A method to prepare Sera-Mag SpeedBeads for purification and size selection of nucleic acids
John B. Ridenour, Rafal Donczew
Solid-phase reversible immobilization (SPRI)
Nucleic acid purification
Nucleic acid size selection
High-throughput sequencing
Abstract
In this protocol, we describe a method to prepare Sera-Mag Speedbeads for purification and size selection of nucleic acids. We additionally describe a method to validate speedbead preparations and a general method for purification of nucleic acids using speedbeads. This protocol is based on previously described methods (DeAngelis et al., 1995; Rohland and Reich, 2012; Glenn et al., 2019; Jolivet and Foley, 2020; and Möller et al., 2023). We use homebrewed speedbeads as a cost-effective substitute for commercial solid-phase reversible immobilization (SPRI) products (e.g., Mag-Bind TotalPure NGS Beads, AMPure XP) during the preparation of samples for high-throughput sequencing and other applications. The amounts of polyethylene glycol (PEG) and sodium chloride (NaCl), which drive SPRI activity, have been optimized to meet our needs.
REFERENCES
DeAngelis MM, Wang DG, Hawkins TL (1995). Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Research , 23 (22), 4742. https://doi.org/10.1093/nar/23.22.4742
Glenn TC, Nilsen RA, Kieran TJ, Sanders JG, Bayona-Vásquez NJ, Finger JW, Pierson TW, Bentley KE, Hoffberg SL, Louha S, Garcia-De Leon FJ, Del Rio Portilla MA, Reed KD, Anderson JL, Meece JK, Aggrey SE, Rekaya R, Alabady M, Belanger M, Winker K, Faircloth BC (2019). Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ , 7 , e7755. https://doi.org/10.7717/peerj.7755
Jolivet P, Foley JW (2020). SPRI bead mix. protocols.io https://dx.doi.org/10.17504/protocols.io.bnz4mf8w
Möller M, Ridenour JB, Wright DF, Martin FA, Freitag M (2023). H4K20me3 is important for Ash1-mediated H3K36me3 and transcriptional silencing in facultative heterochromatin in a fungal pathogen. PLOS Genetics 19 (9), e1010945. https://doi.org/10.1371/journal.pgen.1010945
Rohland, N., & Reich, D. (2012). Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Research , 22 (5), 939-946. https://doi.org/10.1101/gr.128124.111
Steps
Preparation of 10% (v/v) Tween 20
Place a 50 ml conical tube on a balance and tare the balance.
Slowly add 5.475 g of Tween 20 to the conical tube using a serological pipette.
Remove the conical tube from the balance and add 45 ml of MilliQ water.
Thoroughly mix the solution with shaking until homogenous (approximately 20 min). Store 10% (v/v) Tween 20 at room temperature.
Preparation of speedbead DNA buffer
Prepare the following buffer in a 50 ml conical tube:
Speedbead DNA buffer
| A | B | C |
|---|---|---|
| Reagent | Volume | Final conc. |
| 1 M Tris-HCl (pH 8.0) | 500 μl | 10 mM |
| 0.5 M EDTA (pH 8.0) | 100 μl | 1 mM |
| 10% (v/v) Tween 20 | 250 μl | 0.05% |
| Nuclease-free water | 49.15 ml | – |
| Total | 50 ml | – |
Preparation of speedbead DNA binding mix
Equilibrate Sera-Mag Speedbeads to room temperature and mix thoroughly by vortexing.
Immediately after mixing, transfer 1 ml of bead slurry to a 1.5 ml microcentrifuge tube. The speedbeads will settle quickly. The bead slurry must be thoroughly mixed immediately before transferring the 1 ml to a 1.5 ml microcentrifuge tube.
Place samples on a magnetic stand until solution is clear.
Carefully remove and discard cleared supernatant without disturbing the bead pellet.
Remove beads from magnetic stand. Add 1 ml of speedbead DNA buffer and vortex beads on setting 4 for 15 seconds. Very briefly centrifuge samples after mixing. Stop the centrifuge before beads settle.
Repeat Steps 8-10 twice. Keep bead slurry at room temperature.
Prepare the following mixture in a 50 ml conical tube as described in Steps 13-21:
Speedbead DNA binding mix
| A | B | C |
|---|---|---|
| Reagent | Volume | Final conc. |
| PEG-8000 | 9 g | 18% (w/v) |
| 5 M NaCl | 100 μl | 2 M |
| 1 M Tris-HCl (pH 8.0) | 500 μl | 10 mM |
| 0.5 M EDTA (pH 8.0) | 100 μl | 1 mM |
| 10% (v/v) Tween 20 | 250 μl | 0.05% |
| Prepared speedbeads | 1 ml | – |
| Nuclease-free water | up to total | – |
| Total | 50 ml | – |
Add 9 g of PEG-8000 to a 50 ml conical tube.
Add 20 ml of 5 M NaCl to the conical tube.
Add 500 μl of 1.0 M Tris-HCl (pH 8.0) to the conical tube.
Add 100 μl 0.5 M EDTA (pH 8.0) to the conical tube.
Add nuclease-free water (up to 45 ml) to the 50 ml conical tube.
Mix the solution with shaking until PEG-8000 goes into solution (approximately 5-10 min).
Add 250 μl of 10% (v/v) Tween 20 to the conical tube and mix with gentle shaking until Tween 20 goes into solution (approximately 5 min).
Ensure beads (prepared in Steps 6-11) are fully resuspended in speedbead DNA buffer and add the bead suspension to the conical tube.
Add nuclease-free water up to 50 ml to the conical tube and mix the speedbead DNA binding mix thoroughly by vortexing. Nuclease-free water can be added up to 50 ml by eye using a 1000 μl pipet.
Wrap the speedbead DNA binding mix in foil and store at 4°C. Speedbead DNA binding mix can be stored at 4°C for several months.
Validate speedbeads after preparation to ensure they are working as expected (see Step 25).
Preparation of speedbead DNA binding buffer
Prepare the following buffer in a 50 ml conical tube:
Speedbead DNA binding buffer
| A | B | C |
|---|---|---|
| Reagent | Volume | Final conc. |
| PEG-8000 | 9 g | 18% (w/v) |
| 5 M NaCl | 100 μl | 2 M |
| 1 M Tris-HCl (pH 8.0) | 500 μl | 10 mM |
| 0.5 M EDTA (pH 8.0) | 100 μl | 1 mM |
| 10% (v/v) Tween 20 | 250 μl | 0.05% |
| Nuclease-free water | up to total | – |
| Total | 50 ml | – |
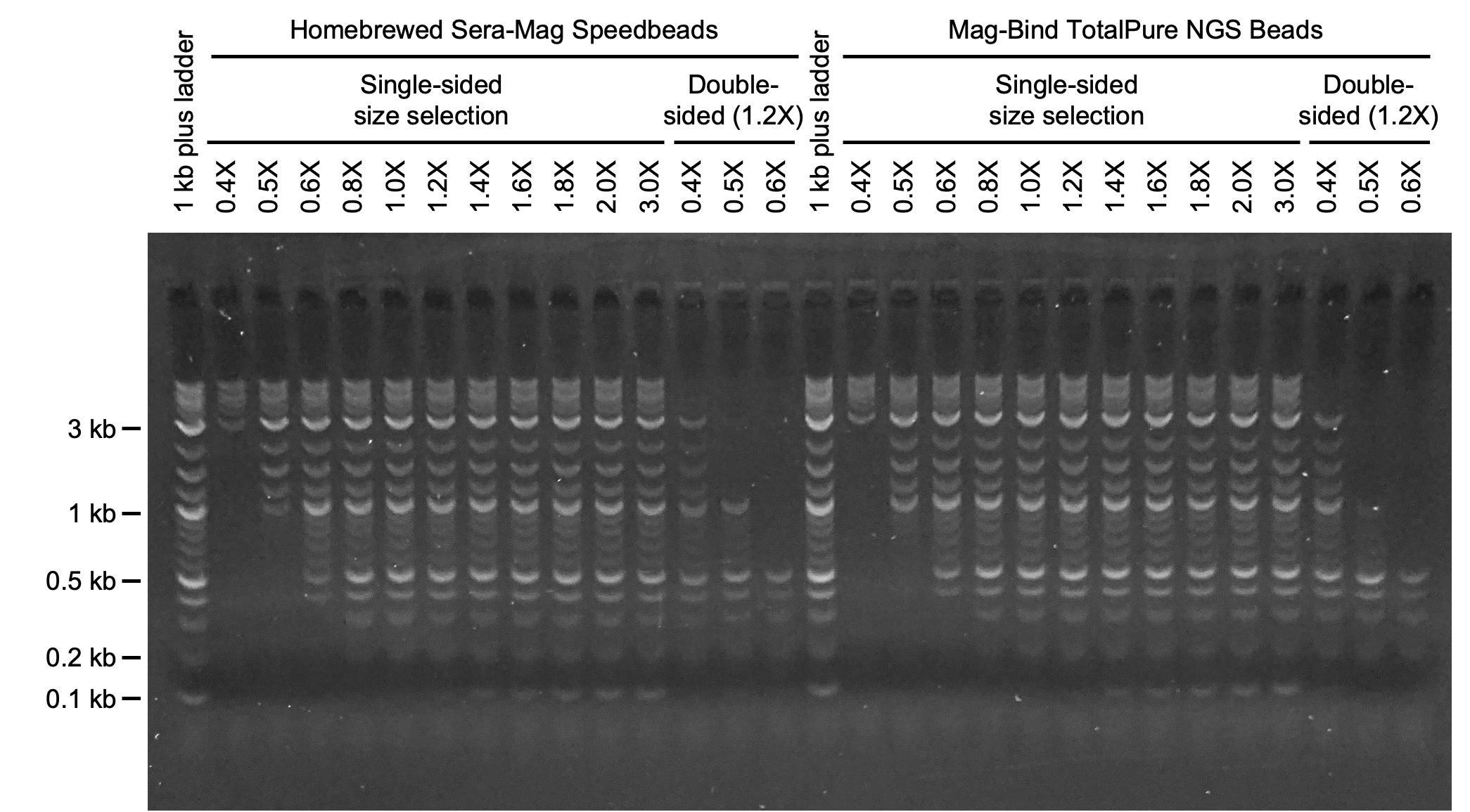
Validation of speedbeads
Prepare 10 aliquots of DNA ladder in 0.2 ml PCR tubes. Combine 3 μl of a 1:10 dilution of DNA ladder and 22 μl of 10 mM Tris-HCl (pH 8.0) for each aliquot if using Quick-load purple 1 kb plus DNA ladder.
Add 0.4X, 0.5X, 0.6X, 0.8X, 1.0X, 1.2X, 1.4X, 1.6X, 1.8X, and 2.0X volumes of speedbeads to the aliquots of DNA ladder. Careful and consistent pipetting of speedbeads is essential to ensure accurate size selection. Additional ratios of beads to sample can be tested as needed.
Purify the aliquots of DNA ladder as described in the general method for speedbead purification .
Elute DNA in 21 μl of 10 mM Tris-HCl (pH 8.0) prewarmed to 50°C.
Combine 4 μl of 6X DNA loading dye with each purified sample (20 μl). Run 8 μl of the mixture on a 1.0% agarose gel at 150 V for 60 min. As a control, run 8 μl of a mixture of DNA ladder (3 μl of a 1:10 dilution), 10 mM Tris-HCl (pH 8.0) (17 μl), and 6X DNA loading dye (4 μl). Conditions for gel electrophoresis (e.g., percent agarose, voltage, time) can vary depending setup.
General method for speedbead purification
Equilibrate speedbeads to room temperature and mix thoroughly.
Carefully add indicated amount of speedbeads to the sample.
Mix well by pipetting up and down a minimum of 10 times. Alternatively, vortex samples on setting 4 for 3-5 seconds. If samples require centrifugation after mixing, stop the centrifuge before beads settle.
Incubate samples at room temperature for 5 min. Prolonged incubation or incubation at low temperature will increase binding of small DNA fragments (e.g., adapter dimers).
Place samples on a magnetic stand for 5 min (or until solution is clear) to collect beads. Keep samples on the magnetic stand for Steps 34-38.
Carefully remove and discard cleared supernatant without disturbing bead pellet.
Add 200 μl of 80% ethanol (freshly prepared with MilliQ water) and incubate for 30 s. Carefully remove ethanol and discard without disturbing bead pellet.
Repeat Step 36 once. Carefully inspect samples and remove remaining ethanol using a 10 μl pipet.
Air dry samples for 2 min with lid open. Do not over-dry samples. Speedbeads (and other SPRI products) will clump and can not be fully resuspended during elution if samples are over-dried.
Remove samples from the magnetic stand. Add the indicated volume of appropriate solution for elution and mix well by pipetting or vortexing (see Step 32).
Incubate mixture at room temperature for 2-5 min.
Place samples on a magnetic stand for 2 min (or until solution is clear) to collect beads.
Transfer cleared supernatant (volume of solution added for elution less 1 μl) to a fresh PCR tube.

