Tutorial: methods for three-dimensional visualization of archival tissue material
Tariq Sami Haddad, Peter Friedl, Navid Farahani, Darren Treanor, Inti Zlobec, Iris Nagtegaal
sample preparation
three-dimensional visualization
archival tissue
microscopy techniques
disease mechanisms
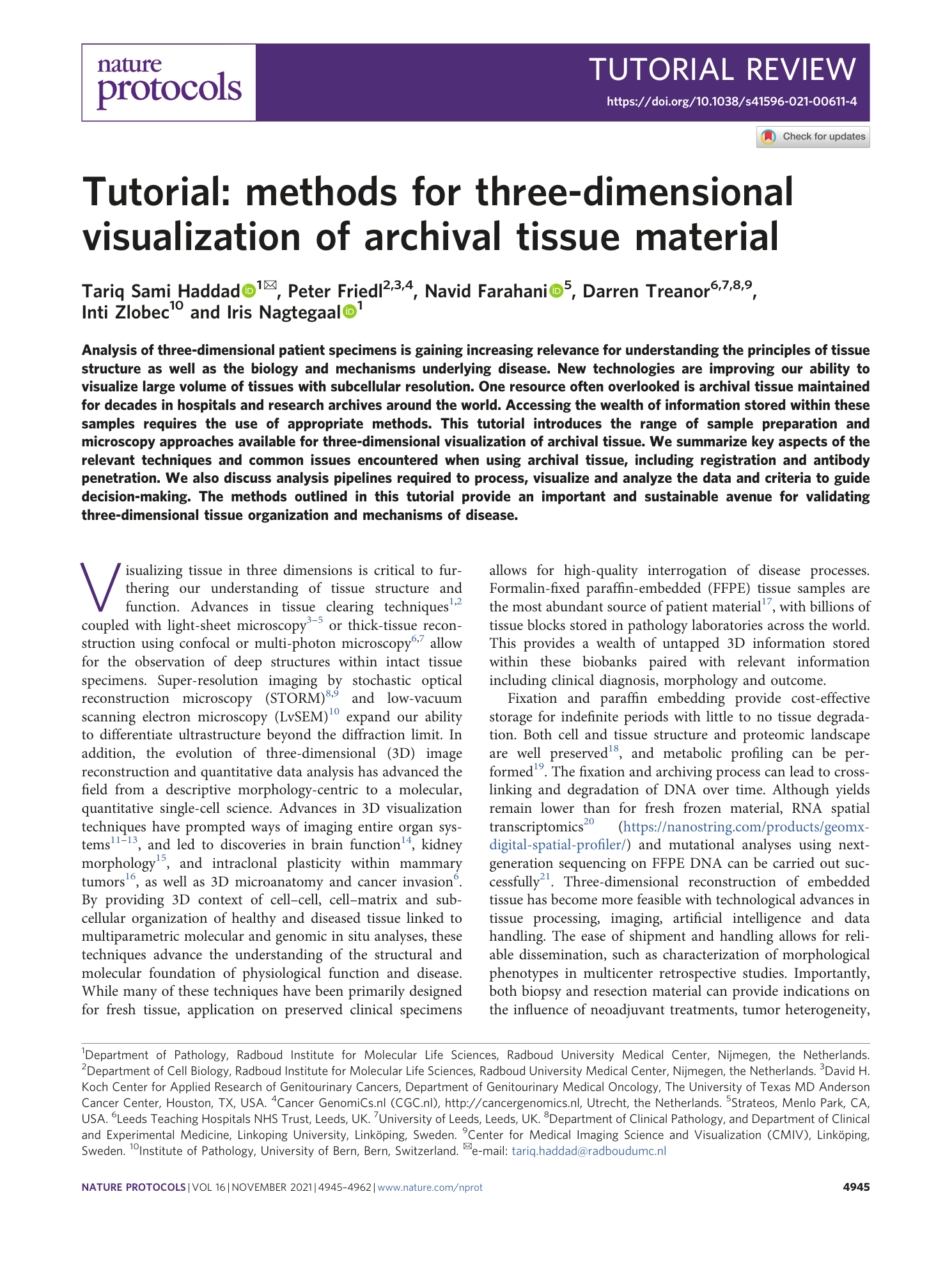

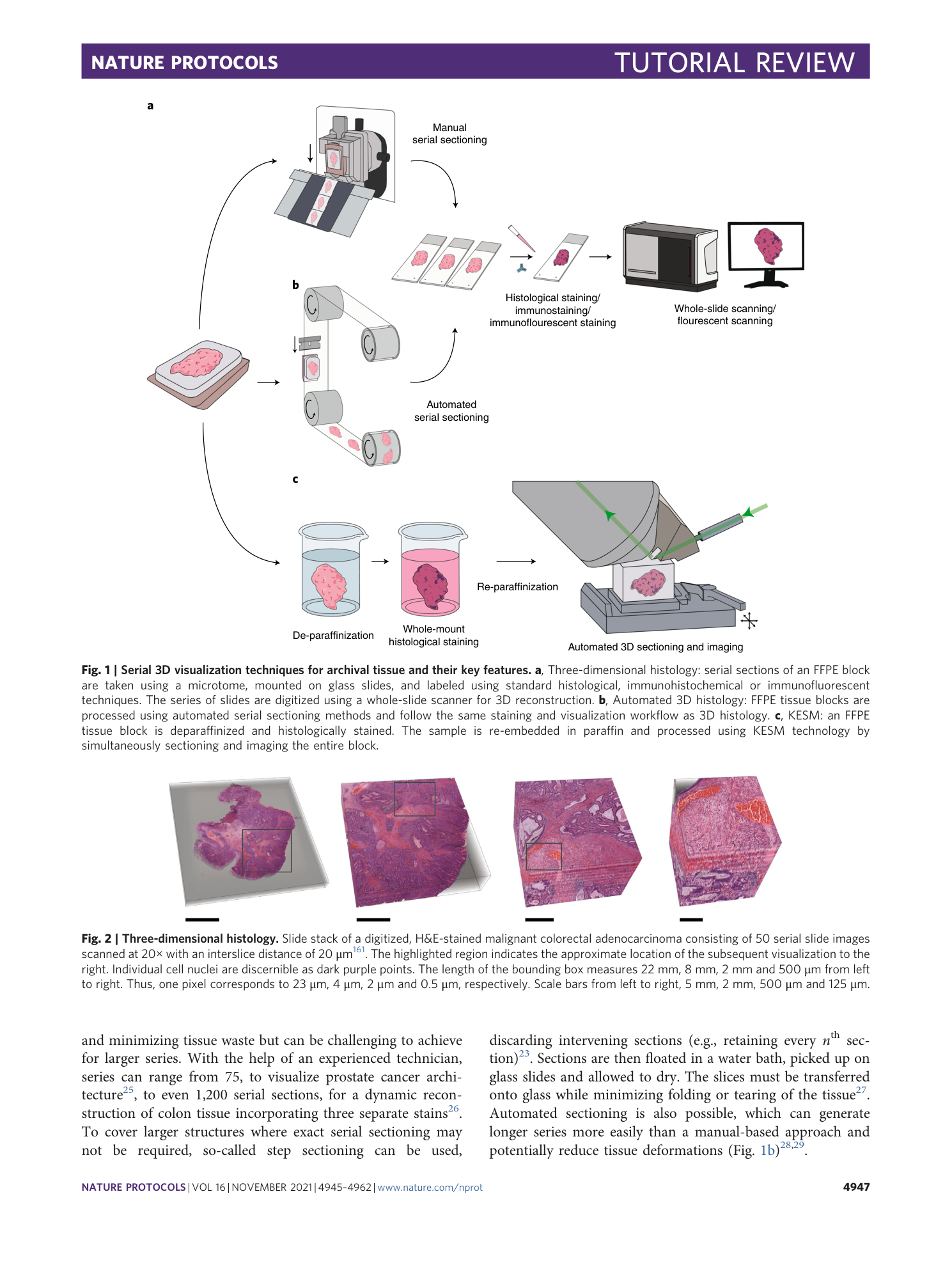

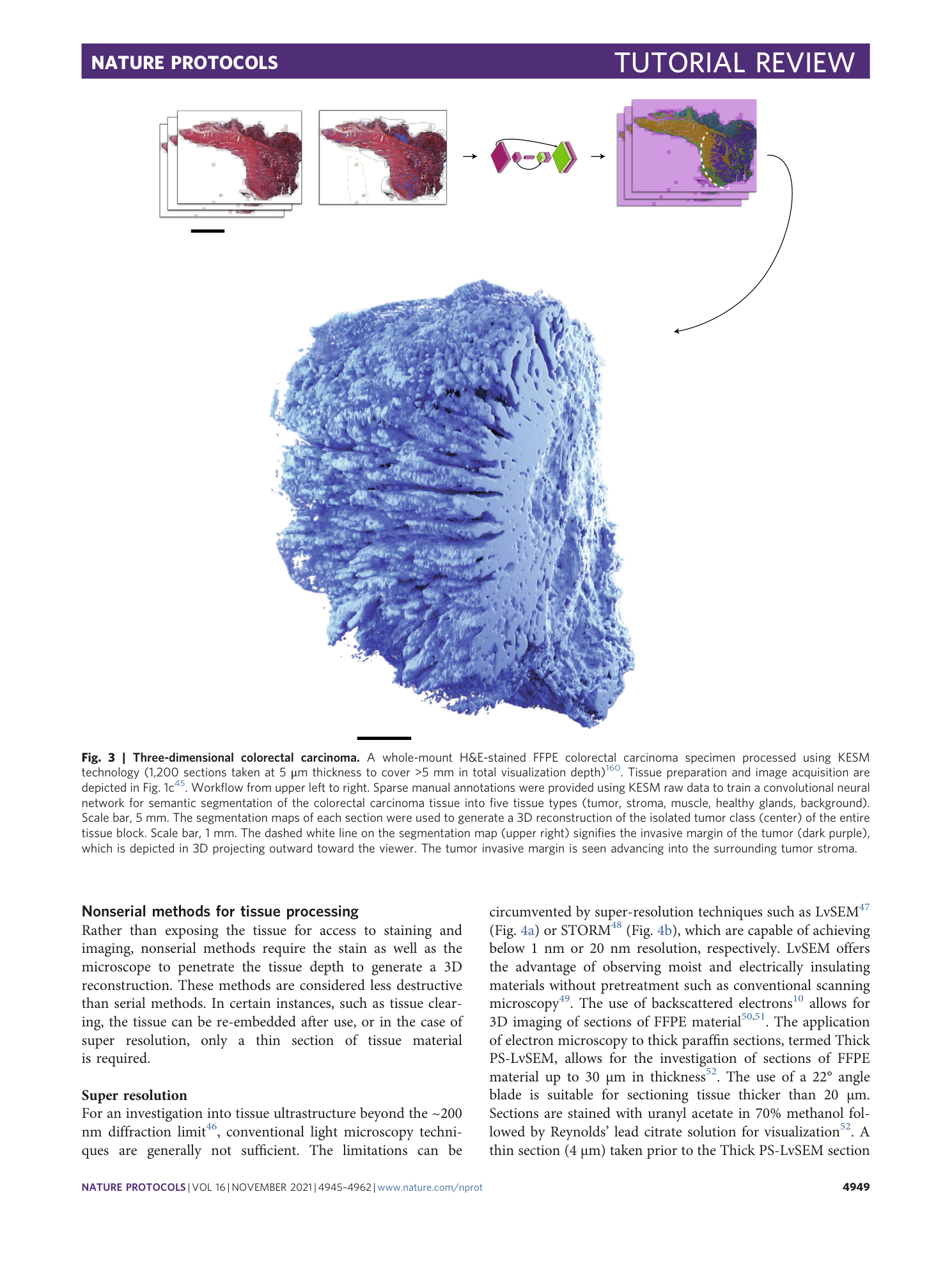
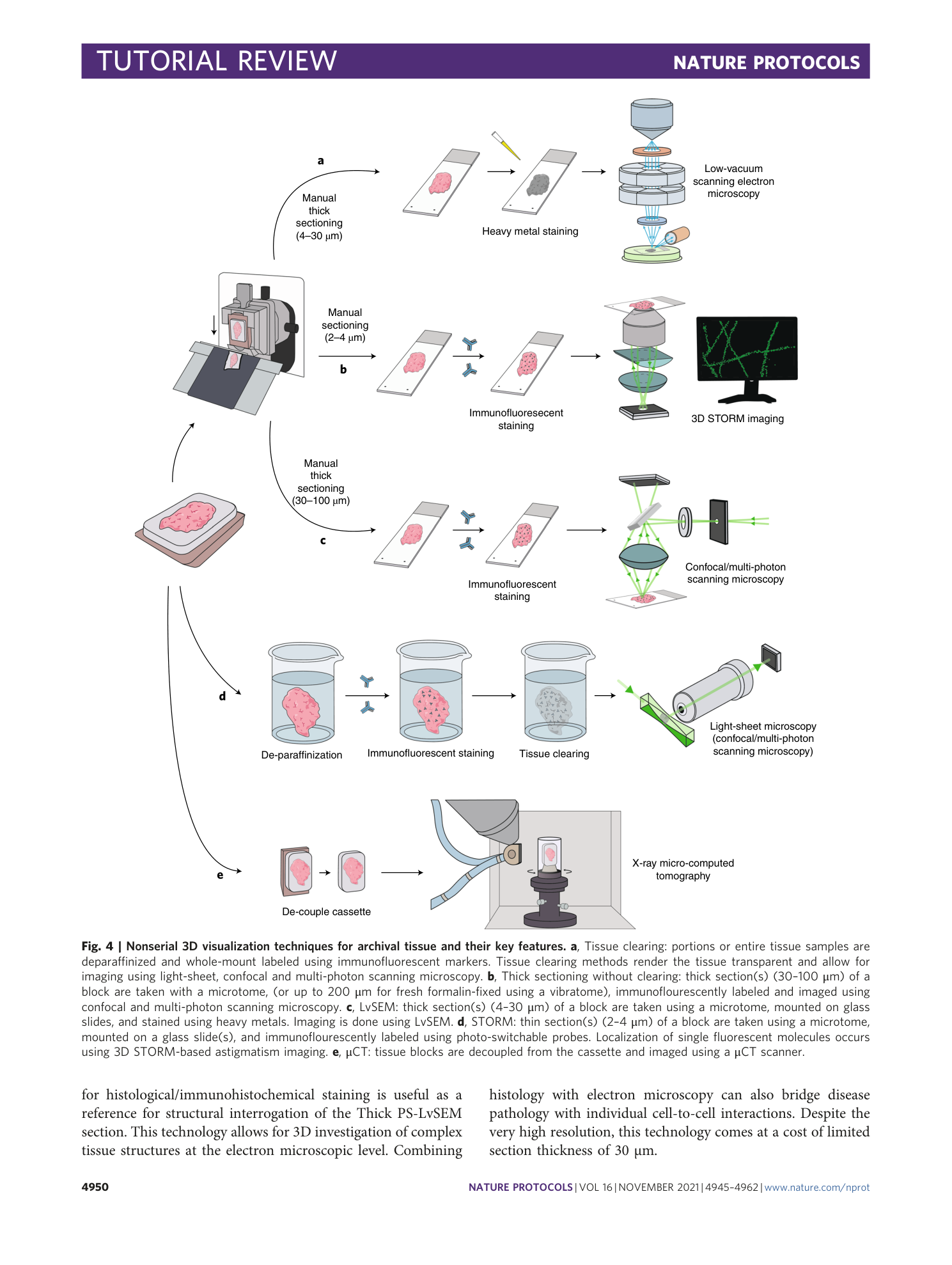








Supplementary information
Supplementary Video 1
3D malignant colorectal adenocarcinoma consisting of 50 slide images with an interslice distance of 20 μm digitized twice at 20x, once with an H&E stain and a second time with a Pan-cytokeratin immunohistochemical stain to reveal the epithelium in brown. Data related to Figure 2.
Supplementary Video 2
3D reconstruction of the isolated tumor class of a formalin-fixed paraffin-embedded colorectal carcinoma specimen processed using Knife-Edge Scanning Microscopy (KESM) technology and segmented using deep learning. Data related to Figure 3.
Supplementary Video 3
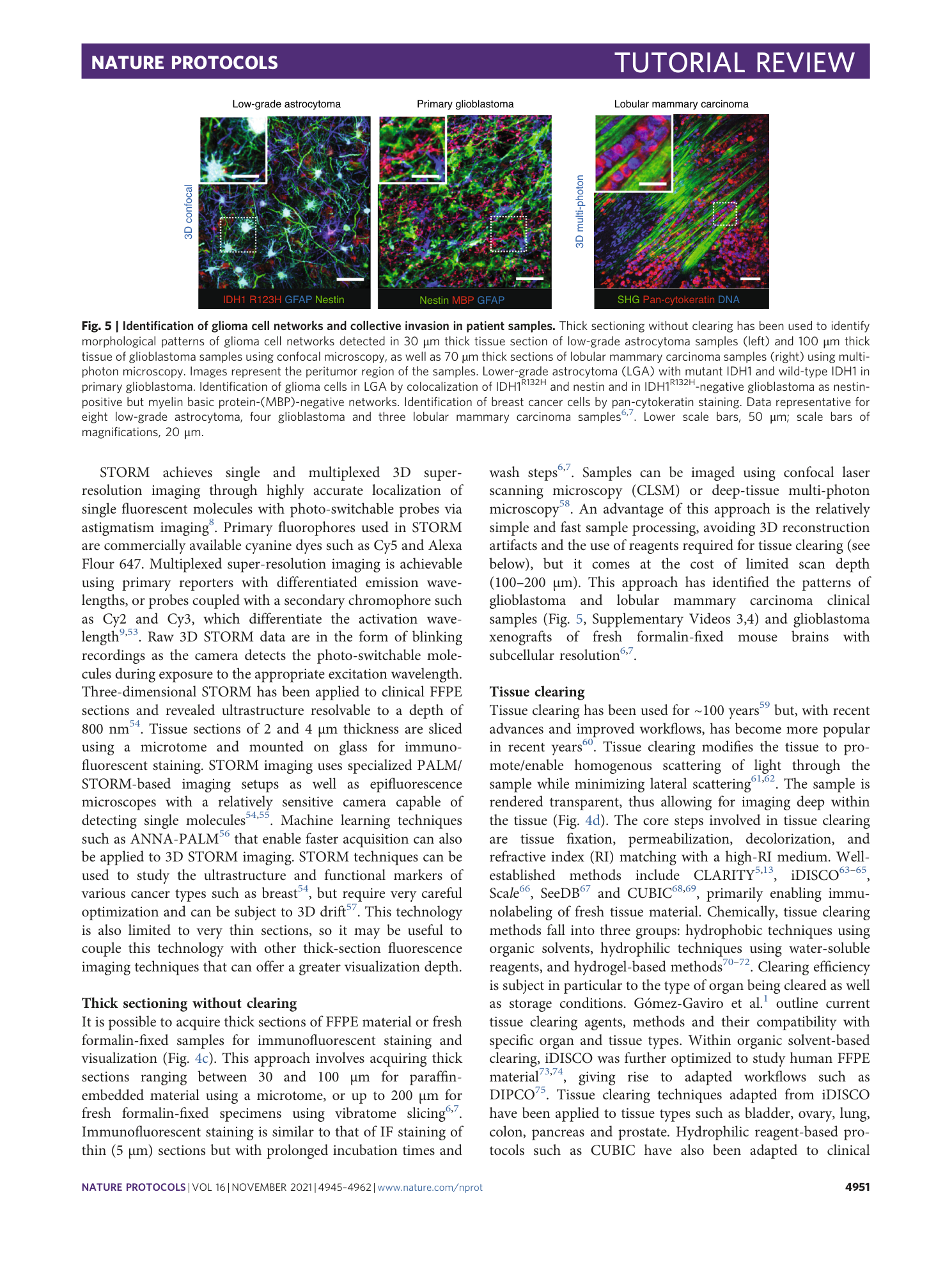
3D reconstruction of invasive ductal carcinoma lesion in a patient sample (30 µm Z-stack, 5 µm step). Collective strands of carcinoma cells were detected by positive E-cadherin staining; collagen bundles (SHG); nuclei (DAPI). The video is representative for 7 independent samples. Data related to Figure 5.
Supplementary Video 4
3D reconstruction of invasive lobular carcinoma lesion in a patient sample (60 µm Z-stack, 5 µm step). Collective strands of carcinoma cells were detected by positive staining for pan-cytokeratin; collagen bundles (SHG); nuclei (DAPI). The video is representative for 3 independent samples. Data related to Figure 5.

