A Mouse Model of Celiac Disease
Valérie Abadie, Valérie Abadie, Chaitan Khosla, Chaitan Khosla, Bana Jabri, Bana Jabri
Abstract
The design and use of mouse models that reproduce key features of human diseases are critical to advance our understanding of the pathogenesis of autoimmune diseases and to test new therapeutic strategies. Celiac disease is a unique organ-specific autoimmune-like disorder occurring in genetically susceptible individuals carrying HLA-DQ2 or HLA-DQ8 molecules who consume gluten. The key histological characteristic of the disease in humans is the destruction of the lining of the small intestine, a feature that has been difficult to reproduce in immunocompetent animal models. This unit describes the DQ8-Dd-villin-IL-15 transgenic mouse model of CeD, which was engineered based on the knowledge acquired from studying CeD patients’ intestinal samples, and which represents the first animal model that develops villous atrophy in an HLA- and gluten-dependent manner without administration of any adjuvant. We provide detailed protocols for inducing and monitoring intestinal tissue damage, evaluating the cytotoxic properties of intraepithelial lymphocytes that mediate enterocyte lysis, and assessing the activation of the enzyme transglutaminase 2, which contributes to the generation of highly immunogenic gluten peptides. Detailed protocols to prepare pepsin-trypsin digested gliadin (PT-gliadin) or chymotrypsin-digested gliadin (CT-gliadin), which allow antibody detection against native or deamidated gluten peptides, are also provided in this unit. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Induction of celiac-like disease in DQ8-Dd-villin-IL-15tg mice
Basic Protocol 2 : Histological assessment of villous atrophy
Support Protocol 1 : Morphometric assessment of villous/crypt ratio
Support Protocol 2 : Evaluation of epithelial cells renewal
Support Protocol 3 : Evaluation of the density of intraepithelial lymphocytes
Basic Protocol 3 : Analysis of cytotoxic intraepithelial lymphocytes
Basic Protocol 4 : Transglutaminase 2 activation and measurement of antibodies against native and deamidated gluten peptides
Support Protocol 4 : Preparation of CT-gliadin
Support Protocol 5 : Preparation of PT-gliadin
INTRODUCTION
Celiac disease (CeD) is a complex intestinal disorder with autoimmune features, which develops in genetically predisposed individuals carrying the major histocompatibility complex (MHC) class II human leukocyte antigen (HLA)–DQ2 or HLA-DQ8 variants (Abadie, Sollid, Barreiro, & Jabri, 2011; Ludvigsson et al., 2013). The driving antigens are dietary gluten proteins that encompass prolamins (gliadin from wheat and related prolamin proteins from barley and rye) and glutenins (Shewry, Tatham, Forde, Kreis, & Miflin, 1986). The histological hallmarks of CeD are an infiltration of the epithelium by intraepithelial lymphocytes (IELs), crypt hyperplasia, and the development of villous atrophy in the later stages of the disease that leads to malabsorption (Khan et al., 2019; Lebwohl, Sanders, & Green, 2018; Lindfors et al., 2019; Rashtak & Murray, 2009; Sollid & Jabri, 2005). In addition, CeD patients produce disease-specific antibodies directed against deamidated gluten peptides and the enzyme transglutaminase 2 (TG2) (Dieterich et al., 1998; Dieterich, Neurath, & Zopf, 2020; Husby et al., 2020). The knowledge of the disease-driving antigen and access to the tissue targeted by the pathogenic immune responses at different stages of disease in patients and in healthy controls have enabled the characterization of key immunological features of the disease in humans. More specifically, CeD is characterized by the development of gluten-specific HLA-DQ2- or -DQ8-restricted CD4+ T cells that produce interferon-gamma (IFN-γ) and interleukin (IL)-21 (Bodd et al., 2010; Molberg et al., 1997; Nilsen et al., 1998), expansion of gluten and TG2-specific plasma cells in the intestinal mucosa (Di Niro et al., 2012; Steinsbo et al., 2014), and expansion and activation of cytotoxic intraepithelial CD8+ lymphocytes that mediate intestinal epithelial cells destruction based on the recognition of stress signals (Jabri & Sollid, 2009; Setty et al., 2015).
To gain a deeper understanding of the immunopathogenesis of CeD, early studies have focused on mouse strains expressing one of the HLA molecules known to be associated with genetic susceptibility to CeD (Black, Murray, & David, 2002; de Kauwe et al., 2009; Du Pre et al., 2020; Du Pre et al., 2011; Lindstad et al., 2021; Marietta et al., 2004; Verdu et al., 2008). Following peripheral or oral gluten immunization with adjuvants, these mouse models displayed gluten-specific HLA-DQ8- or HLA-DQ2-restricted T cells, yet none of them developed villous atrophy. Small intestine tissue damage was only observed in non-gluten- and non-HLA-dependent models after the transfer of gliadin-primed CD4+ CD45RBlow CD25– T cells into lymphopenic mice (Freitag et al., 2009) or in ovalbumin-fed IL-15 transgenic (tg) mice in which the pro-inflammatory cytokine IL-15, known to be chronically upregulated in CeD small intestine (Jabri et al., 2000; Maiuri et al., 2000; Mention et al., 2003), was overexpressed in the epithelium under the T3b promoter (Korneychuk et al., 2014; Yokoyama et al., 2009).
The generation of the DQ8-Dd-villin-IL-15tg mouse model of CeD with villous atrophy was based on the concept that tissue destruction in organ-specific autoimmune disorders and CeD requires not only the development of an antigen-specific CD4 T cell response, but also expression of stress signals by the targeted tissue that are essential to license cytotoxic T cells to induce tissue destruction (Jabri & Abadie, 2015; Jabri & Sollid, 2006). In particular, the following observations in humans led to the development of DQ8-Dd-villin-IL-15tg mice (Abadie et al., 2020; Jabri & Sollid, 2009): (i) more than two-thirds of CeD patients display significant upregulation of IL-15 both in the epithelium and the lamina propria (Abadie & Jabri, 2014; Jabri & Abadie, 2015; Jabri et al., 2000; Maiuri et al., 2000; Mention et al., 2003), with IL-15 acting in a cell-contact dependent manner (Waldmann, 2006); (ii) the development of villous atrophy requires the concomitant presence of epithelial stress and anti-gluten adaptive immunity (Setty et al., 2015); and (iii) IELs mediate destruction of epithelial cells based on recognition of stress-induced non-classical MHC class I molecules upon activation with IL-15 (Meresse et al., 2004; Roberts et al., 2001). DQ8-Dd-villin-IL-15tg mice were obtained by crossing Dd-IL-15tg mice (Fehniger et al., 2001) onto DQ8-villin-IL-15tg mice to obtain the first-generation DQ8-Dd-villin-IL-15tg mice. Next generations were obtained by backcrossing DQ8-Dd-villin-IL-15tg mice with DQ8-villin-IL-15tg mice. These mice developed villous atrophy in a gluten-, HLA-, and TG2-dependent manner (Abadie et al., 2020). Furthermore, the expansion of IELs that have acquired a cytolytic phenotype is a hallmark of CeD (Hue et al., 2004; Meresse et al., 2004; Meresse et al., 2006) that was reproduced in the DQ8-Dd-villin-IL-15tg CeD mouse model. In this model, villous atrophy is dependent on CD4+ and CD8+ T cells and requires IFN-γ. These features of the CeD mouse model are in agreement with observations reported from CeD patients and with the current understanding of CeD pathogenesis (Abadie et al., 2011; Hue et al., 2004; Iversen & Sollid, 2020; Jabri & Sollid, 2009, 2017; Meresse et al., 2004; Meresse et al., 2006; Nilsen et al., 1998; Nilsen et al., 1995; Setty et al., 2015; Voisine & Abadie, 2021).
To study the relative contribution of IL-15 over-expression in the gut epithelium and in the lamina propria, we took advantage of the fact that the MHC class I H-2Dd promoter used to drive IL-15 expression in Dd-IL-15tg mice is inactive in intestinal epithelial cells in C57BL/6 mice (DePaolo et al., 2011). Thus, studying DQ8-Dd-IL-15tg mice that express IL-15 in hematopoietic but not epithelial cells, and DQ8-villin-IL-15tg mice (DePaolo et al., 2011; Meisel et al., 2017) that express IL-15 selectively in epithelial cells, allowed us to define the distinct contribution of IL-15 in these different compartments. Similar to first-degree family members of CeD patients who overexpress IL-15 in epithelial cells in the absence of an adaptive anti-gluten immune response (Setty et al., 2015), DQ8-villin-IL-15tg mice have an expansion of intraepithelial lymphocytes with upregulation of granzyme B and perforin, but no tissue destruction (Abadie et al., 2020; DePaolo et al., 2011). Conversely, and similar to potential CeD patients, who despite a pathogenic anti-gluten immune response with accompanying anti-TG2 antibodies have a normal intestinal architecture, DQ8-Dd-IL-15tg develop a T helper 1 (TH1) response against gluten but no tissue destruction (DePaolo et al., 2011). Licensing of IELs to kill, and the development of villous atrophy, only occur in gluten-fed DQ8-Dd-villin-IL-15tg mice, in which IL-15 is upregulated both in the epithelial and hematopoietic compartments (Abadie et al., 2020), as seen in active CeD (Setty et al., 2015). The observation in these models that IL-15 acts in a tissue compartment–specific manner, even though the IL-15 gene is preceded by the IL-2 leader oligonucleotide, is likely explained by the fact that IL-15Rα expression is required to see the functional effect of IL-15 (Liu et al., 2012). Finally and importantly, villous atrophy did not develop in Dd-villin-IL-15 transgenic mice, reflecting the requirement for the presence of a predisposing HLA molecule for the development of villous atrophy (Koning, 2012; Sollid et al., 1989). Nevertheless, while HLA-DQ8 was required for the development of villous atrophy, it was not required for the development of a TH1 response against gluten (Abadie et al., 2020). This is in line with observations in mice showing that gluten is immunogenic in diverse genetic backgrounds (Kagnoff et al., 1982), and in humans where DR or DP-restricted gluten-specific T cells can be found in the blood (Jensen et al., 1995; Molberg et al., 1998). Finally, TG2, which was shown to be inactive in the intestine under physiological conditions (Siegel et al., 2008), becomes activated upon gluten feeding in the DQ8-Dd-villin-IL-15tg mouse model (Abadie et al., 2020). Taken together, and in line with observations reported from CeD patients, villous atrophy develops in an HLA-dependent manner in DQ8-Dd-villin-IL-15tg mice upon oral administration of gluten in the absence of any adjuvant, and in a gluten-dependent manner, as tissue injury occurs only upon gluten feeding and recedes upon gluten exclusion (Abadie et al., 2020). The HLA dependency and the observation that TG2 is activated upon gluten feeding makes DQ8-Dd-villin-IL-15g mice a unique model to study the requirement for TG2 in the induction of a pathogenic anti-gluten CD4 T cell response.
This article provides detailed protocols for inducing CeD in the DQ8-Dd-villin-IL-15tg mouse model and for characterizing the key features of CeD, including small intestinal tissue destruction, infiltration of cytotoxic intraepithelial CD8 T cells, production of antibodies against deamidated gluten peptides, and TG2 activation. Basic Protocol 1 describes the administration of gluten by oral gavage and the timeline of gluten exposure that can be used to study the initial phase of the disease or relapse. Basic Protocol 2 outlines methods for the histologic assessment of tissue damage and intraepithelial lymphocytosis. Basic Protocol 3 describes methods to study the cytotoxic properties of intraepithelial lymphocytes. Basic Protocol 4 outlines the steps required to analyze TG2 activation and the antibody responses to native or deamidated gluten peptides.
NOTE : All experiments using live animals must be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must conform to government regulations for the care and use of laboratory animals.
Basic Protocol 1: INDUCTION OF CELIAC-LIKE DISEASE IN DQ8-Dd-villin-IL-15tg MICE
To promote the development of CeD, DQ8-Dd-villin-IL-15tg mice must be exposed to gluten via the oral route (Abadie et al., 2020). Mice that are born and raised on a gluten-free diet are transferred to a gluten-containing diet. Although rodent regular chow might provide sufficient gluten exposure, intragastric administration of gliadin—a major component of wheat gluten protein—by oral gavage ensures that all the animals receive a consistent amount of antigen. This protocol details the preparation of gliadin for intragastric administration and the regimen of gluten administration used to study the initiation of CeD, remission upon gluten withdrawal, and relapse following gluten re-exposure.
Materials
-
Gliadin from wheat (Sigma-Aldrich, cat. no. G3375)
-
Dulbecco's phosphate-buffered saline (DPBS), sterile (Corning Cellgro)
-
10-week-old DQ8-Dd-villin-IL-15tg mice (Jabri and Abadie laboratories, available upon request)
-
Teklad Global 18% Protein Rodent Diet (Envigo, 2918)
-
AIN-76A Purified Diet (Envigo, cat. no. 170481)
-
70% ethanol
-
Balance
-
Weighing dish
-
Cell culture dish (100 mm × 20 mm, polystyrene, BD Falcon, cat. no. 353003)
-
Microcentrifuge tubes (DOT Scientific Inc, cat. no. 609-GMTB)
-
Wood toothpicks (Fisher Scientific, cat. no. NC1678836)
-
Razor blades (Fisher Scientific, cat. no. 12-640)
-
Animal feeding/oral gavage needles, 22 × 1ʺ W/1-1/4 (Cadence Science, cat. no. 7901)
-
1-ml syringes, sterile (BD, cat. no. 329654)
-
Additional reagents and equipment for euthanasia of mice (see Current Protocols article Donovan & Brown, 2006)
Model to study anti-gluten immune responses upon the initial gluten introduction, gluten withdrawal, and gluten challenge
Utilizing DQ8-Dd-villin-IL-15tg mice, establish a mouse study in an appropriate mouse holding room according to one of the study regimens described in Figure 1.
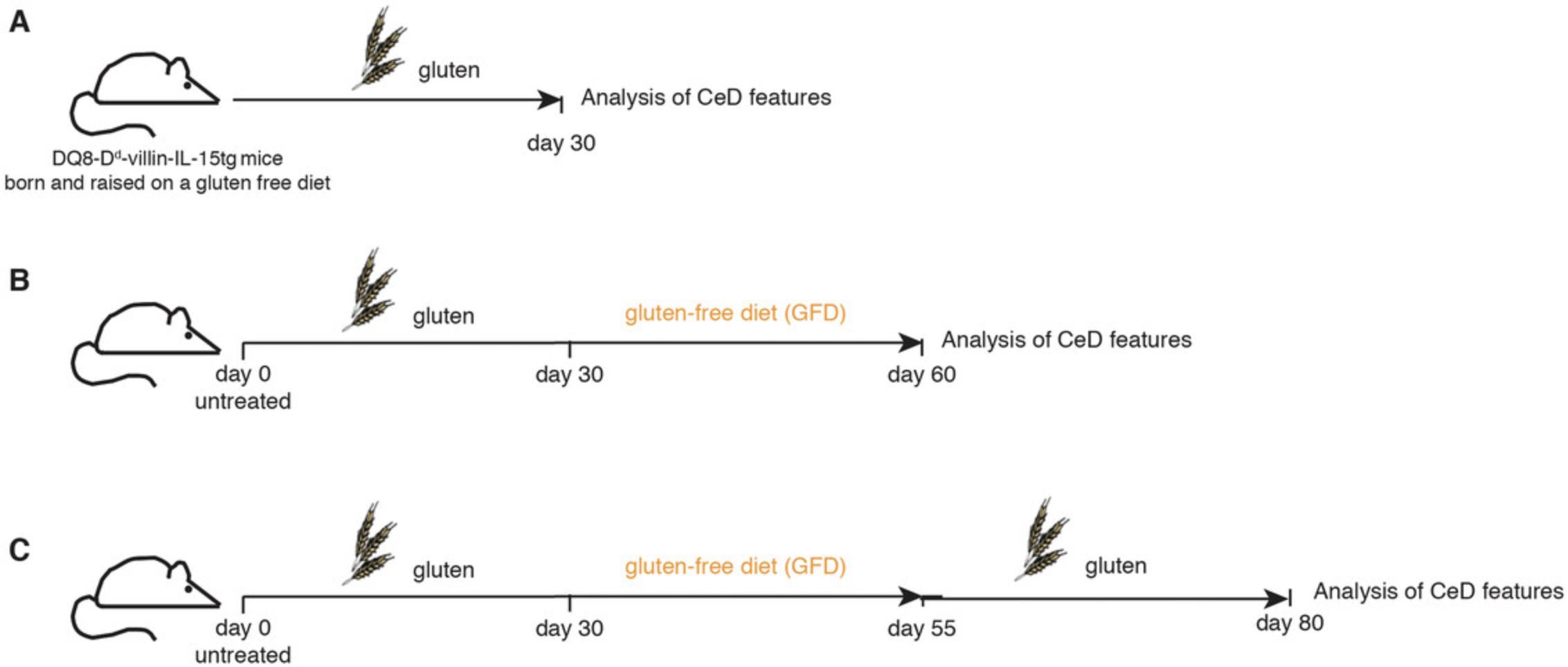
To study the immunological and histological features developing during the initial gluten-induced activation phase (with patent villous atrophy typically observed after 4 weeks of gluten exposure), set up the regimen in Figure 1A. To study remission and gut healing after gluten withdrawal, set up the regimen in Figure 1B. To study disease reactivation with a regimen that mimics clinical trials in patients with established CeD who are on a gluten-free diet and are then challenged with gluten, set up the regimen in Figure 1C.
Preparation of gliadin balls
The studies outlined above require administration of gliadin balls by oral gavage; this is accomplished as follows:
1.Weigh 200 mg of gliadin in a weighing dish.
2.Add to a microcentrifuge tube, and then add 1 ml of 1× DPBS. Mix using a toothpick and let sit for a couple of minutes.
3.Extract from the microcentrifuge tube using the toothpick, pressing against the tube's walls to remove any excess fluid, and place in a dish. Using a razor blade, cut the preparation in 10 equal pieces so as to obtain 10 “balls” each containing ∼20 mg of crude gliadin.
Oral gavage
4.Ear tag the mice for identification.
5.Transfer mice that are to be exposed to gluten into a cage containing regular rodent chow. Maintain control mice on a gluten-free diet.
6.Fit an animal feeding needle on a 1-ml syringe, and place a gliadin ball on the tip of the needle. For controls, fill a syringe with 100 μl of DPBS, avoiding formation of any air bubbles.
7.Remove the first mouse from the cage by the base of the tail. Using the thumb and index finger, grasp the nape of the neck. Lift the mouse and hold it in an upright position, securing the tail between the palm and little finger of the hand.
8.Once the mouse is properly restrained, insert the feeding needle into the mouse's mouth, over its tongue, and deposit a gliadin ball into the back of the mouth. The mouse will either swallow it directly or chew it first before swallowing it. For DPBS administration (control), slowly inject the 100 μl of DPBS into the esophagus using the gavage needle. In both cases, slowly remove the needle following the same angle used for its insertion.
9.Monitor the animal for normal breathing pattern before returning it to the cage.
10.Clean the work area with appropriate disinfectant. Discard the syringes. Wash feeding needle with soap and disinfect with 70% ethanol before sterilizing.
11.Euthanize mice (Donovan & Brown, 2006) at 30 days or at later time points (see Fig. 1 regarding the timeline of gluten administration) following the first gluten exposure to assess the development of villous atrophy (Basic Protocol 2), determine the activation of cytotoxic intraepithelial lymphocytes (Basic Protocol 3), and analyze TG2 activation and the levels of anti-gluten or anti-gliadin antibodies (Basic Protocol 4).
Basic Protocol 2: HISTOLOGICAL ASSESSMENT OF VILLOUS ATROPHY
The DQ8-Dd-villin-IL-15tg mouse model phenocopies key features of active CeD following gluten exposure, including intraepithelial lymphocytosis and the development of villous atrophy. However, the site of these changes in the mouse model differs from that in humans with CeD. While in humans the lesions are classically found in the proximal small bowel, in gluten-fed DQ8-Dd-villin-IL-15tg mice, the ileum is the major site of histopathological changes. This may stem from differences in gut physiology between mice and humans, such as the mouse stomach being less acidic and mice having a significantly faster transit time (Hugenholtz & de Vos, 2018). In support of this explanation, in gluten-dosed rodents, the ileum contained the highest concentration of gluten, followed by the jejunum, and gluten was almost absent in the duodenum (Gass, Vora, Bethune, Gray, & Khosla, 2006). In addition, we have found that while the primary site of tissue damage at 4 weeks after gluten exposure is the distal ileum, after prolonged exposure to gluten (for up to 11 weeks), alterations in the proximal ileum and jejunum are also present. Yet another difference between CeD in humans and the DQ8-Dd-villin-IL-15tg mouse model is that crypt hyperplasia is not consistently observed in this model.
Basic Protocol 2 describes how to process small intestine biopsies, as well as methods required to perform quantitative histology, i.e., measurements of villous height and crypt depth presented as a villous height–to–crypt depth ratio, and intraepithelial lymphocyte counts that allow the most accurate and reliable determination of mucosal injury in CeD (Adelman et al., 2018; Arguelles-Grande, Tennyson, Lewis, Green, & Bhagat, 2012; Cummins et al., 2011; Daveson et al., 2019; Picarelli et al., 2014; Taavela et al., 2013).
Materials
-
Gluten-fed mice (Basic Protocol 1)
-
70% ethanol
-
DPBS, sterile (Corning Cellgro)
-
Formalin solution, neutral-buffered, 10%, histological tissue fixative (Sigma-Aldrich, cat. no. HT501128)
-
Dissecting board
-
Sterile dissection instruments including dissecting pins, scissors (eye scissors, straight, sharp/sharp; Harvard Apparatus, cat. no. 72-8426), forceps (curved, smooth; Harvard Apparatus, cat. no. 72-8685)
-
Paper towels (Fisher Scientific, cat. no. 06-666-114)
-
Tissue-processing histology cassettes (Fisher Scientific, cat. no. 22-272416)
-
Additional reagents and equipment for euthanasia of mice (see Current Protocols article Donovan & Brown, 2006) and paraffin embedding, sectioning, and hematoxylin/eosin staining (see Current Protocols article Hofman & Taylor, 2013)
Collection and processing of the small intestine
1.Euthanize the mouse using CO2 inhalation followed by cervical dislocation in accordance with your Institutional Animal Care and Use Committee guidelines.
2.Place the mouse on its back on a dissecting board and spray 70% ethanol on its chest and abdomen to prevent contamination by hair.
3.Perform a midline incision using sharp straight scissors and retract the skin from the peritoneal wall underneath using forceps. Pin the skin on both sides of the abdomen.
4.Using sharp straight scissors and forceps, perform an incision through the peritoneal wall to reveal the abdominal cavity.
5.Cut the small intestine 0.5 cm from the stomach, below the pyloric sphincter and at the lowest extent of the distal small intestine above the cecum. Slowly pull out the whole small intestine and place it on a PBS-moistened paper towel.
6.Cut a ∼0.8 cm segment of the distal ileum 0.5 cm from the cecum. Gently push the luminal content out of the gut segment using the back of the curved tips of forceps.
7.Place the segment in a tissue-processing histology cassette (identified by marking with a pencil) and immerse the cassette in 10% (v/v) neutral-buffered formalin solution for 24-48 hr.
8.Transfer the cassette to 70% ethanol until the tissue can be processed and embedded in paraffin. Cut 5 μm transverse section, apply the tissue section onto a slide, and stain with hematoxylin and eosin.
9.Analyze the sections under a standard light microscope to assess gluten-mediated small intestinal histopathological changes (see Support Protocol 1).
Support Protocol 1: MORPHOMETRIC ASSESSMENT OF VILLOUS/CRYPT RATIO
Morphometric analyses of small intestinal biopsies in well-oriented sections represent the best way of assessing the extent of tissue damage and remodeling in CeD (Adelman et al., 2018; Arguelles-Grande et al., 2012; Daveson et al., 2019; Picarelli et al., 2014; Taavela et al., 2013). Intestinal damage is the hallmark of CeD, yet CeD can be heterogeneous in its histological presentation, with various degrees of villous atrophy ranging from mild villous blunting up to partial or total villous atrophy (Husby et al., 2012; Marsh, 1990).
Villous height–to–crypt depth ratio measurement
1.Under a light microscope equipped with a high-resolution camera and imaging system, take pictures of the whole intestinal section.
2.Using image processing and analysis software allowing distance measurements, measure the villous height and crypt depth of five well-oriented villi starting with the shortest ones for any sample analyzed. The villous height is measured from the tip to the shoulder of the villus or up to the top of the crypt of Lieberkuhn. The crypt depth is measured as the distance from the top of the crypt of Lieberkuhn to the deepest level of the crypt (Fig. 2).
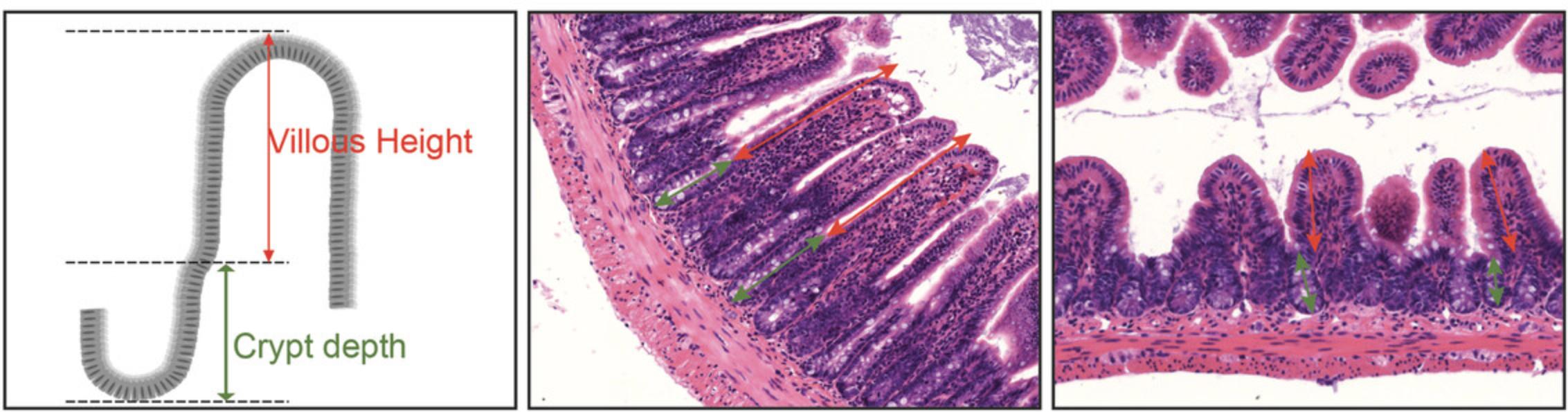
3.The villous height/crypt depth ratio is calculated by dividing the villous height by the corresponding crypt depth.
Support Protocol 2: EVALUATION OF EPITHELIAL CELLS RENEWAL
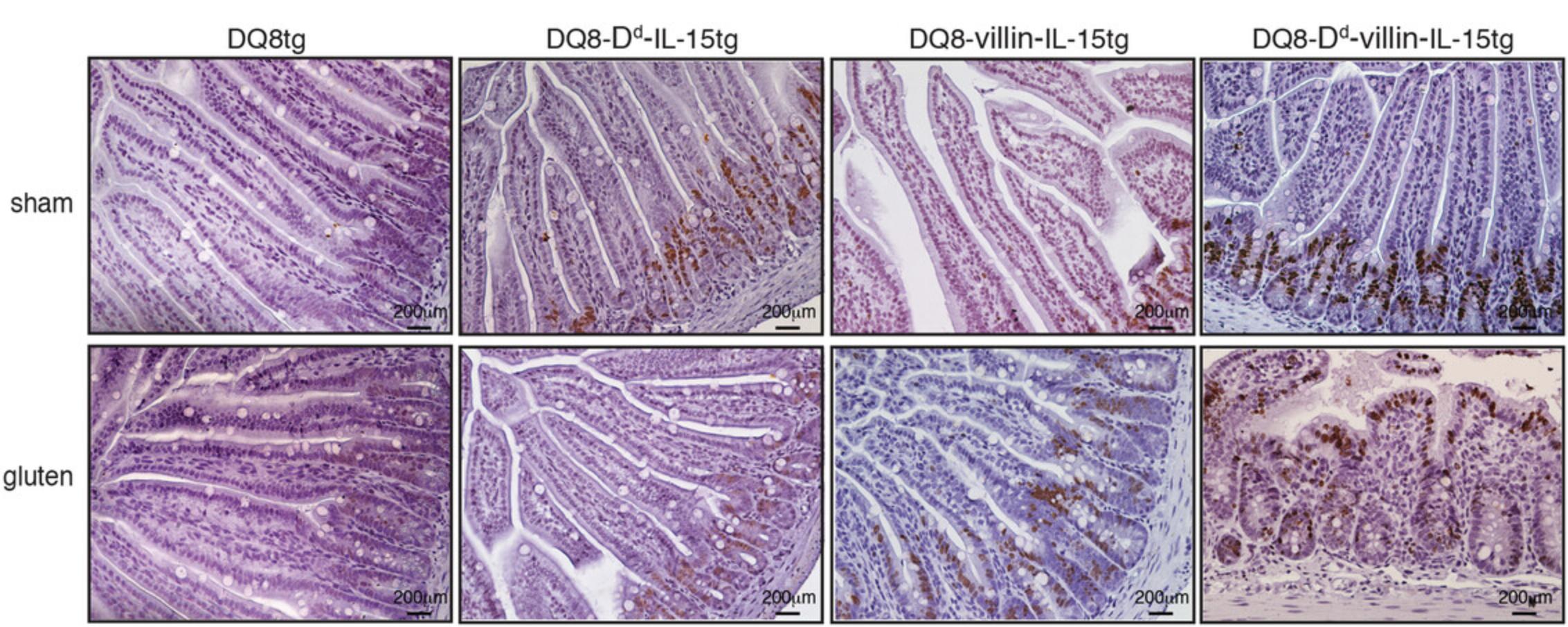
BrdU incorporation analyses have demonstrated that the decrease in villous height to crypt depth ratio in DQ8-Dd-villin-IL-15tg mice are associated with increased epithelial cells renewal. In DQ8-Dd-villin-IL-15tg mice with villous atrophy, BrdU incorporation is seen at the tip of villi, i.e., at a site where epithelial cells are proliferating to compensate for epithelial cells loss. In the other DQ8 and IL-15 tg mouse strains that do not have mucosal injury upon gluten administration, BrdU incorporation is only visible at the bottom of the villi and in crypt cells (Fig. 3).
Materials
-
BrdU solution (BD Biosciences, cat.no. 550891)
-
BrdU In-Situ Detection kit (BD Pharmingen, cat. no. 550803)
-
Xylene (Sigma-Aldrich)
-
Ethanol (Sigma-Aldrich)
-
Hydrogen peroxide (H2O2; Sigma-Aldrich)
-
1× phosphate-buffered saline (PBS), sterile (see recipe)
-
Gill's hematoxylin (Sigma-Aldrich)
-
Aquamount mounting medium
-
Coplin jars
-
Coverslips
1.24 hr before euthanasia of mice subjected to one of the studies outlined in Basic Protocol 1, inject mice intraperitoneally with 100 µl (1 mg) of BrdU solution.
2.Euthanize mice and collect and process the small intestine as described in Basic Protocol 2.
3.Deparaffinize slides in two changes of xylene for 5 min each. Transfer the slides to 100% ethanol. Perform two changes for 3 min each time. Transfer once into 95% ethanol for 3 min
4.Block endogenous peroxidase activity by incubating the slides for 10 min in 3% H2O2 in PBS.
5.Rinse the slides in PBS, three times for 5 min each time.
6.Proceed to antigen retrieval by immersing the slides in a Coplin jar containing the working solution of BD Retrievagen A (from the BrdU In-Situ Detection kit) and heat in a microwave oven to 193°F (89°C). Hold the slides at that temperature for 10 min. BD Retrievagen A is prepared per manufacturer's recommendations by mixing 9 ml of Solution 1 with 41 ml of Solution 2 and bringing up the volume to 500 ml with distilled water.
7.Remove the Coplin jar with the slides, cover the jar tightly, and allow the solution to slowly cool down to room temperature for 20 min.
8.Rinse the slides three times in PBS, each time for 5 min.
9.Dilute the biotinylated anti-BrdU antibody 1:10 in the Diluent buffer. Apply the antibody to the sections on the slide (100 μl/slide) and incubate for 1 hr in a humidified chamber.
10.Rinse slides three times in PBS, each time for 2 min.
11.Apply the ready-to-use Streptavidin-HRP from the kit to each slide and allow to incubate for 30 min, at room temperature.
12.Rinse the slides three times in PBS, each time for 2 min.
13.Prepare the DAB substrate solution by adding 1 drop of DAB chromogen to 1 ml of DAB Buffer, according to the kit manufacturer's recommendations. Apply DAB substrate solution to cover the tissue sections and incubate for 5 min. Monitor the development of the color and the color intensity.
14.Rinse the slides three times in distilled water, 2 min each time.
15.Stain with Gill's hematoxylin, for ∼30 s.
16.Rinse thoroughly in water, changing the water a few times.
17.Dehydrate through four changes of ethanol (95%, 95%, 100%, 100%) for 5 min each.
18.Clear in three changes of xylene, air dry, apply Aquamount mounting medium, and coverslip.
19.Observe the brown staining indicative of BrdU incorporation under a light microscope, as seen in Figure 3.
Support Protocol 3: EVALUATION OF THE DENSITY OF INTRAEPITHELIAL LYMPHOCYTES
Another hallmark of CeD is a sharp increase in the density of intraepithelial lymphocytes, a factor that correlates with the grade of villous atrophy (Kutlu et al., 1993). The amount of intraepithelial lymphocytes per one hundred intestinal epithelial cells is determined on hematoxylin & eosin–stained ileal sections (Basic Protocol 2) from sham and gluten-fed mice.
Materials
- Sample: Hematoxylin and eosin stained small intestine tissue sections (Basic Protocol 2)
- Bal supply cell counter (Fisher Scientific, cat. no. 02-670-12)
1.Under a light microscope equipped with high-magnification objective, count the number of intraepithelial lymphocytes throughout the surface epithelium using a two-key differential counter.
2.Express results as the number of intraepithelial cells per 100 intestinal epithelial cells. In sham-fed DQ8-Dd-villin-IL-15tg mice, the density of intraepithelial lymphocytes ranges from 15 to 25 per 100 intestinal epithelial cells, while gluten-fed DQ8-Dd-villin-IL-15tg mice have, on average, 40 intraepithelial lymphocytes per 100 intestinal epithelial cells
Basic Protocol 3: ANALYSIS OF CYTOTOXIC INTRAEPITHELIAL LYMPHOCYTES
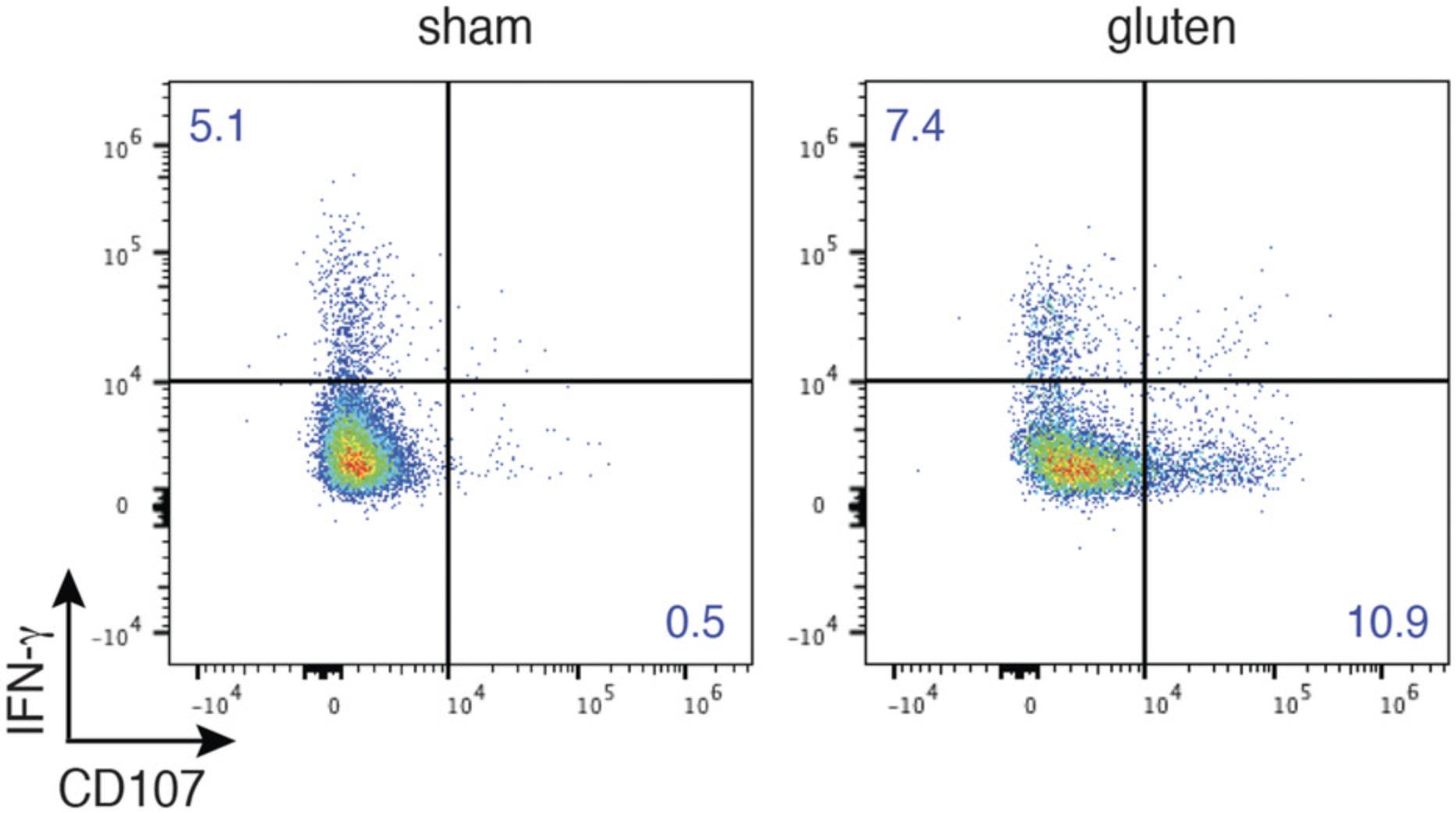
In CeD, intraepithelial cytotoxic CD8 T lymphocytes are the key cell type mediating tissue destruction (Jabri & Sollid, 2006; Meresse et al., 2006). Enterocyte lysis by intraepithelial CD8 lymphocytes is not mediated through gluten-specific mechanisms, but rather involves recognition of stress-induced non-classical MHC class I molecules expressed by intestinal epithelial cells by activating natural killer receptors (NKR) such as NKG2D and CD94/NKG2A, whose expression is upregulated by IL-15 on the surface on IELs (Meresse et al., 2004). The pathways employed by cytotoxic CD8+ T lymphocytes to mediate the killing of target cells rely on the release of cytolytic granules. These granules are composed of a dense core containing perforin, various granzymes, and granulysin complexed with proteoglycan, surrounded by an outer membrane containing various lysosomal membrane proteins including CD107a, CD107b, and CD63.During the process of degranulation, the membrane of the granule fuses with the plasma membrane, and the granule contents are released into the immunological synapse between the activated cell and the target cell (Betts & Koup, 2004; Betts et al., 2003). Degranulation is required for perforin-granzyme mediated killing, and hence the detection of CD107a and CD107b proteins is the best indicator of the functional cytotoxic activity of activated T cells.
In the DQ8-Dd-villin-IL-15tg mouse model, the critical role for IELs in mediating intestinal epithelial cells destruction was supported by the finding that CD8 T cell depletion prevented the induction of villous atrophy, and that the expression of activating NK receptors, granzyme B, and CD107 by intraepithelial lymphocytes strongly correlated with the development of villous atrophy (Abadie et al., 2020). Basic Protocol 3 details the flow cytometric assay used to detect intraepithelial CD8 T lymphocytes degranulation through the measurement of the expression of CD107.
Materials
- Sham and gluten-fed mice (Basic Protocol 1)
- Harvest IEL medium (see recipe)
- FACS wash buffer: DPBS containing 2% fetal bovine serum (FBS)
- 40% Percoll solution (see recipe)
- Culture medium (see recipe)
- Dulbecco's phosphate-buffered saline (DPBS) without Ca2+ and Mg2+ (Hyclone, cat. no. SH30028.03)
- BD GolgiPlug™ (Brefeldin A, BD Biosciences, cat. no 555029)
- BD GolgiStop™ (monensin, BD Biosciences, cat. no 554724)
- Phorbol 12-myristate 13-acetate (PMA; see recipe)
- Ionomycin (see recipe)
- Fluorochrome-labeled antibodies (see Table 1)
- FcR blocking reagent (anti-CD16/CD32 diluted 1:300 in FACS wash buffer)
- Fluorescent fixable viability dye (LIVE/DEADTM Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation; Life Technologies, cat. no. L34957)
- Cell-surface staining cocktail (CD45, TCRb, CD4, CD8a, CD8b listed in Table 1)
- Intracellular staining cocktail (IFN-γ, listed in Table 1)
- Fixation/permeabilization solution (Cytofix/Cytoperm 1× solution, BD Biosciences, cat. no. 2092KZ) or 4% paraformaldehyde solution (Alfa Aesar, cat. no. J19943-K2)
- Permeabilization solution (Perm Wash Buffer, BD Biosciences, cat. no. 2091KZ)
| Marker | Antibody clone |
|---|---|
| CD16/CD32 | 2.4G2 |
| CD107a | 1D4B |
| CD107b | ABL-93 |
| CD45 | 30-F11 |
| TCRb | H57-597 |
| CD4 | GK1.5 |
| CD8a | 53-6.7 |
| CD8b | H35-17.2 |
| IFN-γ | XMG1.2 |
- a
Antibodies can be purchased labeled with fluorochrome of choice.
-
Sterile dissection instruments including dissecting pins, scissors (eye scissors, straight, sharp/sharp; Harvard Apparatus, cat. no. 72-8426), forceps (curved, smooth, Harvard Apparatus; cat. no. 72-8685)
-
25-ml syringes
-
18-G or 20-G oral gavage needle (Cadence Science)
-
Cell culture dishes (100 mm × 20 mm, polystyrene; BD Falcon, cat. no. 353003)
-
15-ml and 50-ml conical polypropylene centrifuge tubes (Corning)
-
Shaker-incubator
-
Falcon cell strainers, 70 μm (BD Falcon, cat. no. 352350) and 40 µm nylon (BD Falcon cat. no. 352340)
-
Refrigerated centrifuge
-
Round bottom 96-well cell culture plate, sterile (BD Falcon, cat. no. 353077)
-
Multichannel pipettor
-
FalconTM round-bottom polystyrene test tubes with cell strainer snap cap, 5 ml (Falcon, cat. no. 352235)
-
Additional reagents and equipment for removing the small intestine (Basic Protocol 1) and counting cells using a hemacytometer (see Current Protocols article Phelan & May, 2015)
A. Isolation of epithelial cells and intraepithelial lymphocytes (IELs) from the small intestine
The intestinal epithelial compartment comprises both epithelial cells and IELs. For IEL purification, this compartment is first dissociated from the subjacent lamina propria and is then processed by Percoll centrifugation to separate/isolate the IELs from the epithelial cells as follows:
1.Remove the small intestine as described in Basic Protocol 2, and place the intestine on a PBS-moistened paper towel. Remove any fat and connective tissue by teasing with two sets of forceps. Excise all the Peyer's patches by gently positioning them in the tips of forceps and pulling them out. Gently push the feces out of the gut segment using the back of the curved tips of forceps.
2.Remove any remaining fecal matter or luminal content. To do so, fill a 25-ml syringe equipped with 18- or 20-G gavage needle with 20 ml of cold PBS. Hold the intestine with forceps on top of a large beaker, insert the needle into the gut lumen, and inject the cold PBS to flush out the luminal content.
3.Place the intestine back on the PBS-moistened paper towel. Using fine sharp straight scissors, cut the small intestine longitudinally. Transfer tissue to a cell culture dish containing PBS and wash carefully by moving the tissue around, held by forceps, to remove any remaining fecal matter.
4.Cut the small intestine into ∼1 cm pieces on top of a 50-ml conical centrifuge tube containing 15 ml of pre-warmed (37°C) harvest IEL medium, marked with the sample name.
5.Shake the 50-ml conical centrifuge tubes at 250 rpm, 37°C, for 20 min in a shaker-incubator. The tubes must be placed either on a tube rack that can be inclined, or horizontally into a plastic box that is secured to the shaker with platform clamps.
6.Following incubation, pour the contents of the 50-ml conical centrifuge tube into another labeled 50-ml conical centrifuge tube through a 70-μm cell strainer. Spin the harvested sample 10 min at 450 × g , 4°C. Resuspend the pellet with 1 ml of PBS containing 2% FBS.
7.Add 15 ml of pre-warmed (37°C) harvest IEL medium to the first tube containing the tissue pieces. Close the tube tightly and shake the 50-ml conical centrifuge tubes at 250 rpm, 37°C, for 20 min in a shaker-incubator.
8.Pour the contents of the 50-ml conical centrifuge tube into another labeled 50-ml conical centrifuge tube through a 70-μm cell strainer. Spin the harvested sample 10 min at 450 × g , 4°C. Resuspend the pellet with 1 ml of PBS containing 2% FBS and pool together the cells recovered after each shake.
9.Centrifuge the cell suspension 5 min at 510 × g , 4°C. Discard the supernatant.
10.Resuspend the pellet in 2 ml of a pre-warmed (room temperature) 40% Percoll solution and transfer the cell solution into a labeled 15-ml conical centrifuge tube that has been pre-filled with 8 ml of the 40% Percoll solution.
11.Centrifuge the gradient 20 min at 1800 × g , room temperature, with minimal acceleration and without brake.
12.Following centrifugation, the gradient is composed of a layer that contains unwanted cells and debris, and a pellet that contains the lymphocytes. Gently aspirate and discard the gradient and top layer, being careful not to disrupt the pellet, and rinse the pellet with 10 ml of PBS containing 2% FBS.
13.Centrifuge 5 min at 650 × g , 4°C.
14.Discard the supernatant and resuspend intraepithelial lymphocytes with 1 ml of culture medium.
15.Count viable cells using trypan blue dye exclusion, a hemacytometer, and a microscope. Keep cells on ice until further use.
B. Assay of IEL cytotoxic potential
The functional potential of cytotoxic T lymphocytes (in this case IELs) can be determined ex vivo by cellular expression of CD107a, the best indicator of cell degranulation. In this part of Basic Protocol 3, the main steps involved in cell stimulation, cell-surface staining, and intracellular detection of CD107 are described.
Intraepithelial lymphocytes stimulation in vitro
16.Dispense 100 μl of cell suspension (2 × 106 cells, from step 15) in culture medium into appropriate wells of a 96-well round-bottom cell culture plate.
17.Add 100 μl of culture medium supplemented with GolgiPlug (1:500 dilution from stock in culture medium), GolgiStop (1:500 dilution from stock in culture medium), diluted PMA (prepared as indicated in the Reagents and Solutions; 50 ng/ml final), and diluted ionomycin (prepared as indicated in Reagents and Solutions; 1000 ng/ml final).
18.Add 10 μl of CD107a/CD107b-FITC cocktail (prepared as indicated in Reagents and Solutions) to each well. Mix wells by pipetting up and down several times.
19.Cover plate and incubate for 4 hr in a cell culture incubator (5% CO2, 37°C).
20.Centrifuge the plate 5 min at 450 × g , 4°C.
Cell-surface antigen staining
21.Following centrifugation, discard supernatant by inverting the plate above a waste tray.
22.Add 50 μl FcR blocking reagent (anti-CD16/CD32 diluted 1:300 in FACS wash buffer) to each well and mix by pipetting up and down. Incubate the plate 10 min at 4°C in the dark.
23.Wash the cells with 150 μl of FACS wash buffer. Centrifuge the plate 5 min at 450 × g , 4°C.
24.Following centrifugation, discard supernatant by inverting the plate above a waste tray. Add 50 μl per well of the viability dye prepared at the appropriate dilution (determined in advance by titration) in DPBS. Mix by pipetting up and down. Incubate the plate for 15 min at 4°C in the dark.
25.Wash the cells with 150 μl of FACS wash buffer. Centrifuge the plate 5 min at 450 × g , 4°C.
26.Following centrifugation, discard supernatant by inverting the plate above a waste tray. Add 50 μl of cell-surface staining cocktail to the appropriate wells. Mix gently by pipetting up and down with a multichannel pipettor. Incubate the plate for 20 min, at 4°C, in the dark.
27.Wash the cells twice, each time with 150 μl of FACS wash buffer. Centrifuge the plate 5 min at 450 × g , 4°C, following each wash.
Intracellular antigen staining
28.Following centrifugation, discard supernatant by inverting the plate above a waste tray. Following the BD Cytofix/Cytoperm “Alternative Fixation and Permeabilization Protocol” steps, resuspend cells in 100 µl of 4% paraformaldehyde solution. Incubate the plate for 20 min, at 4°C, in the dark.
29.Wash the cells with 100 μl of FACS wash buffer. Centrifuge the plate 5 min at 650 × g , 4°C.
30.Following centrifugation, discard supernatant by inverting the plate above a waste tray. Wash the cells with 200 μl of FACS wash buffer. Centrifuge the plate 5 min at 650 × g , 4°C.
31.Resuspend the cells in 200 µl of FACS wash buffer and place the plate covered with aluminum foil at 4°C (in the refrigerator) overnight.
32.Centrifuge the plate 5 min at 650 × g , 4°C, and resuspend cells in 200 µl of 1× Perm/Wash buffer. Incubate the plate for 15 min, at 4°C, in the dark.
33.Centrifuge the plate 5 min at 650 × g , 4°C.
34.Following centrifugation, discard supernatant by inverting the plate above a waste tray. Add 50 μl of intracellular staining cocktail to the appropriate wells. Mix gently by pipetting up and down with a multi-channel pipettor. Wrap plate in aluminum foil to protect from light and incubate for 45 min on ice.
35.Wash the cells with 150 μl of Perm/Wash buffer. Centrifuge the plate 5 min at 650 × g , 4°C.
36.Discard supernatant by inverting the plate above a waste tray. Wash two times the cells with 200 μl of Perm/Wash buffer. Centrifuge the plate 5 min at 650 × g , 4°C.
37.Resuspend cells in 200 μl of FACS wash buffer. Filter cells using round-bottom polystyrene test tubes with cell-strainer snap cap right before flow cytometric analysis.
Basic Protocol 4: TRANSGLUTAMINASE 2 ACTIVATION AND MEASUREMENT OF ANTIBODIES AGAINST NATIVE AND DEAMIDATED GLUTEN PEPTIDES
Gluten proteins are the source of the immunogenic peptides that are the proximal triggers driving CeD pathogenesis. Following ingestion, these proteins are generally resistant to proteolytic degradation (Shan et al., 2002). Yet, they are good substrates for transglutaminase 2 (TG2, a ubiquitous enzyme that catalyzes the conversion of selected glutamine residues in gluten peptides into glutamate; Arentz-Hansen et al., 2000; Molberg et al., 1998; van de Wal et al., 1998), and thus promote the generation of immunogenic deamidated peptides that bind to HLA-DQ2 or HLA-DQ8 with much stronger affinity than native gluten peptides (Abadie et al., 2011; Tollefsen et al., 2006). Generation of highly disease-specific antibodies against deamidated gluten peptides and autoantibodies against TG2 itself is a hallmark of CeD (Dieterich et al., 1997; Dieterich et al., 1998; Husby et al., 2020). Although TG2 is activated upon gluten and gliadin exposure in the DQ8-Dd-villin-IL-15tg mouse model, and one can measure the production of anti-deamidated gluten and gliadin peptide antibodies, the detection of circulating anti-TG2 antibodies has proven inconsistent, perhaps reflecting the absence of a mouse homolog of the human VH5-51 gene segment that recognizes TG2 in its germline configuration (Abadie et al., 2020). This protocol describes how to detect and quantify TG2 activation and antibodies against both native and deamidated gliadin peptides in the DQ8-Dd-villin-IL-15tg mouse model of CeD.
Materials
-
Mice (including sham-fed and gluten-fed; see Basic Protocol 1)
-
5–(biotinamido)-pentylamine (5-BP; Sigma-Aldrich, cat. no. 914134)
-
Tissue-Tek OCT embedding compound (Sakura Finetek)
-
4% Paraformaldehyde (PFA) solution (Alfa Aesar, cat. no. J19943-K2)
-
1× phosphate-buffered saline (PBS; see recipe)
-
IHC blocking solution (see recipe)
-
Tween-20
-
Antibodies:
- Rabbit polyclonal anti-TG2 antibody (stock 1.1 mg/ml; custom produced by Pacific Immunology)
- Alexa Fluor 488 F(ab′)2-Goat anti-Rabbit IgG (H+L) (stock 2 mg/ml; Invitrogen A11070)
- Alexa Fluor 594 streptavidin (stock 2 mg/ml; Thermo Fisher S32356)
- Goat anti-mouse IgA-HRP (Southern Biotech, cat. no. 1040-05)
- Goat anti-mouse IgG-HRP (Southern Biotech, cat. no. 1030-05)
- Standard mouse IgA-UNLB (Southern Biotech, cat. no. 0106-01)
- Standard mouse IgG-UNLB (Southern Biotech, cat. no. 0107-01)
-
CT-gliadin (see Support Protocol 3)
-
PTD-gliadin (see Support Protocol 4)
-
Na2HPO4 solution (see recipe)
-
ELISA wash buffer (see recipe)
-
ELISA blocking solution (see recipe)
-
TMB Blue Substrate System (Agilent Dako, cat. no. S1599)
-
2 N H2SO4 (Fluka Analytical)
-
Lab rocker platform
-
Pap pen (Dako, cat. no. S2002)
-
Kimwipe disposable wipers (Kimberly Clark)
-
Confocal microscope
-
High-binding ELISA 96-well plates (Nunc MaxiSorp flat-bottom, Thermo Scientific)
-
Microplate reader
-
Tween-20
-
Additional reagents and equipment for euthanasia of mice (see Current Protocols article Donovan & Brown, 2006) and basic immunohistochemical techniques (see Current Protocols article Hofman & Taylor, 2013)
Transglutaminase 2 activation
TG2 activation can be visualized in vivo upon administration of the TG2 substrate, 5-(biotinamido)-pentylamine (5-BP) (Siegel et al., 2008). TG2 enzymatic activity is measured using 5-BP cross-linking on small intestine sections co-stained with fluorescently labeled streptavidin to visualize 5-BP, and with an anti-TG2 antibody as follows:
1a. At 6 and 3 hr prior to euthanasia of mice to be subjected to one of the studies in Basic Protocol 1, inject the mice intraperitoneally with 100 mg/kg of 5-BP. Proceed with Basic Protocol 1.
2a. Collect small intestinal pieces as described in Basic Protocol 2 and freeze them in optimum cutting temperature (OCT) compound (see Hofman & Taylor, 2013).
3a. Cut 5-μm thick sections from OCT-embedded tissue samples and air-dried overnight at room temperature (Hofman & Taylor, 2013).
4a. To fix the section, immerse the slides in a Coplin jar or staining dish containing 1% PFA diluted in DPBS from the 4% stock solution for 30 min at room temperature.
5a. Wash the slides three times in PBS, each time for 10 min under agitation using a rocker platform.
6a. Draw a hydrophobic circle around the tissue section using a Pap pen.
7a. Add IHC blocking solution and incubate for 1 hr at room temperature.
8a. Tilt the slides to remove the solution and remove the remaining solution using Kimwipes.
9a. Apply primary antibody against TG2 diluted at 1 μg/ml in IHC blocking solution in sufficient quantity to cover the tissue (∼ 300 μl per slide).
10a. Wash the slides three times in PBS/0.1% Tween-20, each time for 10 min under agitation using a rocker platform.
11a. Add secondary antibodies:
- a. Alexa Fluor 488 F(ab′)2-Goat anti-Rabbit IgG (H+L) (stock 2 mg/ml) diluted 1:400 in IHC blocking solution.
- b. Alexa Fluor 594 streptavidin (stock 2 mg/ml) diluted 1:400 in IHC blocking solution. Apply in sufficient quantity to cover the tissue (∼ 300 μl per slide).
12a. Wash the slides three times in PBS/0.1% Tween-20, each time for 10 min under agitation using a rocker platform.
13a. Dry slides carefully around the tissue using a Kimwipe and mount coverslips using VectaShield with DAPI mounting medium (2 drops per slide, enough to cover the stained tissue; push out all air bubbles).
14a. Analyze the sections under a confocal microscope.
Measurement of antibodies against native and deamidated gliadin peptides
Both antibodies against native gliadin peptides and deamidated gliadin peptides can be measured by ELISA using different preparations of gliadin as antigen coating. Pepsin-trypsin digestion of gluten induces chemical deamidation (Molberg et al., 2001). Hence, an ELISA assay using pepsin-trypsin digested gliadin (PT-gliadin) will detect anti-gliadin antibodies directed against both native and deamidated gluten peptides, while TG2-treated chymotrypsin-digested gliadin (CT-gliadin) should be used to solely detect anti-deamidated gliadin antibodies.
1b. Collect blood at the desired time points from sham-fed and gluten-fed mice. Allow the blood sample to clot and centrifuge 5 min at 1800 × g to further separate the serum from the clot and blood cells.
2b. Coat ELISA 96-well plates with 50 μl of 100 μg/ml deamidated CT-gliadin or PTD-gliadin in 100 mM Na2HPO4 overnight at 4°C.
3b. Wash the plates three times, each time with 250 μl of ELISA wash buffer.
4b. Add 200 μl of ELISA blocking solution per well, cover the plate with aluminum foil, and incubate for 2 hr at room temperature.
5b. Discard the blocking solution by inverting the plate above a waste container. Add 100 μl of serum sample per well. Incubate overnight at 4°C.
6b. Wash the plates three times with 250 μl of ELISA wash buffer.
7b. Add 100 μl per well of anti-mouse Ig-horseradish peroxidase (HRP) prepared in IHC blocking solution (1:500 dilution from stock). Cover the plate with aluminum foil, and incubate for 1 hr at room temperature. Use anti-mouse IgG-HRP or anti-mouse IgA-HRP to detect one or the other isotype.
8b. Wash the plates five times with 250 μl of ELISA wash buffer.
9b. Add 50 μl of the HRP substrate TMB per well. Monitor the appearance of a blue coloration and stop the reaction by adding 50 μl of 2 N H2SO4 that changes the color of the HRP-TMB byproduct from blue to yellow.
10b. Read the absorbance at 450 nm using a microplate reader.
Support Protocol 4: PREPARATION OF CT-gliadin
Materials
-
Gliadin from wheat (Millipore Sigma, cat. no. G3375)
-
Milli-Q water
-
10× Tris buffer (see recipe)
-
1 M CaCl2 solution (see recipe)
-
Chymotrypsin (Sigma Aldrich, cat. no. C1429)
-
1 mM HCl solution (see recipe)
-
Dialysis buffer (see recipe)
-
Liquid N2
-
10× digest buffer (see recipe)
-
mTG2 (Zedira)
-
1× phosphate-buffered saline (PBS), sterile (see recipe)
-
50-ml polypropylene centrifuge tube (BD Falcon, cat. no. 352070)
-
37°C shaking incubator
-
Boiling water bath
-
500-1000 Da MWCO dialysis tubing (Spectra/Por dialysis tubing, cat. no. 131096T)
-
50-ml polypropylene tubes
-
Lyophilizer
-
NanoDrop spectrophotometer
-
Additional reagents and equipment for dialysis (see Current Protocols article Phillips & Signs, 2005)
1.Weigh out the desired number of 500 mg aliquots of gliadin in individual 50-ml polypropylene tubes.
2.Add 22.5 ml of Milli-Q water to each tube, followed by 2.5 ml of 10× Tris buffer and then 125 µl of 1 M CaCl2 solution (10 mM final concentration). Vortex well until gliadin is suspended (it will not dissolve, but the powder will be dispersed in the liquid and not sitting at the bottom of the tube).
3.Place the tubes in a shaking incubator at 37°C for 30 min.
4.Dissolve the chymotrypsin to a concentration of 4 mg/ml in cold 1 mM HCl solution.
5.After the suspended gliadin has pre-dissolved in the shaking incubator for 30 min, add chymotrypsin to a final concentration of 0.2 mg/ml by adding 1.25 ml of the 4 mg/ml chymotrypsin stock. For control tubes without protease, add 1.25 ml of 1 mM HCl.
6.Vortex tube briefly and return to 37°C shaking incubator for 12 hr.
7.After digestion, quench by boiling (be careful—ensure caps are vented) at 95°–98°C for 10 min.
8.Centrifuge the digests 10 min at 1800 × g to pellet insoluble, undigested gliadin.
9.Dialyze the digests by pipetting 25 ml of the digests into one dialysis tubing. Seal the tubing with dialysis clamps. Dialyze at 4°C for 6–8 hr in dialysis buffer. Repeat the procedure with fresh dialysis buffer three times, with the last incubations lasting 12 hr.
10.Transfer the digests into 50-ml polypropylene tubes that have been weighed beforehand.
11.Flash freeze digests in liquid nitrogen and lyophilize to dryness (∼48 hr).
12.Dissolve the digest in Milli-Q water and measure the absorbance at 280 nm using a NanoDrop spectrophotometer. Calculate the concentration of a 2-µl aliquot using the extinction coefficient below, and dilute to the desired final concentration.
13.To proceed to enzymatic deamidation using TG2, dissolve the CT-gliadin to 5.5 mg/ml in water.
14.Add an appropriate volume of 10× digest buffer to bring the buffer concentration to 1×.
15.Add mTG2 such that the w/w ratio is 1:250. To negative control tubes, add 10× digest buffer alone. Mix by inverting the tubes.
16.Incubate at 37°C for 1 hr with gentle shaking.
17.Quench the digestion by boiling at 95°–98°C for 10 min (again making sure that the tubes are compatible with boiling).
18.After cooling, centrifuge 5 min at 2300 × g in a tabletop centrifuge to pellet any TG2 that precipitated upon boiling. Freeze tubes, or continue directly with dialysis, as described above adjusting volumes depending on the volume that was used for the deamidation and control reactions.
19.Resuspend the peptide solution at the desired concentration in water or PBS based on the mass of peptide that was used for the deamidation reaction. Vortex well and confirm the concentration as described in step 2.
Support Protocol 5: PREPARATION OF PT-gliadin
Materials
-
Gliadin from wheat (Millipore Sigma, cat. no. G3375)
-
Pepsin (Sigma Aldrich, cat. no. P3286)
-
0.2 N HCl solution (see recipe)
-
2 N NaOH (see recipe)
-
Trypsin (Sigma Aldrich, cat. no. T1763)
-
Pancreatin (Sigma Aldrich, cat. no. P1750)
-
Magnetic stir plate and stirrer
-
Centrifuge
1.Incubate 100 g of gliadin with 2 g of pepsin in 1 L of 0.2 N HCl. Stir the mixture magnetically at 37°C for 2 hr.
2.Adjust the pH to 8.0 with 2 N NaOH.
3.Add 2 g of trypsin to the digest and incubate at 37°C for 4 hr.
4.Add 2 g of pancreatin and mechanically stir for 2 hr. During the incubation, periodically check the pH and maintain it to 8.0.
5.Centrifuge the digest 20 min at 6000 × g , 20°C.
6.Harvest the supernatant and freeze at −20°C.
7.Measure the concentration as described in the previous section.
REAGENTS AND SOLUTIONS
CaCl2, 1 M
- 1.47 g CaCl2⋅2H2O
- 10 ml Milli-Q water
- Store up to 6 months at −20°C
CD107a/CD107b-FITC cocktail
On the day of the experiment, combine 0.1 µg CD107a-FITC with 0.5 µg CD107b-FITC in 1× DPBS (Corning Cellgro). Prepare enough for all required wells such that 10 µl of the cocktail is added to each well. Prepare fresh.
Culture medium
- 444.5 ml RPMI 1640 medium
- 50 ml fetal bovine serum, heat-inactivated
- 5 ml penicillin-streptomycin-glutamine (GIBCO, cat. no. 10378016)
- 0.5 ml β-mercaptoethanol (55 mM)
- Filter on 0.22-mm filter unit
- Store up to 1 month at 4°C
Dialysis buffer (5 mM sodium phosphate)
- 2.83 g Na2HPO4
- 4 L Milli-Q water
- Adjust the pH to 7.4 using concentrated NaOH and HCl
- Store up to 6 months in cool place
Digest buffer, 10×
- Dissolve MOPS (Sigma Aldrich, cat. no. M1254) to 1 M concentration
- Adjust the pH to 7.4
- Add CaCl2 to 50 mM from 1 M stock (see recipe)
- Store until usage at 4°C
ELISA blocking solution
- 479.75 ml PBS (Corning Cellgro)
- 250 μl Tween-20
- 20 ml bovine serum albumin (BSA; Sigma-Aldrich, cat. no. A8806-1G)
- Store up to 6 months at 4°C
ELISA wash buffer
- 121.1 g Trizma base
- 40 ml Tween-20
- 800 ml H2O
- Adjust the pH to 7.3 using ∼60 ml conc. HCl
- Add water to 1 L
- Mix 50 ml of this 20× stock solution and 950 ml distilled H2O to prepare 1× solution of ELISA wash buffer
- Store up to 6 months at room temperature
Harvest IEL medium
- 29.595 ml RPMI 1640 + L-glutamine
- 300 μl FBS, dialyzed overnight at 4°C against PBS (Corning Cellgro) using dialysis membrane tubing 12–14 kDa MWCO (Spectrum, cat. no. S432700)
- 45 μl 0.5 M EDTA (1 mM final)
- 60 μl 1 M MgCl2 (1.5 mM final)
- Prepare fresh
HCl, 1 mM
- 0.5 ml of 1 M HCl
- 499.5 ml Milli-Q water
- Verify that pH ∼ 3
- Store up to 1 year at room temperature, in a cool, dry, well-ventilated area
HCl, 0.2 N
- 17.18 ml 12.1 N HCl
- Milli-Q water to 1 L
- Verify that pH ∼ 3
- Store up to 1 year at room temperature, in a cool, dry, well-ventilated area
IHC blocking solution
- 474.5 ml PBS (Corning Cellgro)
- 500 μl Tween-20
- 25 ml bovine serum albumin (Sigma-Aldrich, cat. no. A8806-1G)
- Store up to 1 month at 4°C
Ionomycin
From the lyophilized ionomycin (Enzo Life Sciences, cat. no. CA-201), prepare a stock solution of 1 mg/ml in DMSO, aliquot, and store at −20°C. This solution is stable at −20°C for at least 6 months. On day of experiment, dilute an aliquot of frozen ionomycin stock 1:10 in PBS (Corning Cellgro) to make 100 μg/ml concentration.
Na2HPO4, 0.1 M
- 7.098 g Na2HPO4 (FW = 141.96 g/mol)
- H2O to 500 ml
- Store up to 6 months at 4°C
Percoll solution
Prepare a 1× stock solution by mixing 90 ml Percoll (GE Healthcare) with 10 ml of 10× PBS (Corning Cellgro). Prepare fresh working 40% solution by mixing 40 ml 1× Percoll stock with 60 ml of 1× PBS containing 2% fetal bovine serum (FBS). Store stock solution up to 1 year at 4°C.
Phorbol 12-myristate 13-acetate (PMA)
From the lyophilized PMA (Enzo Life Sciences, cat. no. PE-160), prepare a stock solution of 1 mg/ml in DMSO, aliquot and store at −20°C. This solution is stable at −20°C for at least 6 months. On day of experiment, serially dilute an aliquot of PMA stock from freezer 1:1000 in DPBS (Corning Cellgro) to make 1 μg/ml concentration.
Phosphate-buffer (PBS), 1×
- 0.2 g KCl
- 0.2 g KH2PO4
- 8.0 g NaCl
- 2.16 g Na2HPO4 -7H2O
- Water to 1 L
- pH 7.2-7.4
Tris buffer (500 mM), 10×
- 3.03 g Tris base
- 45 ml Milli-Q water
- Adjust pH to 7.8 with HCl
- Βring final volume to 50 ml
- Store up to 2 weeks at room temperature
COMMENTARY
Background Information
Mouse models of CeD can help us understand how tissue-derived signals drive tissue destruction and tissue immunity. IL-15 is the main pro-inflammatory cytokine overexpressed in the intestinal tissue in CeD (Jabri et al., 2000; Maiuri et al., 2000; Mention et al., 2003). Unlike other secreted cytokines, IL-15 acts mainly in a cell contact–dependent manner (Waldmann, 2006), suggesting that the location of IL-15 upregulation is critical and will determine its biological effect. The humanized HLA-DQ8 mice strains overexpressing IL-15 in different intestinal compartments, i.e., only in the gut epithelium in the DQ8-villin-IL-15tg mouse, or in the intestinal lamina propria and gut-associated lymphoid organs in the DQ8-Dd-IL-15tg mouse, and both in the gut epithelium and the other tissues and organs in the DQ8-Dd-villin-IL15tg mouse, have made it possible to study the impact of the differential expression of IL-15 on the gluten-induced immune responses and ensuing mucosal injury.
In accordance with earlier studies (Ohta et al., 2002; Yokoyama et al., 2009), IL-15 overexpression in the gut epithelium in DQ8-villin-IL-15tg mice promotes the expansion of intraepithelial lymphocytes and the acquisition of activating NK receptors. This is also in agreement with observations in CeD patients (Setty et al., 2015) and with our view that, in addition to signal 1 (TCR stimulation) and signal 2 (co-stimulation), IL-15 and stress-inducible non-classical MHC class I molecules expressed by distressed tissue cells provide co-stimulatory signals to cytotoxic tissue-resident memory cells that are critical in order to license them to become killer cells and destroy tissue cells (Jabri & Abadie, 2015; Jabri & Sollid, 2006). This concept is demonstrated by the finding that DQ8-Dd-IL-15tg mice overexpressing IL-15 in all compartments except the gut epithelium do not present alterations in the amount and phenotype of IELs despite their ability to develop a TH1 anti-gluten immune responses (DePaolo et al., 2011). Only DQ8-Dd-villin-IL-15tg mice that also express IL-15 in the epithelium show licensing of cytotoxic intraepithelial lymphocytes to kill, and development of villous atrophy. Overall, although the identity of IL-15-producing cells in the context of CeD still needs to be identified, these mouse models have demonstrated the functional impact of the location of IL-15 expression, and more generally are a good example of how inflammatory signals within tissue deeply impact local antigen-driven immune responses.
The requirement of CD4 and CD8 T cells, HLA, and TG2 for the development of CeD in the DQ8-Dd-villin-IL-15tg mouse reflects the complex cellular and immune interactions that characterize autoimmune diseases but not Mendelian disorders. The DQ8-Dd-villin-IL-15tg mouse model constitutes a unique tool to dissect their relative and combined contributions to CeD pathogenesis. Finally, not only are DQ8-Dd-villin-IL-15tg mice a good model to study immune responses underlying complex autoimmune disorders, but they can also be used as a preclinical tool to test novel therapeutic strategies. The inhibition of TG2 was effective in preventing the development of villous atrophy in the DQ8-Dd-villin-IL-15tg mouse model of CeD (Abadie et al., 2020), a therapeutic approach to the treatment of CeD that has proven to be promising in a cohort of CeD patients (Jabri, 2021; Schuppan et al., 2021).
Critical Parameters and Troubleshooting
Several factors need to be taken into consideration when working with the DQ8-Dd-villin-IL-15tg mouse model. Animals between 9 and 11 weeks old are usually used, to ensure experiment reproducibility and to observe consistency in the development of villous atrophy. Mucosal damage is only observed in around 70% of gluten-fed animals and is not sex-dependent. It is essential to transfer the mice from a gluten-free diet to a gluten-containing rodent chow to ensure a daily consumption of gluten. Control mice that are never exposed to gluten, remaining on a gluten-free diet, should be used as a control group to monitor gluten-dependent manifestations. In addition, when using mouse models such as the DQ8-Dd-villin-IL-15tg model to compare different treatments, mice should be co-housed for at least a week, to equilibrate their microbiota.
Overall, the techniques described in this unit are straightforward and provide consistent results. One difficulty that may be encountered is the analysis of villous atrophy in small intestinal sections. This arises because improper tissue orientation and cutting can lead to an erroneous assessment about the presence of atrophy. It is critical to only perform the morphometric assessment of the villous height and crypt depth on specimens where only one longitudinally cut crypt is observed between two adjacent villi, indicative of a good orientation.
Statistical Analysis
Villous atrophy develops in approximately 70% of gluten-fed DQ8-Dd-villin-IL-15tg mice. Therefore, 10–12 mice per group are required to perform significant comparisons between different groups of mice.
Time Considerations
The standard protocol allowing the investigator to observe patent villous atrophy requires 30 days of gluten exposure. From the moment the mice are euthanized, dissection and harvesting of the small intestine should take 10 min per animal. Isolating IELs from the small intestine takes ∼2 hr. The measurements of the villous height and crypt depth, and the enumeration of IELs, may take half an hour per sample. The assessment of degranulation takes ∼8–9 hr from cell stimulation to the analysis by flow cytometry.
Acknowledgements
This work is supported by grants from NIH: R01 DK128352 to V.A, R01 DK063158 and R01 DK126487 to C.K. and B.J., and R01 DK067180 and Digestive Diseases Research Core Center P30 DK42086 to B.J.
Author Contributions
Valérie Abadie : conceptualization, formal analysis, writing original draft, writing review and editing; Chaitan Khosla : methodology, writing review and editing; Bana Jabri : conceptualization, formal analysis, writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data are available from the corresponding authors upon request.
Literature Cited
- Abadie, V., & Jabri, B. (2014). IL-15: A central regulator of celiac disease immunopathology. Immunological Reviews , 260(1), 221–234. doi: 10.1111/imr.12191
- Abadie, V., Kim, S. M., Lejeune, T., Palanski, B. A., Ernest, J. D., Tastet, O., … Jabri, B. (2020). IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature , 578(7796), 600–604. doi: 10.1038/s41586-020-2003-8
- Abadie, V., Sollid, L. M., Barreiro, L. B., & Jabri, B. (2011). Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annual Review of Immunology , 29, 493–525. doi: 10.1146/annurev-immunol-040210-092915
- Adelman, D. C., Murray, J., Wu, T. T., Maki, M., Green, P. H., & Kelly, C. P. (2018). Measuring change in small intestinal histology in patients with celiac disease. American Journal of Gastroenterology , 113(3), 339–347. doi: 10.1038/ajg.2017.480
- Arentz-Hansen, H., Korner, R., Molberg, O., Quarsten, H., Vader, W., Kooy, Y. M., … McAdam, S. N. (2000). The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. Journal of Experimental Medicine , 191(4), 603–612. doi: 10.1084/jem.191.4.603
- Arguelles-Grande, C., Tennyson, C. A., Lewis, S. K., Green, P. H., & Bhagat, G. (2012). Variability in small bowel histopathology reporting between different pathology practice settings: Impact on the diagnosis of coeliac disease. Journal of Clinical Pathology , 65(3), 242–247. doi: 10.1136/jclinpath-2011-200372
- Betts, M. R., Brenchley, J. M., Price, D. A., De Rosa, S. C., Douek, D. C., Roederer, M., & Koup, R. A. (2003). Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. Journal of Immunological Methods , 281(1-2), 65–78. doi: 10.1016/S0022-1759(03)00265-5
- Betts, M. R., & Koup, R. A. (2004). Detection of T-cell degranulation: CD107a and b. Methods in Cell Biology , 75, 497–512. doi: 10.1016/S0091-679X(04)75020-7
- Black, K. E., Murray, J. A., & David, C. S. (2002). HLA-DQ determines the response to exogenous wheat proteins: A model of gluten sensitivity in transgenic knockout mice. Journal of Immunology , 169(10), 5595–5600. doi: 10.4049/jimmunol.169.10.5595
- Bodd, M., Raki, M., Tollefsen, S., Fallang, L. E., Bergseng, E., Lundin, K. E., & Sollid, L. M. (2010). HLA-DQ2-restricted gluten-reactive T cells produce IL-21 but not IL-17 or IL-22. Mucosal Immunology , 3(6), 594–601. doi: 10.1038/mi.2010.36
- Cummins, A. G., Alexander, B. G., Chung, A., Teo, E., Woenig, J. A., Field, J. B., … Roberts-Thomson, I. C. (2011). Morphometric evaluation of duodenal biopsies in celiac disease. American Journal of Gastroenterology , 106(1), 145–150. doi: 10.1038/ajg.2010.313
- Daveson, A. J. M., Popp, A., Taavela, J., Goldstein, K. E., Isola, J., Truitt, K. E., … on behalf of the RESET CeD Study Group. (2019). Baseline quantitative histology in therapeutic trials reveals villus atrophy in most patients with coeliac disease who appear well controlled on gluten-free diet. GastroHep , 2, 22–30. doi: 10.1002/ygh2.380
- de Kauwe, A. L., Chen, Z., Anderson, R. P., Keech, C. L., Price, J. D., Wijburg, O., … McCluskey, J. (2009). Resistance to celiac disease in humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin CD4+ T cells. Journal of Immunology , 182(12), 7440–7450. doi: 10.4049/jimmunol.0900233
- DePaolo, R. W., Abadie, V., Tang, F., Fehlner-Peach, H., Hall, J. A., Wang, W., … Jabri, B. (2011). Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature , 471(7337), 220–224. doi: 10.1038/nature09849
- Di Niro, R., Mesin, L., Zheng, N. Y., Stamnaes, J., Morrissey, M., Lee, J. H., … Sollid, L. M. (2012). High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nature Medicine , 18(3), 441–445. doi: 10.1038/nm.2656
- Dieterich, W., Ehnis, T., Bauer, M., Donner, P., Volta, U., Riecken, E. O., & Schuppan, D. (1997). Identification of tissue transglutaminase as the autoantigen of celiac disease. Nature Medicine , 3(7), 797–801. doi: 10.1038/nm0797-797
- Dieterich, W., Laag, E., Schopper, H., Volta, U., Ferguson, A., Gillett, H., … Schuppan, D. (1998). Autoantibodies to tissue transglutaminase as predictors of celiac disease. Gastroenterology , 115(6), 1317–1321. doi: 10.1016/S0016-5085(98)70007-1
- Dieterich, W., Neurath, M. F., & Zopf, Y. (2020). Intestinal ex vivo organoid culture reveals altered programmed crypt stem cells in patients with celiac disease. Science Reports , 10(1), 3535. doi: 10.1038/s41598-020-60521-5
- Donovan, J., & Brown, P. (2006). Euthanasia. Current Protocols in Immunology , 73, 1.8.1–1.8.4. doi: 10.1002/0471142735.im0108s73
- Du Pre, M. F., Blazevski, J., Dewan, A. E., Stamnaes, J., Kanduri, C., Sandve, G. K., … Sollid, L. M. (2020). B cell tolerance and antibody production to the celiac disease autoantigen transglutaminase 2. Journal of Experimental Medicine , 217(2), e20190860. doi: 10.1084/jem.20190860
- Du Pre, M. F., Kozijn, A. E., van Berkel, L. A., ter Borg, M. N., Lindenbergh-Kortleve, D., Jensen, L. T., … Samsom, J. N. (2011). Tolerance to ingested deamidated gliadin in mice is maintained by splenic, type 1 regulatory T cells. Gastroenterology , 141(2), 610–620.e611–612. doi: 10.1053/j.gastro.2011.04.048
- Fehniger, T. A., Suzuki, K., Ponnappan, A., VanDeusen, J. B., Cooper, M. A., Florea, S. M., … Caligiuri, M. A. (2001). Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. Journal of Experimental Medicine , 193(2), 219–231. doi: 10.1084/jem.193.2.219
- Freitag, T. L., Rietdijk, S., Junker, Y., Popov, Y., Bhan, A. K., Kelly, C. P., … Schuppan, D. (2009). Gliadin-primed CD4+CD45RBlowCD25– T cells drive gluten-dependent small intestinal damage after adoptive transfer into lymphopenic mice. Gut , 58(12), 1597–1605. doi: 10.1136/gut.2009.186361
- Gass, J., Vora, H., Bethune, M. T., Gray, G. M., & Khosla, C. (2006). Effect of barley endoprotease EP-B2 on gluten digestion in the intact rat. Journal of Pharmacology and Experimental Therapeutics , 318(3), 1178–1186. doi: 10.1124/jpet.106.104315
- Hofman, F. M., & Taylor, C. R. (2013). Immunohistochemistry. Current Protocols in Immunology , 103, 21.24.21–21.24.26. doi: 10.1002/0471142735.im2104s103
- Hue, S., Mention, J. J., Monteiro, R. C., Zhang, S., Cellier, C., Schmitz, J., … Caillat-Zucman, S. (2004). A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity , 21(3), 367–377. doi: 10.1016/j.immuni.2004.06.018
- Hugenholtz, F., & de Vos, W. M. (2018). Mouse models for human intestinal microbiota research: A critical evaluation. Cellular and Molecular Life Sciences , 75(1), 149–160. doi: 10.1007/s00018-017-2693-8
- Husby, S., Koletzko, S., Korponay-Szabo, I., Kurppa, K., Mearin, M. L., Ribes-Koninckx, C., … Wessels, M. (2020). European ociety paediatric gastroenterology, hepatology and nutrition guidelines for diagnosing coeliac disease 2020. Journal of Pediatric Gastroenterology and Nutrition , 70(1), 141–156. doi: 10.1097/MPG.0000000000002497
- Husby, S., Koletzko, S., Korponay-Szabo, I. R., Mearin, M. L., Phillips, A., Shamir, R., … Nutrition. (2012). European Society for pediatric gastroenterology, hepatology, and nutrition guidelines for the diagnosis of coeliac disease. Journal of Pediatric Gastroenterology and Nutrition , 54(1), 136–160. doi: 10.1097/MPG.0b013e31821a23d0
- Husby, S., & Murray, J. A. (2014). Diagnosing coeliac disease and the potential for serological markers. Nature reviews Gastroenterology & hepatology, 11(11), 655–663.
- Iversen, R., & Sollid, L. M. (2020). Autoimmunity provoked by foreign antigens. Science , 368(6487), 132–133. doi: 10.1126/science.aay3037
- Jabri, B. (2021). Transglutaminase 2 inhibition for prevention of mucosal damage in celiac disease. New England Journal of Medicine , 385(1), 76–77. doi: 10.1056/NEJMe2107502
- Jabri, B., & Abadie, V. (2015). IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nature Reviews Immunology , 15(12), 771–783. doi: 10.1038/nri3919
- Jabri, B., de Serre, N. P., Cellier, C., Evans, K., Gache, C., Carvalho, C., … Cerf-Bensussan, N. (2000). Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology , 118(5), 867–879. doi: 10.1016/S0016-5085(00)70173-9
- Jabri, B., & Sollid, L. M. (2006). Mechanisms of disease: Immunopathogenesis of celiac disease. Nature Reviews Gastroenterology & Hepatology, 3(9), 516–525.
- Jabri, B., & Sollid, L. M. (2009). Tissue-mediated control of immunopathology in coeliac disease. Nature Reviews Immunology , 9(12), 858–870. doi: 10.1038/nri2670
- Jabri, B., & Sollid, L. M. (2017). T cells in celiac disease. Journal of Immunology , 198(8), 3005–3014. doi: 10.4049/jimmunol.1601693
- Jensen, K., Sollid, L. M., Scott, H., Paulsen, G., Kett, K., Thorsby, E., & Lundin, K. E. (1995). Gliadin-specific T cell responses in peripheral blood of healthy individuals involve T cells restricted by the coeliac disease associated DQ2 heterodimer. Scandinavian Journal of Immunology , 42(1), 166–170. doi: 10.1111/j.1365-3083.1995.tb03640.x
- Kagnoff, M. F., Austin, R. K., Johnson, H. C., Bernardin, J. E., Dietler, M. D., & Kasarda, D. D. (1982). Celiac sprue: Correlation with murine T cell responses to wheat gliadin components. Journal of Immunology , 129(6), 2693–2697.
- Khan, M. R., Nellikkal, S. S., Barazi, A., Larson, J. J., Murray, J. A., & Absah, I. (2019). The risk of autoimmune disorders in treated celiac disease patients in Olmsted county, Minnesota. Journal of Pediatric Gastroenterology and Nutrition , 69(4), 438–442. doi: 10.1097/MPG.0000000000002418
- Koning, F. (2012). Celiac disease: Quantity matters. Seminars in Immunopathology , 34(4), 541–549. doi: 10.1007/s00281-012-0321-0
- Korneychuk, N., Ramiro-Puig, E., Ettersperger, J., Schulthess, J., Montcuquet, N., Kiyono, H., … Cerf-Bensussan, N. (2014). Interleukin 15 and CD4(+) T cells cooperate to promote small intestinal enteropathy in response to dietary antigen. Gastroenterology , 146(4), 1017–1027. doi: 10.1053/j.gastro.2013.12.023
- Kutlu, T., Brousse, N., Rambaud, C., Le Deist, F., Schmitz, J., & Cerf-Bensussan, N. (1993). Numbers of T cell receptor (TCR) alpha beta+ but not of TcR gamma delta+ intraepithelial lymphocytes correlate with the grade of villous atrophy in coeliac patients on a long term normal diet. Gut , 34(2), 208–214. doi: 10.1136/gut.34.2.208
- Lebwohl, B., Sanders, D. S., & Green, P. H. R. (2018). Coeliac disease. Lancet , 391(10115), 70–81. doi: 10.1016/S0140-6736(17)31796-8
- Lindfors, K., Ciacci, C., Kurppa, K., Lundin, K. E. A., Makharia, G. K., Mearin, M. L., … Kaukinen, K. (2019). Coeliac disease. Nature Reviews Disease Primers , 5(1), 3. doi: 10.1038/s41572-018-0054-z
- Lindstad, C. B., Qiao, S. W., Johannesen, M. K., Fugger, L., Sollid, L. M., & du Pre, M. F. (2021). Characterization of T-cell receptor transgenic mice recognizing immunodominant HLA-DQ2.5-restricted gluten epitopes. European Journal of Immunology , 51(4), 1002–1005. doi: 10.1002/eji.202048859
- Liu, R. B., Engels, B., Arina, A., Schreiber, K., Hyjek, E., Schietinger, A., … Schreiber, H. (2012). Densely granulated murine NK cells eradicate large solid tumors. Cancer Research , 72(8), 1964–1974. doi: 10.1158/0008-5472.CAN-11-3208
- Ludvigsson, J. F., Leffler, D. A., Bai, J. C., Biagi, F., Fasano, A., Green, P. H., … Ciacci, C. (2013). The Oslo definitions for coeliac disease and related terms. Gut , 62(1), 43–52. doi: 10.1136/gutjnl-2011-301346
- Maiuri, L., Ciacci, C., Auricchio, S., Brown, V., Quaratino, S., & Londei, M. (2000). Interleukin 15 mediates epithelial changes in celiac disease. Gastroenterology , 119(4), 996–1006. doi: 10.1053/gast.2000.18149
- Marietta, E., Black, K., Camilleri, M., Krause, P., Rogers, R. S. 3rd, David, C., … Murray, J. A. (2004). A new model for dermatitis herpetiformis that uses HLA-DQ8 transgenic NOD mice. Journal of Clinical Investigation , 114(8), 1090–1097. doi: 10.1172/JCI200421055
- Marsh, M. N. (1990). Grains of truth: Evolutionary changes in small intestinal mucosa in response to environmental antigen challenge. Gut , 31(1), 111–114. doi: 10.1136/gut.31.1.111
- Meisel, M., Mayassi, T., Fehlner-Peach, H., Koval, J. C., O'Brien, S. L., Hinterleitner, R., … Antonopoulos, D. A. (2017). Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. The ISME Journal , 11(1), 15–30. doi: 10.1038/ismej.2016.114
- Mention, J. J., Ben Ahmed, M., Begue, B., Barbe, U., Verkarre, V., Asnafi, V., … Cerf-Bensussan, N. (2003). Interleukin 15: A key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology , 125(3), 730–745. doi: 10.1016/S0016-5085(03)01047-3
- Meresse, B., Chen, Z., Ciszewski, C., Tretiakova, M., Bhagat, G., Krausz, T. N., … Jabri, B. (2004). Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity , 21(3), 357–366. doi: 10.1016/j.immuni.2004.06.020
- Meresse, B., Curran, S. A., Ciszewski, C., Orbelyan, G., Setty, M., Bhagat, G., … Jabri, B. (2006). Reprogramming of CTLs into natural killer-like cells in celiac disease. Journal of Experimental Medicine , 203(5), 1343–1355. doi: 10.1084/jem.20060028
- Mino, M., & Lauwers, G. Y. (2003). Role of lymphocytic immunophenotyping in the diagnosis of gluten-sensitive enteropathy with preserved villous architecture. American Journal of Surgical Pathology , 27(9), 1237–1242. doi: 10.1097/00000478-200309000-00007
- Molberg, O., Kett, K., Scott, H., Thorsby, E., Sollid, L. M., & Lundin, K. E. (1997). Gliadin specific, HLA DQ2-restricted T cells are commonly found in small intestinal biopsies from coeliac disease patients, but not from controls. Scandinavian Journal of Immunology , 46(3), 103–109. doi: 10.1046/j.1365-3083.1996.d01-17.x
- Molberg, O., McAdam, S. N., Korner, R., Quarsten, H., Kristiansen, C., Madsen, L., … Sollid, L. M. (1998). Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nature Medicine , 4(6), 713–717. doi: 10.1038/nm0698-713
- Molberg, O., McAdam, S., Lundin, C., Kristiansen, H., Arentz-Hansen, K., & Sollid, L. M. (2001). T cells from celiac disease lesions recognize gliadin epitopes deamidated in situ by endogenous tissue transglutaminase. European Journal of Immunology , 31(5), 1317–1323. doi: 10.1002/1521-4141(200105)31:5<1317::AID-IMMU1317>3.0.CO;2-I.
- Nilsen, E. M., Jahnsen, F. L., Lundin, K. E., Johansen, F. E., Fausa, O., Sollid, L. M., … Brandtzaeg, P. (1998). Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology , 115(3), 551–563. doi: 10.1016/S0016-5085(98)70134-9
- Nilsen, E. M., Lundin, K. E., Krajci, P., Scott, H., Sollid, L. M., & Brandtzaeg, P. (1995). Gluten specific, HLA-DQ restricted T cells from coeliac mucosa produce cytokines with Th1 or Th0 profile dominated by interferon gamma. Gut , 37(6), 766–776. doi: 10.1136/gut.37.6.766
- Ohta, N., Hiroi, T., Kweon, M. N., Kinoshita, N., Jang, M. H., Mashimo, T., … Kiyono, H. (2002). IL-15-dependent activation-induced cell death-resistant Th1 type CD8 alpha beta+NK1.1+ T cells for the development of small intestinal inflammation. Journal of Immunology , 169(1), 460–468. doi: 10.4049/jimmunol.169.1.460
- Phelan, K., & May, K. M. (2015). Basic techniques in mammalian cell tissue culture. Current Protocols in Cell Biology , 66, 1.1.1–1.1.22. doi: 10.1002/0471143030.cb0101s66
- Phillips, A. T., & Signs, M. W. (2005). Desalting, concentration, and buffer exchange by dialysis and ultrafiltration. Current Protocols in Protein Science , 38, 4.4.1–4.4.15. doi: 10.1002/0471140864.ps0404s38
- Picarelli, A., Borghini, R., Donato, G., Di Tola, M., Boccabella, C., Isonne, C., … Tiberti, A. (2014). Weaknesses of histological analysis in celiac disease diagnosis: New possible scenarios. Scandinavian Journal of Gastroenterology , 49(11), 1318–1324. doi: 10.3109/00365521.2014.948052
- Rashtak, S., & Murray, J. A. (2009). Celiac disease in the elderly. Gastroenterology Clinics of North America , 38(3), 433–446. doi: 10.1016/j.gtc.2009.06.005
- Roberts, A. I., Lee, L., Schwarz, E., Groh, V., Spies, T., Ebert, E. C., & Jabri, B. (2001). NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. Journal of Immunology , 167(10), 5527–5530. doi: 10.4049/jimmunol.167.10.5527
- Rubio-Tapia, A., & Murray, J. A. (2013). Response to Remes-Troche et al. American Journal of Gastroenterology , 108(2), 284. doi: 10.1038/ajg.2012.410
- Schuppan, D., Maki, M., Lundin, K. E. A., Isola, J., Friesing-Sosnik, T., Taavela, J., … Group, C. E. C. T. (2021). A randomized trial of a transglutaminase 2 inhibitor for celiac disease. New England Journal of Medicine , 385(1), 35–45. doi: 10.1056/NEJMoa2032441
- Setty, M., Discepolo, V., Abadie, V., Kamhawi, S., Mayassi, T., Kent, A., … Jabri, B. (2015). Distinct and synergistic contributions of epithelial stress and adaptive immunity to functions of intraepithelial killer cells and active celiac disease. Gastroenterology , 149(3), 681–691.e610. doi: 10.1053/j.gastro.2015.05.013
- Shan, L., Molberg, O., Parrot, I., Hausch, F., Filiz, F., Gray, G. M., … Khosla, C. (2002). Structural basis for gluten intolerance in celiac sprue. Science , 297(5590), 2275–2279. doi: 10.1126/science.1074129
- Shewry, P. R., Tatham, A. S., Forde, J., Kreis, M., & Miflin, B. J. (1986). The classification and nomenclature of wheat gluten proteins—a reassessment. Journal of Cereal Science , 4(2), 97–106. doi: 10.1016/S0733-5210(86)80012-1
- Siegel, M., Strnad, P., Watts, R. E., Choi, K., Jabri, B., Omary, M. B., & Khosla, C. (2008). Extracellular transglutaminase 2 is catalytically inactive, but is transiently activated upon tissue injury. PLoS One , 3(3), e1861. doi: 10.1371/journal.pone.0001861
- Sollid, L. M., & Jabri, B. (2005). Is celiac disease an autoimmune disorder? Current Opinion in Immunology , 17(6), 595–600. doi: 10.1016/j.coi.2005.09.015
- Sollid, L. M., Markussen, G., Ek, J., Gjerde, H., Vartdal, F., & Thorsby, E. (1989). Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. Journal of Experimental Medicine , 169(1), 345–350. doi: 10.1084/jem.169.1.345
- Steinsbo, O., Henry Dunand, C. J., Huang, M., Mesin, L., Salgado-Ferrer, M., Lundin, K. E., … Sollid, L. M. (2014). Restricted VH/VL usage and limited mutations in gluten-specific IgA of coeliac disease lesion plasma cells. Nature Communication , 5, 4041. doi: 10.1038/ncomms5041
- Taavela, J., Koskinen, O., Huhtala, H., Lahdeaho, M. L., Popp, A., Laurila, K., … Maki, M. (2013). Validation of morphometric analyses of small-intestinal biopsy readouts in celiac disease. PLoS One , 8(10), e76163. doi: 10.1371/journal.pone.0076163
- Tollefsen, S., Arentz-Hansen, H., Fleckenstein, B., Molberg, O., Raki, M., Kwok, W. W., … Sollid, L. M. (2006). HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. Journal of Clinical Investigation , 116(8), 2226–2236. doi: 10.1172/JCI27620
- van de Wal, Y., Kooy, Y., van Veelen, P., Pena, S., Mearin, L., Papadopoulos, G., & Koning, F. (1998). Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. Journal of Immunology , 161(4), 1585–1588.
- Verdu, E. F., Huang, X., Natividad, J., Lu, J., Blennerhassett, P. A., David, C. S., … Murray, J. A. (2008). Gliadin-dependent neuromuscular and epithelial secretory responses in gluten-sensitive HLA-DQ8 transgenic mice. American Journal of Physiology Gastrointestinal and Liver Physiology , 294(1), G217–225. doi: 10.1152/ajpgi.00225.2007
- Voisine, J., & Abadie, V. (2021). Interplay between gluten, HLA, innate and adaptive immunity orchestrates the development of coeliac disease. Frontiers in Immunology , 12, 674313. doi: 10.3389/fimmu.2021.674313
- Waldmann, T. A. (2006). The biology of interleukin-2 and interleukin-15: Implications for cancer therapy and vaccine design. Nature Reviews Immunology , 6(8), 595–601. doi: 10.1038/nri1901
- Yokoyama, S., Watanabe, N., Sato, N., Perera, P. Y., Filkoski, L., Tanaka, T., … Perera, L. P. (2009). Antibody-mediated blockade of IL-15 reverses the autoimmune intestinal damage in transgenic mice that overexpress IL-15 in enterocytes. Proceedings of the National Academy of Sciences of the United States of America , 106(37), 15849–15854. doi: 10.1073/pnas.0908834106
Citing Literature
Number of times cited according to CrossRef: 2
- Tobias L. Freitag, Leif C. Andersson, Anja Kipar, Concerns about the histological assessment in a mouse model of human celiac disease, Scandinavian Journal of Immunology, 10.1111/sji.13351, 99 , 3, (2024).
- Anton Chaykin, Elena Odintsova`, Andrey Nedorubov, Celiac Disease: Disease Models in Understanding Pathogenesis and Search for Therapy, Open Access Macedonian Journal of Medical Sciences, 10.3889/oamjms.2022.11024, 10 , F, (705-719), (2022).

