SANGER Sequencing EFGL
EagleFish GeneticsLab
Abstract
This protocol describes how the Eagle Fish Genetics Lab (EFGL) prepares extracted DNA samples for the ABI 3500xL Genetic Analyzer, and how we run and collect data from this machine. This process is achieved by PCR amplification, ExoSAP-IT purification, E -Gel confirmation, Qubit quantification, BigDye sequencing, CleanSeq clean up, set up of the 3500xL Genetic Analyzer (3500), and finally the collection of results. This should result in easy-to-read nucleotide peaks that are exported as an ABI file.
Steps
PCR Amplification of Target Sequence
Materials needed:
• Forward and reverse primers
• Heat Seal
• 1.5mL vials
• PCR master mix reagents/kits
• DNA tray(s)
• Unskirted PCR plate(s)
• Pipette and tip
To note: As there are several PCR kits our lab uses for this section, below is a generic protocol for amplifying PCR products for Sanger sequencing. Refer to the Appendix for our working recipes/programs.
Obtain sample list from lead biologist or data coordinator, and print tray maps if necessary.
1. If this is your first attempt, start small (ideally 8-12 samples – as Step 4 in Post cycle
sequencing clean-up can be challenging).
Consult the lead biologist on the master mix recipe and cycling program to use.
1. **Appendix section’s recipes and programs** have been successful at the Eagle Fish Genetics Lab,
but please double check if those are the right ones for your lab.
b. Depending on the species and PCR product size, the PCR kit(s) could vary and/or be used in
combination.
Locate forward and reverse primers stock (usually at 100 µM concentration) located in -20°C freezer and dilute each individually to a working concentration of 10µM).
a. In a 1.5mL vial, add`20µL` of 100µM primer to`180µL` of water.
Repeat for the other primer.
b. Store the 100µM stock back in lab`-20°C` freezer and keep 10µM working stock in lab fridge.
Prep PCR master mix in a 1.5mL vial. All reagents and/or kits are in our lab mini -20°C freezer
In a new unskirted PCR plate, dispense master mix into each well.
Using a multi-channel pipette, add DNA samples into each well.
Heat seal, gently vortex and quick spin ,0h 0m 0s down.
Load the unskirted plate and run the appropriate thermal cycler program.
(In most cases, these PCR runtimes can range between 4h 0m 0s or longer , so plan accordingly)
Run 1% e-gel to confirm the successful amplification of PCR products. Follow e-gel protocol found in:
S:\Eagle Fish Genetics Lab\LAB PROTOCOL BOOK\05 - DNA quantification\E-Gel
or on protocols.io under " E-Gel Protocol EFGL "
1. Save e-gel images in a project folder
b. If successful, and depending on the sample size of the project, consider using double comb gels if
possible (more samples and less time)
Repeat steps 7 to 13 for the remaining samples for your project.
a. It is NOT recommended to PCR all samples at one go, especially for a large project and the
availability of 3500xL Genetic Analyzer is entirely dependent on the Wildlife lab’s schedule.
They have dibs.
b. Stagger the sections, if possible.
SECTION 1 – PCR Purification with ExoSAP-IT
Materials needed:
• ExoSAP-IT Express reagent (pre-aliquot strip tubes in-20°C small freezer)
• PCR product plate
• 1 unskirted PCR plate
• p10 multichannel pipette and 2 boxes of tips
• Ice block-20°C(in small freezer)
Vortex ExoSAP-IT Express reagent and keep on an ice block (ExoSAP-IT doesn’t freeze).
Aliquot 5µL of PCR product into a new unskirted PCR plate.
Add 2µL of ExoSAP-IT Express to each PCR sample (ExoSAP-IT it is quite viscous, pipette slowly).
Heat seal, vortex gently, and quick spin ,0h 0m 0s down
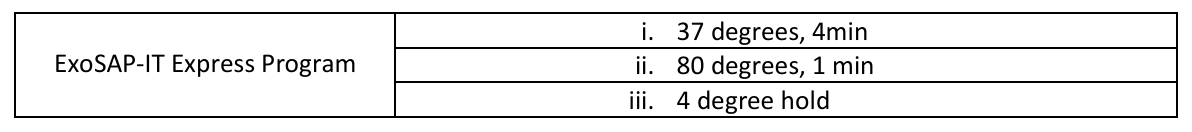
Run ExoSAP-IT program on thermal cycler. Runs for about 0h 5m 0s.
a. User name: General
b. General → Sanger → Exo sapit
c. The program is currently only on our labs machine 1’s and machine 10’s groups as the Eagle Fish
Genetics Lab
Samples are now PCR purified
The unskirted plates are to be placed on ice blocks for downstream steps.
(If not using immediately)
Heat seal and store in -20°C freezer. (Samples are still stable and usable after a month
[tested in May 2022])
SECTION 2 – Quantify Purified PCR Products with Qubit
Materials needed:
• Qubit dsDNA high sensitivity kit
• Purified PCR products
• p10 multichannel pipette and box of tips
• 1 unskirted PCR plate
• Ice block(s)
Note: Remember that you will end up needing to sequence forwards and reverses separately (1µL of
purified PCR product each), and that after Qubit there will only be5µL of purified PCR product left. If the concentration is too low to do a forward and reverse run, you will need to ExoSAP more PCR
product, and qubit them again to make sure you have enough.
If the purified PCR product was previously frozen, it is OK to quick spin,0h 0m 0s to thaw (~0h 0m 5s), gently vortex, and quick spin,0h 0m 0s down again. Remember to place plate back on ice block immediately after .
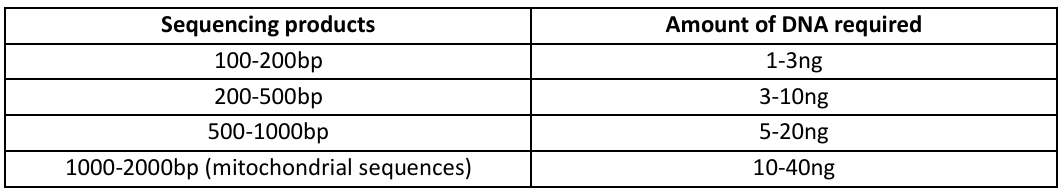
Obtain the average qubit scores for each sample. Depending on the length of your sequencing products,
refer to the table above to determine the required amount of DNA.
In some cases, these purified PCR products are often higher than 60ng/µl. (Qubit machine will read:
“Too high”.)
a. For mitochondrial sequences:
i. Recommended to try a 1:10 dilution prior to Qubit, most samples should stay within
10-30 range after dilution.
ii. 1:10 dilution:`1µL`Add of purified PCR produc`9µL` in of water in a new unskirted
PCR plate.
iii. Keep diluted plate on ice block as well
b. If 1:10 dilution doesn’t fall within the required range, calculate out the actual DNA present,
and either change the dilution factor or proceed with undiluted purified PCR products
Heat seal, and store the original purified PCR product plate back in -20°C freezer, if not using.
1. If you need to go back to these samples, note that the diluted samples may drop in DNA
concentration. These are still viable for sequencing after a month in`-20°C` freezer.
(tested in May 2022)
When proceeding to the next step, purified PCR products should stay on ice block.
SECTION 3 – BigDye Sequencing Cycle
Note: Note: Reach out to Wildlife lab to make sure 3500 machine is available for use, prior to step 3. It is ideal to do STEP 3-5 on the same day. If sequencing within next 1-3 days, it is OK to stop at either the end of step 3 or step 4.
Materials needed:
• Forward and reverse primer (10µM concentration)
• BigDye and BigDye Terminator v3.1 5X sequencing buffer
• 2 of 1.5mL vials to dilute forward and reverse primer to 1.6µM concentration
• 2 of 1.5mL vials for forward and reverse master mix
• 2 unskirted PCR plates (if only running <48 samples, could use same plate)
• p10 multichannel pipette with a box of tips
• Repeater pipette, and 0.1mL combitip
• Ice block(s)
• Ice cooler
Dilute both forward and reverse primers to 1.6µM.
• Full tray:16µL of 10µM primer, with84µL of water =100µL primer at 1.6µM
• Small sample size (up to 40 samples): Ad6.4µL of 10µM primer, wit33.6µL water 40µL primer at 1.6µM
Thaw BigDye and keep in ice cooler.
1. There are usually several aliquots of `20µL` BigDye in 1.5mL vials. Use those first, unless more
is required
b. BigDye is susceptible to multiple freeze/thaw. Aliquot out more in`20µL` (or higher, full tray
need`100µL` ) when require ****Avoid exposure to light during storage.*** .
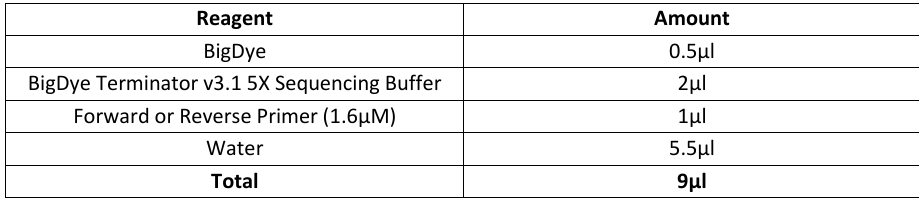
Prep 2 master mixes, one for forward sequencing, and one for reverse sequencing.
We’ll be doing eighth reactions in a total volume of10µL (This is a deviation from the manufacturer’s protocol).
Note: If the purified PCR from ExoSAP-IT does not have sufficient DNA concentration, add more
purified PCR product and remove water. If going this route, you will need a different master mix ratio.
Mix with pipette or vortex gently, and spin down.
Use repeater pipette to aliquot out 9µL of forward Master Mix into each well on new unskirted PCR
plate.
1. Repeat step for reverse Master Mix into separate wells. This can be on the same plate as the
forward sequences if it all fits to a single plate. If not, use a separate unskirted PCR plate.
Add 1µL of purified PCR product into each well that has the forward Master Mix, and repeat for wells
with reverse Master Mix.
Heat seal plate(s), vortex briefly and spin ,0h 0m 0s down (Do not worry about bubbles).
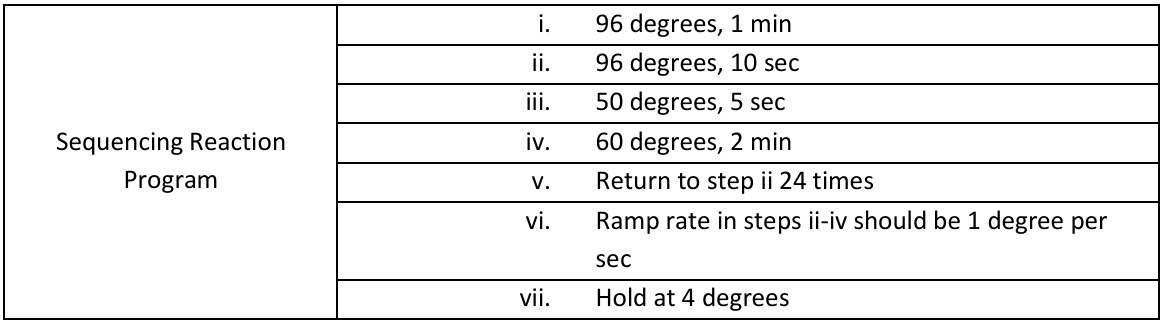
Run BigDye sequencing thermal cycler program. Runs for about 1h 30m 0s.
1. User name: General
b. General → Sanger → BigDye sequencing
i. Go to "Edit", to make sure that it has an asterisk beside step 2-4. This means the
temperature is ramped up by 1 degree per second
3. Program is only on machine 10’s group in our lab
This can be left overnight, but do not delay too long (no more than a week). If delay is needed, store
in-20°C freezer, and protect from light.
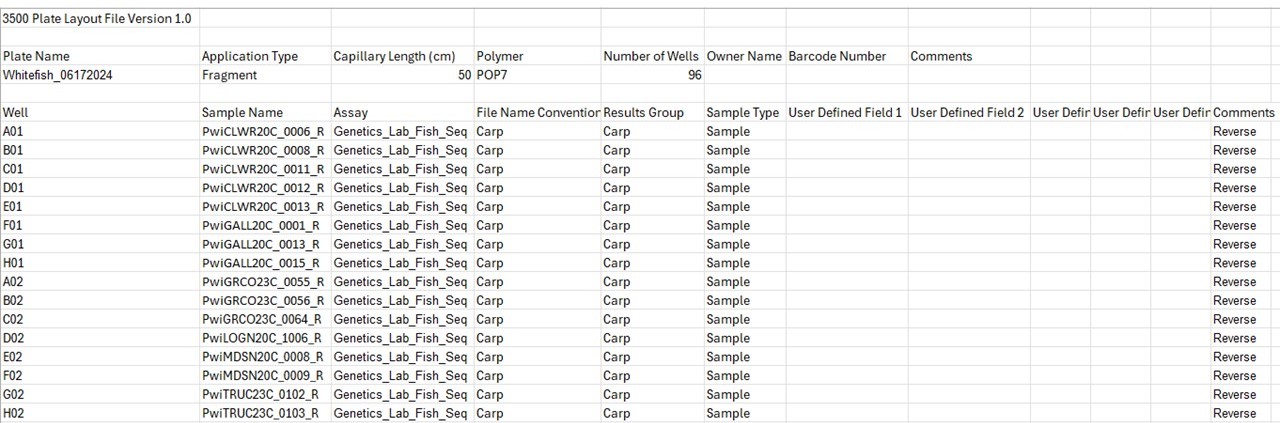
Prep electronic sample sheet for 3500 xL Genetic Analyzer machine while waiting for run to complete
a. Go to LAB PROTOCOL BOOK → 13 – Sanger Sequencing
b. Open Genemapper Plate Template.xlsx
c. Make edits to:
i. Plate Name (cell A4)
ii. Sample Name (cell B6-B102) – leave blank if empty, max 96 samples per
spreadsheet. If more than 1 plate, do each plate separately
d. The following stays the same:
i. Assay: Genetics_Lab_Fish_Seq
ii. File name convention: Carp
iii. Results Group: Carp
e. Save file(s) as Text(tab delimited) on both project folder, and flash drive.
SECTION 4 – Post Cycle Sequencing Clean-up
Note 3500 machine does injections in columns of 3 . If you have a partial tray that the sample
size is not a factor of 3 columns, or a full column → Fill it with water.
For example:
• If you have 4 columns of samples, fill column 5 and 6 with water.
• If you have 2 columns + 7 samples, add water to the 8th well in the 3rd column.
• Each injection takes ~ 2h 0m 0s
CleanSeq <-- Yields better results
Materials needed:
• Agencourt CleanSeq beads
• 85% ethanol
50mL• falcon tube(s)
• ABI plate(s)
• Multichannel and single pipettes, and corresponding tips (~6 boxes for 48 samples)
• Repeater pipette
• 5mL combitip
•25mL reservoir
• Magnetic plate
• Lab grade H2O
(no longer in use, Cleanseq beads cleanup is the preferred choice in EFGL)
X-Terminator
Materials needed:
- BigDye Xterminator, and SAM solution (both in a yellow Ziploc bag in skinny fridge)
- Safety googles
- p10 multichannel pipette, and a box of tips
- p100 pipette, and box of tips
- Wide bore tips
- 5mL tube
- ABI plate(s)
- Water
- Rubber bands
Not recommended to do more than 48 samples at a time, to avoid overdrying of beads mid clean-up
Make 85% ethanol in a 50mL falcon tube. You will need a total of 242µL of 85% ethanol per sample for this section. Multiply the volume per sample and accounting for pipette error.
a. For a full tray -->`21250µL` 100% ethanol `3750µL` water =`25mL`
2. For 16 samples --> (16 (samples) + 3 (pipetting error) ) X `242µL` = `4598µL`of 85%
ethanol
9.`4598µL`of 85% ethanol --> `3908.3µL` pure ETOH and `689.7µL` water
There are 2 separate ways to proceed from here:
Option A might be easier with fewer samples (less than half a tray), and tends to have better results. Option B would be quicker in general, especially if you have a large number of samples.
Add 10µL of CleanSeq beads into each well with a repeater pipette, aim on the high side of the well
to prevent contamination.
Option A
- Obtain a new ABI plate
- Invert Xterminator solution bottle 10x to ensure it is thoroughly mix. Recommended to put on safety googles as Xterminator solution contains Powdex, which can cause serious eye damage.
- Use p100 pipette with wide bore tips to add 10µl of Xterminator solution to each well in ABI plate, mix bottle every 2 columns or so.
- Next, use a p100 pipette to add
45µLof SAM solution to each of those wells. Avoid touching the side of wells to prevent contamination - Use a multichannel p10 pipette and transfer all
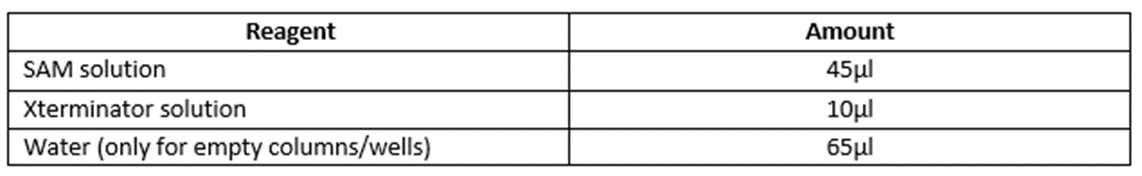
10µLof BigDye sequencing products to ABI plate. Discard tips after each use. - Do not forget! If you’re doing a partial tray, empty wells/columns need to be filled with
65µLof water. - Seal ABI plate with the Nexttec adhesive clear seal.
- Bring ABI plate and flash drive to Wildlife lab.
-
Remove adhesive seal and seal tray with rubber sealer
-
Vortex at high speed for
0h 20m 0s(you may want to bring your own rubber bands for plate to stay on vortex machine) -
Centrifuge for
0h 2m 0sat 1000 rcf (should be pre-programed) -
Load plate to 3500 for sequencing
-
Pipette out the required ethanol (42µL each sample) in a 25mL reservoir, and use a multichannel pipette to add 42µL of 85% ethanol to each well
a. Pipette up and down 5-10 times to mix solution, discard tips after each use.
b. No incubation needed after mixing
Option B
-
Obtain a new ABI plate
-
Invert Xterminator solution bottle 10x to ensure it is thoroughly mix. Recommended to put on safety googles as Xterminator solution contains Powdex, which can cause serious eye damage.
-
Make Xterminator master mix in 5mL/50mL tube. Multiply volumes by number of samples, accounting for pipette error.
10µLof Xterminator solution +45µLof SAM solution per sample
-
Use a p100 pipette with wide bore tip to transfer 55µl of Xterminator master mix into each well. Make sure to mix the
5mL/50mLtube every 1 or 2 columns to ensure the powdex stay in solution (it settles very quickly). -
Use a multichannel p10 pipette and transfer all
10µLof BigDye sequencing products to ABI plate. Discard tips after each use. -
Do not forget! If you’re doing a partial tray, empty wells/columns need to be filled with 65µl of water.
-
Seal ABI plate with the Nexttec adhesive clear seal.
-
Bring ABI plate and flash drive to Wildlife lab.
- Remove adhesive seal and seal tray with rubber sealer
d. Load plate to 3500 for sequencing(you may want to bring your own rubber bands for plate to stay on vortex machine)
-
Centrifuge for
0h 2m 0sat 1000rcf (should be pre-programed) -
Load plate to 3500 for sequencing
Place plate on magnetic plate, and wait 0h 5m 0s at room temperature. Magnetic beads will form a
ring (or 4 lines down the wells, pattern usually alternates on columns depending on the magnetic plate
you’re using)
Note Steps 56- 59 can be challenging, but you must be relatively quick, as stalling can cause beads to dry out which will affect the end result. If just starting out, do 2 wells at a time, and slowly ramp up until you are comfortable with 8 wells.
Set multichannel pipette to ~65µL, and take out all the supernatant. Avoid touching or aspirating the
beads as much as you can.
a. It is easier to do 4 at a time or less, until you are comfortable
b. Lower the pipette tip to the bottom of the well to aspirate all of the ethanol
(Plate stays on magnet)
Once all supernatant is removed, use a 5mL combitip and dispense 100µL of 85% ethanol to each well. Wait 0h 0m 30s.
(Plate stays on magnet) Use a multichannel pipette and set to 110µL to remove all ethanol.
1. Lower the tip all the way to the bottom of well and aspirate
Repeat step 57 and 58 once.
Air dry beads at room temperature for 0h 5m 0s, avoid overdrying (if cracks are seen, it is overdried.
You will want the beads to look matte)
**About into drying** `0h 1m 0s` into drying: Remove the plate from magnet. Check to ensure there is not an
excess of ethanol left in the wells. Pipette out any remaining ethanol. Discard tip after each use.
b. Leave plate off of the magnet for the remaining time to dry.
Use a multichannel pipette to dispense 40µL of water into each well
a. Remove any air pockets formed at the bottom of well(s) with a p10 pipette
b. Tap plate gently on bench a couple of times, so beads will slide down and go back into the
solution for an easier elution.
Incubate plate at room temperature for 0h 5m 0s, while off magnet.
Place plate back on magnet for 0h 5m 0s.
The unskirted PCR plate should sit on the face of the magnet plate inbetween the holes, not down in the holes.
a. This creates bead pellets at the bottom corner of the well instead of a ring, for easier elution.
Use a multichannel pipette, and transfer 35µL of the solution into a new ABI plate. Avoid transferring beads!
a. If there is **more than delay** `24h 0m 0s` delay to sequencing, transfer to non-skirted PCR plate
instead of ABI plate. Heat seal, and store in dark in `-20°C` freezer.
(good up to a month – not tested)
i. If delay is less tha`24h 0m 0s` , store in dark in`4°C` freezer.
b. It is suggested to transfer 2-4 wells at a time by lowering tip to the opposite end of the pellet
c. If beads get into your pipette tip, place the solution back into the well and wait for solution to
clear up.
Seal ABI plate with Nexttec adhesive seal and bring to Wildlife lab for processing in a light blocking container.
Switch out Nexttec seal with the rubber seal and load it on the 3500 xL Genetic Analyzer machine to sequence.
SECTION 5 – 3500 Sequencing Run
Materials needed:
• ABI Plate in light protected container
• USB stick
Make sure both computer and 3500 are turned on.
a. If 3500 is off, and computer is on. The computer needs to be restarted.
i. Log in under Administrator, password is Administrator
b. Follow the instructions (bottom left corner) on 3500 to sync 3500 and computer.
i. Data collection is the software that you launch to start the run. So, donot open the
software on the computer until you see the green check mark.
c. If 3500 is on, but you’re experiencing issues with the 3500 data collection software, reboot
the com HOWEVER! WEVER! Please make sure whoever is using that computer has their GeneMapper results saved! Please check with the techs in the lab.
Once it’s all green, plug in your flash drive (there are more USB slots at back of the tower if the front
ones are occupied)
Open the 3500 data collection software (icon looks like the 3500 machine)
Load plate(s) onto 3500.
a. Press “Tray” button and wait for machine to bring out the tray platform forward for loading.
b. Open the door and load plate(s).
Go back to the computer, you should be on the “Dashboard” tab on top left.
Under "Consumables Information", make sure there’s sufficient samples left to run your samples. If you are about to run out, reach out to the lab manager to get a new one.
Go back to “Dashboard” tab, and select “Create New Plate”
Enter your plate name, it is recommended to use the same plate name in your .txt file.
a. No other changes to settings
b. Click “Assign plate contents
Click “Import” to import your .txt file
a. It might give you some warnings to the import, Click on “Proceed” anyway
b. The screen should now show you a map with all your samples to be run
If you have your template set up right, you should see samples highlighted with a blue dot, and all 3
boxes below the tray map are checked with “Genetics_Lab_Fish_Seq” , “CARP”, CARP”.
a. If not, do the following:
b. In Assay box, if it is empty or not the right assay. Select “Add from library”.
c. Under filter, select “Sequencing” → Select “Genetics_Lab_Fish_Seq”
d. The other 2 boxes, select “Add from library”, and select “CARP”.
e. Highlight all samples, and check the “Genetics_Lab_Fish_Seq” box, along with both “CARP”
boxes. Your samples should now be highlighted in blue. Empty wells should remain
blank/un-highlighted.
f. To un-highlight the empty wells, select them and uncheck all 3 boxes
Click “Link plate for run”
a. If you only have 1 plate → wait`0h 2m 0s` → run window appears → Click on “Start run”
or go to step “b” to add plate B.
b. If you have 2 plates, click on “Assign plate contents” on the left column
i. Repeat steps 9-13
ii. It’ll ask to overwrite plate A, or assign to plate B. Assign it to plate B.
iii. W`0h 2m 0s`it a , then click “Start run”
It will take 0h 2m 0s before the run is initiated, and the estimated runtime is on the top right of the
screen.
a. Each injection takes abou`2h 5m 0s` regardless the number of samples within each
injection.
b. You’ll want to wait for at le`0h 5m 0s`st to make sure the timer continues to countdown
(it refreshes e`0h 0m 5s`ery ). At`1h 58m 0s`the mark, there’ll be movement in the
machine where the first injection is initialized. If the timer continues to countdown after,
the run has successfully begun. You can walk away now.
Make sure to check back on the machine 1h 0m 0s later regardless, as the data collection
software might crash.
a. If it does (timer stops counting down, or the software has quit itself), and no error message
is given: re-start the plate loading process, and start the run again.
i. Start from the beginning if no data present, or till where it stopped last. Updated: 2/2/2024
1. To check: Go to Computer → Data → Applied Biosystems → 3500 → Data
→ Carp → the last run folder → check each injection (successful AB1 file
should be ~300kB in size)
ii. If injection folders are empty, none of the samples sequenced
b. If there is an error message, reach out to Wildlife Lab to troubleshoot if necessary.
i. Note down the error message and timestamp
Things to note/Troubleshooting steps:
• Polymer pouch count goes down each time the run is restarted, even though it has not been
used technically. If you are certain you have sufficient remaining samples to run, proceed to run
anyway – despite the error message.
• There are 2 reasons to why the timer stop might stop. If timer stops at about0h 5m 0s into the run, chances are the oven is not up to temperature (oven needs to be at60°C )
o Go to "Dashboard", "oven temp" should be green, and at abou`60°C`
o Then, check "EPT" tab in the Workflow window, under "Instrument Run Views" and "Flags"
o Check to see if lines are still climbing. Takes abo`0h 10m 0s`t \~ since you’ve clicked
“Start Run”, and timer should countdown at that point.
o There’ll be moveme`0h 2m 2s`t at (switching between buffer A and B, and at about`0h 1m 58s` for first injection)
o Overall, this could take`0h 15m 0s`up to from the time you click “Start Run” even if the oven
is at temp. So, be patient.
• If everything seems to be right, and the timer is still not counting down, and you feel you have given
sufficient time.
o It might be a software issue, there is no known reason for the cause, or error messages
to tell you it is not running. last fixed by defragmentation of the computer 6/21/22
o Click on “Terminate Run”, abort the entire injection list.
o Close the Data collection program, then turn 3500 off.
o Shut down the computer.
o Then follow instructions to start both the computer and the 3500 back up.
o Load the plates on the software again, they should have already been saved. You can
load by clicking on “Existing plate” rather than “Create new plate” on Dashboard.
black_small_square ‱ Filter: Sequencing black_small_square ‱ Type in your plate names, assign plate A (and B)black_small_square
‱ Start run
• If new polymer pouch is required:
o In the Dashboard homepage, go to Wizards. Select “Refresh polymer”
o Go through each step shown on the screen
<sup>black_small_square</sup> ‱ The lever to load/unload the polymer pouch can be “sticky”, so some force is required
<sup>black_small_square</sup> ‱ Use a kimwipe with a bit of distilled water to clean the spout.
<sup>black_small_square</sup> ‱ Check for bubbles, and if all is well, you are set to run the plates
o Close the 3500 machine door
• If you keep getting errors with setting up your assays, and that the oven is not pre-heating.
The 3500 data collection software may have glitched out. You will have to reboot
the computer while leaving the 3500 turned on.
o After it reboots, everything should go as planned.
SECTION 6 – Collect Results
Go to “Computer” → Select “Data (D:)” → Applied Biosystems → 3500 → Data → Carp
The run folder will be listed according to their start date/time, or look for the latest.
Copy the entire run folder to your flash drive, and load to your project folder on S: drive.
Remove your trays from the machine.
a. Remove them from the “jacket” and place the trays in the pile of used trays.
b. Wildlife lab will autoclave the rubber tops to reuse, so do not toss them.
Review quality score on Sequencher. (Ideal is 70% or higher)
a. If lower than 60-70%, and scores do not improve after tweaking the sequences, you may
need to re-run those samples
Appendix – Successful PCR recipes
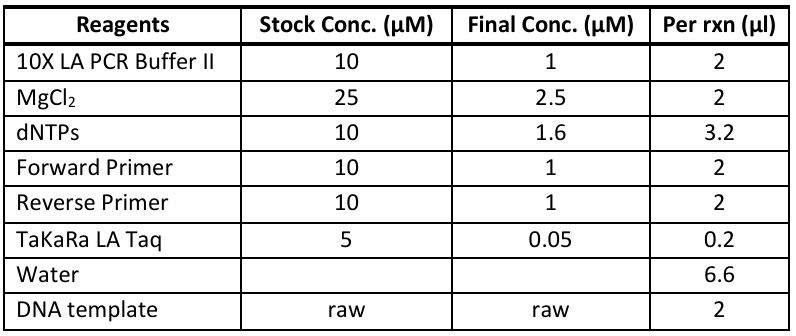
TaKaRa LA Taq (Cat# RR002A) - 20µL reactions
Redband ND1_ND2 region: ND12L and ND12H primers
Redband dloop region: dloop_Brunelli_F and 28RIBa_DL_R
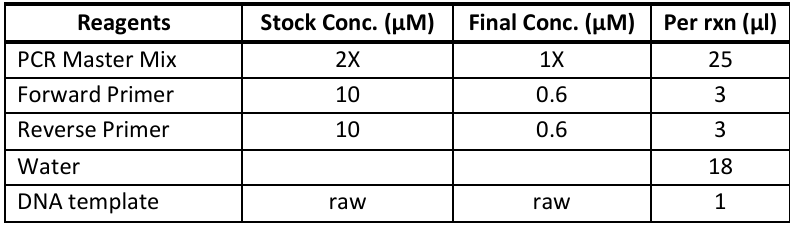
Thermo Scientific PCR Master Mix (2X) (Cat# K0171) - 50µL reactions
Whitefish CytB region: whitefish2_cytb_765F and whitefish2_cytb_765R
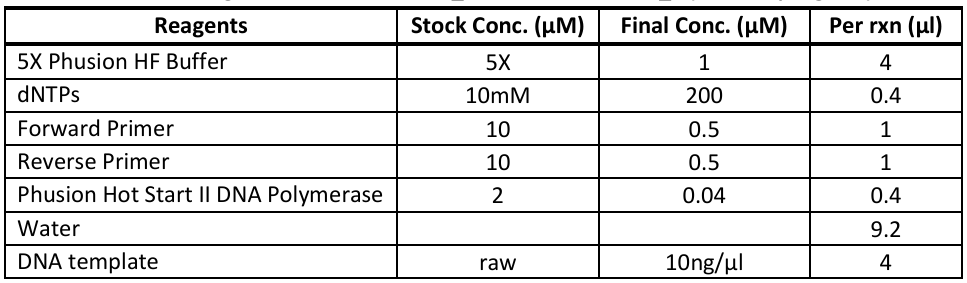
Thermo Scientific Phusion Hot Start II HF DNA Polymerase (Cat# F-549S) - 20µL reactions
Redband mitogenome: EmsermtDNA_F and EmsermtDNA_R (work in progress)
Appendix – Successful PCR cycling programs
ND1_ND2 and dloop
Run ND12NEW (General → Sanger → ND12NEW)
Program located on machines #10-14
Whitefish Cytb
Run pstar60 (General → Sanger → pstar60)
Program located on machines #1-5
Mitogenome
Run Emser_THERMO (General → Sanger → Emser_THERMO) (work in progress)
Program located on machines #10-14