Mn2+-Phos-Tag Polyacrylamide for the Quantification of Protein Phosphorylation Levels
Kasturi Markandran, Kasturi Markandran, Jane Vanetta Lee En Xuan, Jane Vanetta Lee En Xuan, Haiyang Yu, Haiyang Yu, Lim Meng Shun, Lim Meng Shun, Michael A. Ferenczi, Michael A. Ferenczi
Abstract
This paper provides a guideline for optimizing and utilizing Mn2+ Phos-tag gel technology to separate phosphorylated proteins from their unphosphorylated counterparts. It provides key insights into methods for careful sample preparation and experimental directions for determining the appropriate Phos-tag gel compositions and electrophoresis and western blotting conditions. This protocol has been used to successfully resolve proteins extracted from cardiac and skeletal muscles. The guidelines can be extended for optimizing protocols to resolve proteins from other cells or tissue sources. With this, phosphoproteomics and the elucidation of underlying mechanisms of disease progression can be accelerated. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
INTRODUCTION
Much of biology relies on phospho-linkages, as evidenced vy the sugar phosphate backbone of DNA in all living organisms (Schneider, Neidle, & Berman, 1997). Phosphorylation of amino acid residues in eukaryotic cells is the most common post-translational modification, given that 9 of the 20 naturally occurring amino acids can be phosphorylated (Cieśla, Frączyk, & Rode, 2011). About 30% of human proteins are covalently bound to phosphate ions, and about 500 protein kinases and 200 phosphatases are encoded in the human genome (Cohen, 2001; Sacco, Perfetto, Castagnoli, & Cesareni, 2012).
Phosphorylation of amino acids facilitated by kinases creates a negative charge density on protein surfaces, allowing stronger and stable intramolecular and intermolecular interactions. This elicits physical and chemical responses such as structural modifications, alterations in the interactions with surrounding molecules, and regulation of signaling pathways (Hunter, 2012). Phosphatases are responsible for dephosphorylation and complement the role of kinases by regulating the rate and duration of signaling (Heinrich, Neel, & Rapoport, 2002). These quick and reversible mechanisms usually affect kinetics and pathways at a molecular, cellular, and possibly physiological level, over time scales of seconds to days.
Protein phosphorylation plays vital roles in basic cellular processes such as cell-cycle progression, differentiation, organ development, intercellular communication, cytoskeletal arrangements, and apoptosis, which preserves the natural phenotype of living organisms (Johnson, 2009); it can also lead to adverse consequences, such as promoting disease progression (Kristjánsdóttir & Rudolph, 2004; Perluigi, Barone, Di Domenico, & Butterfield, 2016). Thus, protein phosphorylation in biological systems is a subject of intense research.
Existing Methods to Quantify Protein Phosphorylation Levels
Generally, phosphoproteins exist in low abundance, which imposes a major challenge for their detection and measurement (Goshe, 2006). Nonetheless, various methods to study and analyze the phosphorylation levels of proteins have been devised. For example, mass spectrometry and one- and two-dimensional (1D and 2D) gel electrophoresis are necessary steps for obtaining phosphoproteomic data, but these are time consuming and require careful interpretation (Kinoshita-Kikuta, Kinoshita, & Koike, 2009a; Tsunehiro et al., 2013).
Certain gel-electrophoretic methods (e.g., urea-glycerol, Phos-tag, gradient polyacrylamide gels, and 2D gel electrophoresis) and the phospho-specific enzyme-linked immunosorbent assay (ELISA) are useful for analyzing phosphorylation levels and hence the kinase activity of targeted proteins. However, intensive optimization is required to obtain repeatable and reliable results. Although gradient polyacrylamide gels may be useful to resolve large amounts of sarcomeric proteins, they may not be capable of resolving phosphorylated proteins from their unphosphorylated counterparts (Zaremba et al., 2007). In the past, it was common to perform global radioisotope (32P) labeling of phosphorylated proteins before gel electrophoresis. However, the usefulness of this approach is limited by the difficulty of deciphering the phosphorylation levels of protein of interest. Also, it is challenging to identify the labeled proteins using mass spectrometry due to sensitivity limitations (Aponte et al., 2009).
Meanwhile, although phospho-specific ELISA is a promising tool, there are only a few commercially available phospho-specific ELISA kits for targeted proteins. Moreover, the use of phospho-specific ELISA kits and phosphofluoro antibodies will limit the findings to a particular phosphorylation site rather than the total phosphorylation activity of the protein of interest (Jczernik et al., 1991).
Immunohistochemistry can reveal the spatial localization of phosphorylated proteins in cells or tissues. Immunohistochemistry's power depends on the availability of the targeted phosphor-fluorophore and imaging modality (controlling spatial resolution) that can be used to visualize the fluorophore-tagged phosphorylated proteins (Lima et al., 2008; Yaseen et al., 2015).
A summary of commonly used techniques for studying protein phosphorylation is provided in Table 1. The choice of technique will be driven by the abilities of the different techniques to answer research questions, their reliability, their efficiency, and the cost-effectiveness relationship.
| Technique | Quantitative | Qualitative | Specific protein detection | Codetection | Throughput |
|---|---|---|---|---|---|
| Pro-Q Diamond phosphoprotein gel stain | ✓ | Depends on the method to resolve proteins | |||
| Western blotting (WB) using phosphofluoro antibodies | ✓ | ✓ | Depends on the method to resolve proteins | ||
| Urea-glycerol PAGE + WB | ✓ | ✓ | ✓ | Moderate | |
| 2D gel electrophoresis + WB | ✓ | ✓ | ✓ | Low | |
| Phos-tag acrylamide gel + WB | ✓ | ✓ | ✓ | Moderate | |
| Mass spectrometry (MS/MS, LC/MS/MS, MRS-MS) | ✓ | ✓ | ✓ | High | |
| Phospho-specific ELISA | ✓ | ✓ | |||
| Immunohistochemistry and histology | ✓ | ✓ | ✓ |
- a Quantitative: Gives numerical output for protein quantification. Qualitative: Gives non-numerical output; post-analysis of immediate results required for protein quantification. Specific protein detection: identification of phosphorylated protein of interest. Codetection: Phosphorylated and non-phosphorylated protein of interest can be identified concurrently. Throughput: Rate of processing samples.
Significance of Phos-tag Polyacrylamide Technology
To obtain data on the total phosphorylation of a specific protein with respect to its unphosphorylated counterpart, mass spectrometry, 2D sodium dodecyl sulfate (SDS)-PAGE, urea-glycerol PAGE, or Phos-tag SDS-PAGE followed by western blotting is commonly performed. These approaches provide concurrent quantitative and qualitative description of the phosphorylation levels of proteins of interest.
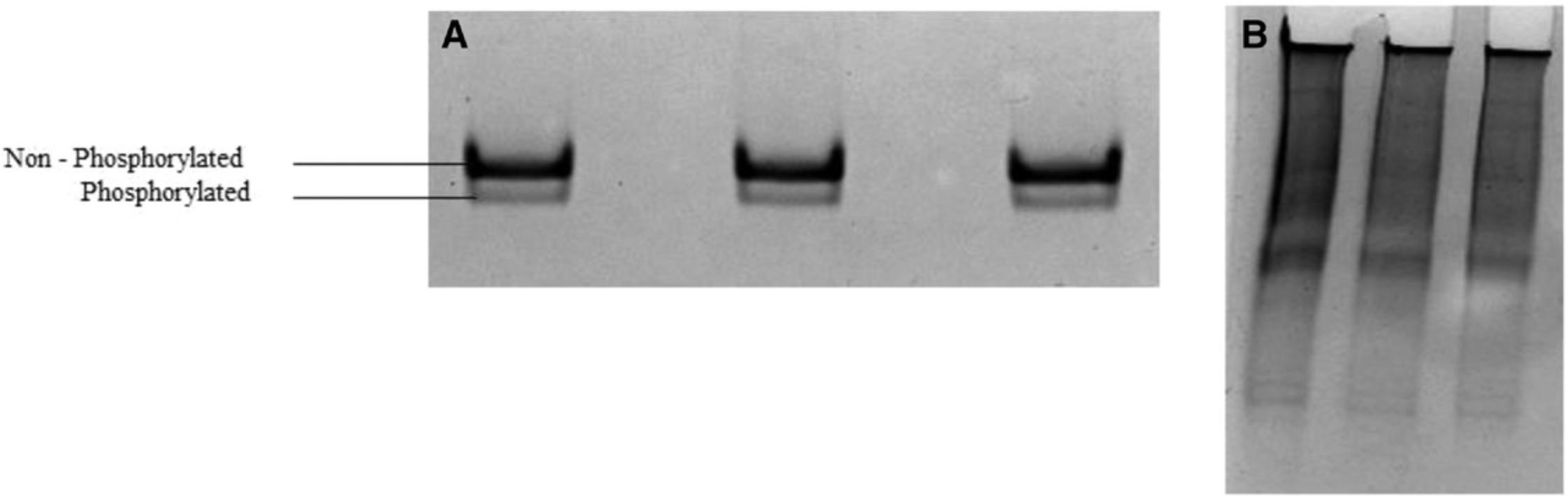
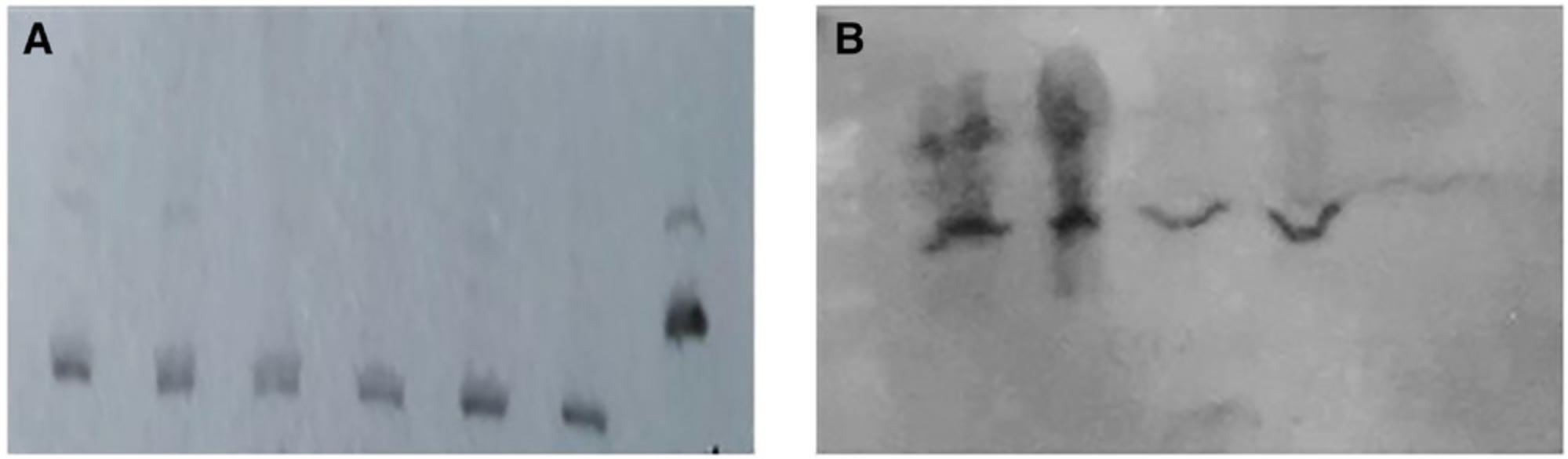
As mentioned earlier, the large data output of mass spectrometry is time consuming and expensive to analyze. On the other hand, 2D gel electrophoresis has low throughput because only one sample can be analyzed per gel. Moreover, it requires phosphor-specific antibodies to detect phosphorylated proteins, as other post-translational modifications (i.e., deamidation) could result in a similar shift of proteins (Scruggs & Solarob, 2011). In our laboratory, we attempted urea-glycerol PAGE, a technique in use since the late 1980s to resolve phosphorylated proteins extracted from smooth and skeletal muscles, along with various troubleshooting steps. But we found it difficult to resolve cell-lysate proteins with this approach (Fig. 1; Sobieszek & Jertschin, 1986). After careful optimization studies, we settled on Phos-tag technology for the separation, detection, and purification of phosphorylated proteins. This technology indeed provides new opportunities for the systematic collection of high-quality phosphoproteomic data.
Phos-tag is a divalent metal (Mn2+ or Zn2+) complex with a molecular weight of 594.7 Daltons (Da). It specifically binds to the phenyl phosphomonoester dianion of the phosphorylated residues, increasing the overall molecular weight of the phosphorylated protein (Kinoshita et al., 2009a). This results in the electrophoretic retardation of the phosphorylated protein, separating the phosphorylated protein from its unphosphorylated counterpart and thus facilitating the study of phosphorylation and related processes of proteins in vitro.
There are two types of Phos-tag molecules: the Mn2+-Phos-tag and the Zn2+-Phos-tag (Kinoshita, Kinoshita-Kikuta, & Koike, 2009b). Because the Mn2+-Phos-tag molecule is compatible with the well-known Laemmli SDS-PAGE protocol, we preferred the Mn2+-Phos-tag over the Zn2++-Phos-tag, as the gel composition used in Laemmli's system can be conveniently transferred to making the Mn2+-Phos-tag gel.
Nonetheless, the Phos-tag SDS-PAGE assay requires meticulous optimization. In our experience, Phos-tag SDS-PAGE is particularly sensitive to temperature, pH, and the presence of chelating agents. Care is required during sample preparation and in the optimization of polyacrylamide gel content and electrophoresis and western blotting conditions to optimally resolve proteins. There are papers with brief methodology descriptions for using Mn2+-Phos-tag gel for protein phosphorylation quantification (Ito et al., 2016; Sutherland & Walsh, 2012; Sutherland, MacDonald, & Walsh, 2016; Toepfer et al., 2013). However, published procedures include many variations in the experimental factors, and clearly there is no one-size-fits-all protocol, as summarized in Table 2. Hence, we present here a guide to optimize the factors required for success in Mn2+-Phos-tag polyacrylamide gel electrophoresis, along with a protocol that we have optimized to quantify protein phosphorylation levels from cardiomyocytes specifically.
| S/N | Title | Phos-tag conc. (µM) | Target MW (kDa) | Lysis buffer | Extraction procedure | Protein loaded/lane | Protein source | Resolving gel content | Electrophoresis current, duration | EDTA conc., duration | Blotting voltage, duration | Blotting, method, type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mobility shift detection of phosphorylation on large proteins using a Phos-tag SDS-PAGE gel strengthened with agarose (Kinoshita, Kinoshita-Kikuta, Ujihara, & Koike, 2009) | 20 | 200-350 | Laemmli buffer: 65 mM Tris·Cl (pH 6.8), 1.0% (w/v) SDS, 5.0% (v/v) 2-ME | NR (107 cells/0.5 ml) | 20 μg | HeLa cells | 3% (w/v) PAM, 0.5% (w/v) agarose | 15 mA, gel, 2 hr | 1 mM, 10 min | 4 V/cm, 16 hr | Wet transfer, PVDF |
| 2 | Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE (Kinoshita et al., 2009b) | 20 | >200 | Laemmli buffer: 1.0% (w/v) SDS and 5% (w/v) 2-ME | NR (107 cells/0.5 ml) | 20 μg | HeLa cells | 3% (w/v) PAM, 0.5% (w/v) agarose | 15 mA/gel, 2 hr | 1 mM, 10 min | 3.5 V/cm, 16 hr | Wet transfer, PVDF |
| 3 | Analysis of phosphorylation of the myosin-targeting subunit of myosin light chain phosphatase by Phos-tag SDS-PAGE (Sutherland et al., 2015) | 30 | 115-130 | Laemmli buffer: 65 mM Tris·Cl, pH 6.8, 2% (w/v) SDS, 100 mM DTT | Tissues were dehydrated and lyophilized. Dried tissues were immersed in 1 ml Laemmli buffer, heated to 95°C for 2 min, cooled to RT, and rotated O/N at 4°C. | 30 μl | Sprague-Dawley rats vascular smooth muscle tissue | 6% (w/v) acrylamide | 15 mA (ice bath), 3.5 hr | 2 mM, 15 min | 28 V (4°C), O/N | Wet transfer, NC |
| 4 | A highly sensitive technique to measure myosin regulatory light chain phosphorylation: the first quantification in renal arterioles (Takeya et al., 2008) | 30 | 20 | Laemmli buffer: 65 mM Tris·Cl, pH 6.8, 4% (w/v) SDS, 100 mM DTT | 20 μl Laemmli buffer/arteriole. Samples were subjected to constant shaking in a microcentrifuge tube for 2 hr at RT, sonicated 10 min, and heated to 95°C for 5 min before electrophoresis. | NR | Wistar rats renal afferent arterioles tissue | 9.7% (w/v) acrylamide | 20 mA, until BPB front leaves gel | 2 mM, 15 min | 27 V (4°C), O/N | Wet transfer, NC |
| 5 | Myosin regulatory light chain diphosphorylation slows relaxation of arterial smooth muscle (Sutherland & Walsh, 2012) | 50 | 20 | Laemmli buffer: 60 mM Tris·Cl, pH 6.8, 2% (w/v) SDS, 100 mM DTT | Tissues were immersed in cold 10% TCA, acetone, 10 mM DTT, washed three times (1 min each) with acetone/DTT, and lyophilized for 36 hr. Dried tissues were immersed in 1 ml of SDS gel sample buffer. Heated to 95°C for 2 min, cooled to RT and rotated O/N at 4°C. | 40 μl | Sprague-Dawley rat caudal arterial smooth muscle tissue | 12.5% acrylamide | 30 mA/gel, 70 min | 2 mM , 15 min (based on citation) | 27 V (4°C), O/N | Wet transfer, PVDF |
| 6 | Addition of urea and thiourea to electrophoresis sample buffer improves efficiency of protein extraction from TCA/acetone-treated smooth muscle tissues for Phos-tag SDS-PAGE (Takeya et al., 2018) | 40 | 20 | LDS buffer: 2% (w/v) LDS, 6 M urea, 2 M thiourea, 65 mM Tris·Cl (pH 6.8), 100 mM DTT, 10% glycerol, 0.01% (w/v) BPB | Muscles were treated in a series of buffers ending with TCA. 30 μl LDS buffer was added and samples were heated 5 min at 95°C. Proteins were then extracted by vigorous mixing on a vortex mixer for 2 hr at RT. Samples were centrifuged 5 min at 18,000 × g (to precipitate insoluble fraction) before electrophoresis. | NR | Mesenteric artery smooth muscle tissue | 11.6% (w/v) acrylamide, 0.4% (w/v) N,N-methylene bisacrylamide | 50 V, 30 min; then 140 V until BPB reached the bottom of gel | 5 mM, 30 min | 27 V (4°C), 16 hr | Wet transfer, PVDF |
| 7 | Determining the phosphorylation status of Hippo components YAP and TAZ using Phos-tag (Chen et al, 2019) | 14.9 | 44, 70 | Laemmli buffer: 50 mM Tris·Cl, pH 6.8, 2% SDS, 5% 2-ME | Cells harvested by adding 150 μl 1× SDS sample buffer per 12-well plate (70-80% confluency) and shaking on a plate shaker. Samples were then heated 5-10 min at 95°C, vortexed, and centrifuged. | 12 μl | Cultured cells | 7.5% PAM | 60-70 V, 30 min, until all samples entered Phos-tag separating gel; then 110-120 V until 55-kDa marker was ∼2 cm from bottom of gel | NR | ∼400 mA (4°C), 1.5-2 hr | Wet transfer, PVDF |
| 8 | Specific glutamic acid residues in targeted proteins induce exaggerated retardations in Phos-tag SDS-PAGE migration (Kinoshita et al., 2017) | 25 | 15, ∼46 | Hypotonic lysis buffer | Cells were washed with TBS solution (10 mM Tris·Cl, pH 7.5, 0.10 M NaCl) and collected in a conical tube. Hypotonic lysis buffer was used for protein extraction. | 1 μg | HeLa cells | 10%-16% (w/v) PAM | 30 mA/gel, RT, until BPB dye reached bottom of separating gel | NR | NR | NR |
| 9 | Phos-tag analysis of Rab10 phosphorylation by LRRK2: a powerful assay for assessing kinase function and inhibitors (Ito et al., 2016) | 75 | ∼23 | 4× Laemmli buffer: 250 mM Tris·Cl (pH 6.8), 8% (w/v) SDS, 4% (v/v) 2- ME | Cell/tissue lysates were mixed with 4 × SDS/PAGE sample buffer and heated 5 min at 95°C . | 10-30 µg | Mouse embryonic fibroblasts, lung and spleen-derived B-cells | 12% (w/v) acrylamide | 70 V for stacking part, 150 V for separating part | 10 mM, 30 min | 100 V, 180 min (on ice bath) | Wet transfer, NC |
| 10 | Phos-tag SDS-PAGE resolves agonist- and isoform-specific activation patterns for PKD2 and PKD3 in cardiomyocytes and cardiac fibroblasts (Qiu & Steinberg, 2016) | 5 | ∼80 | NR | NR | NR | Cardio-myocytes and fibroblasts isolated from Wistar rats | 6% (w/v) acrylamide | 80 V (4°C), 16 hr | 1 mM, 10 min | NR | PVDF |
| 11 | Detection of phosphorylated T and B cell antigen receptor species by Phos-tag SDS- and Blue Native-PAGE (Deswal et al., 2010) | 5.20, 100 | ∼17-20 | Cell lysis buffer: 20 mM Tris·Cl (pH 8), 10% glycerol, 10 mg/ml leupeptin, 10 mg/ml aprotinin, 1 mM PMSF, 500 μM Na3VO4, 1 mM NaF, 0.3% Brij96 (NaCl and EDTA excluded as they may functionally interfere with Phos-tag reagent) | Cells were lysed for protein extraction. TCR-CD3 complex were immunopurified and boiled in Laemmli buffer 5 min at 95°C. | NR |
Murine M.mζSBP T cells |
10% acrylamide | NR | 10 mM, 15 min | 1.6 mA, cm2, 2 hr | Semidry transfer, PVDF |
| 12 | Myosin regulatory light chain (RLC) phosphorylation change as a modulator of cardiac muscle contraction in disease (Toepfer et al., 2013) | NR | ∼19 |
SDS-PAGE sample buffer |
10 mg tissue was pulverized in dry ice, added to SDS-PAGE sample buffer, and heated 2 min at 95°C. | 10 µl | Left ventricular myocardium from Sprague-Dawley rats | 15% acrylamide | NR, 120 min | NR | NR, 60 min | Wet transfer, PVDF |
| 13 | The use of phosphate-affinity SDS-PAGE to measure the cardiac troponin I phosphorylation site distribution in human heart muscle (Messer et al., 2009) | 50 | ∼24 |
Laemmli buffer: 8 M urea, 2 M thiourea, 0.05 M Tris·Cl, pH 6.8, 75 mM DTT, 3% SDS, 0.05% BPB |
∼20-30 mg tissue was homogenized in 1 ml relax buffer (recipe in paper), then in rigor buffer and K-60 buffer, and dissolved in Laemmli buffer. | NR | Left ventricular human tissue | 10% acrylamide | NR |
1 mM, 10 min |
350 mA, 100 V, 60 min |
Wet transfer, PVDF |
- BPB, bromophenol blue; DTT, dithiothreitol; 2-ME, 2-mercaptoethanol; MW, molecular weight; NC, nitrocellulose; NR, not reported; O/N, overnight; PAM, polyacrylamide; PVDF, polyvinylidene fluoride; RT, room temperature; S/N, serial number.
STRATEGIC PLANNING
Initial Questions
To make it possible to perform successful Phos-tag SDS-PAGE, it is necessary to first answer the following questions:
- 1.What method of sample preparation will be used? This will depend on the type of samples used.
- 2.What polyacrylamide percentage will be used for the gels? This depends on the molecular weight of the protein to be resolved.
- 3.What Phos-tag concentration will be used for the gels? This will affect the separation of phosphorylated and unphosphorylated proteins, as well as subsequent transfer efficiency during immunoblotting.
- 4.How much protein will be added to each well? This is important so as to achieve an appropriate intensity for detection and measurement via western blotting. It is also important to avoid overloading wells, which could result in smearing.
- 5.What electrophoresis conditions (temperature, duration) will you use? This will affect the resolution of protein bands.
- 6.What conditions will be used for treatment of Phos-tag gels and polyvinylidene fluoride (PVDF) before western blotting? The conditions chosen are important as they affects protein transfer efficiency from Phos-tag gels to PVDF membranes.
- 7.Should SDS be added to the transfer buffer for immunoblotting? The inclusion of SDS in the transfer buffer has been suggested to increase transfer efficiency but may not be suitable for proteins of lower molecular sizes.
These questions are especially relevant to optimize the various parameters of concern summarized in Table 3.
| Steps | Processes to be optimized |
|---|---|
| Step 1: Sample preparation |
Frozen tissue pulverization method Protein extraction buffer |
| Step 2: Phos-tag gel preparation |
Percentage of polyacrylamide in resolving gel Protein amount to be loaded Phos-tag concentration |
| Step 3: Gel electrophoresis |
Electrophoresis condition Electrophoresis duration for optimal resolution |
| Step 4: Western blotting |
Treatment of gels and polyvinylidene fluoride membrane Transfer buffer Immunostaining and Imaging |
Step 1: Sample Preparation
Muscle pulverization
Although monolayer cells can be simply lysed by adding lysis buffer followed by gentle agitation, tissue samples, due to their intricacy, must be effectively pulverized by physical methods such as mechanical, sonication, and manual grinding methods. Though sonication reduces protein loss during the procedure, the proteins in the lysate are at risk of degradation or denaturation because of the high-energy sound waves and increase in temperature (Pchelintsev, Adams, & Nelson, 2016). Mechanical pulverization using blenders are not suitable for small volumes of muscles (e.g., a portion of the left ventricle from a mouse). On the other hand, pulverizing muscles using mortar and pestle may reduce protein yield due to the loss of muscle particles during transfer from the mortar to a tube. Each method possesses its own advantages and disadvantages. Ultimately, the final choice of pulverization method will depend on factors such as the cell type, the abundance of the protein of interest, and the stability of the proteins.
Manual pulverization carried out in the cold reduces damage to labile proteins. Snap-frozen muscle should not be allowed to thaw before manual grinding, in order to prevent repeated cycles of freeze-thaw that may produce ice-crystal stresses on the proteins, resulting in degradation (Cao, Chen, Cui, & Foster, 2003). Protein degradation results in multiple bands that will be detected on western blot images, making analysis complex or even inaccurate (Zilliges et al., 2011). A precalculated volume of lysis buffer should be added quickly to the pulverized muscle and the sample immediately placed in a 100°C heat block. This rapid heating of pulverized muscle particles in buffer denatures proteases and phosphatases that can alter protein post-translational modifications.
Lysis buffer for protein extraction
Appropriate buffers must be chosen based on the experimental design. Two commonly used lysis buffers are radioimmunoprecipitation assay and Laemmli buffer. Radioimmunoprecipitation assay buffer cannot be used for this method of phosphorylation analysis because of the incompatibility between ethylenediaminetetraacetic acid (EDTA) and Phos-tag gel: EDTA chelates Mn2+ ions in the gel, making the Phos-tag molecule ineffective and thus possibly leading to distorted (curved) protein bands on Phos-tag gels. On the other hand, Laemmli buffer is effective in extracting proteins, even of low abundance, from pulverized striated muscle tissues (Kinoshita, 2016).
Laemmli buffer—62.5 mM Tris, (pH 6.8), 2% SDS, 10% (v/v) glycerol, 50 mM dithiothreitol, 0.004% (w/v) bromophenol blue, 6.7 M urea—is commonly used as lysis buffer for endogenous protein extraction. There are slight variations in the buffer composition across different published papers (Table 2), but the above-mentioned mixture worked well for extracting insoluble proteins from skeletal and cardiac muscle and denaturing them to unfolded primary structures for optimal resolution of the protein bands on the gel (Cho et al., 2008).
Addition of a high concentration of urea (6-8 M) is crucial to unfold the hydrophobic proteins (Zangi, Zhou, & Berne, 2009). Lysate proteins experience carbamylation when heated with urea. However, studies have shown that carbamylation has no significant effect on Phos-tag gel results (Takeya, Kaneko, Miyazu, & Takai, 2018).
Also, the proteins must not be treated with phosphate-buffered saline (PBS) or any other chemicals containing phosphates as this may alter the phosphorylation level. In some studies, additives such as hypotonic solution have been used to facilitate cell lysis (Qi, Yang, & Chen, 2011). However, to extract proteins from muscle tissues for Phos-tag gel electrophoresis, Laemmli buffer works effectively.
Cell lysis and protein extraction
Again, there are variations in the ratios of lysis buffer volume to muscle mass reported (Table 2). Based on our experience, to successfully extract the low-abundance, insoluble proteins from left ventricular cardiac muscles, Laemmli buffer (abovementioned constituents) of a ratio of 20 µl per milligram of muscle is suitable. In practice, the total volume of solution prepared at this stage varied from 100 to 300 µl. After the samples have been heated in lysis buffer for 5 min, ensuring thorough denaturation of proteins, the lysate must be centrifuged at maximum speed (or 20,000 × g) for 2 min at room temperature to collect cell debris as a pellet, leaving the solubilized proteins in the supernatant. The centrifugation step is crucial to pellet nucleic acids and membranes (with high lipid content), as loading these into the gel can cause smearing of proteins during electrophoresis (Caprette, 1996a; Kurien & Scofield, 2012). Additional steps can be taken to shear the nucleic acid materials. However, this is not discussed here as the genetic material and lipid contents did not pose any problems with our protocol.
The protein concentration can be conveniently measured by colorimetry using a protein assay kit (Micro BCA Protein Assay Kit, Thermo Fisher Scientific). However, some colorimetry kits are incompatible with bromophenol blue, as it interferes with the output reading. In such situations, bromophenol blue should not be added before protein concentration measurement. It is a good practice to divide the proteins into smaller-volume aliquots and store them at –80°C. This minimizes freeze-thaw cycles and allows the samples to retain its form for a longer duration (Sitaramamma, Shivaji, & Rao, 1998).
Step 2: Phos-tag Polyacrylamide Gel Preparation
Optimizing polyacrylamide percentage
The percentage of polyacrylamide in the gel depends on the molecular weight of the protein to be resolved. Published protocols are useful for gauging a suitable polyacrylamide percentage. For example, protocols have recommended that resolving gels should comprise ∼15% polyacrylamide (a relatively high percentage, resulting in smaller pores in the gel matrix) for proper separation of low-molecular-weight proteins (19-25 kDa) and ∼8% for high-molecular-weight proteins (90-200 kDa; Abcam, 2020; Novus Biologicals, 2019a; Rath, Cunningham, & Deber, 2013).
The ratio of acrylamide to N,N ′-methylene bisacrylamide (the cross-linking agent) also affects the pore size. A smaller ratio results in a smaller pore size of the gel matrix. To resolve low-molecular-weight cardiac lysate proteins, gels made from a 30% (w/v) polyacrylamide stock solution with N,N ′-methylene bisacrylamide at a 29:1 ratio is suitable.
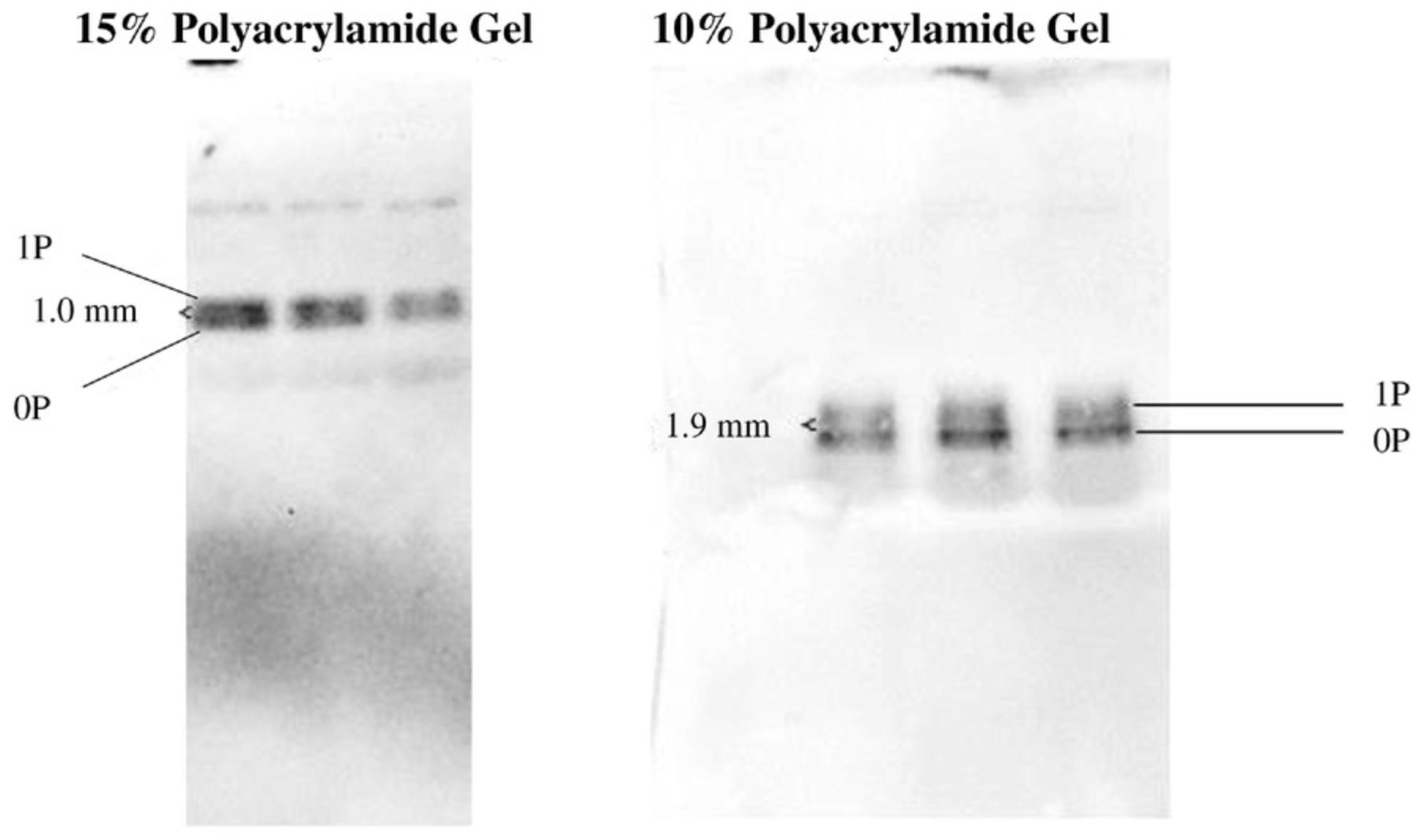
In our experiments, we observed that resolving proteins of the same molecular weight but of a different muscle type may require a different polyacrylamide concentration in the gel (Fig. 2). To be specific, a different polyacrylamide concentration was required to resolve and detect regulatory light-chain isoforms (∼19 kDa) from skeletal and cardiac muscle, respectively. A lower percentage of polyacrylamide (10%) was needed to resolve regulatory light chains from New Zealand white rabbit psoas muscle, whereas 15% worked well for resolving cardiac regulatory light chains from C57BL/6 mice (Fig. 2). Though the reason why a different percentage of polyacrylamide was needed for cardiac and skeletal isoforms is unclear, it is possibly due to tissue-specific protein sequences, species-related isoforms, or differences in intracellular ion interactions with the gel (Lindskog et al., 2015). Thus, obtaining the best protein resolution requires optimization of the polyacrylamide percentage adapted to each protein of interest.
Optimizing Phos-tag concentration
The concentration of Phos-tag molecules is important as it affects the separation of proteins and the transfer efficiency during western blotting.
Increasing Phos-tag concentration improves the separation between the non-phosphorylated and phosphorylated lysate proteins, but decreases the protein transfer efficiency, especially for phosphorylated proteins (Fig. 3). The same pattern was observed for purified recombinant regulatory light chains of Homo sapiens origin (Fig. 3). Thus, it is crucial to find a balance between Phos-tag concentration and transfer efficiency, as evidenced by the fact that such optimization has been described in other studies (Bekešová et al., 2015; Kinoshita-Kikuta, Kinoshita, Matsuda, & Koike, 2014). In our optimization, 25 µmol/L Phos-tag molecules in the resolving gel was found to be sufficient to separate the striated muscle lysate proteins of molecular weights between 18 and 150 kDa from their phosphorylated counterparts with an acceptable transfer efficiency (data not shown).
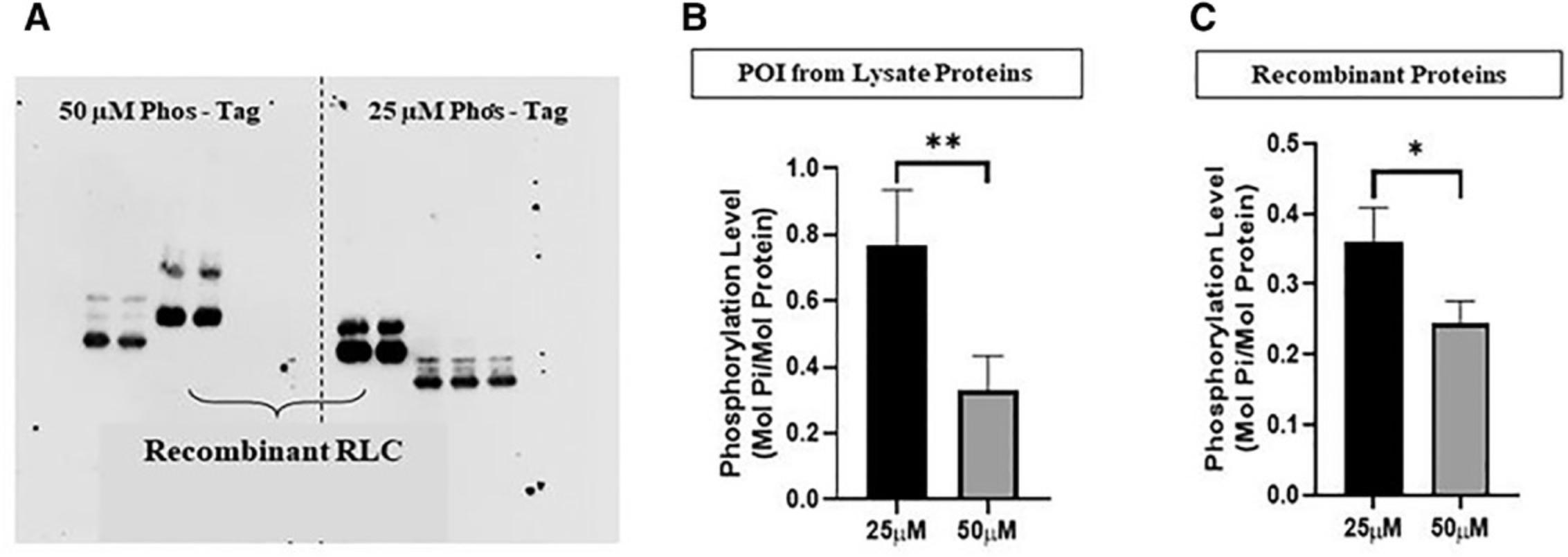
On a separate note, the difference in the molecular weights between the recombinant (Homo sapiens origin) and lysate regulatory light chains (Mus musculus origin) in Figure 3 may be attributable to the His-tag component in the recombinant proteins or the differences in the nucleotide sequences (Zhao, 2016).
Optimizing protein amount to be loaded
The amount of protein loaded per well is a key factor for effective resolution of protein bands and for achieving an appropriate intensity for detection and measurement via western blotting.
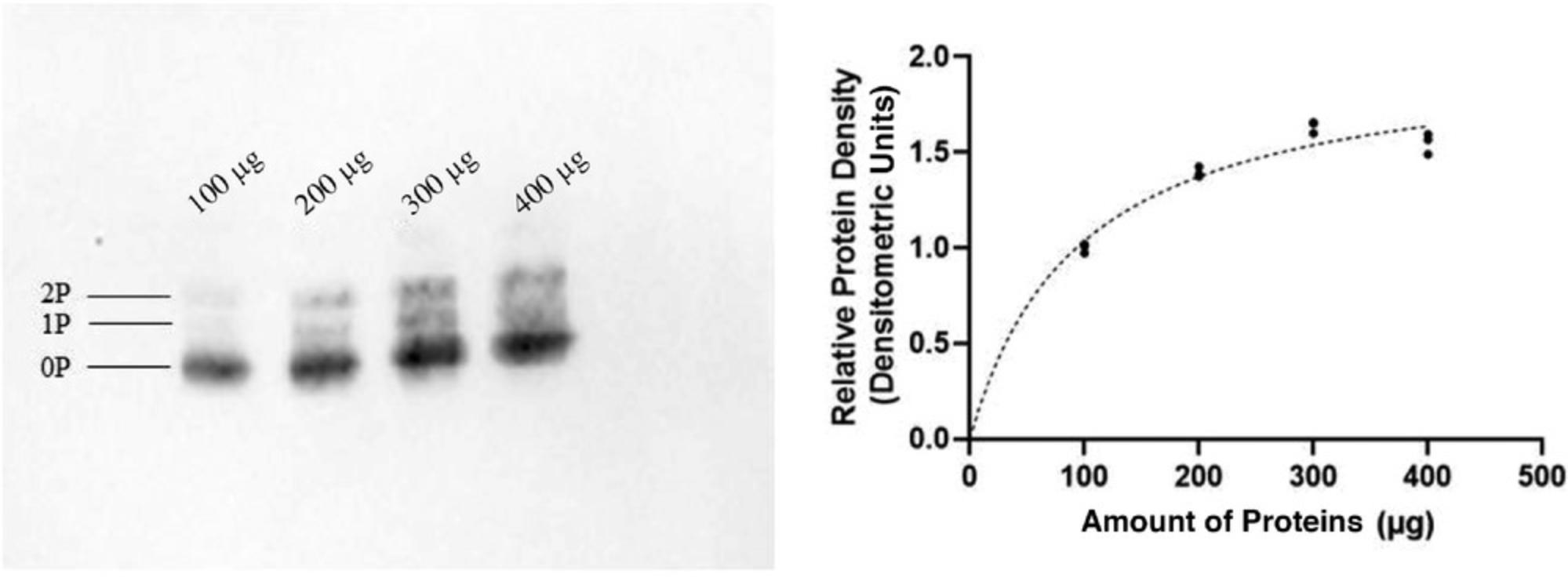
Protein-overloaded wells result in gels with smeared proteins (Caprette, 1996a). Scientific manuals recommend a total load of protein from muscle extract of ∼0.5 µg protein per band for the gel dimensions described above (1-mm-thick gels; Caprette, 1996b). However, the number of protein bands resolved from a crude lysate varies among tissues of different species, and the abundance of individual proteins varies. Thus, protein amount optimization is required to answer different research questions. This can be done by loading a range of protein amounts and then determining the best amount to use through visual interpretation and software detection.
We used such an optimization procedure to elucidate the amount of lysate protein to be added per well for the detection of regulatory light chains, which we determined to be 300 µg (Fig. 4). The antibody signal tends to drop as the amount of protein increases beyond a certain value (Fig. 4). This could arise if, when overloaded proteins are transferred, the antibodies bind only to the surface layer of the proteins, resulting in the collection of underestimate results (Taylor, Berkelman, Yadav, & Hammond, 2013).
Step 3: Phos-tag Gel Electrophoresis
Electrophoretic conditions
Heating during electrophoresis leads to diffused, distorted, or poorly resolved protein bands. Gel temperature is affected by room temperature, electrophoresis voltage, number of gels in a tank, and electrophoresis duration. Thus, a balance among these factors is needed to maintain appropriate gel temperature. The temperature of the running buffer during gel electrophoresis should be kept below 25°C for optimal lysate protein resolution. “Smiling” bands can occur when gels experience excess or uneven distribution of heat (Fig. 5; Anonymous, 2017).
Gel electrophoresis can be conducted in the cold room to reduce the effect of heat on the gel. However, during protocol optimization, we found that the separation was not distinct even after 3 hr of gel electrophoresis at 4°C (Fig. 5), as low temperatures decrease the conductivity of the electrolytes in the buffer, decreasing the rate of protein migration (Rogacs & Santiago, 2013). As a result, the protein bands became compact, with poor resolution. Poor resolution of proteins can also occur due to protein diffusion as a result of prolonged gel electrophoresis.
From our experience, it is best to subject only one Phos-tag gel to electrophoresis per tank to reduce gel heating, as more gels result in more electric current flow. If it is necessary to run two gels at once, it is advisable to wrap the tank with a wet towel to conduct the heat away and maintain the temperature below 25°C to prevent protein distortion (Anonymous, 2017).
Our experience and that of others converges on the use of a constant voltage (V) in the range of 140-160 V at room temperature to resolve lower-molecular-weight proteins (Copeland et al., 2010; Kampourakis, Sun, & Irving, 2016; Nagy, Comer, & Smolenski, 2018). As the voltage remains constant, the current decreases during electrophoresis as the resistance (R) increases due to the depletion of electrolytes in the buffer (Kelly, Altria, & Clark, 1997). The power (P), the product of current and voltage, is proportional to the heat generated. This is the heat that must be dissipated by conduction to the running buffer and that is then dissipated to the ambient air. Given the relation P = V 2/R , as resistance increases under constant voltage conditions, power gradually decreases, reducing the heating of the gel over the course of electrophoresis. The higher the electrophoresis voltage, the number of gels, and the electrophoresis duration, the greater the heat dissipated. Use of a constant voltage limits heating of the buffer and gel.
Electrophoresis duration
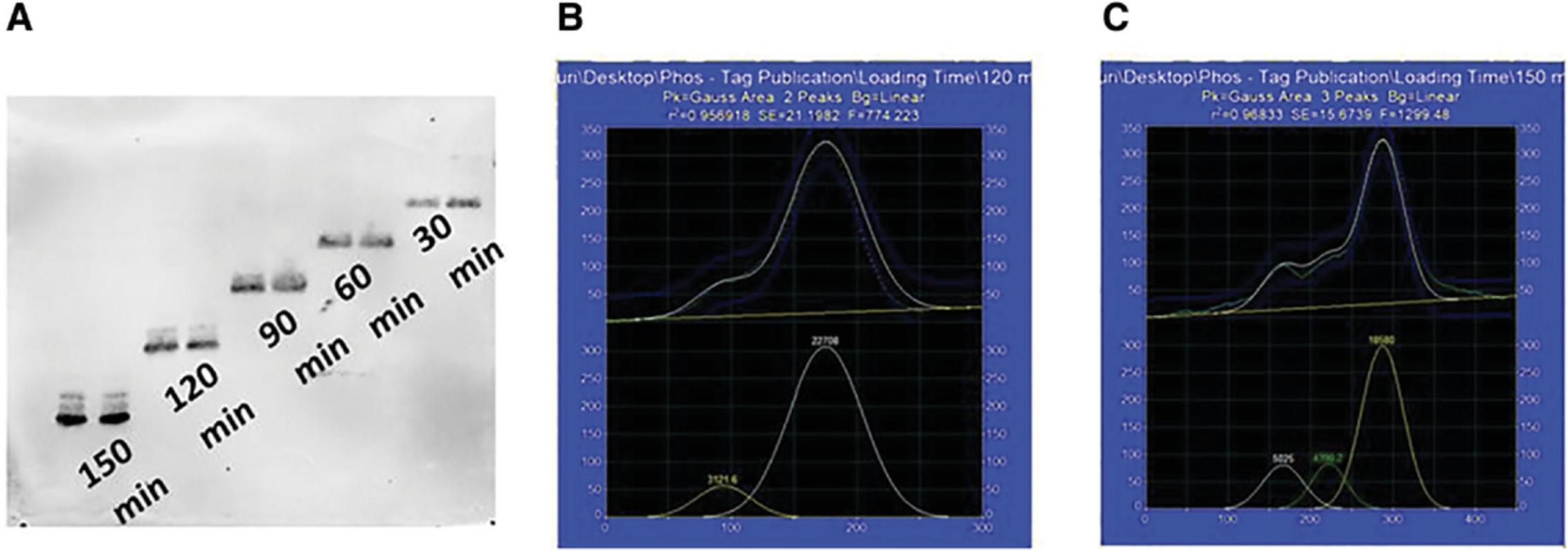
Even though it is ideal to keep the duration of electrophoresis short to reduce heating and protein diffusion, it must be sufficient to clearly resolve the proteins. Optimization experiments are required to determine the shortest possible duration of electrophoresis for optimal protein resolution. This involves loading equal amount of proteins into the lanes at different time-points such that the proteins resolve for varying length of time on the same gel.
There are two steps to determine the ideal duration for optimal protein resolution. First, a visual observation for obvious separation of protein bands (based on the possible number of phosphorylation sites) must be conducted on the scanned blots. Second, quantitative measurement of the scanned peaks, for example using PeakFit v4.12 software, gives objective measures of resolution. The software calculates peak parameters according to peak-fitting constraints. The densitometric pixel values can be fitted to a normal distribution (Gaussian) curve at a 95% confidence interval with the PeakFit software. A good fit can be verified based on the coefficient of determination (r 2) value.
Our optimization experiment identified the ideal duration for resolution of low-molecular-weight proteins to be 150 min (Fig. 6). This was determined from the clear resolution of protein bands and the r 2 value (calculated by PeakFit) near 1 (r 2 = 0.96l Fig. 6).
Step 4: Western Blotting
The western blotting procedure presented here was optimized based on published protocols to achieve good protein transfer efficiency from Phos-tag gels to PVDF membrane (Hycult Biotech, 2010; Mahmood & Yang, 2012) and protein detection.
Treatment of gels and PVDF membrane
Our experience and that of others has established that phosphorylated proteins do not readily transfer to PVDF membrane, possibly because they bind to the Phos-tag molecules. Treating the gel with EDTA to chelate the Mn2+ ions in the Phos-tag metal ion complex improved the transfer efficiency of phosphorylated proteins (Kinoshita-Kikuta et al., 2014). According to the literature, a range of 1-10 mM EDTA is generally used to treat the gels of similar size. The gel is then washed with EDTA-free transfer buffer to remove any excess EDTA present. The ideal duration of the washes is dependent on the thickness of the gel. For 1.0-mm-thick Phos-tag gels, 30-min washes with 10 mM EDTA followed by 30 min without EDTA were suitable. Additional details are presented in the supplementary section (Kinoshita et al., 2009b; Komis, Takáč, Bekešová, Vadovič, & Samaj, 2014). It is important to minimize the duration of washes to avoid protein diffusion or protein migrating out of the gel.
Before protein transfer is performed, the hydrophobic PVDF membrane must be treated with 100% methanol to displace the air trapped in the membrane, which makes it much less hydrophobic (Roche Laboratories, 2019). After this, the membrane is swirled for a few seconds in transfer buffer to remove excess methanol. This allows water-based transfer buffers to diffuse into the membrane and proteins to be adsorbed onto it. When the gel and membrane are ready, they are tightly sandwiched in the cassette holder to remove all air bubbles between the gel and membrane and ensure close contact, which facilitates protein transfer. The PVDF membrane comes in different pore sizes. Thus, a membrane of appropriate pore size must be chosen depending on the molecular weight of the protein of interest. For proteins between 18 and 150 kDa, PVDF membranes with a pore size of 0.2 µm are suitable.
Transfer buffer
The addition of SDS to the transfer buffer increases the ionic strength and thus conductivity of the buffer, which improves protein transfer. However, whether to include SDS must be decided based on the molecular weight of the protein of interest and the polyacrylamide and Phos-tag concentrations in the gel. Generally, SDS is recommended for the transfer of proteins with a large molecular weight, as they migrate out of the gel very slowly. Also, larger proteins tend to precipitate, and the presence of SDS reduces precipitation (Abcam, n.d.).
The binding affinity between the dinuclear manganese (II) complex and phenyl phosphate di-anion in aqueous solution has not been reported. Assuming it is similar to that for Zn2+ complex binding to the phenyl phosphate di-anion, the binding affinity would be ∼ K d = 2.5 × 10−8 M under neutral pH. The high binding affinity and compromised protein transfer efficiency from Phos-tag gels prompted the exploration of adding SDS in the transfer buffer (Kinoshita et al., 2009b; Kinoshita, Kinoshita-Kikuta, & Koike, 2007; Kinoshita-Kikuta et al., 2014).
Our experiments showed that the amount of proteins detected in blots transferred with 0.08% (w/v) SDS is significantly lower than in the absence of SDS (Fig. 7). Results of previous studies involving non-Phos-tag gels support these experimental data, emphasizing that the addition of SDS is essential to increase transfer efficiency of only high molecular weight proteins, as smaller proteins (∼19 kDa) easily migrate out of the gel to be adsorbed on the PVDF membrane even in the absence of SDS. The addition of SDS leads the proteins to acquire additional negative charges, which could cause them to migrate through the positively charged PVDF membrane, resulting in the loss of proteins (Novus Biologicals, 2019b; Thermo Fisher Scientific, n.d.). Smaller proteins are not retained on the blot even at low concentrations of SDS in the transfer buffer (Abcam, n.d.; Bolt & Mahoney, 1997) and thus can be eliminated.
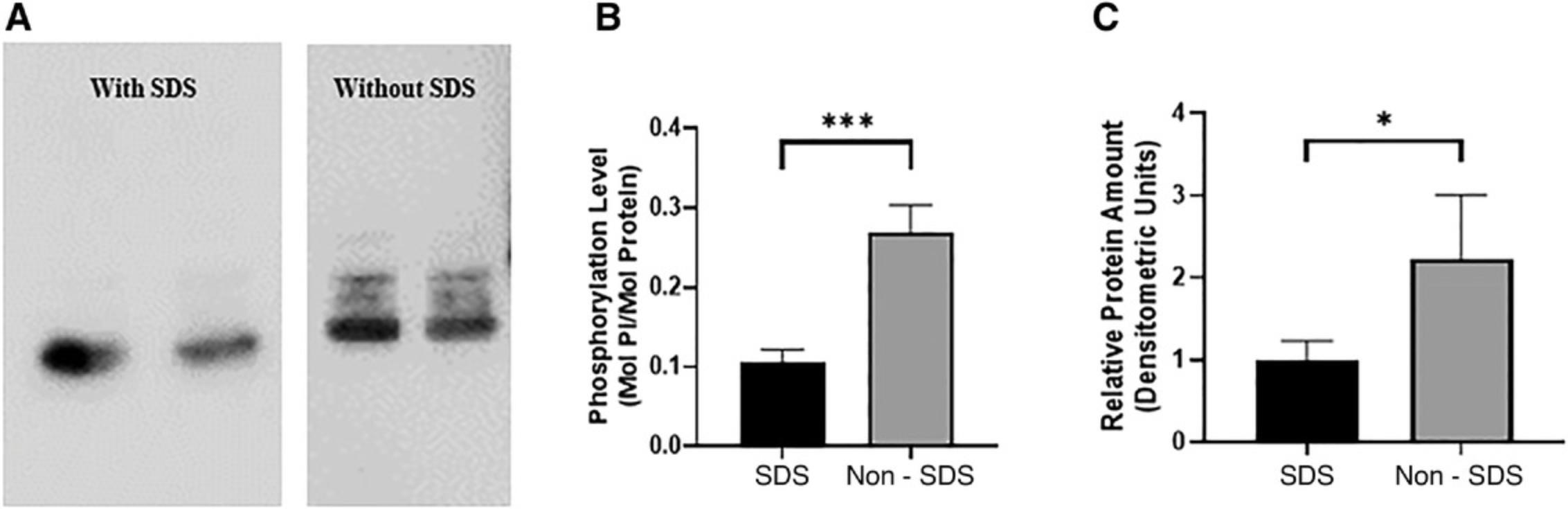
Methanol present in the transfer buffer removes SDS (from lysis buffer) bound to the proteins and swells the gel. This promotes protein binding to the PVDF membrane and the migration of proteins out of the gel, respectively. However, methanol also precipitates proteins and changes the charges of basic proteins to neutral or positively charged, which can inhibit efficient protein transfer to the PVDF membrane. Thus, methanol should be limited to 20% of the buffer. Methanol should be omitted from buffers used to transfer large-molecular-weight proteins (Abcam, 2020; LI-COR, 2020).
The pH of the transfer buffer, which is temperature dependent, also affects protein transfer to the PVDF membrane. Thus, if the transfer is taking place in the cold room (∼4°C), the pH of the buffer must be adjusted for that temperature, as Tris buffers have high temperature sensitivity (AppliChem, 2008). Also, it is important for the pH to be close to 8.3, which is higher than the isoelectric point of most proteins, so that proteins carry a net negative charge and migrate toward the anode (positively charged electrode) for adsorption on the positively charged PVDF membrane (Egger & Bienz, 1994).
Fixation of proteins on PVDF
Subsequent treatment of PVDF membranes with buffers containing Tween 20 (addressed in the next section) may strip proteins, reducing accuracy in the quantification of phosphorylation. To reduce the loss of proteins, fixation of proteins with glutaraldehyde treatment of the PVDF membranes after blotting can be introduced. Although we do not explore the differences arising from including versus omitting such a protein fixation step, this has been successfully used in other protocols and is worth considering (Fiesel, Hudec, & Springer, 2016; Takeya et al., 2018; Takeya, Loutzenhiser, Shiraishi, Loutzenhiser, & Walsh, 2008). The fixation is typically done before blocking of the blotted membrane, using 0.25%-0.5% glutaraldehyde for 20-45 min with shaking, and is followed by a washing step. Depending on the abundance of the protein of interest, this step must be optimized accordingly.
Immunostaining
5% bovine serum albumin (BSA) dissolved in Tris-buffered saline/2% Tween 20 buffer can be used to block all potential antibody binding sites (antigens) of proteins on the PVDF membrane. This prevents nonspecific binding of antibodies and thus increases the signal-to-noise ratio. When antibodies are added, they displace BSA and bind to their respective antigen of interest as they have a higher affinity for the binding sites than for BSA (Jensen, 2012). The recommended dilution factor for antibodies is usually provided in the product specifications. Using the range provided as a guide, an optimization procedure can be performed to determine the most appropriate dilution factor to detect the protein of interest (data not shown) and avoid signal saturation, which would be deleterious to quantitative analysis. The membranes are washed in Tris-buffered saline/2% Tween 20 to remove excess antibodies.
Secondary antibodies with different types of labels (for example, Alexa Fluor and horseradish peroxidase) are used to observe the protein of interest, depending on the availability of instrumentation for quantitative assays. Here, secondary antibodies conjugated with horseradish peroxidase were used together with chemiluminescent reagents to visualize the protein bands.
Imaging and quantification
As mentioned in the previous section, chemiluminescent reagents are useful for visualizing protein bands on blots as the reagents become oxidized by horseradish peroxidase. The oxidation emits a signal (light) that is detected with the Bio-Rad ChemiDoc MP-Imaging system and captured using the ImageLab5.2 software. Commercially available chemiluminescent reagents eact with proteins even in the picogram and femtogram concentration ranges. To obtain results that can be compared from membrane to membrane, the scanning duration must be kept constant among the different blots.
If film-based methods are used to detect the chemiluminescent signals, the films must be scanned using transmission mode instead of reflection mode to capture an image for densitometric analysis. That is, the light source and detector must be on the opposite sides of the scanner. Also, the scanner must provide a linear intensity response. These two factors are essential to obtain accurate densitometric values. Thus, office scanners are not reliable for scanning and interpreting the results from film-based methods (Tiago Ferreira, 2012). Again, to obtain results that can be compared between blots, the duration of exposure of the blots to the films must be kept constant.
To ensure reproducibility among different blots, the image-processing steps must be done the same way for each blot image. The area (aspect ratio) of the region of interest analyzed must be equal, as the densitometric values are computed from the product of the region of interest area and the gray values of all pixels (Tiago Ferreira, 2012). In our analysis, the only modification done to the image was to convert it to an 8-bit image using ImageJ before measuring the area density of bands.
Attention to the parameters described above, together with systematic optimization processes, will improve the usability and reliability of Phos-tag gels for phosphorylation studies.
SAFETY CONSIDERATIONS
Chemicals
CAUTION : The monomer of polyacrylamide used for gel casting is probably carcinogenic and can result in peripheral neuropathy. It is important to handle polyacrylamides in a fume hood while wearing personal protective equipment such as goggles, nitrile gloves, and lab coats. In the case of splashes, immediately replace gloves. The polymerized gel is not toxic and can be disposed of in biosafety bags.
Hydrochloric acid and sodium hydroxide solutions of high concentrations may be used to alter the pH of various solutions. Methanol is also used to activate PVDF membranes and in the transfer buffer. Spillage of solutions containing these chemicals can result in skin irritation and organ damage. Furthermore, they produce fumes that can be dangerous upon inhalation. Care should be taken to handle these chemicals with personal protective equipment in a well-ventilated area such as fume hoods.
Experimental waste (e.g., used chemicals) must be disposed of appropriately according to institution's policies.
Electrical Safety
When performing electrophoresis and western blotting, the equipment is able to supply up to 300 V, which can result in lethal shocks. Do not use other plugs or banana jacks which may not be safety compliant for use with the power supply. Ensure that the power supply is connected to a grounded three-prong AC outlet. When handling the power supply, do not grasp both voltage leads at the same time. Conduct frequent checks of the equipment to ensure that there are no exposed wires. When removing gels or blots after runs, ensure that the power supply is turned off beforehand.
Risk of Burns
Heating must be performed before protein samples are loaded and during gel staining. Temperatures can reach as high as 100°C, which can result in burns. Care must be taken when handling any heated samples and equipment as there is a possibility of burns.
Basic Protocol: Mn2+-PhOS-TAG GEL ELECTROPHORESIS TO RESOLVE PHOSPHORYLATED PROTEINS FROM CARDIOMYOCYTES
By considering the parameters that have been discussed in this paper, we have optimized a Phos-tag protocol for detecting phosphorylated low-molecular-weight (∼20 kDa) proteins from C57BL/6 mouse cardiac tissues. Although this protocol has been written specifically for cardiac tissues, it can be easily optimized for proteins from other tissue sources by taking into consideration the factors presented in Table 4.
| Day 0 (sample preparation) | ||
|---|---|---|
| Step | Key optimized conditions | Timea |
| Muscle pulverization |
Mechanical pulverization using mortar and pestle for small volumes of muscle tissue. Rapid heating of pulverized particles on 100°C heat block for 5 min to ensure thorough denaturation of proteins. |
Dependent on the number of samples to process |
| Protein extraction |
Avoid any treatment with PBS or phosphate-containing chemicals. Laemmli buffer is suitable and effective for muscle tissues (use 20 µl/mg for muscles). Centrifugation of lysate at maximum speed (or 20,000 × g) for 2 min at room temperature to remove nucleic acids and membranes as pellet to prevent smearing of proteins. |
|
| Day 1 (total duration: ∼9.5 hr) | ||
| Step | Key optimized conditions | Time |
| Gel casting |
Polyacrylamide percentage of 10%-15%. Phos-tag concentration of 25 µM in resolving gel to balance resolution of separation and transfer efficiency. Lysate proteins loaded per well of 20-300 µg, depending on protein abundance, for sufficient signal without overloading. |
Preparations for casting gel: 30 min Casting resolving and stack gels: 135 min |
| Gel electrophoresis and preparation for western blotting |
Ideally only one gel at a time should be run in each electrophoresis tank. If two must be gels in a single tank, ensure it is wrapped with wet towels throughout. 140-160 V constant voltage to limit heating of buffer and gel. Electrophoresis duration of 150 min. Gel treatment with EDTA to chelate Mn2+ ions for increased transfer efficiency. Subsequent washes needed to remove EDTA. |
Running gel: 150 min Gel treatment and washes: 70 min |
| Western blotting and primary antibody incubation |
Polyvinylidene fluoride membrane of pore size 0.2 µM suitable for proteins of 15-150 kDa Transfer buffer without sodium dodecyl sulfate for smaller proteins (∼19 kDa). Methanol limited to 20% of transfer buffer, and eliminated for high-molecular-weight proteins. Blocking with 5% BSA in Tris-buffered saline/2% Tween 20 buffer. Optimize dilution factor for antibodies based on range specified in product specification. |
Western blotting: 60 min Blocking: 60 min Buffer washing time: 60 min Primary antibody incubation: Overnight or ≥14 hr |
| Day 2 (total duration: ∼3 hr) | ||
| Step | Key optimized conditions | Time |
| Washes and secondary antibody incubation | Use secondary antibodies with appropriate labels based on instruments available in the laboratory (e.g., secondary antibodies conjugated with horseradish peroxide with chemiluminescent reagents). |
Washes post primary antibody: 30 min Secondary antibody incubation: 60 min Washes post secondary antibody: 30 min |
| Staining and incubation |
For film-based methods, transmission mode must be used to detect chemiluminescent signals. Scanners must provide linear intensity response. Scanning duration and area of the region of interest analyzed must be equal for comparison between membranes. |
60 min (including a 30-min buffer time) |
- a
Timing may vary depending on the number of gels and membranes handled. This timing is applicable when experimenting with one gel and membrane.
Materials
-
70% (v/v) ethanol
-
Milli-Q deionized water (0.55 nS/cm conductivity)
-
30% (w/v) 29:1 acrylamide/bisacrylamide solution
-
1.5 M Tris/0.4% SDS, pH 8.8 (see recipe)
-
10% (w/v) SDS
-
10 mM MnCl2
-
5 mM Phos-tag in 3% (v/v) methanol
-
Tetramethylethylenediamine (TEMED)
-
10% (w/v) ammonium persulfate (see recipe)
-
100% ethanol
-
0.5 M Tris/0.4% SDS, pH 6.8 (see recipe)
-
1× electrophoresis running buffer (see recipe)
-
Protein sample extracted for analysis
-
Protein ladder
-
1× loading buffer (Laemmli buffer; see recipe)
-
100% all-purpose bleach
-
1× transfer buffer (see recipe)
-
1× transfer buffer with EDTA (see recipe)
-
Primary antibodies (e.g., regulatory light chain–targeted antibodies, Abcam ab92721) and secondary antibodies
-
1× Tris-buffered saline/2% Tween 20 (see recipe)
-
1× blocking buffer (see recipe)
-
Chemiluminescent reagent (e.g., Bio-Rad 1705061)
-
Blotting paper
-
Electrophoresis equipment (e.g., using Bio-Rad Mini-PROTEAN vertical electrophoresis cell module and Bio-Rad PowerPac Basic Electrophoresis Power Supply, assembled with short plates, spacer plates, integrated spacers, cell bugger dams, combs, casting stand gaskets, casting stand, casting frame, buffer tank, cell lid with power cables, and gel holder cassette)
-
Western blotting equipment: foam pads, thick blot filter paper, central core
-
Imaging system (such as ChemiDoc MP from Bio-Rad)
-
100°C heating block, for denaturing pulverized proteins
-
Microwave
-
Software: ImageLab 5.2 (Bio-Rad), PeakFit v.4.12 (SeaSolve Software), and ImageJ 1.51j8 (National Institutes of Health, USA)
NOTE : Besides the abovementioned companies, electrophoresis and imaging equipment is available from other suppliers such as Thermo Fisher, Bio-Rad, Analytik-Jena, Amersham, or Licor.
Day 1: Gel casting
1.Clean short and spacer plates thoroughly with 70% ethanol and then with deionized water (Milli-Q; 0.55 nS/cm conductivity). Using clean plates reduces the risk of distortions in the protein bands.
2.Assemble the plates on a casting stand and fill with deionized water to the brim. Check that the glass plates are assembled properly. Improper assembly results in water leakage and the level of water in between the glass plates gradually drops.
3.Once the gel casting setup is ready, prepare the resolving gel with 25 µM Phos-tag, as shown in Table 5.
| Component | Volume |
|---|---|
| Deionized water | 1.08 ml |
| 30% (w/v) 29:1 acrylamide/bisacrylamide solution | 2.475 ml |
| 1.5 M Tris/0.4% SDS, pH 8.8 | 1.25 ml |
| 10% SDS | 50 µl |
| 10 mM MnCl2 | 25 µl |
| 5 mM Phos-tag in 3% (v/v) methanol | 25 µl |
| Tetramethylethylenediamine | 7.15 µl |
| 10% ammonium persulfate | 35.75 µl |
4.Add the resolving gel constituents and ensure they are thoroughly mixed before pouring 4.5 ml into the gel casting setup. Add 100% ethanol to the gel casting setup to the brim and gently shake the gel cast from side to side, making sure that the denser gel mixture settles and forms a straight front line. Leave the gel to polymerize for 90 min. Tip: The ratio of Phos-tag concentration to MnCl2 concentration is 1:2, as 2 mol of Mn2+ ions bind to 1 mol of Phos-tag molecule.
5.About 75 min into the polymerization of the resolving gel, prepare the stacking gel, as shown in Table 6.A stacking gel of ∼25 mm depth is a gel of low acrylamide content into which proteins migrate rapidly. It improves the sharpness of the protein bands.
| Component | Volume |
|---|---|
| Deionized water | 1.5 ml |
| 30% (w/v) 29:1 acrylamide/bisacrylamide solution | 0.35 ml |
| 0.5 M Tris/0.4% SDS, pH 6.8 | 0.625 ml |
| Tetramethylethylenediamine | 10 µl |
| 10% ammonium persulfate | 12.5 µl |
6.Decant the ethanol, and rinse the gel front with deionized water three times. Remove as much water as possible by tilting the casting stand, and absorb the small amount of water remaining on the edges using a clean tissue.
7.Pour the stacking gel solution to the brim of the plates and insert a clean 15-well comb into the gap. Leave the stacking gel to polymerize for 45 min.
8.While waiting for polymerization, clean the tanks and tank lid for electrophoresis and prepare the electrophoresis running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS; see recipe). Thaw the aliquoted protein samples on ice.
9.After the 45 min are over, carefully remove the gel comb from the stacking gel, and rinse the top of the gel with water to remove bubbles and any impurities.
10.Position the plates in the vertical electrophoresis apparatus and place them into the electrophoresis tank. Gently pour the electrophoresis running buffer into the tank up till the recommended level marked on it.
Day 1: Gel electrophoresis
11.Load the proteins and protein ladder for calibrating molecular weights into the wells. Empty wells must be loaded with loading buffer (Laemmli buffer) to prevent inter-lane buffer concentration gradients, which may lead to distorted protein bands.
12.Set the PowerPac™ Basic Power Supply to operate for 2 hr 30 min at 140 V.
13.After electrophoresis, transfer the gel to a container containing 15 ml of transfer buffer with 10 mM EDTA. Agitate the gel for 15 min on a seesaw rocker at 25 rpm to remove as many Mn2+ ions as possible. Repeat this step once more.
14.Wash the gel with transfer buffer for 15 min at 25 rpm on a seesaw rocker. Repeat this step once more.
15.Ten minutes before the end of the gel wash in transfer buffer, prepare the equipment for western blotting. Two bleach-cleaned containers are required to contain methanol and transfer buffer, respectively. Thorough washing of the transfer tanks, cassette, foam pad and filter paper, roller, and forceps with deionized water is necessary.
Day 1: Western blotting
16.The PVDF membrane is shielded from dirt or impurities by being sandwiched between two protective papers. Cut the membrane (while it is still in between protective papers) to an appropriate size such that it fully covers the protein band region of the gel.
17.Assemble the clean cassette, foam pads, and wet filter paper as shown in Figure 8.
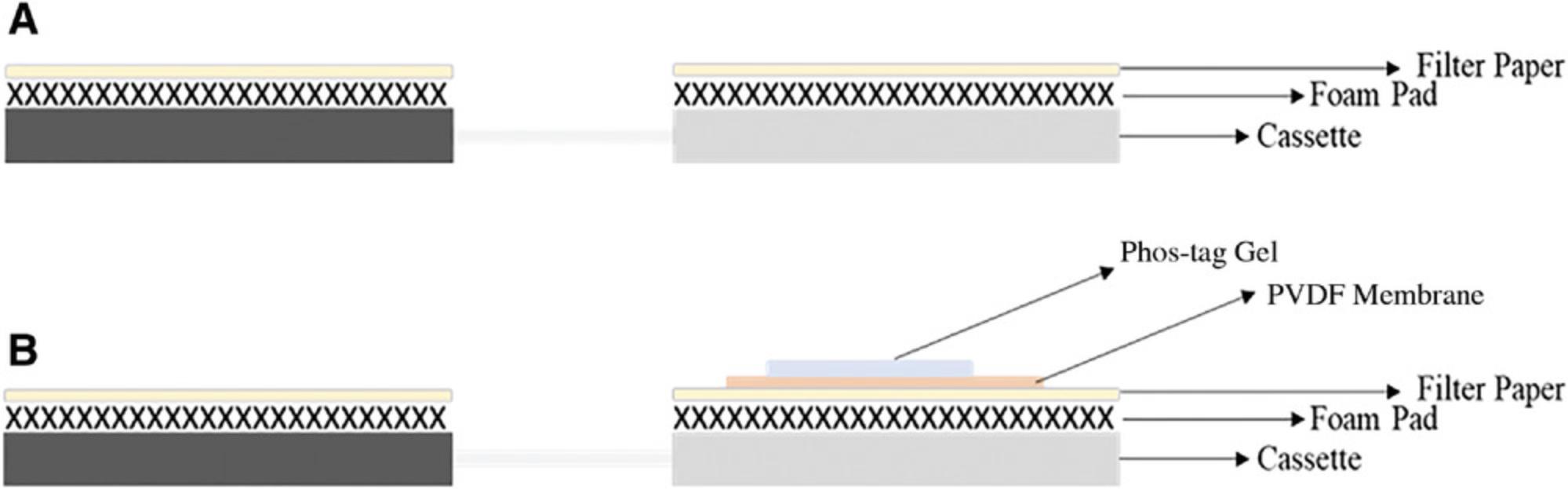
18.When the gel washes (step 14) are completed, activate the PVDF membrane by submerging it in 100% methanol for 20 s. Gently swirl the container to ensure that the air pockets in the membrane are fully displaced.
19.Transfer the activated membrane to the container containing transfer buffer to remove methanol from the membrane pockets, as methanol may inhibit the elution of proteins from the gel and/or adhesion of proteins to the membrane. This can be achieved by gently swirling the container.
20.Place the PVDF membrane on filter paper on the transparent/grey part of the cassette.
21.Place the gel on the PVDF membrane and roll out any air bubbles again. The setup will look like the schematic diagram in Figure 8.
22.Carefully bring together the two sides of the cassette and clamp them together. A tight clamp will increase the contact between the gel and membrane and release trapped air bubbles, if any.
23.The orientation of the cassette inserted into the electrode assembly is crucial as it affects the direction of protein transfer. For the Bio-Rad assembly, the correct orientation is achieved when the black portion of the cassette faces the black wall (cathode) of the electrode assembly.
24.After setting up the tank, electrode assembly, and cassette, place an ice pack in the tank and quickly pour the transfer buffer into the tank to prevent the PVDF membrane from drying. Ensure that the transfer buffer is adjusted to pH 8.3 at 4°C prior to the blotting step.
25.Transport the setup to the cold room (∼4°C) and place the buffer tank lid. Adjust the settings of the Bio-Rad PowerPac Basic to run at a constant current of 400 mA for 60 min to start the blotting process.
26.After the proteins are blotted into the PVDF membrane, transfer the membrane to a bleached container containing blocking buffer. Block the membrane for an hour at room temperature on a seesaw rocker at a speed of 25-30 rpm.
27.Dilute primary antibodies in blocking buffer just before use. Prepare a total volume of 10 ml of diluted antibodies in a 50-ml centrifuge tube (6250× dilution for regulatory light chain–targeted antibodies, Abcam ab92721). This volume is required to fully cover the membrane to ensure that it remains hydrated.
28.Roll the PVDF membrane and place it in the 50-ml centrifuge tube containing the primary antibodies. Then incubate it on a rod-rotating shaker at 20 rpm in the cold room (∼4°C) overnight or for at least 14 hr.
Day 2: Washes and secondary antibody incubation
29.Add Tris-buffered saline/2% Tween 20 to one of the bleached containers (from the day before) to cover the membrane entirely.
30.Transfer the PVDF membrane incubated in primary antibody to the container containing Tris-buffered saline/2% Tween 20.Wash the membrane on a see-saw rocker at 70 rpm for 10 min. Repeat this step twice more to remove as much nonspecific primary antibody binding as possible.
31.During the last 10 min of the third wash, add the secondary antibodies. They are reconstituted in a total of 10 ml of blocking buffer (1429× dilution) in a 50-ml centrifuge tube. Incubate the membrane as for the primary antibodies (step 28), but for 1 hr at room temperature.
32.Wash the membrane with Tris-buffered saline/2% Tween 20 on a see-saw rocker at 70 rpm for 10 min. Repeat this step twice more to remove as much nonspecific secondary antibody binding as possible.
Day 2: Staining and imaging
33.After the washes, rinse the membrane in water, submerge it in the chemiluminescent reagent (contained in a bleached container) for 10 s, and immediately image using the Bio-Rad ChemiDoc MP Imaging system.
34.Capture the image and export it in both TIFF and JPEG format using the ImageLab5.2 software.
35.Measure phosphorylation from the images using ImageJ and peak-fitting software such as PeakFit v4.12.
REAGENTS AND SOLUTIONS
Ammonium persulfate, 10%
- 1 g ammonium persulfate
- 10 ml water
CAUTION : 10% (w/v) ammonium persulfate must be prepared on the day of experiment (no more than 24 hr before use; store at 4°C once prepared).
Blocking buffer, 1×
- 1× Tris-buffered saline (see recipe), pH 8.1
- 2% Tween 20
- 5% bovine serum albumin (BSA)
- Stable for at least 1 month when stored at 4°C.
Electrophoresis running buffer, 10×
- 250 mM Tris⋅Cl, pH 8.1
- 1.92 M glycine
- 10% (w/v) sodium dodecyl sulfate (SDS)
- Store at room temperature; functional for at least 6 months.
Laemmli buffer, 1× (loading buffer)
- 62.5 mM Tris⋅Cl, pH 6.8
- 2% (w/v) SDS
- 10% (v/v) glycerol
- 50 mM dithiothreitol
- 0.004% (w/v) bromophenol blue
- 6.7 M urea
- Store at room temperature; stable for only ∼10 hr
CAUTION : Because reducing agents such as dithiothreitol have a short half-life due to oxidation in air, that must be added to the buffer minutes before the protein extraction steps to optimally reduce the disulfide bonds between cysteine residues.
MnCl2, 10 mM
- 125.84 mg MnCl2
- Dilute to 100 ml with water
- Store at room temperature; stable for at least 6 months
Transfer buffer with EDTA, 1×
- 25 mM Tris, pH 8.3
- 192 mM glycine
- 20% (v/v) methanol
- 10 mM ethylenediaminetetraacetic acid (EDTA)
- Store at room temperature; stable for at least 2 months
Transfer buffer, 1×
- 25 mM Tris, pH 8.3
- 192 mM glycine
- 20% (v/v) methanol
- Store at room temperature; stable for at least 2 months
0.5 M Tris/0.4% SDS, pH 6.8
- 30.285 g Tris base
- 2 g SDS
- Dilute to 500 ml with water and adjust pH to 6.8
- Store at room temperature; stable for at least 6 months
1.5 M Tris/0.4% SDS, pH 8.8
- 90.855 g Tris base
- 2 g SDS
- Dilute to 500 ml with water and adjust pH to 8.8
- Store at room temperature; stable for at least 6 months
Tris-buffered saline
- 20 mM Tris, pH 8.1
- 150 mM NaCl
- Store at room temperature; stable for at least 1 month
Tris-buffered saline/2% Tween 20, 1×
- 20 mM Tris, pH 8.1
- 150 mM NaCl
- 2% Tween 20
- Store at room temperature; stable for at least 1 month
COMMENTARY
Critical Parameters and Troubleshooting
Variations in the protocol are inevitable due to the sensitivity of Phos-tag gels in response to different samples being used. Hence, this article was written to attend to these variations and enable scientists to easily follow the guidelines to optimize a protocol so as to answer their unique scientific questions.
Table 7 presents a list of problems that may arise with this procedure along with their possible causes and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Distortion of bands | Blank lanes leading to a concentration gradient causing diffusion | Add equal volume of Laemmli buffer into the blank lanes to eliminate the concentration gradient. |
| Smearing of bands (“smiling”- shaped bands) | Uneven heat distribution or excess heat | Ensure that only one gel is run in each electrophoresis tank at a time. If it is necessary to run more than one gel at a time, wrap the electrophoresis tank with wet towels and replace at least once during the entire run to remove excess heat. |
| Poor transfer to membrane | Poor contact between the gel and membranes | Ensure that there are no air bubbles between the gel and membrane before western blotting. |
| Mn2+ ions in the Phos-tag metal ion complex reducing the transfer efficiency of phosphorylated proteins | Slightly increase the duration of EDTA washes to chelate Mn2+ ions. | |
| Poor resolution | Gel- or electrophoresis-related problems | Perform optimization of (i) polyacrylamide concentration, (ii) Phos-tag concentration, and (iii) electrophoresis conditions, as detailed in Strategic Planning. |
Understanding Results
Phos-tag gel electrophoresis detects low-abundance phosphoproteins at high throughput and hence enables rapid data collection. It is relatively low in cost compared to tools such as Phospho-ELISA kits. The dynamic changes in proteins’ phosphorylation levels during disease progression or in response to various conditions can be identified through longitudinal studies using Phos-tag gel to map out potential pathophysiological pathways. For example, Mn2+ Phos-tag gels were used to detect and analyze multiple phosphorylatable residues of myosin targeting subunit 1 of myosin light chain phosphatase (a key subfragment regulatory protein) from rat caudal arterial smooth muscle, under varying chemical perturbations, as shown in Figure 9 (Sutherland et al., 2016).
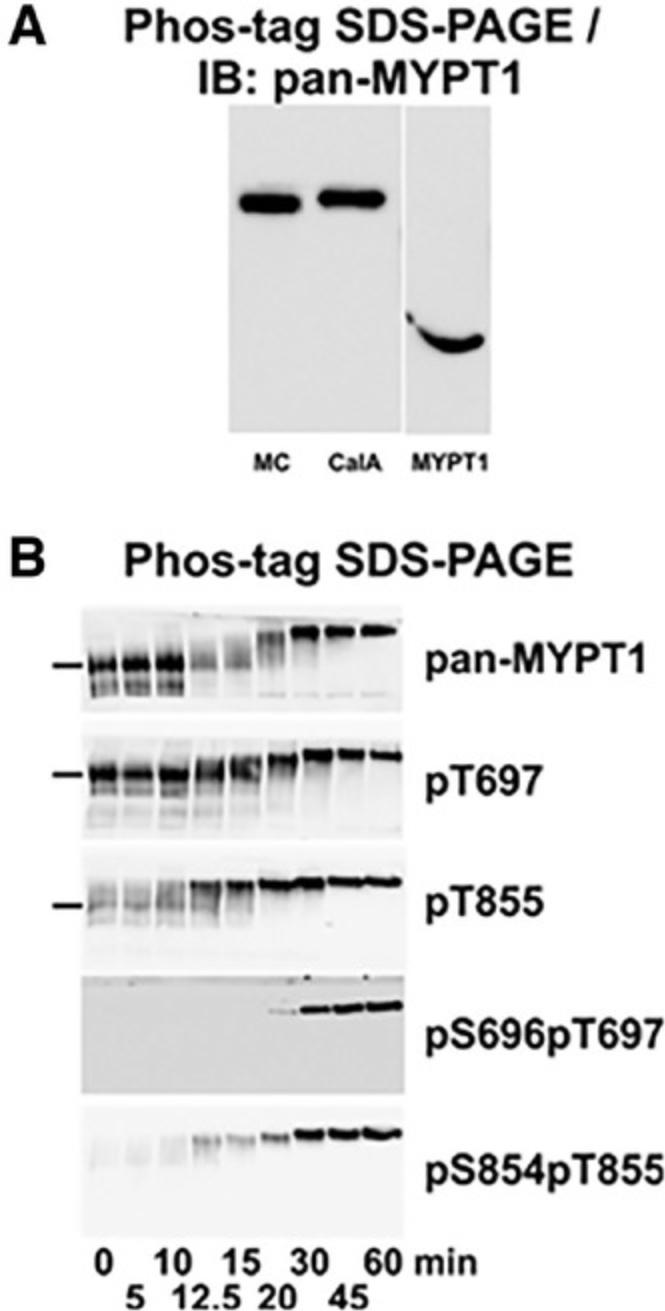
Kinase/phosphatase activity (kinomics) plays an important role in pathological mechanisms. Phos-tag gels are also useful for uncovering activities of these enzymes during disease progression (Barbieri & Stock, 2008; Hosokawa, Saito, Asada, Fukunaga, & Hisanaga, 2010; Ito et al., 2016; Kinoshita et al., 2009a). Phos-tag gels enable the identification of novel or altered signaling pathways, which can lead to the identification of therapeutic targets for diseases (Berard, Kroeker, McQueen, & Coombs, 2018).
Total phosphorylation level
The key data that can be obtained from Phos-tag gels are total phosphorylation levels. Phosphorylated proteins band(s) appear above unphosphorylated proteins due to their slower rates of migration. It is also reported that Phos-tag polyacrylamide gels are able to separate proteins based on the phosphorylation site, rather than the overall molecular weight (Kinoshita et al., 2008).
In proteins with multiple phosphorylation sites, more than two bands will be seen due to the presence of multiple possible phosphorylation states (mono- and diphosphorylated proteins, etc.). In some images, the separation of protein bands is not very distinct, but curve fitting (PeakFitv4.12) provides the ability to detect the density of individual bands and to reliably measure protein abundance.
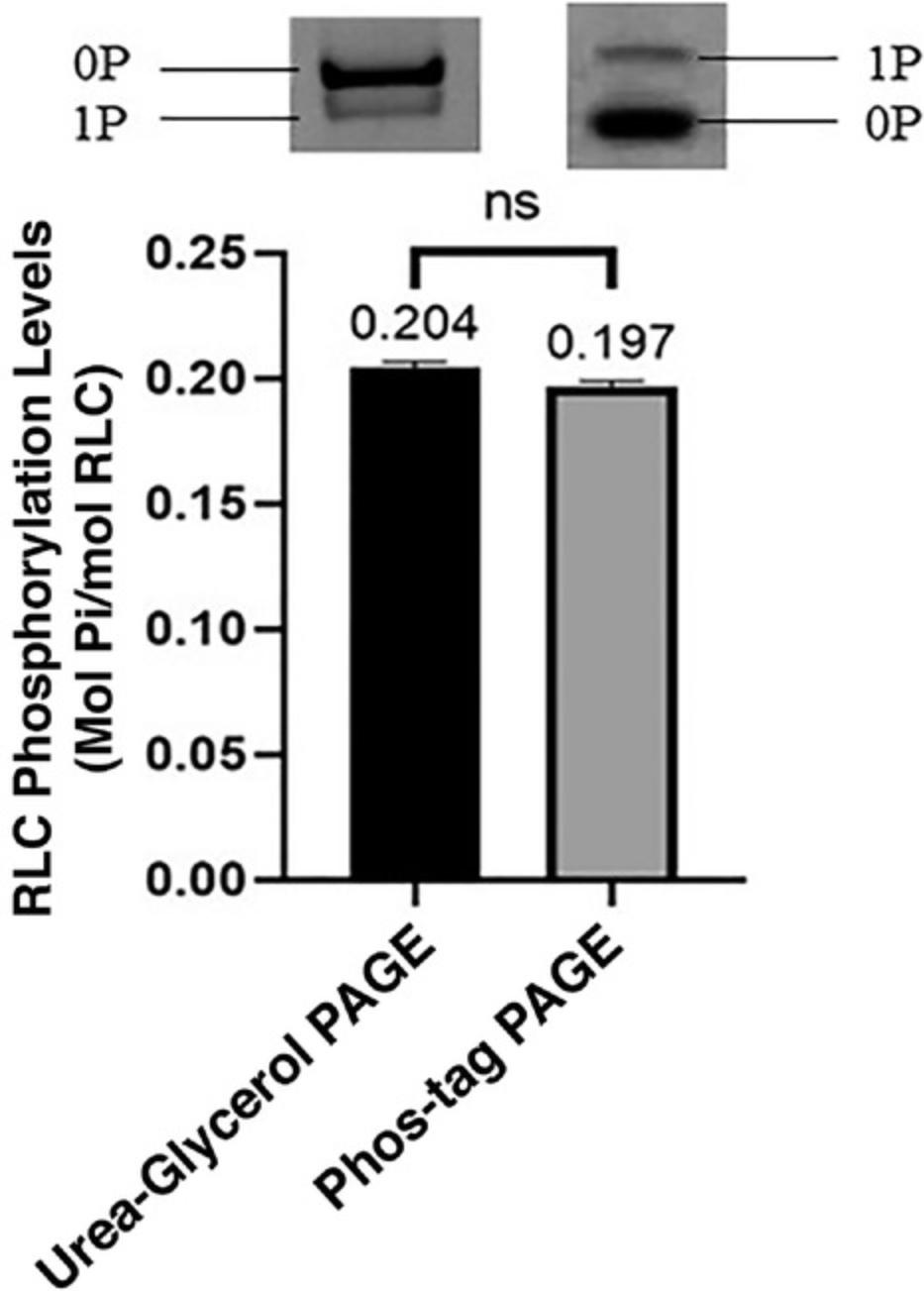
To quantify phosphorylation levels, calculate the ratio of the intensity of the sum of phosphorylated bands (mono- [represented as 1P] and/or diphosphorylated [represented as 2P] proteins) to the total proteins (unphosphorylated [lowest band] + phosphorylated bands). The intensity of the protein bands are calculated based on pixel values, given in arbitrary units, by ImageJ. A higher value implies greater intensity. The steps to calculate phosphorylation levels of proteins are shown below. A working example with mouse cardiac regulatory light chains (RLC) resolved with 50 µM Phos-tag (Fig. 3) is presented below. The formula to calculate phosphorylation level is:
=(intensityof2P∗2)+intensityof1Pintensityof2P+1P+0P=(4476.3∗2)+2139.5)4476.3+2139.5+26367=0.34molPi/molRLC
Phosphorylation levels calculated using Phos-tag gels, and urea-glycerol polyacrylamide gels (the traditional method for resolving phosphorylated proteins from their counterparts) showed no significant differences, suggesting that Phos-tag gels are as reliable as urea-glycerol gels (Fig. 10). Urea-glycerol and Phos-tag gels separate proteins using different mechanisms, causing phosphorylated proteins to appear differently in faster and slower migration bands, respectively. In Phos-tag gels, the binding of SDS to proteins occurs at a consistent charge-to-mass ratio. This masks the intrinsic charge of proteins, allowing the separation of proteins by weight (Gallagher, 2012) and causing in the tagged phosphorylated proteins to appear to have a higher molecular weight and migrate more slowly than their unphosphorylated counterparts (Kinoshita et al., 2009a). In urea-glycerol gels, phosphorylated and unphosphorylated myosin light-chain species are separated based on their charge differences at the pH value of the buffer (Perrie, Smillie, & Perry, 1973), where the more negatively charged proteins migrate faster (Persechini, Kamm, & Stull, 1986). Our experimental conditions allowed for faster migration of the phosphorylated proteins.
PeakFit v4.12
PeakFit is a software that semi-automatically resolves protein bands on gel or blot images read by ImageJ. The coordinates of intensity peaks generated by ImageJ are obtained via the macro function codes. The coordinates can be saved as a MS-DOS file to run compatibly in PeakFit software. Constraints can be adjusted in PeakFit to fit the peaks as accurately as possible. The macro function codes (https://forum.image.sc/t/x-and-y-duplicates-when-exporting-coordinates-of-an-outline/7420) are provided below:
h = getHeight(); run("Make Binary"); run("Skeletonize"); run("Invert LUT"); doWand(0, 0.5*h); getSelectionCoordinates(x, y); tabDelText = "" for (i = 0; i < ×.length; i++) {tabDelText += "" + ×[i] + "\t" + y[i] + "\n";} path = getDirectory("Choose a Directory"); File.saveString(tabDelText, path + "myTabDelText.txt");
Further research
The principle behind the separation of phosphoproteins on the basis of their phosphorylation sites is unclear, but it may be related to the effect that gives rise to different mobilities for cardiac and skeletal regulatory light chains, as mentioned earlier. Further experiments should be designed to uncover the cause of such results. This will expand the use of Phos-tag polyacrylamide gels.
Alternatively, 2D-Phos-tag gel electrophoresis (isoelectric focusing + Phos-tag gel electrophoresis) separates phosphoproteins isotypes better. The blots need to be stained with different primary and secondary antibodies to detect the proteins phosphorylated at specific sites. The gels can also be analyzed using mass spectrometry to verify the phosphoisoproteins. This strategy has been applied in a previous study to determine the phosphorylation sites of intracellular β-catenin (Kinoshita, Kinoshita-Kikuta, & Koike, 2012).
Acknowledgments
The work presented herein was supported by the Singapore Ministry of Education under its Academic Research Fund Tier 2 (Project No. MOE2016-T2-1-106). The APC was funded by the Ministry of Education Research Funding. The authors would like to thank the Research Administration and Support Services of Lee Kong Chian School of Medicine and Professor Philip Ingham, FRS, for their support in this publication.
Author Contributions
Kasturi Markandran : Conceptualization, data curation, formal analysis, investigation, methodology, validation, writing—original draft, writing—review and editing; Jane Vanetta Lee En Xuan : data curation, investigation, validation, writing—original draft, writing—review and editing; Haiyang Yu : supervision; Lim Meng Shun Darren : project administration, supervision; Michael A. Ferenczi : funding acquisition, supervision, writing—original draft, writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Abcam. (2020). Preparation of PAGE gels. Electrophoresis for western blot. Retrieved from https://www.abcam.com/protocols/electrophoresis-for-western-blot.
- Abcam. (n.d.). Transfer and staining of proteins in western blot. Retrieved from https://www.abcam.com/protocols/transfer-and-staining-of-proteins-in-western-blot.
- Anonymous, A. (2017). Electrophoresis. Retrieved from https://openwetware.org/wiki/Electrophoresis.
- Aponte, A. M., Phillips, D., Harris, R. A., Blinova, K., French, S., Johnson, D. T., & Balaban, R. S. (2009). 32P labeling of protein phosphorylation and metabolite association in the mitochondria matrix. Methods in Enzymology , 457, 63–80. doi: 10.1016/S0076-6879(09)05004-6.
- AppliChem. (2008). Biological BuffersAppliChem. Retrieved from https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.221/attachments/BioBuffer.pdf.
- Barbieri, C. M., & Stock, A. M. (2008). Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag- based reagents. Analytical Biochemistry , 376(1), 73–82. doi: 10.1016/j.ab.2008.02.004.
- Bekešová, S., Komis, G., Křenek, P., Vyplelová, P., Ovečka, M., Luptovčiak, I., … Šamaj, J. (2015). Monitoring protein phosphorylation by acrylamide pendant Phos-Tag™ in various plants. Frontiers in Plant Science , 6, 336. doi: 10.3389/fpls.2015.00336.
- Berard, A., Kroeker, A., McQueen, P., & Coombs, K. M. (2018). Methods and approaches to disease mechanisms using systems kinomics. Synthetic and Systems Biotechnology , 3(1), 34–43. doi: 10.1016/j.synbio.2017.12.004.
- Bolt, M. W., & Mahoney, P. A. (1997). High-efficiency blotting of proteins of diverse sizes following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Analytical Biochemistry , 247(2), 185–192. doi: 10.1006/abio.1997.2061.
- Bryan, P. M., Smirnov, D., Smolenski, A., Feil, S., Feil, R., Hofmann, F., … Potter, L. R. (2006). A sensitive method for determining the phosphorylation status of natriuretic peptide receptors: cGK-Iα does not regulate NPR-A. Biochemistry , 45(4), 1295–1303. doi: 10.1021/bi051253d.
- Cao, E., Chen, Y., Cui, Z., & Foster, P. R. (2003). Effect of freezing and thawing rates on denaturation of proteins in aqueous solutions. Biotechnology and Bioengineering , 82(6), 684–690. doi: 10.1002/bit.10612.
- Caprette, D. R. (1996a). SDS-PAGE "Hall of Shame". Experimental Biosciences. Retrieved from https://www.ruf.rice.edu/~bioslabs/studies/sds-page/sdsgoofs.html.
- Caprette, D. R. (1996b). Preparing protein samples for electrophoresis. Experimental Bioscience. Retrieved from https://www.ruf.rice.edu/~bioslabs/studies/sds-page/denature.html.
- Chen, R., Plouffe, S. W., & Guan, K.-L. (2019). Determining the phosphorylation status of Hippo components YAP and TAZ using Phos-tag. Methods in Molecular Biology , 1893, 281–287. https://doi.org/10.1007/978-1-4939-8910-2_21.
- Cho, J.-H., Hwang, H., Cho, M.-H., Kwon, Y.-K., Jeon, J.-S., Bhoo, S. H., & Hahn, T.-R. (2008). The effect of DTT in protein preparations for proteomic analysis: Removal of a highly abundant plant enzyme, ribulose bisphosphate carboxylase/oxygenase. Journal of Plant Biology , 51, 297–301. doi: 10.1007/BF03036130.
- Cieśla, J., Frączyk, T., & Rode, W. (2011). Phosphorylation of basic amino acid residues in proteins: Important but easily missed. Acta Biochimica Polonica , 58(2), 137–148. doi: 10.18388/abp.2011_2258.
- Cohen, P. (2001). The role of protein phosphorylation in human health and disease. European Journal of Biochemistry , 268(19), 5001–5010. doi: 10.1046/j.0014-2956.2001.02473.x.
- Colburn, J., Michnoff, C., Hsu, L., Slaughter, C. A., Kamm, K., & Stull, J. (1988). Sites phosphorylated in myosin light chain in contracting smooth muscle. Journal of Biological Chemistry , 263(35), 19166–19173.
- Copeland, O'.N., Sadayappan, S., Messera, A. E., Steinen, G. J. M., van der Velden, J., & Marston, S. B. (2010). Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. Journal of Molecular and Cellular Cardiology , 49(6), 1003–1011. doi: 10.1016/j.yjmcc.2010.09.007.
- Deswal, S., Beck-García, K., Blumenthal, B., Dopfer, E. P., & Schamel, W. W. Detection of phosphorylated T and B cell antigen receptor species by Phos-tag SDS- and Blue Native-PAGE. Immunology Letters , 130(1-2), 51–56. https://doi.org/10.1016/j.imlet.2009.12.012.
- Egger, D., & Bienz, K. (1994). Protein (western) blotting. Molecular Biotechnology , 1, 289–305. doi: 10.1007/BF02921696.
- Fiesel, F. C., Hudec, R., & Springer, W. (2016). Non-radioactive in vitro PINK1 kinase assays using ubiquitin or parkin as substrate. Bio-protocol , 6(19), e1946. doi: 10.21769/BioProtoc.1946.
- Gallagher, S. R. (2012). SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Current Protocols Essential Laboratory Techniques , 6(1), 7–3.
- Goshe, M. B. (2006). Characterizing phosphoproteins and phosphoproteomes using mass spectrometry. Briefings in Functional Genomics , 4(4), 363–376. doi: 10.1093/bfgp/eli007.
- Heinrich, R., Neel, B. G., & Rapoport, T. A. (2002). Mathematical models of protein kinase signal transduction. Molecular Cell , 9(5), 957–970. doi: 10.1016/S1097-2765(02)00528-2.
- Hosokawa, T., Saito, T., Asada, A., Fukunaga, K., & Hisanaga, S. (2010). Quantitative measurement of in vivo phosphorylation states of Cdk5 activator p35 by Phos-tag SDS-PAGE. Molecular & and Cellular Proteomics, 9(6), 1133–1143. doi: 10.1074/mcp.M900578-MCP200.
- Hunter, T. (2012). Why nature chose phosphate to modify proteins. Philosophical Transactions of The Royal Society , 367, 2513–2516. doi: 10.1098/rstb.2012.0013.
- Hycult Biotech. (2010). Western blotting. Retrieved from https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.221/attachments/Protocol_Western_Blotting.pdf.
- Ito, G., Katsemonova, K., Tonelli, F., Lis, P., Baptista, M. A., Shpiro, N., … Alessi, D. R. (2016). Phos-tag analysis of Rab10 phosphorylation by LRRK2: A powerful assay for assessing kinase function and inhibitors. The Biochemical Journal , 473(17), 2671–2685. doi: 10.1042/BCJ20160557.
- Jczernik, A., Girault, J.-A., Nairn, A. C., Chen, J., Snyder, G., Kebabian, J., & Greengard, P. (1991). Production of phosphorylation state-specific antibodies. Methods in Enzymology , 201, 264–283.
- Jensen, E. C. (2012). The basics of western blotting. The Anatomical Record , 295(3), 369–371. doi: 10.1002/ar.22424.
- Johnson, L. N. (2009). The regulation of protein phosphorylation. Biochemical Society Transactions , 37(4), 627–641. doi: 10.1042/BST0370627.
- Kampourakis, T., Sun, Y-B., & Irving, M. (2016). Myosin light chain phosphorylation enhances contraction of heart muscle via structural changes in both thick and thin filaments. Proceedings of the National Academy of Sciences of the United States of America , 113(21), E3039–E3047. doi: 10.1073/pnas.1602776113.
- Kelly, M. A., Altria, K. D., & Clark, B. J. (1997). Approaches used in the reduction of the buffer electrolysis effects for routine capillary electrophoresis procedures in pharmaceutical analysis. Journal of Chromatography A , 768(1), 73–80. doi: 10.1016/S0021-9673(97)00054-X.
- Kinoshita, E. (2016). Phos-tag™ SDS-PAGE guidebook. doi: 10.13140/RG.2.2.11535.18084.
- Kinoshita, E., Kinoshita-Kikuta, E., & Koike, T. (2007). Specific recognition and detection of phosphorylated proteins using characteristics of metal ions. Yakugaku Zasshi: Journal of the Pharmaceutical Society of Japan , 127(12), 1897–1913. doi: 10.1248/yakushi.127.1897.
- Kinoshita, E., Kinoshita-Kikuta, E., & Koike, T. (2009a). Phosphate-affinity gel electrophoresis using a Phos-tag molecule for phosphoproteome study. Current Proteomics , 6(2), 104–121. doi: 10.2174/157016409788680965.
- Kinoshita, E., Kinoshita-Kikuta, E., & Koike, T. (2009b). Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nature Protocols , 4(10), 1513–1521. doi: 10.1038/nprot.2009.154.
- Kinoshita, E., Kinoshita-Kikuta, E., & Koike, T. (2012). Phos-tag affinity electrophoresis for protein kinase profiling. In Protein kinase technologies (pp. 13–34). Totowa, NJ: Humana Press.
- Kinoshita, E., Kinoshita-Kikuta, E., Matsubara, M., Yamada, S., Nakamura, H., Shiro, Y., … Koike, T. (2008). Separation of phosphoprotein isotypes having the same number of phosphate groups using phosphate-affinity SDS- PAGE. Proteomics , 8(15), 2994–3003. doi: 10.1002/pmic.200800243.
- Kinoshita, E., Kinoshita-Kikuta, E., Ujihara, H., & Koike, T. (2009). Mobility shift detection of phosphorylation on large proteins using a Phos-tag SDS-PAGE gel strengthened with agarose. Proteomics , 9(16), 4098–4101. https://doi.org/10.1002/pmic.200900020.
- Kinoshita-Kikuta, E., Kinoshita, E., & Koike, T. (2009). Phos-tag beads as an immunoblotting enhancer for selective detection of phosphoproteins in cell lysates. Analytical Biochemistry , 389(1), 83–85. doi: 10.1016/j.ab.2009.03.026.
- Kinoshita-Kikuta, E., Kinoshita, E., Matsuda, A., & Koike, T. (2014). Tips on improving the efficiency of electrotransfer of target proteins from Phos-tag SDS-PAGE gel. Proteomics , 14(21-22), 2437–2442. doi: 10.1002/pmic.201400380.
- Kinoshita, E., Kinoshita-Kikuta, E., Karata, K., Kawano, T., Nishiyama, A., … Koike, T. (2017). Specific glutamic acid residues in targeted proteins induce exaggerated retardations in Phos-tag SDS-PAGE migration. Electrophoresis , 38(8), 1139–1146. https://doi.org/10.1002/elps.201600520.
- Komis, G., Takáč, T., Bekešová, S., Vadovič, P., & Samaj, J. (2014). Affinity-based SDS PAGE identification of phosphorylated Arabidopsis MAPKs and substrates by acrylamide pendant Phos-Tag™ (Vol. 1171). Totowa, NJ: Humana Press.
- Kristjánsdóttir, K., & Rudolph, J. (2004). Cdc25 phosphatases and cancer. Chemistry & Biology, 11(8), 1043–1051. doi: 10.1016/j.chembiol.2004.07.007.
- Kurien, B., & Scofield, R. (2012). Common artifacts and mistakes made in electrophoresis. Methods in Molecular Biology , 869, 633–640. doi: 10.1007/978-1-61779-821-4_58.
- LI-COR, Inc. (2020). Transfer options. Optimizing wet tank transfer. Retrieved from https://www.licor.com/bio/guide/westerns/transfer_options.
- Lima, R., Wada, S., Tanaka, S., Takeda, M., Ishikawa, T., Tsubota, K-i., … Yamaguchi, T. (2008). In vitro blood flow in a rectangular PDMS microchannel: Experimental observations using a confocal micro-PIV system. Biomedical Microdevices , 10(2), 153–167. doi: 10.1007/s10544-007-9121-z.
- Lindskog, C., Linné, J., Fagerberg, L., Hallström, B. M., Sundberg, C. J., Lindholm, M., … Uhlén, M. (2015). The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody-based profiling. BMC Genomics , 16(1), 475. doi: 10.1186/s12864-015-1686-y.
- Mahmood, T., & Yang, P.-C. (2012). Western blot: Technique, theory, and trouble shooting. North American Journal of Medical Sciences , 4(9), 429–434. doi: 10.4103/1947-2714.100998.
- Messer, A. E., Gallon, C. E., McKenna, W. J., Dos Remedios, C. G., & Marston, S. B. (2009). The use of phosphate-affinity SDS-PAGE to measure the cardiac troponin I phosphorylation site distribution in human heart muscle. Proteomics Clinical Applications , 3(12), 1371–1382. https://doi.org/10.1002/prca.200900071.
- Nagy, Z., Comer, S., & Smolenski, A. (2018). Analysis of protein phosphorylation using phos-tag gels. Current Protocols in Protein Science , 93(1), e64. doi: 10.1002/cpps.64.
- Novus Biologicals. (2019a). Loading and running the gel. Western blot SDS-PAGE. Retrieved from https://www.novusbio.com/support/support-by-application/western-blot-sds-page.
- Novus Biologicals. (2019b). Protein transfer. Protein transfer from gel to membrane in western blot. Retrieved from https://www.novusbio.com/support/support-by-application/protein-transfer-gel.
- Pchelintsev, N. A., Adams, P. D., & Nelson, D. M. (2016). Critical parameters for efficient sonication and improved chromatin immunoprecipitation of high molecular weight proteins. PLoS One , 11(1), e0148023. doi: 10.1371/journal.pone.0148023.
- Perluigi, M., Barone, E., Di Domenico, F., & Butterfield, D. A. (2016). Aberrant protein phosphorylation in Alzheimer disease brain disturbs pro-survival and cell death pathways. Biochimica et Biophysica Acta , 1862(10), 1871–1882. doi: 10.1016/j.bbadis.2016.07.005.
- Perrie, W. T., Smillie, L. B., & Perry, S. V. (1973). A phosphorylated light-chain component of myosin from skeletal muscle. Biochemical Journal , 135(1), 151–164.
- Persechini, A., Kamm, K., & Stull, J. (1986). Different phosphorylated forms of myosin in contracting tracheal smooth muscle. The Journal of Biological Chemistry , 261(14), 6293–6299.
- Qi, L., Yang, L., & Chen, H. (2011). Detecting and quantitating physiological endoplasmic reticulum stress. Methods in Enzymology , 490, 137–146. doi: 10.1016/B978-0-12-385114-7.00008-8.
- Qiu, W., & Steinberg, S. F. (2016). Phos-tag SDS-PAGE resolves agonist- and isoform-specific activation patterns for PKD2 and PKD3 in cardiomyocytes and cardiac fibroblasts. Journal of Molecular and Cellular Cardiology , 99, 14–22. https://doi.org/10.1016/j.yjmcc.2016.08.005.
- Rath, A., Cunningham, F., & Deber, C. M. (2013). Acrylamide concentration determines the direction and magnitude of helical membrane protein gel shifts. Proceedings of the National Academy of Sciences of the United States of America , 110(39), 15668–15673. doi: 10.1073/pnas.1311305110.
- Roche Laboratories. (2019). PVDF western blotting membranes. PVDF western blotting membranes. Retrieved 14 August 2021 from https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-assays-analysis/western-blotting/transfer-proteins-western-blot/membranes-transfer-buffers-western-blotting.html. (pp. 1–2). Germany: Roche Diagnostics GmbH.
- Rogacs, A., & Santiago, J. G. (2013). Temperature effects on electrophoresis. Analytical Chemistry , 85(10), 5103–5113. Retrieved from https://pubs.acs.org/doi/10.1021/ac400447k.
- Sacco, F., Perfetto, L., Castagnoli, L., & Cesareni, G. (2012). The human phosphatase interactome: An intricate family portrait. FEBS Letters , 586(17), 2732–2739. doi: 10.1016/j.febslet.2012.05.008.
- Schneider, B., Neidle, S., & Berman, H. M. (1997). Conformations of the sugar–phosphate backbone in helical DNA crystal structures. Biopolymers , 42(1), 113–124. doi: 10.1002/(SICI)1097-0282(199707)42:13.0.CO;2-O.
- Scruggs, S. B., & Solarob, R. J. (2011). The significance of regulatory light chain phosphorylation in cardiac physiology. Archives of Biochemistry and Biophysics , 510(2), 129–134. doi: 10.1016/j.abb.2011.02.013.
- Sitaramamma, T., Shivaji, S., & Rao, G. N. (1998). Effect of storage on protein concentration of tear samples. Current Eye Research , 17(10), 1027–1035. doi: 10.1076/ceyr.17.10.1027.5241.
- Sobieszek, A., & Jertschin, P. (1986). Urea-glycerol-acrylamide gel electrophoresis of acidic low molecular weight muscle proteins: Rapid determination of myosin light chain phosphorylation in myosin, actomyosin and whole muscle samples. Electrophoresis , 7(9), 417–425. doi: 10.1002/elps.1150070906.
- Sutherland, C., & Walsh, M. P. (2012). Myosin regulatory light chain diphosphorylation slows relaxation of arterial smooth muscle. The Journal of Biological Chemistry , 287(29), 24064–24076. doi: 10.1074/jbc.M112.371609.
- Sutherland, C., MacDonald, J. A., & Walsh, M. P. (2016). Analysis of phosphorylation of the myosin-targeting subunit of myosin light chain phosphatase by Phos-tag SDS-PAGE. American Journal of Physiology-Cell Physiology , 310(8), C681–C691. doi: 10.1152/ajpcell.00327.2015.
- Takeya, K., Kaneko, T., Miyazu, M., & Takai, A. (2018). Addition of urea and thiourea to electrophoresis sample buffer improves efficiency of protein extraction from TCA/acetone-treated smooth muscle tissues for phos-tag SDS-PAGE. Electrophoresis , 39(2), 326–333. doi: 10.1002/elps.201700394.
- Takeya, K., Loutzenhiser, K., Shiraishi, M., Loutzenhiser, R., & Walsh, M. P. (2008). A highly sensitive technique to measure myosin regulatory light chain phosphorylation: The first quantification in renal arterioles. American Journal of Physiology-Renal Physiology , 294(6), F1487–F1492. doi: 10.1152/ajprenal.00060.2008.
- Taylor, S. C., Berkelman, T., Yadav, G., & Hammond, M. (2013). A defined methodology for reliable quantification of Western blot data. Molecular Biotechnology , 55(3), 217–226. doi: 10.1007/s12033-013-9672-6.
- Thermo Fisher Scientific. (n.d.). Western blotting transfer methods. Retrieved from https://www.thermofisher.com/sg/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/western-blot-transfer-methods.html.
- Tiago Ferreira, W. R. (2012). ImageJ User Guide—IJ 1.46r. Retrieved 2 March 2020 from https://imagej.nih.gov/ij/docs/guide/user-guide.pdf.
- Toepfer, C., Caorsi, V., Kampourakis, T., Sikkel, M. B., West, T. G., Leung, M. C., … Ferenczi, M. A. (2013). Myosin regulatory light chain (RLC) phosphorylation change as a modulator of cardiac muscle contraction in disease. Journal of Biological Chemistry , 288(19), 13446–13454. doi: 10.1074/jbc.M113.455444.
- Tsunehiro, M., Meki, Y., Matsuoka, K., Kinoshita-Kikuta, E., Kinoshita, E., & Koike, T. (2013). A Phos-tag-based magnetic-bead method for rapid and selective separation of phosphorylated biomolecules. Journal of Chromatography B , 925, 86–94. doi: 10.1016/j.jchromb.2013.02.039.
- Yaseen, M. A., Srinivasan, V. J., Gorczynska, I., Fujimoto, J. G., Boas, D., & Sakadzic, S. (2015). Multimodal optical imaging system for in vivo investigation of cerebral oxygen delivery and energy metabolism. Biomedical Optics Express , 6(12), 4994–5007. doi: 10.1364/BOE.6.004994.
- Zangi, R., Zhou, R., & Berne, B. J. (2009). Urea's action on hydrophobic interactions. Journal of American Chemical Society , 131(4), 1535–1541. doi: 10.1021/ja807887g.
- Zaremba, R., Merkus, D., Hamdani, N., Lamers, J. M., Paulus, W. J., Dos Remedios, C., … van der Velden, J. (2007). Quantitative analysis of myofilament protein phosphorylation in small cardiac biopsies. Proteomics. Clinical Applications , 1(10), 1285–1290. doi: 10.1002/prca.200600891.
- Zhao, D., & Huang, Z. (2016). Effect of His-tag on expression, purification, and structure of zinc finger protein, ZNF191(243-368). Bioinorganic Chemistry and Applications , 2016, 8206854. doi: 10.1155/2016/8206854.
- Zilliges, Y., Kehr, J.-C., Meissner, S., Ishida, K., Mikkat, S., Hagemann, M., … Dittmann, E. (2011). The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One , 6, e17615. doi: 10.1371/journal.pone.0017615.
Citing Literature
Number of times cited according to CrossRef: 1
- Kasturi Markandran, Haiyang Yu, Weihua Song, Do Thuy Uyen Ha Lam, Mufeeda Changaramvally Madathummal, Michael A. Ferenczi, Functional and Molecular Characterisation of Heart Failure Progression in Mice and the Role of Myosin Regulatory Light Chains in the Recovery of Cardiac Muscle Function, International Journal of Molecular Sciences, 10.3390/ijms23010088, 23 , 1, (88), (2021).
