Ribosome Profiling in the Model Diatom Thalassiosira pseudonana
Monica Pichler, Monica Pichler, Andreas Meindl, Andreas Meindl, Markus Romberger, Markus Romberger, Annemarie Eckes-Shephard, Annemarie Eckes-Shephard, Carl-Fredrik Nyberg-Brodda, Carl-Fredrik Nyberg-Brodda, Claudia Buhigas, Claudia Buhigas, Sergio Llaneza-Lago, Sergio Llaneza-Lago, Gerhard Lehmann, Amanda Hopes, Gunter Meister, Jan Medenbach, Thomas Mock
Abstract
Diatoms are an important group of eukaryotic microalgae, which play key roles in marine biochemical cycling and possess significant biotechnological potential. Despite the importance of diatoms, their regulatory mechanisms of protein synthesis at the translational level remain largely unexplored. Here, we describe the detailed development of a ribosome profiling protocol to study translation in the model diatom Thalassiosira pseudonana , which can easily be adopted for other diatom species. To isolate and sequence ribosome-protected mRNA, total RNA was digested, and the ribosome-protected fragments were obtained by a combination of sucrose-cushion ultracentrifugation and polyacrylamide gel electrophoresis for size selection. To minimize rRNA contamination, a subtractive hybridization step using biotinylated oligos was employed. Subsequently, fragments were converted into sequencing libraries, enabling the global quantification and analysis of changes in protein synthesis in diatoms. The development of this novel ribosome profiling protocol represents a major expansion of the molecular toolbox available for diatoms and therefore has the potential to advance our understanding of the translational regulation in this important group of phytoplankton. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Ribosome profiling in Thalassiosira pseudonana
Alternate Protocol : Ribosome profiling protocol for diatoms using sucrose gradient fractionation
INTRODUCTION
Diatoms are unicellular, eukaryotic microalgae, which comprise over 100,000 species throughout all aquatic environments. Their mosaic genomes have been shaped by secondary endosymbiosis and horizontal gene transfer, providing them with diverse regulatory mechanisms to adapt their protein synthesis in response to the changing environmental conditions (Mock et al., 2022). Due to their key role as primary producers in aquatic systems and their biotechnological potential, gene regulatory mechanisms in diatoms have been extensively studied using transcriptomic and proteomic data (e.g., Dong et al., 2016; Mock et al., 2008). The globally distributed species Thalassiosira pseudonana CCMP1335 was chosen for the first diatom genome sequencing project (Armbrust et al., 2004). Subsequently, it became a model organism due to the development of genetic manipulation tools such as the incorporation of recombinant DNA via transformation (Poulsen et al., 2006) and CRISPR/Cas-based genome editing (e.g., Belshaw et al., 2023; Hopes et al., 2016). However, despite these developments, regulation of protein synthesis at the translational level is largely unexplored in T. pseudonana and diatoms in general. The development of ribosome profiling or Ribo-Seq in 2009 (Ingolia et al., 2009) spurred the transcriptome-wide monitoring and quantification of translation in diatoms. This technique is based on the analysis of ∼30 nucleotide (nt) long mRNA fragments which are enclosed by translating ribosomes and protected from nuclease digestion (Steitz, 1969) (Fig. 1A). Deep sequencing of these generated footprints or ribosome-protected fragments (RPFs) thus provides a genome-wide and high-resolution snapshot of translation (Ingolia et al., 2009). Ribosome profiling was first developed in Saccharomyces cerevisiae (Ingolia et al., 2009) and has since been successfully applied to study translation in a wide range of organisms, such as bacteria (Li et al., 2012), fishes (Bazzini et al., 2012), mammals (Guo et al., 2010; Ingolia et al., 2011) and plants (Hsu et al., 2016; Juntawong et al., 2014). Ribosome profiling has revolutionized our understanding of gene expression by unveiling the full complexity and regulation of translation in both prokaryotes and eukaryotes. It has provided novel mechanistic insights into the translation mechanism, such as ribosomal pausing sites (Brar & Weissman, 2015; Ingolia et al., 2011; Karlsen et al., 2018; Shalgi et al., 2013) or previously unannotated (small) open reading frames (ORFs) (Aspden et al., 2014; Bazzini et al., 2014; Calviello et al., 2016; Hsu et al., 2016; Ji et al., 2015; Wu et al., 2019). However, to date, ribosome profiling has only been performed on one algal species, the green alga Chlamydomonas reinhardtii (Chung et al., 2015; Trosch et al., 2018). Here, we report the development of a ribosome profiling protocol for the model diatom T. pseudonana CCMP1335, representing the first application of this technique for any marine algae. Previously developed protocols were adapted (McGlincy & Ingolia, 2017; Meindl et al., 2023; Mohammad et al., 2019), which involved optimizations of harvesting strategy and cell lysis conditions, the amount of nuclease for mRNA digestion, and the implementation of a subtractive hybridization step for rRNA removal. Our optimized strategy ensures the generation of high-quality footprint data for this important class of organisms. Thus, the application of ribosome profiling in diatoms provides a powerful tool for studying their translational regulation. Moreover, this protocol can be adapted for use with other diatom species and may also facilitate the study of other marine algae.
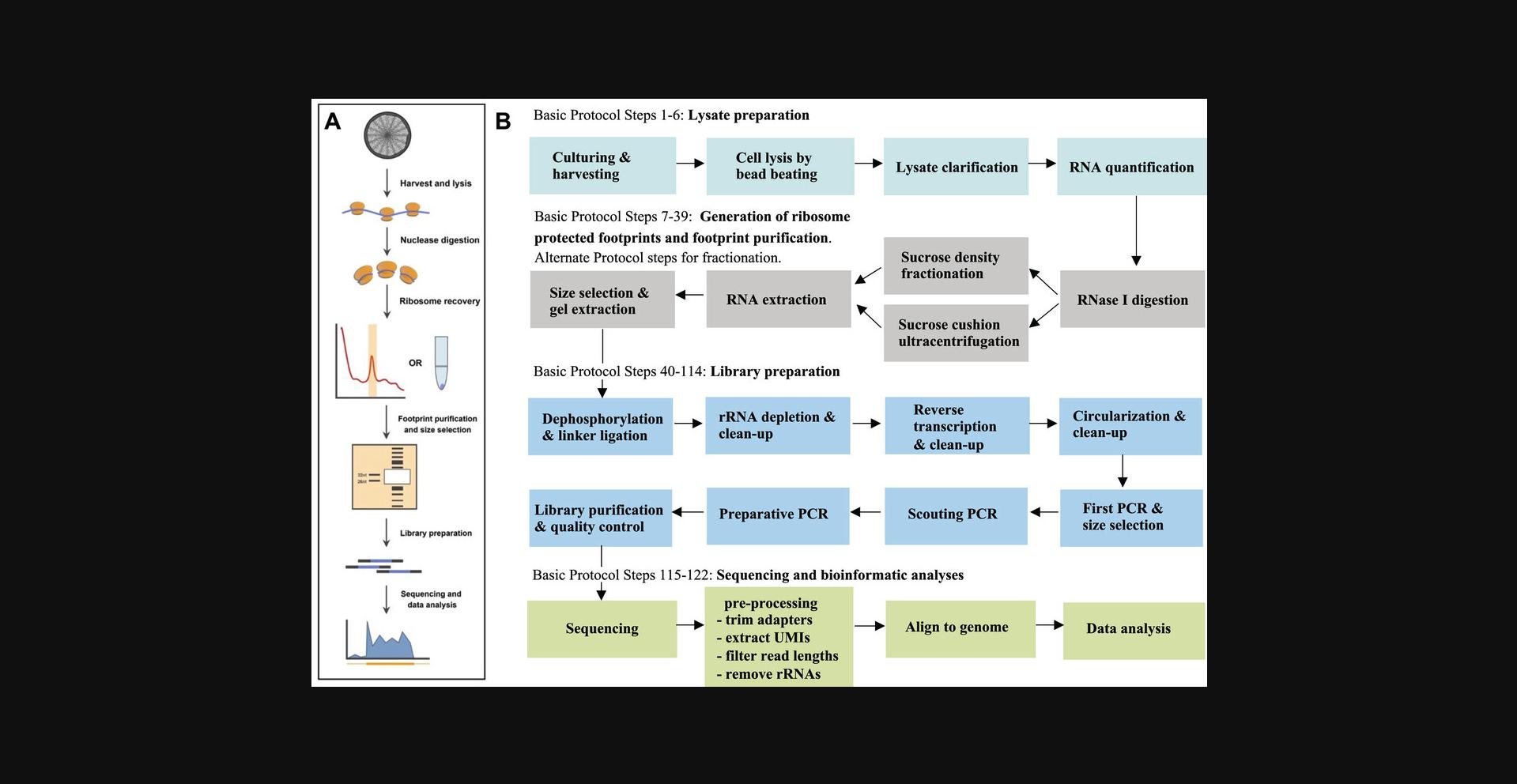
The Basic Protocol involves a series of steps for monosome preparation, isolation of ribosome-protected fragments, and subsequent sequencing library preparation (Fig. 1B). We have also developed an Alternate Protocol that yields comparable results but utilizes a sucrose gradient instead of a sucrose cushion, therefore requiring additional instrumentation. This Alternate Protocol enables quality control of the samples prior to sequencing library preparation, facilitating the adaption of ribosome profiling to other diatom species, which typically requires the optimization of reaction conditions, such as those used for limited nucleolytic digestion.
STRATEGIC PLANNING
To ensure optimal results, ribosome profiling should be carried out using exponentially growing T. pseudonana cells. For all experiments, we recommend a minimum of three biological replicates per condition to enhance data robustness. Additionally, to limit ribosomal run-off and RNA degradation, several steps of the protocol must be carried out in a cold room or on ice. As this protocol utilizes potentially hazardous chemicals such as cycloheximide and organic solvents, careful handling and access to a fume hood is crucial. Following monosome purification, it is imperative to maintain an RNase-free environment, using dedicated workspaces and RNase-free reagents. To minimize variation due to batch effects, it is advised to process samples from different conditions together by the same individual and by using the same lot of reagents. We suggest a sequencing depth of 20 million reads per sample for standard gene expression analyses.
Basic Protocol: RIBOSOME PROFILING IN Thalassiosira pseudonana
The following protocol is designed to generate high-quality ribosome profiling data to quantify translation in T. pseudonana CCMP1335.Initially, cells are promptly harvested and flash-frozen in liquid nitrogen. After mechanical disruption of the cells, optimized RNase digestion conditions are employed to create ribosome-protected footprints. Monosomes are subsequently isolated using sucrose-cushion ultracentrifugation. Alternatively, ribosomes can be purified via sucrose density gradient fractionation as detailed in the Alternate Protocol. This protocol typically yields ∼10 to 50 ng of 26 to 31 nt RNA fragments (corresponding to ∼1 to 5 pmol of RNA), which serves as the optimal amount of starting material for sequencing library preparation. We have incorporated an rRNA removal step via subtractive hybridization with the aim to increase the fraction of informative reads that map to mRNAs.
Materials
- Thalassiosira pseudonana clone CCMP1335 (Bigelow; see Internet Resources)
- Aquil medium (Price et al., 1989; see Internet Resources)
- Polysome resuspension buffer (PRB) (see recipe)
- Sucrose cushion solution (see recipe)
- Qubit RNA BR assay kit (ThermoFisher Scientific, cat. no. Q10210)
- 100 U/µl RNase I (ThermoFisher Scientific, cat. no. AM2294)
- 20 U/µl SUPERase-In RNase inhibitor (ThermoFisher Scientific, cat. no. AM2694 or AM2696)
- TRizol reagent (ThermoFisher Scientific, cat. no. 15596026)
- Direct-zol RNA MiniPrep kit (Zymo Research, cat. no. R2050)
- 3 M sodium acetate (NaOAc), pH 5.5 (ThermoFisher Scientific, cat. no. AM9740)
- GlycoBlue (ThermoFisher Scientific, cat. no. AM9516)
- Isopropanol (Carl Roth, cat. no. 9781)
- 1 M Tris·HCl, pH 8 (ThermoFisher Scientific, cat. no. 15568025)
- RNaseZap (ThermoFisher Scientific, cat. no. AM9780)
- MilliQ H2O
- 15% urea-PAGE gel (see recipe)
- 1× TBE prepared from 10× TBE stock solution (Promega, cat. no. V4251)
- 2× denaturing sample loading buffer (see recipe)
- NI-800 and NI-801 size marker oligos (see Table 1)
- 10,000× SYBR Gold (ThermoFischer Scientific, cat. no. S11494)
- RNA gel extraction buffer (see recipe)
- Ethanol (Carl Roth, cat. no. 9065)
- Qubit microRNA assay kit (ThermoFisher Scientific, cat. no. Q32881)
- T4 polynucleotide kinase (PNK) and buffer (New England Biolabs, cat. no. M0201)
- RNaseOUT recombinant ribonuclease inhibitor (ThermoFisher Scientific, cat. no. 10777019)
- Pre-adenylated rApp-L7 (see Table 2)
- T4 RNA Ligase 2, truncated KQ and buffer (New England Biolabs, cat. no. M0373)
- PEG400 (Sigma Aldrich, cat. no. 202398)
- Dimethylsulfoxide (DMSO) (Sigma Aldrich, cat. no. D8418)
- Depletion oligonucleotides (see Table 3)
- 20× SSC buffer (ThermoFisher Scientific, cat. no. 15557044)
- Dynabeads MyOne streptavidin C1 (ThermoFisher Scientific, cat. no. 65001)
- 1× and 2× binding and washing buffer (see recipe)
- Dynabeads MyOne silane (ThermoFisher Scientific, cat. no. 37002D)
- RLT buffer (Qiagen, cat. no. 79216)
- Phusion High-Fidelity DNA polymerase & dNTP mix (ThermoFisher Scientific, cat. no. F530N)
- RT Oligo P7-circ (see Table 2)
- SuperScript III reverse transcriptase and kit components (ThermoFisher Scientific, cat. no. 18080093 or 18080044)
- 1 M NaOH (Carl Roth, cat. no. 9356)
- 1 M HEPES, pH adjusted to 7.3 with NaOH (Carl Roth, cat. no. 9195)
- CircLigase II ssDNA ligase (Lucigen, cat. no. CL4111K or CL4115K)
- 10 mM ATP solution (ThermoFisher Scientific, cat. no. AM8110G)
- P5Solexa_*s oligonucleotide (see Table 2)
- P7Solexa_*s oligonucleotide (see Table 2)
- ProNex size-selective purification system (Promega, cat. no. NG2001)
- P5Solexa oligonucleotide (see Table 2)
- TrueSeq P7 Index ‘X’ oligonucleotide (see Table 2)
- 6× TBE loading buffer (see recipe)
- 7% PAA-TBE gel (see recipe)
- Ultra low range DNA ladder (ThermoFischer Scientific, cat. no. 10597012)
- Ethidium bromide (Carl Roth, cat. no. 2218)
- 5 mg/ml linear acrylamide (ThermoFisher Scientific, cat. no. AM9520)
- High Sensitivity D1000 reagents (Agilent Technologies, cat. no. 5067-5585)
| Size marker oligos | Sequencea |
|---|---|
| Upper size marker, NI-800: | 5′-AUGUACACUAGGGAUAACAGGGUAAUCAACGCGA/3Phos/ |
| Lower size marker, NI-801: | 5′-AUGUUAGGGAUAACAGGGUAAUGCGA/3Phos/ |
-
aMarker oligonucleotides used in this study are the same as described in McGlingy and Ingolia (2017) and are ordered as RNA oligonucleotides and sourced from Eurofins Genomics.
| Name | Sequencea,b,c |
|---|---|
| rApp-L7 Adapterd | /5rApp/NNNAGATCGGAAGAGCACACGTCTGA/3ddC/ |
| RT Oligo P7 - circe | /5Phos/NNNNNNNAGATCGGAAGAGCGTCGTGT/iSp9/GATTCAGACGTGTGC |
| P5Solexa_*sf | ACACGACGCTCTTCCGATC*T |
| P7Solexa_*sf | GGAGTTCAGACGTGTGCTCTTCCGATC*T |
| P5Solexa | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT |
| TrueSeq_P7_Indexg | CAAGCAGAAGACGGCATACGAGAT-X-GTGACTGGAGTTCAGACGTGTGCTCTTCC |
-
aOligonucleotides used for library preparation are the same as described in Meindl et al. (2023).
-
bAll oligonucleotides are HPLC-purified.
-
cStore and dilute primers in a buffered solution (e.g., 10 mM Tris·HCl, pH 8.0-8.5). To limit the number of freeze-thaw cycles, store as aliquots at −20°C.
-
d5’ ribo-adenylated, 3’ protected by di-deoxy nucleotide (ddC), contains a 3 nt 5’ randomized sequence to minimize ligation bias and to serve as a UMI for the bioinformatic identification of PCR duplicates.
-
eThis RT oligonucleotide serves as a circularization adapter containing a short sequence on its 3’ end that serves as a primer for reverse transcription. Separated by a PEG spacer (denoted iSp9 in the sequence) is a ligation adapter for the P7 side of the amplicon. It contains a 5’ phosphate group followed by seven degenerate nucleotides that minimize ligation bias and that serve as UMIs for the bioinformatic identification of PCR duplicates.
-
fTo prevent degradation by the proof-reading activity of the polymerase, it is recommended to include a phosphorothioate bond at the 3’ terminal end of the primer (highlighted as asterisk).
-
gX marks the position of a 6 nt barcode for experimental multiplexing. See Internet Resources, Illumina TrueSeq single indexes.
| Depletion oligoa,b | Sequence | Target |
|---|---|---|
| #1 | /5biotin-TEG/ATTCACACGGAATTCCACGTGCTCCATGCTACTT | 23S chloroplast |
| #2 | /5biotin-TEG/ACCTCAGCCCGACCCAGGACATGAACAGAAGGCACT | 28S |
| #3 | /5biotin-TEG/TACTTCTCCAGACCACAATTCGATGCAC | 5.8S |
| #4 | /5biotin-TEG/GGCTACTCTCATGCTTACAGTACCACGTTCCTT | 28S |
| #5 | /5biotin-TEG/GGCTAGCTTAGCCTTCTCCGTCCCTCGA | 23S chloroplast |
| #6 | /5biotin-TEG/AAAAGCGCAATGTGCGTTCAAAAGTTTGATGATTCAC | 5.8S |
| #7 | /5biotin-TEG/GACACAATGATAGGAAGAGCCGACATCGA | 28S |
| #8 | /5biotin-TEG/AAAGTTTGAGAATAGGCCGAGGGCA | 28S |
-
aThe primers are used for rRNA depletion via subtractive hybridization.
-
bThe DNA oligonucleotides contain a 5’ biotin tag and are HPLC purified to ensure highest quality.
- Vacuum pump for cell filtration (Welch Vacuum, model 2534C-02)
- Nalgene reusable bottle top filters (ThermoFisher Scientific, cat. no. DS0320-5045)
- 47-mm diameter 1.2-µm isopore filters (Merck, cat. no. RTTP04700)
- Laminar flow hood (Walker Safety Cabinets, model: Class II 1290 Recirc Gen 6 or equivalent)
- Tweezers (ThermoFisher Scientific, cat. no. 15699310)
- 1.5-ml cryogenic vials (Carl Roth, cat. no. AET4)
- Liquid nitrogen
- 425- to 600-µm glass beads (Merck, cat. no. G8772)
- 50-ml centrifuge tubes (ThermoFisher Scientific, cat. no. 339652)
- Bead beater (BioSpec 3110BX Mini-BeadBeater-1, or equivalent)
- Ice
- Refrigerated microcentrifuge 5418R, 4°C (Eppendorf, cat. no. 5401000010)
- Pipettes and tips (ThermoFisher Scientific, cat. no. 05-403-151)
- NanoDrop One (ThermoFisher Scientific, cat. no. ND-ONE-W)
- Qubit 4 fluorometer (ThermoFisher Scientific, cat. no. Q33238)
- Qubit assay tubes (ThermoFisher Scientific, cat. no. Q32856)
- 11 × 34-mm polycarbonate ultracentrifuge tubes (Beckman, cat. no. 343778)
- Optima TLX ultracentrifuge (Beckman, cat. no. 361545)
- TLA 120.2 rotor (Beckman, cat. no. 357656)
- 1.5-ml microcentrifuge tubes (Fisher Scientific, cat. no. 11926955)
- Dry ice
- Heat block (VWR, cat. no. 12621-104)
- Mini-Protean vertical electrophoresis cell (BioRad, cat. no. 1658004)
- Blue light transilluminator, e.g., DarkReader (Clare Chemical Research, cat. no. DR46B)
- Scalpel blades (Merck, cat. no. S2646)
- 18-G needle (VWR, cat. no. BD305195)
- 0.5-ml tubes (ThermoFisher Scientific, cat. no. AB0533)
- Corning Costar Spin-X column centrifuge tube filters (Sigma Aldrich, cat. no. CLS8160)
- DNA LoBind tubes (Eppendorf, cat. no. 0030108051)
- Vortex
- DynaMag-2 magnetic separation rack (Invitrogen, cat. no. 12321D)
- Tube shaker
- 0.2-ml tubes (ThermoFisher Scientific, cat. no. AB0620)
- T100 thermal cycler (BioRad, cat. no. 1861096)
- Automated gel purification system, e.g., BluePippin (optional)
- High Sensitivity D1000 ScreenTape (Agilent Technologies, cat. no. 5067-5584)
- Agilent 2200 TapeStation nucleic acid system (Agilent Technologies, cat. no. G2965AA)
- Illumina sequencing system
- Cutadapt (see Internet Resources)
- UMI-tools (see Internet Resources)
- bowtie2 and genome sequences (see Internet Resources)
- STAR aligner (see Internet Resources)
- R software (see Internet Resources)
- Rstudio software (see Internet Resources)
- riboWaltz (see Internet Resources)
Cell growth
1.Grow T. pseudonana CCMP1335 to mid-exponential phase (5–7 × 105 cells/ml) in Aquil medium under desired experimental conditions.
Cell harvest
2.Collect 2 × 200 ml of culture by gentle vacuum filtration (using a vacuum pump and a reusable bottle top filter) onto a 47-mm filter under a laminar flow hood. Use sterile tweezers to roll up the filter membrane and insert it into a previously labeled 1.5-ml cryogenic vial. Immediately flash-freeze in liquid nitrogen (<1 min).
Cell lysis
3.Add 400 µl of pre-chilled polysome resuspension buffer (PRB) to the tube with slightly thawed cells. Rinse off filter and remove with sterile tweezers. Add two scoops of glass beads (425- to 600-µm).
4.Treat sample in bead beater for 2 min (max speed) with a break on ice after 1 min to prevent overheating.
5.Clarify the lysate by centrifugation for 10 min at 14,000 × g , 4°C using a microcentrifuge. Transfer the supernatant using a pipette in a new 1.5-ml microcentrifuge tube and place on ice.
6.Measure UV absorption at 260 nm of the lysate using a NanoDrop to estimate the RNA concentration.
RNA digestion
7.Take ∼60 µg of total RNA and dilute to 300 µl with PRB if necessary. Add 7 U RNase I (100 U/µl) and incubate for 45 min at room temperature with gentle mixing.
8.Put on ice and add 2 µl SUPERase-In RNase inhibitor (20 U/µl) to stop nuclease digestion.
Ribosome recovery
9.Transfer the sample to a 11 × 34-mm polycarbonate ultracentrifuge tube.
10.Underlay sample with 750 µl 1 M sucrose cushion solution by positioning a pipette tip at the bottom of the tube and gently dispensing the sucrose cushion solution.
11.Pellet ribosomes by centrifugation in an ultracentrifuge using a TLA120.2 rotor, 2 hr at 199,000 × g (75,000 rpm), 4°C.
12.Remove the supernatant and resuspend the pellet in 350 µl of TRizol reagent. Transfer to a 1.5-ml microcentrifuge tube. Replicates of the same sample can be pooled at this point.
13.Purify resuspended RNA by using the Direct-zol RNA MiniPrep kit following the manufacturer's instruction for purification of total RNA. Elute RNA in 50 µl RNase-free H2O provided with the kit.
14.Precipitate eluted RNA by adding 10 µl 3 M NaOAc pH 5.5, 1.5 µl GlycoBlue (to facilitate precipitation) and 38.5 µl RNase-free H2O, followed by 150 µl isopropanol.
15.Carry out precipitation on dry ice >30 min or at −20°C overnight.
16.Pellet RNA by centrifugation for 30 min at 14,000 × g , 4°C using a microcentrifuge. Pipette all liquid from the tube, place sideways and air dry for 10 min.
17.Resuspend pellet in 5 µl 10 mM Tris·HCl, pH 8 prepared from the 1 M stock solution.
Ribosomal footprint fragment purification
It is critical to work in an RNase-free environment from this step onwards. Decontaminate the electrophoresis apparatus and other equipment with RNaseZap. MilliQ H2O can be used to prepare the running buffer.
18.Prepare 15% urea-PAGE gel.
19.Pre-heat a heating block to 80°C.
20.Pre-run gel in the electrophoresis cell at 15 V/cm for 15 min in 1× TBE.
21.Prepare RNA sample from step 17 by adding 5 µl of 2× denaturing sample loading buffer. To prepare marker oligos for two lanes, mix 1 µl lower marker oligo NI-800 (10 µM), 1 µl upper marker oligo NI-801 (10 µM) (see Table 1), 10 µl 2× denaturing sample loading buffer and 8 µl 10 mM Tris·HCl, pH 8.
22.Denature sample and marker oligos for 90 s at 80°C using the heat block. Keep on ice until loading.
23.Load sample onto the gel with marker oligo samples framing the sample lane.
24.Run gel at 15 V/cm until the lower dye (light blue) front has reached the lower third of the gel.
25.Stain gel for 5 min with 1× SYBR gold in 1× TBE with gentle shaking.
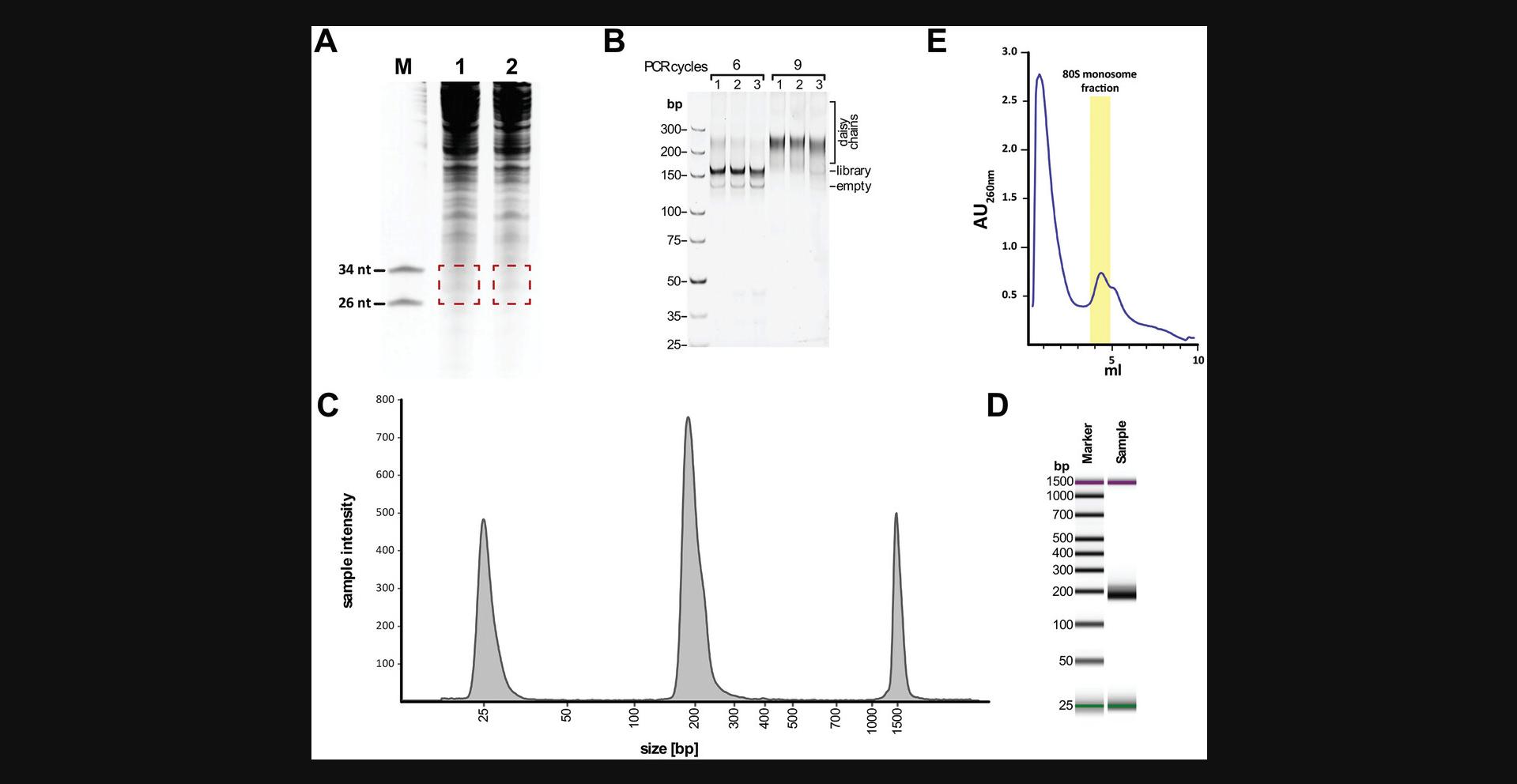
26.Visualize gel on a blue light transilluminator and cut out the 26 to 34 nt region of the gel using a scalpel blade (see Fig. 2A).
Gel extraction
The gel extraction step has been adapted from Mohammad and Buskirk (2019).
27.Using an 18-G needle, poke holes into a 0.5-ml tube. Place the gel cut out into the tube.
28.Place the 0.5-ml tube into a 1.5 ml-tube.
29.Spin 5 min at 14,000 × g , room temperature, in a microcentrifuge until gel extrudes into bottom tube.
30.Add 400 µl RNA extraction buffer and 2 µl SUPERase-In.
31.Incubate overnight at 4˚C with gentle shaking.
32.Spin 5 s at 14,000 × g , room temperature, and transfer gel slurry into a Spin-X column.
33.Spin 3 min at 14,000 × g , room temperature, and transfer the eluate into a new low bind (LoBind) tube.
34.Precipitate RNA by adding 1.5 µl GlycoBlue, mix well and add two volumes 100% EtOH. Vortex briefly.
35.Incubate at −80°C for 1 hr.
36.Pellet by centrifugation for 30 min at 14,000 × g , 4°C.
37.Wash pellet with 300 µl cold 80% EtOH, centrifuge 5 min at 14,000 × g , 4°C, remove all liquid and briefly let air dry (<10 min).
38.Resuspend in 10 µl nuclease-free H2O.
39.Measure concentration using the Qubit microRNA Assay Kit.
Library preparation
For preparation of sequencing libraries, we followed the protocol by Meindl et al. (2023). With this protocol sequencing libraries can be prepared from as little as 0.1 pmol of RNA fragments; an optimal amount of starting material is 1 to 5 pmol of RNA (from step 38). To minimize sample loss, all reactions until PCR amplification of the final library (step 97) should be carried out in low bind reaction tubes.
We additionally implemented a subtractive hybridization step for rRNA removal using eight biotinylated depletion oligonucleotides representing abundant rRNA contaminants in T. pseudonana. The custom depletion step was optimized from Zinshteyn et al., and Green (2020).
RNA dephosphorylation to remove of terminal phosphates
40.For removal of the 2’-3’-cyclic phosphate that is generated by the RNase I cleavage, set up the following reaction:
| Dephosphorylation mix | ||
| x | µl | RNA in H2O (from step 38) |
| 2 | µl | 10 × T4 PNK buffer (supplied with the enzyme) |
| 0.5 | µl | 40 U/µl RNaseOUT |
| 0.5 | µl | T4 PNK (with 3´phosphatase activity) |
| add nuclease-free H2O to 20 µl | ||
41.Mix all components well and incubate for 30 min at 37°C.
42.Inactivate the enzyme for 20 min at 65°C.
3´-adapter ligation
43.Prepare ligation mix (35 µl) using 3´dephosphorylated RNA adapters:
| Ligation of adapter to 3’ end of RNA | ||
| 20 | µl | reaction from previous step |
| 1 | µl | 20 μM pre-adenylated rApp-L7 adapters (see Table 2) |
| 1.5 | µl | 10 × T4 RNA-ligase buffer (supplied with the enzyme) |
| 1 | µl | T4 RNA ligase 2, truncated KQ |
| 6 | µl | 50% PEG400 |
| 4.5 | µl | DMSO |
| 1 | µl | nuclease-free H2O |
44.Incubate overnight at 16°C.
45.Incubate 1 hr at 37°C.
46.Inactivate the enzyme for 15 min at 65°C.
Ribosomal RNA depletion
47.Dilute depletion oligos (see Table 3) in 4× SSC buffer (prepared from 20× stock solution) to concentration of 2.5 µM for each oligo and store at −20˚C.
48.Add 10 µl of depletion oligo mix solution to 35 µl of 3′-adapter ligation product from step 46.
49.Incubate the mixture at 80°C for 2 min to denature RNA.
50.Allow to cool down slowly to 25°C (temperature ramp 3°C/min).
51.Resuspend Dynabeads MyOne streptavidin C1 by vortexing the tube at medium speed.
52.Transfer 80 µl of bead suspension into a new low bind tube.
53.Put on a magnetic rack and wait for 1 min.
54.Aspirate and discard all supernatant.
55.Add 80 µl of 1× binding and washing buffer and agitate tube to resuspend beads. Place on magnetic rack and wait for 1 min.
56.Aspirate and discard all supernatant.
57.Repeat wash step twice.
58.Resuspend beads in 45 µl 2× binding and washing buffer.
59.Add the cooled sample (45 µl) from step 50 to the prepared beads (45 µl) from step 58 and mix well to homogeneously resuspend the beads.
60.Incubate at 25°C for 15 min (with shaking at 500 rpm).
61.Place on magnetic rack for >1 min to attract the beads to the magnet, then carefully transfer supernatant into a new tube.
Clean-up (1)
62.Thoroughly mix the solution of magnetic MyONE Silane beads, using 20 μl beads per sample.
63.Wash the beads with 500 μl RLT buffer. Then, resuspend them in 650 μl RLT buffer per sample and add them to the sample. Mix the solution well.
64.Add 720 μl 100% ethanol and gently mix the solution by pipetting. Incubate the mixture for 5 min, mix again and incubate for another 5 min.
65.Magnetically attract the beads by placing the sample on the magnetic rack and discard the supernatant.
66.Resuspend the beads in 1 ml of freshly prepared cold 80% ethanol and transfer the mix into a new tube.
67.Wash again with 80% ethanol. Incubate 30 s before putting the sample on the magnet. Repeat this wash step.
68.Spin 5 s at 14,000 × g , room temperature, magnetically attract the beads and discard the supernatant. Air-dry the beads for 5 min at room temperature and resuspend them in 12 μl nuclease-free H2O.
69.Incubate the mix for 5 min at room temperature. Magnetically attract the beads and add the eluate to the reverse transcription (RT) mix of the next step (step 70).
Reverse transcription
70.Prepare the following mix in 0.2-ml tubes:
| RNA and primer mix | ||
| 12 | µl | pooled RNA in H2O (from step 69) |
| 1 | µl | 10 mM dNTP mix |
| 1 | µl | 0.5 pmol/µl RT Oligo P7-circ (see Table 2) |
71.In a thermal cycler, heat the sample to 70°C for 5 min and incubate at 25°C until RT mix added, mix by pipetting. Do not put on ice to prevent unspecific annealing of the oligonucleotide used to prime reverse transcription.
72.Prepare the following RT mix and add it to the sample from the previous step (resulting in a total volume of 20 µl):
| RT mix | ||
| 4 | µl | 5× first strand buffer (supplied with the enzyme) |
| 1 | µl | 0.1 M DTT (supplied with the enzyme) |
| 0.5 | µl | 200 U/µl Superscript III RT |
| 0.5 | µl | 40 U/μl RNaseOUT |
73.Incubate as follows:
| 25 | °C | 5 | min |
| 42 | °C | 20 | min |
| 50 | °C | 40 | min |
| 4 | °C | ∞ |
74.To hydrolyze the RNA, add 1.65 μl 1 M NaOH and incubate at 98°C for 20 min. Then add 20 μl 1 M HEPES, pH 7.3 to neutralize the solution.
Clean-up (2)
75.Thoroughly mix the solution of magnetic MyONE Silane beads, using 10 μl of beads per sample.
76.Wash beads with 500 μl RLT buffer. Then, resuspend them in 650 μl RLT buffer per sample and add them to sample. Mix the solution well.
77.Add 720 μl 100% ethanol and gently mix the solution by pipetting. Incubate the mixture for 5 min, mix again and incubate another 5 min.
78.Magnetically attract the beads by placing the sample on the magnetic rack and discard the supernatant.
79.Resuspend the beads in 1 ml of freshly prepared cold 80% ethanol and transfer the mix into a new tube.
80.Wash again with 80% ethanol. Incubate 30 s before putting the sample on the magnet. Repeat this wash step.
81.Spin the mix 5 s at 14,000 × g , room temperature, in a microcentrifuge, magnetically attract the beads and discard the supernatant. Air-dry the beads for 5 min at room temperature and resuspend them in 14 μl nuclease-free H2O.
82.Incubate the mix for 5 min at room temperature. Magnetically attract the beads and add the eluate to the reactions mix of the next step (step 83).
Circularization of the RT product
83.Prepare the following ligation mix (amounts given per sample):
| Ligation mix | ||
| 14 | µl | cDNA (from step 82) |
| 2 | µl | 10 × CircLigase buffer (supplied with the enzyme) |
| 1 | µl | 10 mM ATP |
| 1 | µl | 50 mM MnCl2 (supplied with the enzyme) |
| 2 | µl | CircLigase II |
84.To ensure homogeneity, mix the ligation master-mix by vigorous stirring, pipetting, and flicking. Centrifuge the mix 5 s at 14,000 × g , room temperature, in a microcentrifuge to collect all drops.
85.Incubate 2 hr at 60°C in a thermocycler.
86.Incubate at 80°C for 10 min to inactivate the CircLigase.
Clean-up (3)
87.Thoroughly mix the solution of magnetic MyONE Silane beads, using 10 μl MyONE Silane beads per sample.
88.Wash beads with 500 μl RLT buffer. Then, resuspend them in 650 μl RLT buffer per sample and add them to sample. Mix the solution well.
89.Add 720 μl of 100% ethanol and gently mix the solution by pipetting. Incubate the mixture for 5 min, mix again and incubate another 5 min.
90.Magnetically attract the beads by placing the sample on the magnetic rack and discard the supernatant.
91.Resuspend the beads in 1 ml of freshly prepared cold 80% ethanol and transfer the mix into a new tube.
92.Wash again with 80% ethanol. Incubate 30 s before putting the sample on the magnet. Repeat this wash step.
93.Briefly spin the mix 5 s at 14,000 × g , room temperature, in a microcentrifuge, magnetically attract the beads and discard the supernatant. Air-dry the beads for 5 min at room temperature and resuspend them in 23 μl nuclease-free H2O.
94.Incubate the mix for 5 min at room temperature. Magnetically attract the beads and transfer the solution to a fresh tube.
First PCR (cDNA pre-amplification)
95.Prepare 2× Phusion HF master mix on ice (25 µl required per reaction, volumes provided are for 100 µl):
| 2× Phusion HF master mix | ||
| 40 | µl | 5× Phusion HF buffer |
| 4 | µl | 10 mM dNTPs |
| 54 | µl | nuclease-free H2O |
| 2 | µl | Phusion polymerase |
96.Prepare the following PCR mix:
| PCR mix | ||
| 15 | µl | circularized RT product (from step 94) |
| 7.5 | µl | nuclease-free H2O |
| 2.5 | µl | primer mix of P5Solexa_*s and P7Solexa_*s, 10 μM each (see Table 2) |
| 25 | µl | 2× Phusion HF master mix |
97.Run the following PCR:
| 98 | °C | 30 | s | ||
| 98 | °C | 10 | s | ||
| 65 | °C | 30 | s | perform amplification (steps 2 to 4) 6× | |
| 72 | °C | 30 | s | ||
| 72 | °C | 3 | min | ||
| 16 | °C | ∞ |
Size selection to remove primer-dimers
To remove excess primer dimers, size-select your samples with ProNex size-selective purification system.
98.Equilibrate the ProNex size-selective chemistry (beads) to room temperature for 30 min and resuspend by vigorous vortexing.
99.For 50 μl of sample, add 147.5 μl ProNex size-selective chemistry. This is a 1:2.95 (v/v) ratio of sample to beads. Mix by pipetting 10× up and down.
100.Incubate the mixture at room temperature for 10 min.
101.Place the samples on a magnetic stand for 2 min. Discard the supernatant.
102.Leave the tube on the magnetic stand so that the beads remain attracted to the side of the tube, then add 300 μl ProNex wash buffer. If necessary, add more ProNex wash wuffer to cover all beads on the magnet. While the beads are magnetically attracted, incubate the ProNex wash buffer for 30 to 60 s before removal.
103.Repeat the last wash of the magnetically attracted beads with another 300 μl ProNex wash buffer for 40 to 60 s. Discard the supernatant and allow the samples to air-dry for ∼8 to 10 min (<60 min) until cracks are visible in the bead pellet.
104.Remove the beads from the magnetic stand and elute samples in 23 μl ProNex elution buffer. Resuspend all samples by pipetting and let them stand for 5 min at room temperature.
Second PCR amplification-cycle optimization
Try two different cycle numbers for amplification of the sample, as described in Buchbender et al. (2020). A good starting point for cycle optimization is a range of 6-12 cycles. Ideally, you should observe enough product without overamplification. Overamplification is indicated by the appearance of large assemblies of improperly annealed, partially double-stranded, heteroduplex DNA migrating above the library (known as 'daisy chains', Fig. 2B; see also Huppertz et al., 2014).
105.Prepare the following master mix (10 µl per cycle number to be scouted):
| PCR mix (per reaction) | ||
| 3.7 | µl | nuclease-free H2O |
| 0.3 | µl | primer mix of P5Solexa and TrueSeq P7 Index ‘X’, 10 μM each (see Table 2) |
| 1 | µl | cDNA (from step 104) |
| 5 | µl | 2× Phusion HF master mix (see step 95) |
106.Split into 10 µl reactions and run the following PCR with different cycle numbers:
| 98 | °C | 30 | s | ||
| 98 | °C | 10 | s | ||
| 65 | °C | 30 | s | perform amplification (steps 2 to 4) 6-12× * | |
| 72 | °C | 30 | s | ||
| 72 | °C | 3 | min | ||
| 16 | °C | ∞ |
107.Combine 10 µl PCR product with 2 µl 6× TBE loading buffer. Run 6 μl of the amplified product on a small (∼7 × 10 cm ) 7% PAA-TBE gel for 30 min at 200 V. Use 3 µl ultra low range DNA ladder (diluted 1:4 in loading dye), as a marker.
108.Stain gel for 10 min with ethidium bromide and visualize gel (see Fig. 2B).
Preparative PCR
From your cycle optimization PCR results, estimate the minimum number of PCR cycles to use to amplify the library. Consider that the template for the reaction will be 2.5-times more concentrated (see PCR mix below), therefore one cycle less is needed than in the scouting PCR (steps 105 to 108: PCR cycle optimization).
For experimental multiplexing during sequencing, barcodes are introduced with the P7 primer. For additional information please refer to the Illumina TrueSeq Single Indexes (see Internet Resources). Ensure usage of compatible indices for all libraries to be sequenced on the same lane.
109.Prepare the following PCR mix:
| PCR mix (per reaction) | ||
| 6.5 | µl | nuclease-free H2O |
| 1 | µl | primer mix of P5Solexa and TrueSeq P7 Index ‘X’, 10 μM each (see Table 2) |
| 7.5 | µl | cDNA (from step 104) |
| 15 | µl | 2× Phusion HF master mix (see step 95) |
110.Run the same PCR program as in step 106, but with the adjusted cycle number.
Purification of the sequencing library
111.Purify the PCR reaction using the automated agarose gel electrophoresis system (PippinPrep, or similar) using a 3% agarose cassette according to the manufacturer's instructions with the following settings: target fragment length 168 bp with the setting ‘tight’.
112.Precipitate the purified library (30 µl) by the addition of 3 µl 3 M NaOAc pH 5.5, 2 µl linear acrylamide, and 90 µl ethanol. Incubate at least 30 min at −20°C, then centrifuge 30 min at 14,000 × g , 4°C. speed in a microcentrifuge. Aspirate supernatant, air dry pellet, and resuspend the pellet in 12 µl H2O.
Quantitative and qualitative analysis of the purified sequencing library
For quality control and determination of the concentration of the libraries, a screen tape assay using the High Sensitivity D1000 ScreenTape and the High Sensitivity D1000 Reagents is employed on a TapeStation system.
113.Determine library concentration by measuring UV absorbance at 260 nm using the Nanodrop System.
114.Analyze an appropriate amount of the purified library using a high sensitivity screen tape assay on a TapeStation according to the manufacturer's instructions (Fig. 2C and D).
Sequencing and data analysis
115.Pool libraries as desired and subject to sequencing on an appropriate machine (e.g., Illumina NextSeq2000, P3 Reagents (50 Cycles), 80-85 cycles, single end read using Read1 primer, plus 6 nt Indexread).
116.After de-multiplexing, pre-process the sequencing reads by trimming of adapters using Cutadapt (version 2.8, adapter=AGATCGGAAGAGCACACGTCT, overlap=10, minimum-length=10, discard-untrimmed).
117.Extract unique molecular identifiers (UMIs) and append them to the read name using UMI-tools (version 1.0.1, extract-method=regex).
118.Filter reads by lengths and keep reads between 16 and 40 nt.
119.Align trimmed and filtered reads to an rRNA reference (e.g., from RNAcentral database) using bowtie2 (version 2.3.5.1).
120.Align all non-rRNA aligned reads to the Joint Genome Institutes (JGI) T. pseudonana reference genome (Thaps3_chromosomes_ assembly_chromosomes_repeatmasked.fasta) with genome annotations (Thaps3.filtered_ proteins.FilteredModels2.gff3) using STAR (version, –quantMode TranscriptomeSAM GeneCounts, --outSAMattributes All, outFilterMismatchNmax 2).
121.Remove PCR duplicates using UMI-tools (extract-umi-method, read_id-method unique).
122.Analyze and interpret ribosome profiling data in R (version 3.5.1) and Rstudio using the riboWaltz package (version 1.2.0).
Alternate Protocol: RIBOSOME PROFILING PROTOCOL FOR DIATOMS USING SUCROSE GRADIENT FRACTIONATION
This protocol includes the same steps as the Basic Protocol, but with the addition of a sucrose density gradient instead of a sucrose cushion. This allows users to optimize nuclease digestion conditions to suit a wider range of diatom species as monitored by the collapse of polysomes into monosomes after nucleolytic digestion.
Additional Materials (also see Basic Protocol)
-
10% sucrose solution (see recipe)
-
50% sucrose solution (see recipe)
-
Sucrose chase solution (see recipe)
-
Phenol/chloroform/isoamyl alcohol 25:24.1 (Carl Roth, cat. no. A156)
-
Gradient Master gradient forming device and tube holders (Biocomp Instruments, cat. nos. B108 and 105-914A)
-
Ultracentrifuge (e.g., Beckman Optima L-100 K Ultracentrifuge) with SW41 rotor (Beckman, cat. no. 331362)
-
14 × 89-mm tubes for SW41 rotor (Beckman, cat. no. 331372)
-
siFractor (siTOOLs, cat. no. eq-F001-F0SW41)
-
ÄKTA FPLC (or similar)
Cell growth and harvesting, lysate preparation, and nucleolytic digestion
1.Follow steps 1 to 8 of the Basic Protocol.
Preparation of sucrose gradient
We recommend using a gradient master device (Biocomp Instruments) for rapid and reproducible formation of sucrose gradients. Here, 12 ml gradients are used with a SW41 rotor (Beckman). Conditions for monosome purification using different rotors are described in Meindl et al. (2023).
2.Pre-cool the ultracentrifuge and cool down the rotor and the buckets. Thaw all necessary reagents.
3.Prepare 10% and 50% sucrose solutions.
4.Prepare linear sucrose density gradients using the Biocomp gradient master according to the manufacturer's instructions. Use the ‘long caps’ provided by the manufacturer with the following program: SW41 rotor, sucrose, long caps, 10%-50%, 11 steps. If required, the density gradients can be stored at 4°C for several hours.
Sucrose gradient centrifugation
5.Remove the caps from the gradient tubes and gently overlay the solution with the sample. Pool replicates of the same sample at this point. Balance the tubes carefully with PRB buffer.
6.Carefully transfer tubes into the buckets, close lids and insert buckets into the rotor.
7.Insert rotor into the centrifuge and spin 3 hr at 151,000 × g (35,000 rpm) at 4°C.
Density gradient fractionation
This protocol uses a gradient fractionation system which employs a tube piercing unit to deliver a dense chase solution to displace the gradient. It is connected to an FPLC system for analysis and fractionation. Further details are described in Meindl et al. (2023).
8.Connect the siFractor (siTOOLs Biotech) to an FPLC machine according to the manufacturer's instructions and thoroughly rinse the system with H2O. Fill a super loop (or similar) with sucrose chase solution and connect it to the FPLC system. Prime the tubes with the chase solution.
9.Follow the manufacturer's instructions for the fractionation of the samples from the gradient tubes while continuously monitoring conductivity and UV absorbance at 260 nm. Collect fractions of 1 ml each. Figure 2E depicts a typical UV profile obtained with nuclease treated T. pseudonana cell extract.
RNA isolation from density gradient fractions
10.Select the gradient fractions that contain your complexes of interest (e.g., 80S monosomes) and dilute 1:1 (v/v) with nuclease-free H2O to prevent phase inversion during organic extraction (due to the high sugar concentration of the sample).
11.For organic extraction, transfer the samples to a fume hood and add an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) and mix thoroughly by vortexing.
12.Separate the phases by centrifugation in a bench-top centrifuge for 10-30 min, 14,000 × g , room temperature, then transfer the upper aqueous phase into new tube. Make sure not to transfer any of the organic solution.
13.Precipitate nucleic acids by adding 1/10 volume 3 M NaOAc (pH 5.2), 2.5 µl linear acrylamide, and 0.8 volumes of isopropanol. Mix by inverting the tubes several times. Incubate samples at −20°C for at least 30 min, then centrifuge in a microcentrifuge 30 min at 14,000 × g , 4°C.
14.Aspirate supernatant and wash the pellet with cold 80% ethanol. Aspirate the ethanol, air-dry the pellet and resuspend the RNA in 5 µl 10 mM Tris, pH 8.
15.Continue with footprint fragment purification as outlined in the Basic Protocol step 18.
REAGENTS AND SOLUTIONS
Binding and washing buffer, 1×
- 5 mM Tris·HCl, pH 8 (ThermoFisher Scientific, cat. no. 15568025)
- 0.5 mM EDTA (Carl Roth, cat. no. 8043)
- 1 M NaCl (Carl Roth, cat. no. 3957)
- Double the concentrations for a 2× buffer
- Store indefinitely at room temperature
Denaturing sample loading buffer, 2×
- 98% (v/v) formamide (Carl Roth, cat. no. P040)
- 10 mM EDTA (Carl Roth, cat. no. 8043)
- 300 µg/ml bromophenol blue (Carl Roth, cat. no. A512)
- Store indefinitely at −20°C
PAA-TBE gel, 7%
- Mix in a fume hood:
- 3.5 ml 30% acrylamide/bis-acrylamide solution (19:1) (BioRad, cat. no. 1610154)
- 1.5 ml 10× TBE (Promega, cat. no. V4251)
- 10 ml MilliQ H2O
- For polymerization add:
- 120 µl 10% APS (prepare fresh) (Carl Roth, cat. no. 9592)
- 12 µl TEMED (Carl Roth, cat. no. 2367)
- Mix gently and pour gel
- Insert well comb and allow 30 min for gel to polymerize before use
Polysome resuspension buffer (PRB)
- 50 mM Tris·HCl, pH 8 (ThermoFisher Scientific, cat. no. 15568025)
- 100 mM NH4Cl (Carl Roth, cat. no. K298)
- 200 mM sucrose (Carl Roth, cat. no. 9097)
- 10.5 mM magnesium acetate (Carl Roth, cat. no. P026)
- 0.5 mM EDTA (Carl Roth, cat. no. 8043)
- 1 mM DTT (Carl Roth, cat. no. 6908)
- 0.01% cycloheximide (Carl Roth, cat. no. 8682)
- 0.1% Triton X-100 (Carl Roth, cat. no. 3051)
- 25 U/ml Turbo DNase (ThermoFisher Scientific, cat. no. AM2238 or AM2239)
- Mix all reagents in a fume hood
- Prepare fresh before every use and keep on ice
RNA gel extraction buffer
- 300 mM NaOAc, pH 5.5 (ThermoFisher Scientific, cat. no. AM9740)
- 1 mM EDTA (Carl Roth, cat. no. 8043)
- 0.25% (w/v) SDS (Carl Roth, cat. no. CN30)
- Store indefinitely at room temperature
Sucrose chase solution
- 60 mM Tris·HCl, pH 7.4 (Carl Roth, cat. no. 4855)
- 450 mM NaCl (Carl Roth, cat. no. 3957)
- 15 mM MgCl2 (ThermoFisher Scientific, cat. no. R0971)
- 0.0001% Ponceau S (Carl Roth, cat. no. 5938)
- 60% sucrose (w/v) (Carl Roth, cat. no. 9097)
- Store up to 3 days at 4°C
50 ml of solution are sufficient for four gradients
Sucrose cushion solution
- 1 M sucrose (Carl Roth, cat. no. 9097) in PRB (see recipe)
- 20 U/ml SUPERase-In (ThermoFisher Scientific, cat. no. AM2694, AM2696)
- Prepare fresh before every use and keep on ice
Sucrose solution, 10%
- 20 mM Tris·HCl, pH 7.4 (Carl Roth, cat. no. 4855)
- 150 mM NaCl (Carl Roth, cat. no. 3957)
- 5 mM MgCl2 (ThermoFisher Scientific, cat. no. R0971)
- 10% sucrose (w/v) (Carl Roth, cat. no. 9097)
- Store up to 3 days at 4°C
- Prior to use, add:
- 1 mM DTT (Carl Roth, cat. no. 6908)
- 100 µg/µl cycloheximide (Carl Roth, cat. no. 8682)
25 ml of solution is sufficient for four gradients
Sucrose solution, 50%
- 20 mM Tris·HCl, pH 7.4 (Carl Roth, cat. no. 4855)
- 150 mM NaCl (Carl Roth, cat. no. 3957)
- 5 mM MgCl2 (ThermoFisher Scientific, cat. no. R0971)
- 50% sucrose (w/v) (Carl Roth, cat. no. 9097)
- Store up to 3 days at 4°C
- Prior to use, add:
- 1 mM DTT (Carl Roth, cat. no. 6908)
- 100 µg/µl cycloheximide (Carl Roth, cat. no. 8682)
25 ml of solution is sufficient for four gradients
TBE loading buffer, 6×
In 6.7 ml nuclease-free H2O, dissolve 25 mg bromophenol blue (Carl Roth, cat. no. A512) and 25 mg xylene cyanole (Carl Roth, cat. no. A513). Add 3.3 ml glycerol (Carl Roth, cat. no 3783) and mix. Store indefinitely at −20°C.
Urea-PAGE gel, 15%
- Mix in a fume hood:
- 3.75 ml 40% acrylamide/bis-acrylamide solution (19:1) (BioRad, cat. no. 1610144)
- 4.8 × g urea (Carl Roth, cat. no. 3941)
- 1 ml 10× TBE (Promega, cat. no. V4251)
- Βring up to 10 ml with MilliQ H2O
- Warm at 37°C until dissolved before adding the following:
- 120 µl 10% APS (prepare fresh) (Carl Roth, cat. no. 9592)
- 12 µl TEMED (Carl Roth, cat. no. 2367)
- Mix gently and pour gel
- Insert well comb and allow 30 min for gel to polymerize before use
COMMENTARY
Background Information
Ribosome profiling provides comprehensive snapshots of cellular translation based on the sequencing of mRNA fragments that are protected by translating ribosomes from nucleolytic digestion (Ingolia et al., 2009). These ribosome-protected fragments (RPFs) yield positional information of ribosomes on mRNA and their quantification allows the determination of protein synthesis rates. Moreover, by normalization to RNA abundance, ribosome loading scores can be derived that reflect the translation efficiency of mRNA species. Ribosome profiling monitors the final step of gene expression and therefore allows detection of gene expression changes that occur at different levels (including transcriptional regulation) (Ingolia, 2016). Changes in protein synthesis rates (as measured by changes in RPFs from a locus) can hence be driven, e.g., by transcriptional regulation, RNA turnover, differential RNA processing, or regulated translation. This allows to detect changes in gene expression programs, e.g., induced by a changing environment.
A wide range of ribosome profiling protocols are available for different species. Due to the different nature of cells and ribosomes, these protocols vary in their methods of harvesting, lysis conditions, and the selected nuclease and digestion conditions. In eukaryotic organisms, ribonuclease (RNase) I, A, T1 and micrococcal S7 are commonly used but their cutting efficiencies are species-dependent (Gerashchenko & Gladyshev, 2017). In this protocol, we used RNase I which performed well for T. pseudonana (Fig. 2E).
Different methods have been used in ribosome profiling studies to isolate digested monosomes, the most prevalent ones being ultracentrifugation via sucrose gradient and sucrose cushion (e.g., Ingolia et al., 2012; Meindl et al., 2023). As described in the Alternate Protocol, density gradient centrifugation was performed to identify optimal nuclease digestion conditions for T. pseudonana which had not been established before. Moreover, density gradient centrifugation allows the purification of ribosomal monosomes without carrying over a large portion of messenger ribonucleoproteins (mRNPs) to the next steps in the protocol (McGlincy & Ingolia, 2017). However, it requires additional instrumentation that may not be readily available in all labs.
Ribosome profiling sequencing libraries are typically dominated by rRNA fragments. Despite measures to limit these contaminations, e.g., precise excision of RNA in the right size after denaturing poly-acrylamide gel electrophoresis (PAGE) using appropriate markers, contaminations typically represent ∼90% of the reads. Hence, different measures have been implemented for their depletion. To specifically target the contaminants, they are experimentally identified typically by shallow sequencing of ribosome profiling libraries prepared under the same conditions from the respective organism/sample. Several commercially available rRNA depletion kits have been tested in a recent study and were found generally unsuitable for use with ribosome footprint libraries (Zinshteyn et al., 2020), especially kits based on targeted nuclease cleavage can lead to ribosome footprint degradation, reduction of mappable reads, interference with global gene expression measurements and blurred nucleotide resolution. Moreover, commercial kits that target T. pseudonana rRNA contaminations is not available. Thus, for our protocol we designed biotinylated antisense oligonucleotides that target the common contaminants found in sequencing libraries generated from ribosomal profiling of T. pseudonana and implemented a subtractive hybridization step in the protocol.
Various protocols for library preparation from ribosome profiling samples have been described that differ, e.g., in aspects of linker sequence and ligation, or the purification of the reaction intermediates. More recently, single-pot reactions have been introduced that exploit the template switching activity of reverse transcriptase (Ferguson et al., 2023; Ozadam et al., 2023). Here, we employ a recently developed protocol for sequencing library preparation that is particularly suited for low input samples (Meindl et al., 2023) and hence for the processing of samples derived from T. pseudonana that can have a low yield. In brief, for sequencing library preparation, an adapter sequence is ligated to the ribosome protected fragments, followed by reverse transcription, circularization of the cDNA and PCR amplification. Purification of the reaction intermediates occurs via solid phase reversible immobilization. Degenerate nucleotides on the ends of the adapters used in the ligation and circularization reactions reduce ligation bias and are employed for the identification of PCR duplicates during bioinformatic analyses.
Bioinformatic analysis of ribosome profiling data has been described in detail elsewhere (e.g., McGlincy & Ingolia, 2017). We mapped the sequencing reads against the T. pseudonana genome instead of the transcriptome due to its higher quality.
Taken together, the adjustments made to previously existing protocols result in high-quality sequencing data of actively translating ribosomes from T. pseudonana. Our protocol is robust and can now be used for future gene expression analysis in this organism, as well as a starting point for the adaption of the protocol to other related species.
Critical Parameters
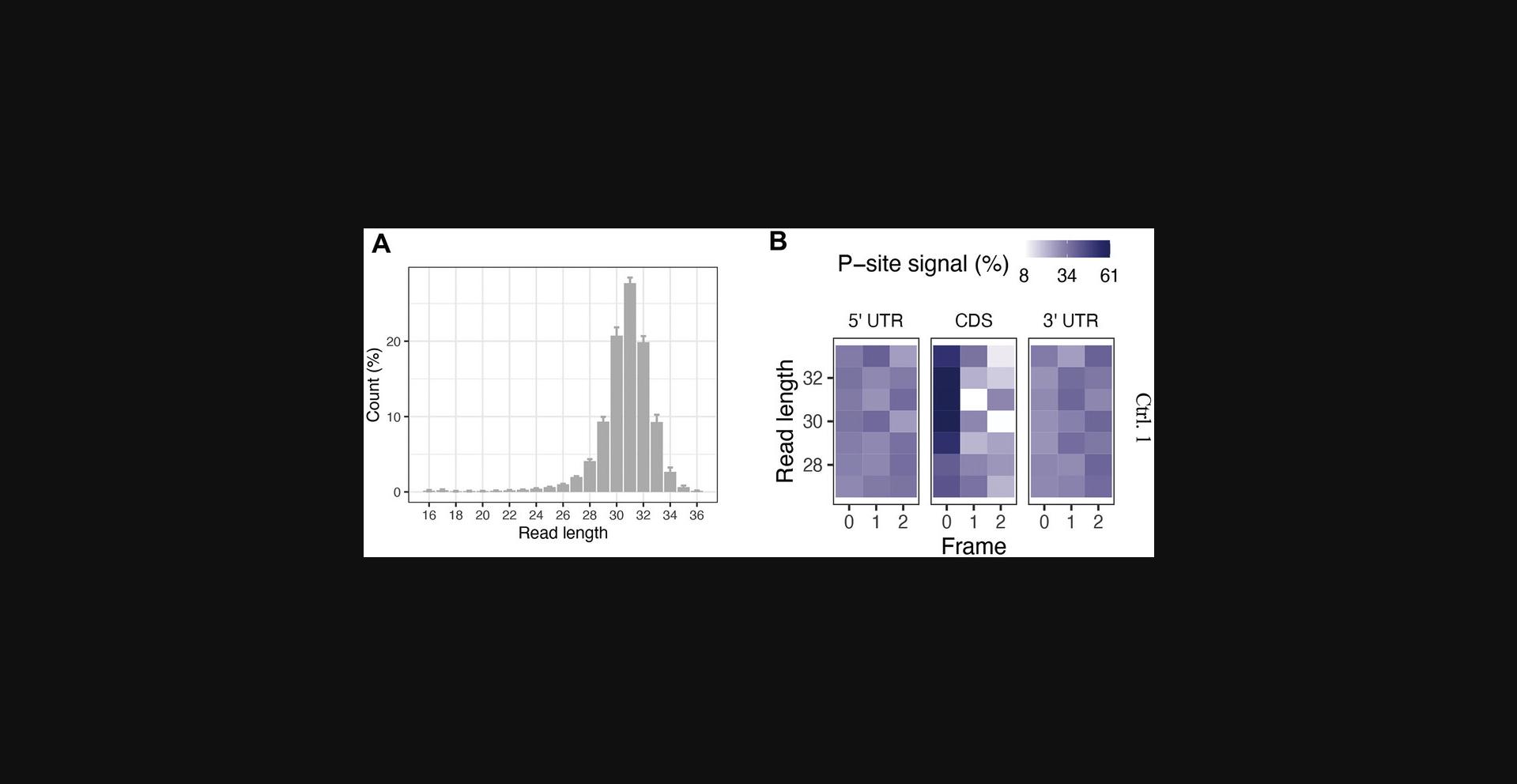
For robust and reproducible results from ribosome profiling experiments several parameters are important, e.g., the way cells are harvested and lysed can have a profound effect on ribosome distribution by triggering of cellular stress responses (e.g., Mohammad et al., 2019). Also, the use of translation inhibitors has been debated since it can introduce artefacts in particular species (Gerashchenko & Gladyshev, 2014; Sharma et al., 2021). Furthermore, the nuclease(s) and conditions for the generation of ribosome protected fragments need to be carefully chosen. Nucleases that work well in one species, might result in a complete loss of the sample in others (Gerashchenko & Gladyshev, 2017). Hence, the conditions under which ribosome profiling is performed must be carefully chosen and adapted to the model system and research question pursued. Our protocol has been optimized for T. pseudonana and yields high-resolution footprints with decent frame information (see Fig. 3B). For other species, conditions suitable for ribosome profiling need to be experimentally established.
In all cases, ribosome profiling experiments need to be performed under conditions that prevent nucleolytic degradation of the samples, since shortening of the RPFs during library preparation compromises data quality. This includes working at low temperatures whenever possible, as well as the use of nuclease-free reagents and RNase inhibitors. Moreover, since the amount of input material for sequencing library preparation is typically rather limited, the use of low bind reaction tubes and low retention tips is highly recommended, once the RPFs have been excised from the gel. Experimental precision during critical steps, e.g., the size selection of the RPFs by denaturing page, limits contaminations and yields high quality data.
Proper amplification of the sequencing library is crucial for obtaining good results, since over-amplification increases the fraction of PCR duplicates in the final library. Yet, insufficient amplification of the sequencing library will produce only minute amounts of material that are often not sufficient for sequencing, the protocol provides a simple and robust step (scouting PCR) to identify optimal PCR conditions for the amplification of the library.
Troubleshooting
Since ribosome profiling is a complex technique with numerous factors contributing to success, troubleshooting of the experiments is complex. In the following, we provide a list of frequently encountered problems and how to deal with them.
Little yield after cell lysis
Incomplete recovery of sample from the filter: ensure to recover as much material as possible from the filter prior to cell lysis. Incomplete lysis of the cells: check for complete lysis of the cells under the microscope, adjust lysis conditions, if required by, e.g., adding additional cycles in the bead beater. Partial degradation of the sample during cell lysis: make sure to keep the sample cooled during cell lysis to prevent unwanted degradation.
Loss of sample or incomplete digestion after nuclease treatment of the extract
Amount of RNase used for digestion: we frequently observe batch to batch variation of the RNase I. Carefully titrate the enzyme to achieve optimal conditions. RNase activity in the extract: supplement the extract with RNase inhibitors such as RNaseOUT (which does not affect RNase I) to inhibit endogenous nucleases. Lysate too concentrated or too dilute: to ensure optimal nucleolytic digestion, ensure a total reaction volume of 300 µl when digesting 60 µg of total RNA. Incorrect RNA quantification: quantify extracts as described in the protocol, adjust amount enzyme when required.
Loss of sample during sequencing library preparation or too many PCR cycles required to produce the sequencing library
Sample has been degraded by nucleases: use nuclease-free reagents, use nuclease inhibitors as indicated in the protocol, work on ice whenever possible, use filter tips to prevent nuclease carry-over from other reactions. Sample loss due to unspecific adsorption onto surfaces: use low bind reaction tubes and low retention tips during library preparation. Sample loss during clean-up: try not to lose beads when aspirating supernatant during the clean-up steps, do not remove beads from magnet during some of the washing steps (as indicated in the protocol); for ProNex size exclusion ensure the correct 1:2.95 v/v ratio of beads to sample as outlined in the protocol. Low quality oligonucleotides: we observe batch to batch variation of the oligonucleotides used for library preparation; ensure that the oligonucleotides are of good quality (e.g., by running them on a denaturing PAGE), order new oligonucleotides if necessary. Enzymatic reaction not efficient: test activity of the enzymes used for library preparation, order new batches if required.
Sequencing library contains too many empty amplicons
Input material for the library preparation was too low. Use more input material, and/or gel-purify the final library as outlined in the protocol.
Sequencing library dominated by a single experimental barcode
Contamination from a previous experiment: we highly recommend to physically separate pre-PCR and post-PCR work in two separate labs or use laminar flow hoods to prevent carryover between experiments. Pooling of samples for sequencing: prior to pooling, quantify the amplicons as precisely as possible using a Qubit fluorimeter, or TapeStation analysis.
Understanding Results
Yield
The protocol provided provides instructions for the processing of a crude cell lysate containing ∼60 µg of total RNA. After RNA extraction, typically 10 to 15 µg of RNA can be recovered for gel purification, resulting in a yield of ∼10 to 50 ng of ∼30 nt RNA fragments (∼1 to 5 pmol of RNA), an ideal starting quantity for library preparation.
PCR cycles required for library amplification
The number of PCR cycles needed for the final amplification can be used as a crude indicator for the yield after sequencing library preparation. Low cycle numbers are observed when using large amounts of starting material, or they indicate a good overall performance during library preparation. Typically, libraries are obtained after 6 to 9 PCR cycles (Fig. 2B); higher PCR cycle numbers can result in increased fractions of PCR duplicates and empty amplicons.
Non-mRNA reads in the libraries (contamination)
Despite the depletion of contaminating rRNA sequences, it is expected to observe ∼60%-80% of total reads mapping to rRNA.
Data processing
We used the riboWaltz R package to demonstrate the quality of the ribosome profiling data and to visualize it. Typically, eukaryotic ribosome profiling data exhibits a read length of 28 to 32 nt; in our results we also found a peak of ribosome-protected fragments at 31 nt across all tested samples (Fig. 3A). By identifying the ribosomal P-site in each ribosome-protected fragment (RPF), riboWaltz allows for visual inspection of triplet periodicity, a distinct feature of ribosome profiling data due to the ribosome's movement along the mRNA 3-nt at a time. As expected for high-quality data, we observed a strong enrichment of sequencing reads in canonical coding sequences (CDSs). Our footprint fragments of 30 to 32 nt in length have a strong frame preference for one of the three sub-codon positions on the CDS, but not on the 5’ and 3’ UTRs (Fig. 3B).
Time Considerations
The protocol (from harvest to library preparation) is expected to take ∼5 days. This could vary depending on the number of samples analyzed and the individuals experience with the techniques involved.
Acknowledgments
This work was supported by the UKRI-BBSRC Norwich Research Park Biosciences Doctoral Training Partnership (grant BB/M011216/1 to MP) and the German Research Foundation (grant SFB960 TP B11 to JM). We thank EMBL GeneCore for excellent support with sequencing.
Author Contributions
Monica Pichler : Data curation, methodology, resources, validation, writing—original draft; Andreas Meindl : Data curation, investigation, methodology; Markus Romberger : Data curation, investigation, methodology; Annemarie Eckes-Shephard : Data curation, investigation, methodology; Carl-Fredrik Nyberg-Brodda : Data curation, investigation, methodology; Claudia Buhigas : Data curation, investigation, methodology; Sergio Llaneza-Lago : Data curation, investigation, methodology; Gerhard Lehmann : Data curation, investigation, methodology; Amanda Hopes : Data curation, investigation, methodology, supervision; Gunter Meister : Funding acquisition; Jan Medenbach : Conceptualization, investigation, methodology, supervision, writing—review and editing; Thomas Mock : Conceptualization, funding acquisition, project administration, supervision, writing - review and editing.
Conflict of Interest
G.M. is a founder of, and J.M. a consultant to siTOOLs Biotech GmbH, Martinsried.
Open Research
Data Availability Statement
The data that support the protocol are openly available on figshare at https://doi.org/10.6084/m9.figshare.22717591 and https://doi.org/10.6084/m9.figshare.23635512.
Literature Cited
- Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., Zhou, S. G., Allen, A. E., Apt, K. E., Bechner, M., Brzezinski, M. A., Chaal, B. K., Chiovitti, A., Davis, A. K., Demarest, M. S., Detter, J. C., Glavina, T., Goodstein, D., Hadi, M. Z., … Rokhsar, D. S. (2004). The genome of the diatom Thalassiosira pseudonana : Ecology, evolution, and metabolism. Science , 306, 79–86. https://doi.org/10.1126/science.1101156
- Aspden, J. L., Eyre-Walker, Y. C., Phillips, R. J., Amin, U., Mumtaz, M. A., Brocard, M., & Couso, J. P. (2014). Extensive translation of small open reading frames revealed by poly-ribo-seq. Elife , 3, e03528. https://doi.org/10.7554/eLife.03528
- Bazzini, A. A., Johnstone, T. G., Christiano, R., Mackowiak, S. D., Obermayer, B., Fleming, E. S., Vejnar, C. E., Lee, M. T., Rajewsky, N., Walther, T. C., & Giraldez, A. J. (2014). Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO Journal , 33(9), 981–993. https://doi.org/10.1002/embj.201488411
- Bazzini, A. A., Lee, M. T., & Giraldez, A. J. (2012). Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science , 336(6078), 233–237. https://doi.org/10.1126/science.1215704
- Belshaw, N., Grouneva, I., Aram, L., Gal, A., Hopes, A., & Mock, T. (2023). Efficient gene replacement by CRISPR/Cas-mediated homologous recombination in the model diatom Thalassiosira pseudonana. New Phytologist , 238, 438–452. https://doi.org/10.1111/nph.18587
- Brar, G. A., & Weissman, J. S. (2015). Ribosome profiling reveals the what, when, where and how of protein synthesis. Nature Reviews Molecular Cell Biology , 16(11), 651–664. https://doi.org/10.1038/nrm4069
- Buchbender, A., Mutter, H., Sutandy, F. X. R., Kortel, N., Hanel, H., Busch, A., Ebersberger, S., & Konig, J. (2020). Improved library preparation with the new iCLIP2 protocol. Methods (San Diego, Calif.) , 178, 33–48. https://doi.org/10.1016/j.ymeth.2019.10.003
- Calviello, L., Mukherjee, N., Wyler, E., Zauber, H., Hirsekorn, A., Selbach, M., Landthaler, M., Obermayer, B., & Ohler, U. (2016). Detecting actively translated open reading frames in ribosome profiling data. Nature Methods , 13(2), 165–170. https://doi.org/10.1038/nmeth.3688
- Chung, B. Y., Hardcastle, T. J., Jones, J. D., Irigoyen, N., Firth, A. E., Baulcombe, D. C., & Brierley, I. (2015). The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-seq data analysis. RNA , 21(10), 1731–1745. https://doi.org/10.1261/rna.052548.115
- Dong, H. P., Dong, Y. L., Cui, L., Balamurugan, S., Gao, J., Lu, S. H., & Jiang, T. (2016). High light stress triggers distinct proteomic responses in the marine diatom Thalassiosira pseudonana. BMC Genomics , 17(1), 994. https://doi.org/10.1186/s12864-016-3335-5
- Ferguson, L., Upton, H. E., Pimentel, S. C., Mok, A., Lareau, L. F., Collins, K., & Ingolia, N. T. (2023). Streamlined and sensitive mono- and diribosome profiling in yeast and human cells. BioRxiv , https://doi.org/10.1101/2023.02.01.526718
- Gerashchenko, M. V., & Gladyshev, V. N. (2014). Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Research , 42(17), e134. https://doi.org/10.1093/nar/gku671
- Gerashchenko, M. V., & Gladyshev, V. N. (2017). Ribonuclease selection for ribosome profiling. Nucleic Acids Research , 45(2), e6. https://doi.org/10.1093/nar/gkw822
- Guo, H., Ingolia, N. T., Weissman, J. S., & Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature , 466(7308), 835–840. https://doi.org/10.1038/nature09267
- Hopes, A., Nekrasov, V., Kamoun, S., & Mock, T. (2016). Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods , 12, 49. https://doi.org/10.1186/s13007-016-0148-0
- Hsu, P. Y., Calviello, L., Wu, H. L., Li, F. W., Rothfels, C. J., Ohler, U., & Benfey, P. N. (2016). Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America , 113(45), E7126–E7135. https://doi.org/10.1073/pnas.1614788113
- Huppertz, I., Attig, J., D'Ambrogio, A., Easton, L. E., Sibley, C. R., Sugimoto, Y., Tajnik, M., Konig, J., & Ule, J. (2014). iCLIP: Protein-RNA interactions at nucleotide resolution. Methods , 65(3), 274–287. https://doi.org/10.1016/j.ymeth.2013.10.011
- Ingolia, N. T. (2016). Ribosome footprint profiling of translation throughout the genome. Cell , 165(1), 22–33. https://doi.org/10.1016/j.cell.2016.02.066
- Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M., & Weissman, J. S. (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature Protocols , 7(8), 1534–1550. https://doi.org/10.1038/nprot.2012.086
- Ingolia, N. T., Ghaemmaghami, S., Newman, J. R., & Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science , 324(5924), 218–223. https://doi.org/10.1126/science.1168978
- Ingolia, N. T., Lareau, L. F., & Weissman, J. S. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell , 147(4), 789–802. https://doi.org/10.1016/j.cell.2011.10.002
- Ji, Z., Song, R., Regev, A., & Struhl, K. (2015). Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife , 4, e08890. https://doi.org/10.7554/eLife.08890
- Juntawong, P., Girke, T., Bazin, J., & Bailey-Serres, J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America , 111(1), E203–212. https://doi.org/10.1073/pnas.1317811111
- Karlsen, J., Asplund-Samuelsson, J., Thomas, Q., Jahn, M., & Hudson, E. P. (2018). Ribosome profiling of synechocystis reveals altered ribosome allocation at carbon starvation. mSystems , 3(5), https://doi.org/10.1128/mSystems.00126-18
- Li, G. W., Oh, E., & Weissman, J. S. (2012). The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature , 484(7395), 538–541. https://doi.org/10.1038/nature10965
- McGlincy, N. J., & Ingolia, N. T. (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods , 126, 112–129. https://doi.org/10.1016/j.ymeth.2017.05.028
- Meindl, A., Romberger, M., Lehmann, G., Eichner, N., Kleemann, L., Wu, J., Danner, J., Boesl, M., Mesitov, M., Meister, G., Koenig, J., Leidel, S. A., & Medenbach, J. (2023). A rapid protocol for ribosome profiling of low input samples. Nucleic Acids Research , gkad459. Advance online publication. https://doi.org/10.1093/nar/gkad459
- Mock, T., Hodgkinson, K., Wu, T., Moulton, V., Duncan, A., van Oosterhout, C., & Pichler, M. (2022). Structure and evolution of diatom nuclear genes and genomes. In A. Falciatore & T. Mock (Eds.), The molecular life of diatoms (pp. 111–145). Springer International Publishing.
- Mock, T., Samanta, M. P., Iverson, V., Berthiaume, C., Robison, M., Holtermann, K., Durkin, C., Bondurant, S. S., Richmond, K., Rodesch, M., Kallas, T., Huttlin, E. L., Cerrina, F., Sussman, M. R., & Armbrust, E. V. (2008). Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proceedings of the National Academy of Sciences of the United States of America , 105(5), 1579–1584. https://doi.org/10.1073/pnas.0707946105
- Mohammad, F., & Buskirk, A. R. (2019). Protocol for ribosome profiling in bacteria. Bio-Protocol , 9(24), e3468. https://doi.org/10.21769/BioProtoc.3468
- Mohammad, F., Green, R., & Buskirk, A. R. (2019). A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife , 8, e42591. https://doi.org/10.7554/eLife.42591
- Ozadam, H., Tonn, T., Han, C. M., Segura, A., Hoskins, I., Rao, S., Ghatpande, V., Tran, D., Catoe, D., Salit, M., & Cenik, C. (2023). Single-cell quantification of ribosome occupancy in early mouse development. Nature , 618(7967), 1057–1064. https://doi.org/10.1038/s41586-023-06228-9
- Poulsen, N., Chesley, P. M., & Kröger, N. (2006). Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). Journal of Phycology , 42(5), 1059–1065. https://doi.org/10.1111/j.1529-8817.2006.00269.x
- Price, N. M., Harrison, G. I., Hering, J. G., Hudson, R. J., Nirel, P. M., Palenik, B., & Morel, F. M. (1989). Preparation and chemistry of the artificial algal culture medium aquil. Biological Oceanography , 6, 443–461. https://doi.org/10.1080/01965581.1988.10749544
- Shalgi, R., Hurt, J. A., Krykbaeva, I., Taipale, M., Lindquist, S., & Burge, C. B. (2013). Widespread regulation of translation by elongation pausing in heat shock. Molecular Cell , 49(3), 439–452. https://doi.org/10.1016/j.molcel.2012.11.028
- Sharma, P., Wu, J., Nilges, B. S., & Leidel, S. A. (2021). Humans and other commonly used model organisms are resistant to cycloheximide-mediated biases in ribosome profiling experiments. Nature Communications , 12(1), 5094. https://doi.org/10.1038/s41467-021-25411-y
- Steitz, J. A. (1969). Polypeptide chain initiation: Nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature , 224(5223), 957–964. https://doi.org/10.1038/224957a0
- Trosch, R., Barahimipour, R., Gao, Y., Badillo-Corona, J. A., Gotsmann, V. L., Zimmer, D., Muhlhaus, T., Zoschke, R., & Willmund, F. (2018). Commonalities and differences of chloroplast translation in a green alga and land plants. Nature Plants , 4(8), 564–575. https://doi.org/10.1038/s41477-018-0211-0
- Wu, H. L., Song, G., Walley, J. W., & Hsu, P. Y. (2019). The Tomato Translational Landscape Revealed by Transcriptome Assembly and Ribosome Profiling. Plant Physiology , 181(1), 367–380. https://doi.org/10.1104/pp.19.00541
- Zinshteyn, B., Wangen, J. R., Hua, B. Y., & Green, R. (2020). Nuclease-mediated depletion biases in ribosome footprint profiling libraries. RNA , 26(10), 1481–1488. https://doi.org/10.1261/rna.075523.120
Internet Resources
A source of the Thalassiosira pseudonana clone CCMP1335.
A source for the full recipe of Aquil medium.
Manual for Cutadapt.
Manual for UMI-tools.
Manual for bowtie2.
Data source of the T. pseudonana reference genome and genome annotations.
Manual for STAR aligner.
R software download source.
Rstudio software download source.
Manual for riboWaltz.
Illumina TrueSeq single indexes online resource.

