Scalable dual-omics profiling with single-nucleus chromatin accessibility and mRNA expression sequencing 2 (SNARE-seq2)
Nongluk Plongthongkum, Song Chen, Kun Zhang, Dinh Diep, Blue B. Lake
chromatin accessibility
single-nucleus sequencing
RNA expression
cellular heterogeneity
scalable omics profiling
































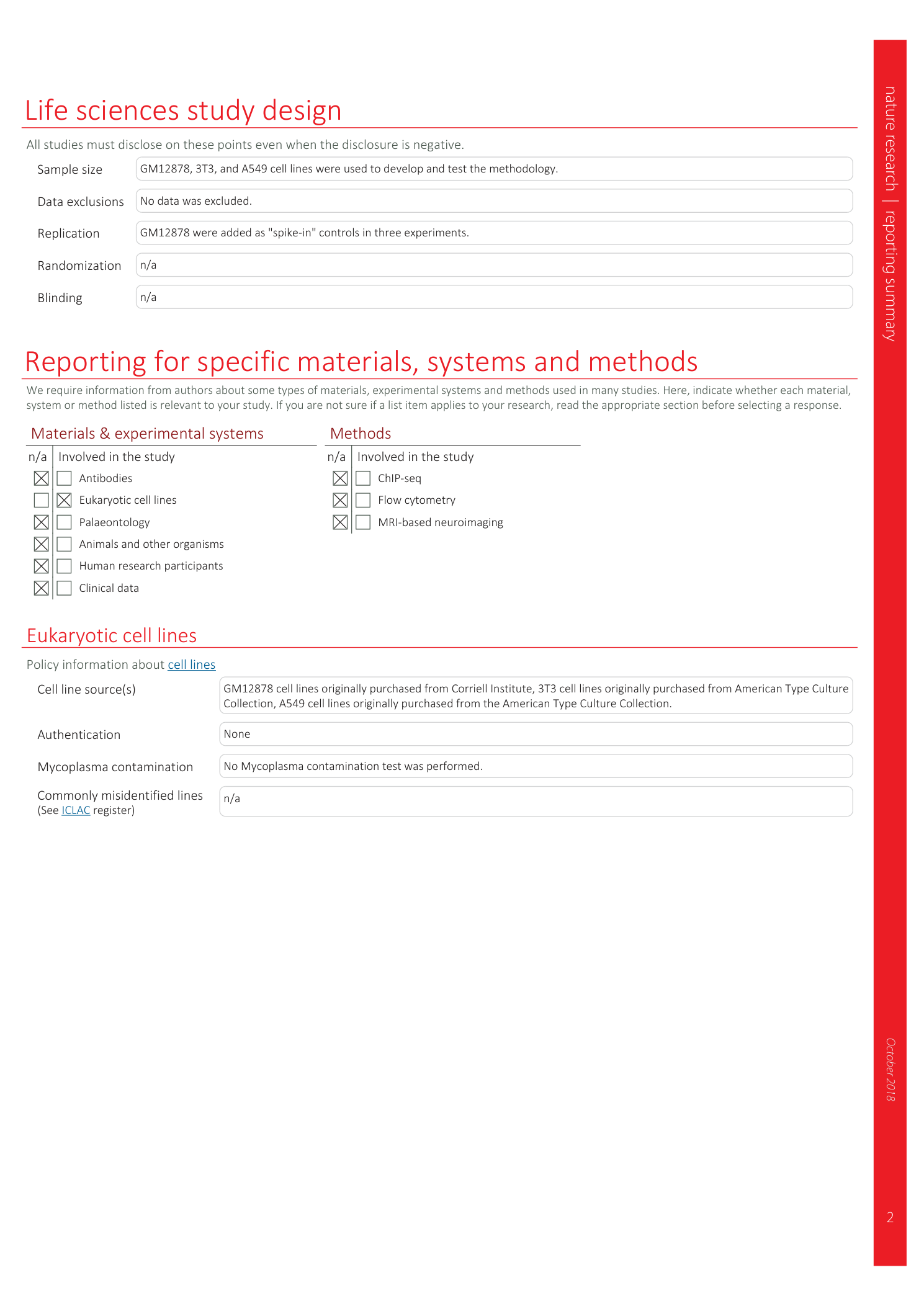
Extended
Extended Data Fig. 1 Schematics of SNARE-seq2 sequencing library generation.
a , The sequences of barcoded oligos and linkers in SNARE-seq2 protocol. b , The sequences of tagmented accessible DNA and mRNA molecules that are available for capture. c , The sequence of tagmented accessible DNA after being ligated to Round 1 barcodes. d , Round 1 barcoded oligos prime polyadenylated and truncated mRNA, respectively, followed by reverse transcription. e and f , The sequences of AC DNA and mRNA after Round 2 and Round 3 barcoding ligation, respectively. g , AC DNA and cDNA are captured with streptavidin beads. h , The sequence of AC DNA after binding to Nextera adapter 1 blocker. i , The sequence of cDNA after reverse transcription is complete and a PCR handle is added to the 3′ end of the cDNA by template-switching oligos. j , The sequence of AC DNA after gap-filling and priming sites for the first PCR. k , The sequence of AC DNA and priming sites for the second (indexing) PCR. l and m , The sequence of AC DNA sequencing library, with priming sites for sequencing primers. n , The sequence of RNA and priming sites for the first PCR. o , The sequence of RNA and priming sites for the second PCR. p , The sequence of RNA after end tagmentation and priming sites for indexing PCR. q and r , The sequence of the RNA sequencing library, with priming sites for sequencing primers. ‘/Phos/’ denotes phosphorylated, ‘/Biosg/’ denotes a 5′ biotin, a lower case ‘x’ denotes a genomic or transcriptomic sequence, ‘//’ denotes genomic or transcriptomic sequences of variable lengths, a dashed arrow denotes the direction of reverse transcription, ‘#’ denotes either DNA or cDNA sequences, ‘…’ denotes only partial sequences shown and ‘/3InvdT/’ denotes a 3′ inverted thymine base. The bolded numbers in brackets denote the order in which Illumina sequencing data are generated. BC, barcode; UMI, unique molecular identifier.

