K., Zhang, Y., Liang, C., Carrie, C., Whelan, J., … Lim, B. L. (2012). A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield. New Phytologist , 194(1), 206–219. doi: 10.1111/j.1469-8137.2011.04026.x.
- Tamoi, M., Miyazaki, T., Fukamizo, T., & Shigeoka, S. (2005). The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. The Plant Journal , 42(4), 504–513. doi: 10.1111/j.1365-313X.2005.02391.x.
- Zhang, Y., Beard, K. F., Swart, C., Bergmann, S., Krahnert, I., Nikoloski, Z., … Fernie, A. R. (2017). Protein-protein interactions and metabolite channelling in the plant tricarboxylic acid cycle. Nature Communications , 8(1), 1–11.
- Zhang, Y., & Fernie, A. R. (2018). On the role of the tricarboxylic acid cycle in plant productivity. Journal of Integrative Plant Biology , 60(12), 1199–1216. doi: 10.1111/jipb.12690.
- Zhang, Y., Sun, F., Fettke, J., Schöttler, M. A., Ramsden, L., Fernie, A. R., & Lim, B. L. (2014). Heterologous expression of AtPAP2 in transgenic potato influences carbon metabolism and tuber development. FEBS Letters , 588(20), 3726–3731. doi: 10.1016/j.febslet.2014.08.019.
- Zhang, Y., Swart, C., Alseekh, S., Scossa, F., Jiang, L., Obata, T., … Fernie, A. R. (2018). The extra-pathway interactome of the TCA cycle: Expected and unexpected metabolic interactions. Plant Physiology , 177(3), 966–979. doi: 10.1104/pp.17.01687.
- Zhang, Y., Yu, L., Yung, K.-F., Leung, D. Y., Sun, F., & Lim, B. L. (2012). Over-expression of AtPAP2 in Camelina sativa leads to faster plant growth and higher seed yield. Biotechnology for Biofuels , 5(1), 19. doi: 10.1186/1754-6834-5-19.
- Zhang, Y., Zhang, Y., McFarlane, H. E., Obata, T., Richter, A. S., Lohse, M., … Giavalisco, P. (2018). Inhibition of TOR represses nutrient consumption, which improves greening after extended periods of etiolation. Plant Physiology , 178(1), 101–117. doi: 10.1104/pp.18.00684.
- Zhu, J., Lau, K., Puschmann, R., Harmel, R. K., Zhang, Y., Pries, V., … Hothorn, M. (2019). Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. Elife , 8, e43582. doi: 10.7554/eLife.43582.
- Zhu, J., Loubéry, S., Broger, L., Zhang, Y., Lorenzo-Orts, L., Utz-Pugin, A., … Hothorn, M. (2019). A genetically validated approach to detect inorganic polyphosphates in plants. The Plant Journal , 102(3), 507–516. doi: 10.1111/tpj.14642.
Citing Literature
Number of times cited according to CrossRef: 11
- Stefan Timm, Nicole Klaas, Janice Niemann, Kathrin Jahnke, Saleh Alseekh, Youjun Zhang, Paulo V. L. Souza, Liang‐Yu Hou, Maike Cosse, Jennifer Selinski, Peter Geigenberger, Danilo M. Daloso, Alisdair R. Fernie, Martin Hagemann, Thioredoxins o1 and h2 jointly adjust mitochondrial dihydrolipoamide dehydrogenase‐dependent pathways towards changing environments, Plant, Cell & EnvironmentPlant, Cell & EnvironmentPlant, Cell & Environment, 10.1111/pce.14899, 47 , 7, (2540-2558), (2024).
- Jonas Giese, Jürgen Eirich, Dirk Walther, Youjun Zhang, Ines Lassowskat, Alisdair R. Fernie, Marlene Elsässer, Veronica G. Maurino, Markus Schwarzländer, Iris Finkemeier, The interplay of post‐translational protein modifications in Arabidopsis leaves during photosynthesis induction, The Plant Journal, 10.1111/tpj.16406, 116 , 4, (1172-1193), (2023).
- Tomasz Piechowiak, Bartosz Skóra, Edible coating enriched with cinnamon oil reduces the oxidative stress and improves the quality of strawberry fruit stored at room temperature, Journal of the Science of Food and Agriculture, 10.1002/jsfa.12463, 103 , 5, (2389-2400), (2023).
- Chengyang Li, Patrick Duckney, Tong Zhang, Yanshu Fu, Xin Li, Johan Kroon, Geert De Jaeger, Yunjiang Cheng, Patrick J. Hussey, Pengwei Wang, TraB family proteins are components of ER-mitochondrial contact sites and regulate ER-mitochondrial interactions and mitophagy, Nature Communications, 10.1038/s41467-022-33402-w, 13 , 1, (2022).
- Tomasz Piechowiak, Katarzyna Grzelak-Błaszczyk, Michał Sójka, Maciej Balawejder, One-time ozone treatment improves the postharvest quality and antioxidant activity of Actinidia arguta fruit, Phytochemistry, 10.1016/j.phytochem.2022.113393, 203 , (113393), (2022).
- Ruchanok Tinikul, Duangthip Trisrivirat, Wachirawit Chinantuya, Thanyaporn Wongnate, Pratchaya Watthaisong, Jittima Phonbuppha, Pimchai Chaiyen, Detection of cellular metabolites by redox enzymatic cascades, Biotechnology Journal, 10.1002/biot.202100466, 17 , 6, (2022).
- James William Allwood, Alex Williams, Henriette Uthe, Nicole M. van Dam, Luis A. J. Mur, Murray R. Grant, Pierre Pétriacq, Unravelling Plant Responses to Stress—The Importance of Targeted and Untargeted Metabolomics, Metabolites, 10.3390/metabo11080558, 11 , 8, (558), (2021).
- Luis Cervela-Cardona, Takuya Yoshida, Youjun Zhang, Masaaki Okada, Alisdair Fernie, Paloma Mas, Circadian Control of Metabolism by the Clock Component TOC1, Frontiers in Plant Science, 10.3389/fpls.2021.683516, 12 , (2021).
- Nina M. Scheurer, Yogeswari Rajarathinam, Stefan Timm, Christin Köbler, Joachim Kopka, Martin Hagemann, Annegret Wilde, Homologs of Circadian Clock Proteins Impact the Metabolic Switch Between Light and Dark Growth in the Cyanobacterium Synechocystis sp. PCC 6803, Frontiers in Plant Science, 10.3389/fpls.2021.675227, 12 , (2021).
- Edward N. Smith, Markus Schwarzländer, R. George Ratcliffe, Nicholas J. Kruger, Shining a light on NAD- and NADP-based metabolism in plants, Trends in Plant Science, 10.1016/j.tplants.2021.06.010, 26 , 10, (1072-1086), (2021).
- Simone Landi, Giorgia Capasso, Sergio Esposito, Different G6PDH isoforms show specific roles in acclimation to cold stress at various growth stages of barley (Hordeum vulgare) and Arabidopsis thaliana, Plant Physiology and Biochemistry, 10.1016/j.plaphy.2021.11.017, 169 , (190-202), (2021).

