Supervised Learning of Gene Regulatory Networks
Zahra Razaghi-Moghadam, Zahra Razaghi-Moghadam, Zoran Nikoloski, Zoran Nikoloski
Abstract
Identifying the entirety of gene regulatory interactions in a biological system offers the possibility to determine the key molecular factors that affect important traits on the level of cells, tissues, and whole organisms. Despite the development of experimental approaches and technologies for identification of direct binding of transcription factors (TFs) to promoter regions of downstream target genes, computational approaches that utilize large compendia of transcriptomics data are still the predominant methods used to predict direct downstream targets of TFs, and thus reconstruct genome-wide gene-regulatory networks (GRNs). These approaches can broadly be categorized into unsupervised and supervised, based on whether data about known, experimentally verified gene-regulatory interactions are used in the process of reconstructing the underlying GRN. Here, we first describe the generic steps of supervised approaches for GRN reconstruction, since they have been recently shown to result in improved accuracy of the resulting networks? We also illustrate how they can be used with data from model organisms to obtain more accurate prediction of gene regulatory interactions. © 2020 The Authors.
Basic Protocol 1 : Construction of features used in supervised learning of gene regulatory interactions
Basic Protocol 2 : Learning the non-interacting TF-gene pairs
Basic Protocol 3 : Learning a classifier for gene regulatory interactions
INTRODUCTION
A genome-wide gene regulatory network (GRN) consists of all transcription factor (TF)–target gene interactions that take place in a biological system. Variation in responsiveness of a target gene to a TF, due to genetic variation, change in the environment, or a combination thereof, can affect target-gene expression. Therefore, computational predictions of TF–target gene interactions based on gene expression (i.e., transcriptomics) data are well established and widely used in modern systems biology (Haury, Mordelet, Vera-Licona, & Vert, 2012; Huynh-Thu, Irrthum, Wehenkel, & Geurts, 2009; Marbach et al., 2012; Margolin et al., 2006; Meyer, Kontos, Lafitte, & Bontempi, 2007; Mordelet & Vert 2008; Petralia, Wang, Yang, & Tu, 2015).
The existing approaches for reconstruction of gene regulatory interactions based on transcriptomics data can be grouped into two categories, i.e., unsupervised and supervised (Maetschke, Madhamshettiwar, Davis, & Ragan, 2014). Unsupervised approaches are most prominently used due to the relatively simple formulation—they rely on application of statistical approaches that make use of the transcriptomics data and thresholding techniques (Omranian, Eloundou-Mbebi, Mueller-Roeber, & Nikoloski, 2016), without consideration of the accumulated knowledge on experimentally verified gene regulatory interactions (Fig. 1). In contrast, supervised approaches use knowledge of known gene regulatory interactions, in addition to transcriptomics profiles, to predict new gene regulatory interactions. A comprehensive comparative study with synthetic and experimentally obtained transcriptomics data sets has indicated the superiority of supervised over unsupervised approaches for GRN reconstruction (Maetschke et al., 2014).
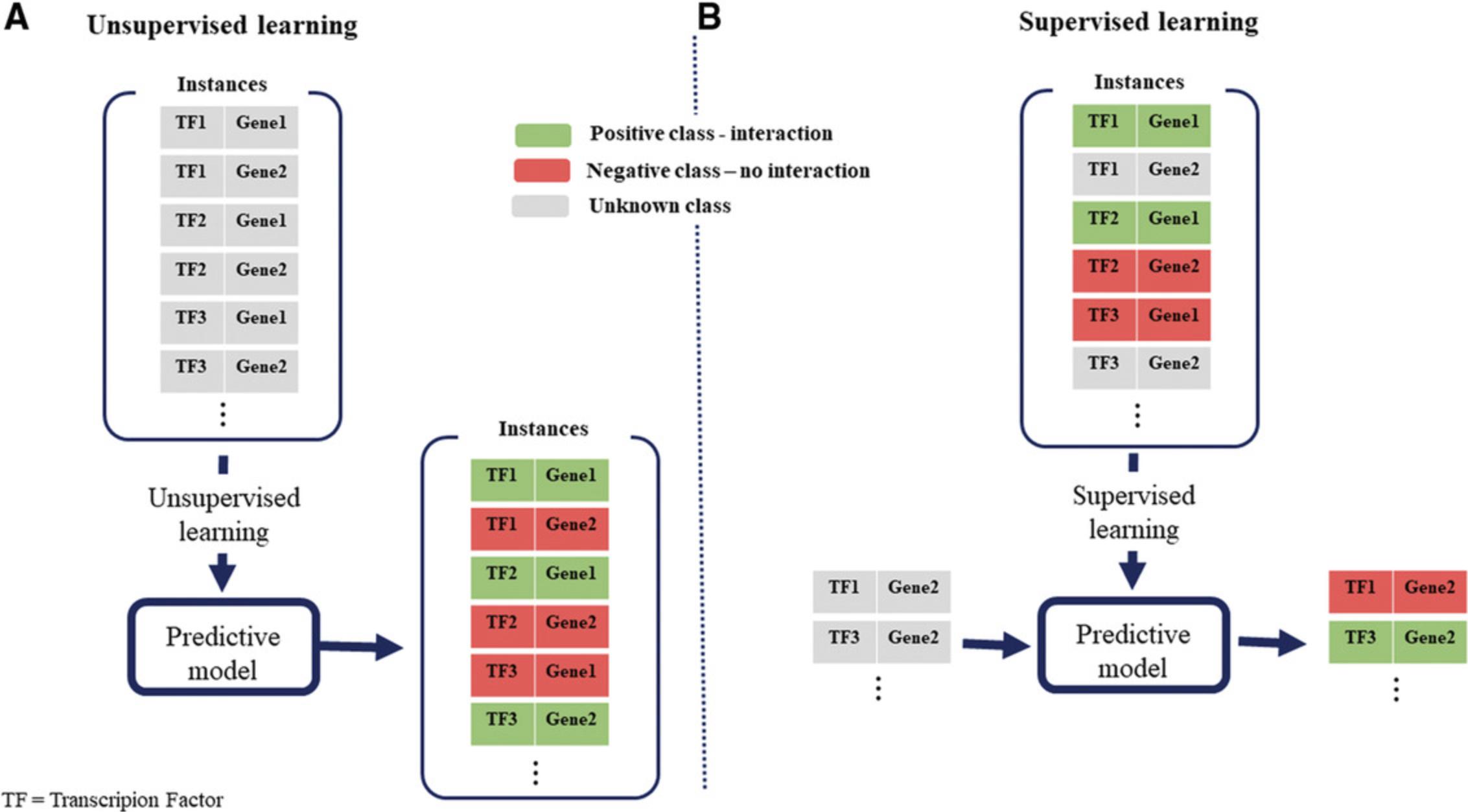
The supervised approaches are based on the idea that if one TF is known to regulate a gene, then all TF-gene pairs with similar features are likely to interact as well. Therefore, supervised approaches necessitate that the expression data profiles for a TF-gene pair be first transformed into feature vectors and then used as input to a supervised learning method. The learning method consists of training a classifier, which is employed to identify whether or not a pair of genes is involved in a regulatory interaction based on the employed features. The key challenges of supervised learning of GRNs are the construction of features used in the learning process, as well as the availability of information that a TF does not have a particular gene as a target, which cannot be readily verified experimentally.
Supervised learning approaches for GRN reconstruction can be further grouped into local and global (Vert, 2010). In local approaches, a classifier is built to discriminate the target of each TF separately. In contrast, global approaches use all TF-target gene pairs to learn a classifier for gene regulatory interactions. The global approaches are better suited for practical applications, since the learned classifier can be used on any TF-gene pair and does not require considerable knowledge of gene regulatory interactions for each TF.
The existing supervised approach for GRN reconstruction, called SIRENE, is local—it builds a binary classifier based on a support vector machine (SVM) which, for each TF, distinguishes target from non-target genes (Mordelet & Vert, 2008). SIRENE overcomes the absence of knowledge that a TF does not directly interact with a given gene roughly, by randomly selecting such pairs. In the following protocols, we describe an improved approach for generation of non-interacting TF-gene pairs that can be used in conjunction with expression-based SVM to improve the prediction accuracy of gene regulatory interactions (Razaghi-Moghadam and Nikoloski, submitted). The code for the following protocols is available at https://github.com/MonaRazaghi/GRADIS/.
Basic Protocol 1: CONSTRUCTION OF FEATURES USED IN SUPERVISED LEARNING OF GENE REGULATORY INTERACTIONS
Supervised learning of gene regulatory interactions is based on features of the TF-gene pair to be classified. To this end, gene-expression profiles provide a plethora of data based on features that can be extracted. A trivial set of features can be obtained by concatenating the gene-expression profiles of the TF and gene in a given pair (Ni et al., 2016). However, such a representation does not consider the relationship between the expression of the putative target and TF in a given experiment. Here, we provide the means to extract transcriptomics features representative for a TF-gene pair.
Materials
- Expression of genes monitored over different developmental and environmental conditions (perturbation experiments) or over time (time-resolved experiments). Gene-expression values are usually represented in a table, Expn ×p , where n denotes the number of genes, p stands for the number of experiments (e.g., conditions or time points), and the entity Expi,j, denotes the expression level of gene i in experiment j.
1.Scale the expression profiles of TFs and genes by their respective maximum expression values (Fig. 2).
The following code snippet scales the expression profile of Exp (Equation 1):
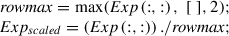
2.Determine the Euclidean distance between every two experiments based on the point representation (Fig. 2).
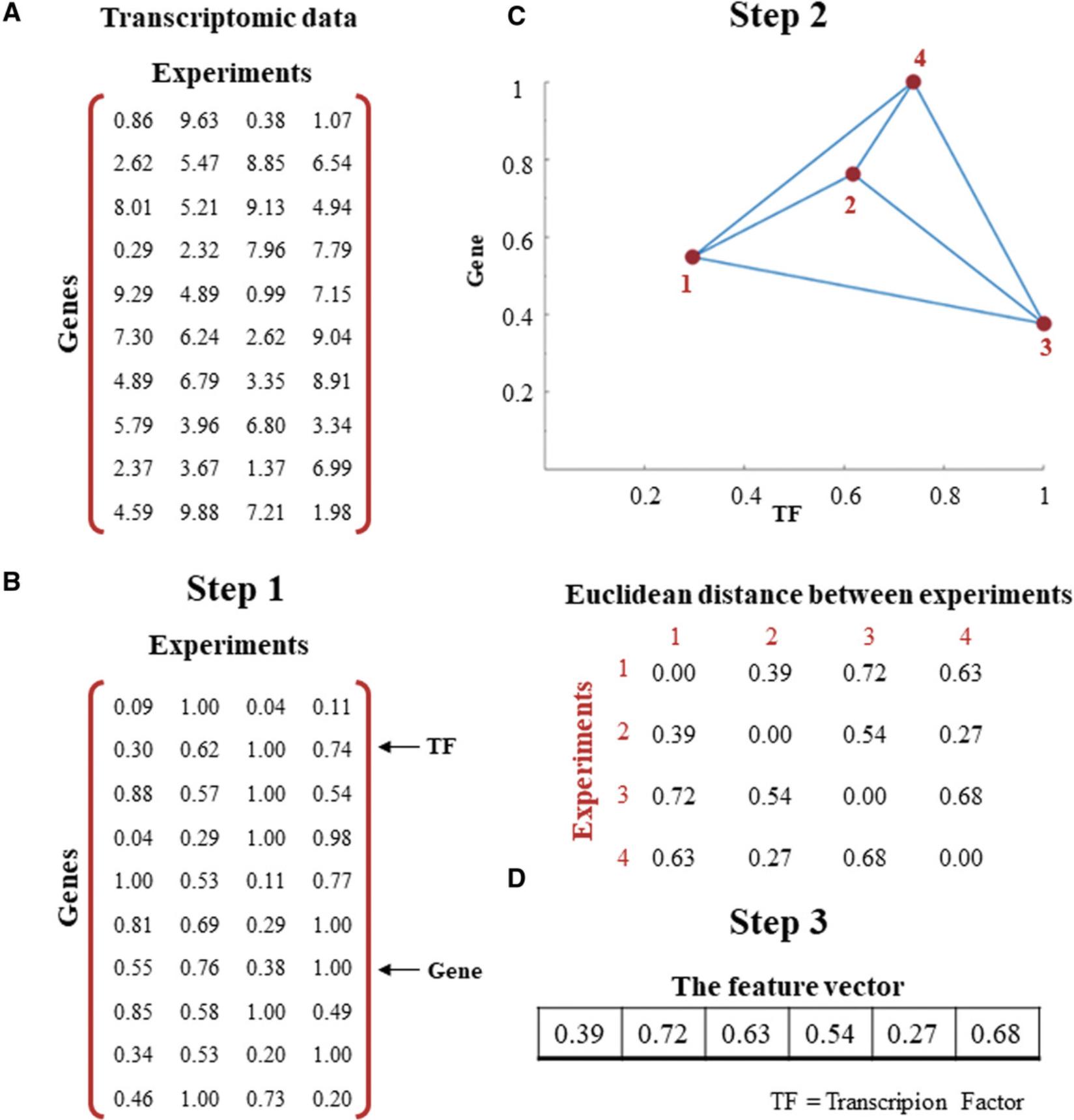
3.Obtain the features of the TF-gene pair by the vectorized form of the resulting Euclidean distance matrix obtained from step 2 (Fig. 2).
Every TF-gene pair is presented by p(p −1)/2 features.
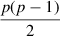
Steps 2 and 3 of Basic Protocol 1 are implemented in lines 98-146 of the code (https://github.com/MonaRazaghi/GRADIS/blob/master/GRADIS_neg.m). These lines determine the Euclidean distance for each TF-gene pair, and vectorize it to form the feature vector.
Basic Protocol 2: LEARNING THE NON-INTERACTING TF-GENE PAIRS
Training a binary classifier requires access to two types of instances, called positive and negative, which in our case correspond to presence or absence of gene regulatory interactions for a given pair of TF and gene. Typically, there is little information available about the absence of gene regulatory interactions between TFs and target genes in real-world datasets. Hence, it is not straightforward to train a classifier, due to the lack of negative instances. We describe a detailed procedure for composing a list of negative instances given knowledge about positive instances, i.e., TF-target gene pairs along with their expression levels.
Materials
Hardware
The approach can be executed on any computer (e.g., i7 processor and 16 GB RAM) with Windows 7 operating system
Software
The only software needed to run the code is Matlab R2017b
Data
- Expression-based features for TF-gene pairs based on the Basic Protocol 1
- A list of pairs of interacting TFs and their target genes. These pairs are referred to as positive instances and are obtained from experimentally verified interactions with different technologies.
- _Positive instance can be obtained from different databases: for instance, DREAM5 challenge (_Marbach et al. , 2012 _), RegulonDB (_Gama-Castro et al. , 2016 _), Yeastract (_Teixeira et al. , 2018 _), and AGRIS (_Yilmaz et al. , 2011 ).
- If TF-target gene interactions are not verified in the organism of interest, consider transfer of interactions according to homology from model organisms.
1.Form the class of positive instances of the training data by collecting the available experimentally verified TF-gene interactions.
2.Consider the remaining TF-gene pairs as uncharacterized, and divide them into subsets of size (almost) equal to that of the positive class (in step 1). Assume that there are k such subsets.
3.Treat one of these subsets, i , 1 ≤ i ≤ k , as a negative class and use it together with the positive class to train an SVM specific to subset i.
4.Treat the uncharacterized TF-gene pairs in all but the i -th subset as test data and assess them by the built SVM classifier.
5.Aggregate the individual classifiers for each of the k subsets to form the set of negative instances.
The implementation for Basic Protocol 2 can be found in the lines 151-189 of the code (https://github.com/MonaRazaghi/GRADIS/blob/master/GRADIS_neg.m).
Basic Protocol 3: LEARNING A CLASSIFIER FOR GENE REGULATORY INTERACTIONS
Having obtained a labeled training set associated with the feature vectors (see Basic Protocol 1, above), an SVM can be trained to find an optimal hyperplane that separates the two classes. The training set consists of m TF-gene pairs p 1, p 2, …, pm , each of which belong to either of the two positive and negative classes, respectively denoted by +1 and −1.Following Basic Protocol 2, the negative class of TF-gene pairs is considerably bigger than the positive. Here we describe a protocol for overcoming this challenge.
Materials
- Expression-based features for TF-gene pairs (see Basic Protocol 1)
- Positive and negative classes of TF-gene pairs: the negative class is obtained based on Basic Protocol 2
1.Form the class of positive instances of the training data by collecting the available experimentally verified TF-gene interactions.
2.Form a class of negative instances of the same size as the class of positive instances by randomly sampling from the negative class instances provided.
3.Train an SVM with the features and classes from steps 1 and 2.
4.Predict the class of the uncharacterized TF-gene pairs based on the SVM.
The implementation for Basic Protocol 3 can be found in lines 191-217 of the code (https://github.com/MonaRazaghi/GRADIS/blob/master/GRADIS_neg.m).
COMMENTARY
Background Information
The class prediction is done by the SVM based on a scoring function of the form: 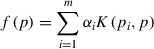
Critical Parameters
The number of experiments for which data are available affect the number of features used. The number of representative experiments should therefore be chosen so as to reduce the redundant information.
Troubleshooting
If positive-class TF-gene pairs are not available in sufficient number, the learned classifier may be either underfitted or overfitted. We recommend inspection of the learning curves for the respective classifiers.
Anticipated Results
The results of the protocol include:
Set of features representing TF-gene pairs Set of negative class TF-gene pairs that are likely not involved in a regulatory interaction Predictions for uncharacterized TF-gene pairs based on whether or not they are involved in a regulatory interaction.
Time Considerations
A timeline depends on the number of TFs and genes in the data set. For a data set that includes 141 TFs and 999 genes (as is the case in Escherichia coli) and the hardware specified in the Basic Protocol 1, the timeline is:
The generation of the features takes approximately 3 min The generation of the negative class instances takes approximately 6 hr The generation of the final SVM take approximately 3 min, while the classification of the uncharacterized TF-gene pairs requires 2 min of computation.
Acknowledgments
This work has been supported by the MELICOMO project 031B0358B of the German Federal Ministry of Science and Education to ZN and ZR-M.
Literature Cited
- Cortes, C., & Vapnik, V. (1995). Support-vector networks. Machine Learning , 20, 273–297. doi: 10.1007/BF00994018.
- Gama-Castro, S., Salgado, H., Santos-Zavaleta, A., Ledezma-Tejeida, D., Muñiz-Rascado, L., García-Sotelo, J. S., … Collado-Vides, J. (2016). RegulonDB version 9.0: High-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Research , 44, D133–D143. doi: 10.1093/nar/gkv1156.
- Haury, A., Mordelet, F., Vera-Licona, P., & Vert, J. (2012). TIGRESS: Trustful inference of gene regulation using stability selection. BMC Systems Biology , 6, 145. doi: 10.1186/1752-0509-6-145.
- Huynh-Thu, V. A., Irrthum, A., Wehenkel, L., & Geurts, P. (2009). Inferring regulatory networks from expression data using tree-based methods. PloS One , 5(9), pii: e12776. doi: 10.1371/journal.pone.0012776.
- Maetschke, S. R., Madhamshettiwar, P. B., Davis, M. J., & Ragan, M. A. (2014). Supervised, semi-supervised and unsupervised inference of gene regulatory networks. Brief Bioinformatics , 15(2), 195–211. doi: 10.1093/bib/bbt034.
- Marbach, D., Costello, J. C., Küffner, R., Vega, N., Prill, R. J., Camacho, D. M., … Stolovitzky, G. (2012). Wisdom of crowds for robust gene network inference. Nature Methods , 9(8), 796–804. doi: 10.1038/nmeth.2016.
- Margolin, A. A., Nemenman, I., Basso, K., Wiggins, C., Stolovitzky, G., Dalla Favera, R., & Califano, A. (2006). ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics , 7, S7. doi: 10.1186/1471-2105-7-S1-S7.
- Meyer, P., Kontos, K., Lafitte, F., & Bontempi, G. (2007). Information-theoretic inference of large transcriptional regulatory networks. EURASIP Journal on Bioinformatics and Systems Biology , 2007, 79879. doi: 10.1155/2007/79879.
- Mordelet, F., & Vert, J. P. (2008). SIRENE: Supervised inference of regulatory networks. Bioinformatics , 24, 76–82.
- Ni, Y., Aghamirzaie, D., Elmarakeby, H., Collakova, E., Li, S., Grene, R., & Heath, L. (2016). A machine learning approach to predict gene regulatory networks in seed development in Arabidopsis. Frontiers in Plant Science , 7, 1936. doi: 10.3389/fpls.2016.01936.
- Omranian, N., Eloundou-Mbebi, J. M. O., Mueller-Roeber, B., & Nikoloski, Z. (2016). Gene regulatory network inference using fused LASSO on multiple data sets. Scientific Reports , 6, 20533. doi: 10.1038/srep20533.
- Petralia, F., Wang, P., Yang, J., & Tu, Z. (2015). Integrative random forest for gene regulatory network inference. Bioinformatics , 31(12), i197–i205. doi: 10.1093/bioinformatics/btv268.
- Razaghi-Moghadam, Z., & Nikoloski, Z. Supervised learning of gene regulatory networks based on graph distance profiles of transcriptomics data. Nature Systems Biology and Applications. Submitted for publication.
- Teixeira, M. C., Monteiro, P. T., Palma, M., Costa, C., Godinho, C. P., Pais, P., … Sá-Correia, I. (2018). YEASTRACT: An upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Research , 46(D1), D348–D353. doi: 10.1093/nar/gkx842.
- Vert, J. P. (2010). Reconstruction of biological networks by supervised machine learning approaches. Elements of Computational Systems Biology , 165–188.
- Yilmaz, A., Mejia-Guerra, M. K., Kurz, K., Liang, X., Welch, L., & Grotewold, E. (2011). AGRIS: The Arabidopsis gene regulatory information server, an update. Nucleic Acids Research , 39, D1118–D1122. doi: 10.1093/nar/gkq1120.
Citing Literature
Number of times cited according to CrossRef: 10
- Yanglan Gan, Jiacheng Yu, Guangwei Xu, Cairong Yan, Guobing Zou, Inferring gene regulatory networks from single-cell transcriptomics based on graph embedding, Bioinformatics, 10.1093/bioinformatics/btae291, 40 , 5, (2024).
- Ankita Abnave, Jerrin John, Erich Grotewold, Andrea I. Doseff, John Gray, Upper level and cross hierarchical regulation of predominantly expressed phenolic genes in maize, Current Plant Biology, 10.1016/j.cpb.2024.100364, 39 , (100364), (2024).
- Heng Dong, Baoshan Ma, Yangyang Meng, Yiming Wu, Yongjing Liu, Tao Zeng, Jinyan Huang, GRNMOPT: Inference of gene regulatory networks based on a multi-objective optimization approach, Computational Biology and Chemistry, 10.1016/j.compbiolchem.2024.108223, 113 , (108223), (2024).
- Meroua Daoudi, Souham Meshoul, Samia Boucherkha, A Semi-Supervised Approach to GRN Inference Using Learning and Optimization, Research Anthology on Bioinformatics, Genomics, and Computational Biology, 10.4018/979-8-3693-3026-5.ch006, (94-118), (2023).
- Jwaone Gaboitaolelwe, Adamu Murtala Zungeru, Abid Yahya, Caspar K. Lebekwe, Dasari Naga Vinod, Ayodeji Olalekan Salau, Machine Learning Based Solar Photovoltaic Power Forecasting: A Review and Comparison, IEEE Access, 10.1109/ACCESS.2023.3270041, 11 , (40820-40845), (2023).
- Vivian Robin, Antoine Bodein, Marie-Pier Scott-Boyer, Mickaël Leclercq, Olivier Périn, Arnaud Droit, Overview of methods for characterization and visualization of a protein–protein interaction network in a multi-omics integration context, Frontiers in Molecular Biosciences, 10.3389/fmolb.2022.962799, 9 , (2022).
- Amar Mujkic, Ena Baralic, Aida Ombasic, Lemana Spahic Becirovic, Lejla Gurbeta Pokvic, Almir Badnjevic, Machine Intelligence in Biomedical Data Modeling, Processing, and Analysis, 2022 11th Mediterranean Conference on Embedded Computing (MECO), 10.1109/MECO55406.2022.9797164, (1-10), (2022).
- Chirag Gupta, Venkategowda Ramegowda, Supratim Basu, Andy Pereira, Using Network-Based Machine Learning to Predict Transcription Factors Involved in Drought Resistance, Frontiers in Genetics, 10.3389/fgene.2021.652189, 12 , (2021).
- Haoze Chen, Zhijie Zhang, Hybrid neural network based on novel audio feature for vehicle type identification, Scientific Reports, 10.1038/s41598-021-87399-1, 11 , 1, (2021).
- Guangzhen Ma, Jirong Chen, Tiantian Wei, Jia Wang, Wenshan Chen, Inhibiting roles of FOXA2 in liver cancer cell migration and invasion by transcriptionally suppressing microRNA-103a-3p and activating the GREM2/LATS2/YAP axis, Cytotechnology, 10.1007/s10616-021-00475-2, 73 , 4, (523-537), (2021).

