Gnotobiotic Plant Systems for Reconstitution and Functional Studies of the Root Microbiota
Paul Schulze-Lefert, Paul Schulze-Lefert, Ka-Wai Ma, Ka-Wai Ma, Jana Ordon, Jana Ordon
Abstract
Healthy plants host a multi-kingdom community of microbes, which is known as the plant microbiota. Amplicon sequencing technologies for microbial genomic markers were a milestone in revealing the taxonomic composition of the microbiota and its variation associated with a plant host in natural environments. However, this method alone does not allow conclusions to be drawn about functions of these microbial assemblages for the plant. The development of culture collections, which recapitulate natural microbial communities in their diversity, and multiple gnotobiotic plant systems therefore represent a breakthrough in plant‒microbiota research such that plants can be inoculated with defined communities to study proposed microbiota functions. These systems provided, for the root microbiota, first insights into mechanisms underlying microbial community establishment and contributions of its microbial members to indirect pathogen protection and mineral nutrition of the host. We argue that the choice of a gnotobiotic system for microbiota reconstitution and subsequent functional analysis depends on the particular plant trait that is influenced by the microbiota. We start by discussing the advantages and limitations of using individual gnotobiotic systems and then describe the general procedures for preparing bacterial cultures from the Arabidopsis thaliana At -R-SPHERE culture collection for inoculation and cocultivation in two gnotobiotic plant growth systems using agar and perlite matrix. Additionally, a protocol for inoculation of plants with opportunistic Pseudomonas pathogens is provided. Lastly, we describe a high-throughput system for visual assessment of roots after inoculation with individual mutants of a transposon library generated from a root-derived bacterial commensal. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Preparation of bacterial cultures from At -R-SPHERE
Support Protocol 1 : Validation of strains by sequencing hypervariable regions of the 16S rRNA gene
Basic Protocol 2 : Coinoculation of plants grown on an agar matrix with microbial elicitor and a defined microbial community
Alternate Protocol : Inoculation of plants cultivated in a perlite-based growth system
Support Protocol 2 : Surface sterilization of Arabidopsis thaliana seeds
Basic Protocol 3 : Inoculation using a Pseudomonas opportunistic pathogen
Basic Protocol 4 : Assessment of commensal-mediated root phenotypes using phytostrips
INTRODUCTION
Healthy above- and below-ground organs of plants grown in natural environments are inhabited by communities of taxonomically diverse microbes known as the phyllosphere and the root microbiota, respectively. The main inoculum source of the root-associated microbial communities is the even more diverse soil biome. Despite the diversity of microbes that can colonize plants, the root and phyllosphere microbiota have a taxonomic structure at the class level, leading to the concept of a core microbiota composed of ubiquitous taxonomic lineages present on different plant species in diverse environments (Bulgarelli et al., 2012; Hacquard et al., 2015; Lundberg et al., 2012; Thiergart et al., 2020; Vorholt, 2012). The study of plant‒microbiota associations is challenging even by today's standard. Staley and Konopka quoted the term “the great plate count anomaly” in 1985 to describe the challenge that up to 99% of bacterial species isolated from the environment cannot be cultured under standard laboratory conditions (Staley & Konopka, 1985). Stepping into the new millennium, short-read next-generation sequencing (NGS) of microbial genomic markers has become standard and enables culture-independent sequencing approaches to reveal the composition of host-associated microbial communities and their variation within and between plant species (Fitzpatrick et al., 2018; Schlaeppi, Dombrowski, Oter, Ver Loren van Themaat, & Schulze-Lefert, 2014; Thiergart et al., 2020; Yeoh et al., 2017).
For community profiling, amplicon reads generated by DNA sequencing the hypervariable region of microbial markers (e.g., the 16S rRNA gene of bacteria and internal transcribed spacer region of fungi) were clustered into operational taxonomic units (OTUs) using an arbitrary threshold (e.g., >97% sequence similarity). Such a clustering approach reduces complication due to sequencing errors but compromises at the expense of detecting microbial diversity. Later, due to the development of statistical denoising methods such as DADA2, analyses of the resultant amplicon sequence variants provided single-nucleotide resolution of the amplicon reads, resulting in higher resolution of plant-associated microbial diversity without great loss of accuracy (Callahan et al., 2016). Despite these improvements in culture-independent sequencing methods coupled with single-nucleotide polymorphism analysis, community profiling of these host-associated microbial assemblages alone does not allow for a valid conclusion to be drawn regarding the cause and effect of the plant microbiota on particular host processes.
The root microbiota is thought to provide beneficial services to plant hosts under adverse environmental conditions (Bulgarelli, Schlaeppi, Spaepen, Ver Loren van Themaat, & Schulze-Lefert, 2013; Trivedi, Leach, Tringe, Sa, & Singh, 2020). These services include indirect protection against soilborne pathogens (Berendsen et al., 2018; Carrion et al., 2019; Duran et al., 2018; Kwak et al., 2018); mobilization of mineral nutrients from soil to alleviate plant mineral malnutrition (Castrillo et al., 2017; Harbort, Hashimoto, Inoue, Niu, et al., 2020; Yu et al., 2021); and improved abiotic stress tolerance in response to drought, salt, or low light (Hou et al., 2021; Kearl et al., 2019; Santos-Medellin et al., 2021). However, it remains a challenge to disentangle the complex interactions compounded by the sheer number of plant‒microbe and microbe‒microbe interactions that ultimately determine whether microbial communities can directly offer beneficial services to the host or not. Deconstruction of these complex interactions is therefore advantageous to identify reduced complexity communities and to study the impact on a given plant trait. Contrary to the expectations of “the great plate count anomaly” (Staley & Konopka, 1985), major advances have been made in generating microbial culture collections that represent the majority of root-associated bacterial and fungal taxa that can be detected in nature using culture-independent community profiling methods (Bai et al., 2015; Duran et al., 2018). This cultivation success on synthetic media in the presence of different soluble organic carbon sources may indicate the metabolic versatility of root-associated microbes.
Individual microbes can then be used to design defined synthetic communities (SynComs), which permit, together with gnotobiotic plant systems, reconstitution experiments to be performed in the laboratory for controlled perturbation of plant‒microbiota associations. For example, plant roots or leaves can recognize seemingly benign microbes (i.e., commensals of the leaf or root microbiota) and respond with induced defenses (Garrido-Oter et al., 2018; Ma et al., 2021; Maier et al., 2021; Stringlis et al., 2018; Teixeira et al., 2021; Vogel, Bodenhausen, Gruissem, & Vorholt, 2016). A systematic mono-association experiment with >100 isolated root commensals from the Arabidopsis thaliana culture collection (At -R-SPHERE; Bai et al., 2015) showed that only a subset of the tested commensals can suppress parts of these plant immune responses (Ma et al., 2021). The composition of defined SynComs based on individual bacterial activities then revealed that commensal-mediated immunosuppression acts dominantly in community contexts to interfere with root growth and defense responses of A. thaliana. (Ma et al., 2021; Teixeira et al., 2021). Therefore, systematic screening of culture collections for specific beneficial microbial traits in binary interactions, followed by the construction of defined SynComs, will help to dissect complex plant‒microbiota interactions as a bottom-up approach.
To facilitate direct comparisons of the results generated from different laboratories, it is essential to develop standard procedures and protocols. A number of gnotobiotic systems were developed over the past few years. However, there is no universal gnotobiotic plant system that can mimic adverse environmental conditions to quantitatively examine all proposed beneficial activities of the microbiota for the plant host. The choice of gnotobiotic system therefore depends on the particular plant trait that is influenced by the microbiota to be tested. For example, an agar-based gnotobiotic growth system is convenient and permits visual inspection and isolation of intact root materials for subsequent analyses. However, the artificial nature of the agar-based growth system with the exposure of surface-grown roots to light are prone to criticism. Peat-based growth systems, such as flowpot and gnotopot (Kremer et al., 2021), were developed by sterilizing peat matrix through repeated autoclaving. However, these systems are mostly incompatible with the application of mineral nutritional stresses due to nutrient leaching, rendering them less useful to assess root microbiota-mediated mineral nutrient mobilization for plant growth under malnutrition conditions. Peat-based gnotobiotic systems also show variable results in experiments involving the application of microbial elicitors to the peat matrix to stimulate chronic immune responses in roots. Readers should familiarize themselves with the individual protocols, and we refer the reader to the Commentary for further discussion on the advantages and disadvantages of each system to make an educated decision.
Basic Protocol 1 describes the general procedure for the preparation of bacterial inocula from the At -R-SPHERE culture collection of bacterial root commensals (Bai et al., 2015). Basic Protocol 2 describes the procedure for the inoculation of individual bacterial strains or SynComs on germ-free plants using an agar-based plant growth system. We also describe an alternative plant growth system that uses inert perlite as expanded volcanic glass matrix for plant growth (Alternate Protocol). To demonstrate the potential application of the agar-based gnotobiotic systems, in Basic Protocol 3 we describe an infection protocol for the inoculation of an opportunistic Pseudomonas pathogen and for monitoring the growth of this pathogen by viable plate counting as a proxy of plant immunity. Finally, Basic Protocol 4 uses a 96-well agar-based phytostrip system to characterize in mono-associations individual mutants of a transposon library generated from a root-derived bacterial commensal for altered root architecture.
CAUTION : Certain bacterial members from the At -R-SPHERE collection are Biosafety Level 2 material. Consult your local safety office, and follow all the appropriate guidelines and safety regulations for the use and handling of these materials.
NOTE : All reactions involving live bacteria and gnotobiotic system should be performed in a laminar flow hood to avoid contamination.
Basic Protocol 1: PREPARATION OF BACTERIAL CULTURES FROM At-R-SPHERE
At -SPHERE (Bai et al., 2015; http://www.at-sphere.com/) is a culture collection of A. thaliana root- and leaf-derived bacteria isolated from plants grown in natural soils. The At -R-SPHERE collection is composed of >200 strains isolated from roots of healthy A. thaliana representing >60% of the OTUs detectable by culture-independent community profiling using 16S rRNA short-read NGS (Bai et al., 2015). The draft genomes of the majority of bacterial isolates in the At -SPHERE collection are available for strain validation and genomic analyses. This protocol summarizes the procedures and quality control measures used to work with these bacteria.
Materials
-
Bacterial strain in glycerol stock
-
Tryptic soy broth (TSB) plate, half strength (see recipe)
-
TSB medium, half strength (see recipe)
-
Glycerol
-
10 mM MgSO4, sterile (e.g., Sigma-Aldrich, cat. no. M7506)
-
25°C incubator with variable shaking
-
1.5-ml microcentrifuge tube
-
Microcentrifuge (e.g., Eppendorf Centrifuge 5424)
-
Spectrophotometer (e.g., Implen) and compatible cuvettes (e.g., Ratiolab, cat. no. 2712120)
1.Request individual strains of interest from the At -SPHERE collection (http://www.at-sphere.com/).
2.Upon arrival of strains, use a sterile pipette tip to streak out an individual strain on a half-strength TSB plate. Incubate at 25°C until single colonies appear.
3.Inoculate a single colony into 2 ml half-strength TSB medium, and incubate at 25°C, 200 rpm, until saturation.
4.Save pure bacterial culture as 25% (v/v) glycerol stocks, and store at ‒80°C.
5.Use a sterile pipette tip or loop to inoculate individual strains from a clean glycerol stock into half-strength TSB medium. Depending on incubation time of the individual strain, allow culture to grow at 25°C for 1 to 4 days.
6.On the day of the inoculation experiment, subculture fully grown bacterial culture in a 1:3 ratio (e.g., 1 ml stock culture to 3 ml fresh medium) in the morning. Allow to grow at 25°C for 3 to 5 hr.
7.Take 500 µl of an actively growing bacterial culture, and transfer to a sterile 1.5-ml microcentrifuge tube. Centrifuge 2 min at 6500 × g , room temperature.
8.Wash pellet with 1 ml of 10 mM MgSO4.
9.Repeat washing step two more times, and then resuspend pellet in 1 ml of 10 mM MgSO4.
10.Measure optical density at 600 nm (OD600): Fill cuvette with 900 µl of 10 mM MgSO4, and use as a blank. Dilute bacterial culture in a 1:9 ratio (e.g., 100 µl resuspended bacterial culture and 900 µl of 10 mM MgSO4), and measure the OD600 value. Dilute stock solution to an OD600 range of 0.2 to 0.5.
Support Protocol 1: VALIDATION OF STRAINS BY SEQUENCING HYPERVARIABLE REGIONS OF THE 16S rRNA GENE
Unlike common laboratory strains, the strains within At -R-SPHERE may not express a known selection marker (i.e., antibiotic resistance) and thus are prone to contamination. It is strongly recommended to perform regular quality control checks to ensure that the original stock is clean from contamination. This protocol provides a method to extract genomic bacterial DNA for subsequent PCR amplification and Sanger sequencing of the 16S rRNA gene for validation.
Materials
-
Fully gown bacterial culture in half-strength TSB medium (see Basic Protocol 1)
-
Buffer 1 (see recipe)
-
Buffer 2 (see recipe)
-
DFS-Taq DNA Polymerase kit (e.g., BIORON) containing:
- 10× Taq incomplete buffer
- 100 mM MgCl2
- 5 U/μl Taq polymerase
-
10 mM dNTPs
-
10 μM forward primer 799F: 5'-AACMGGATTAGATACCCKG-3'
-
10 μM reverse primer 1192R: 5'-ACGTCATCCCCACCTTCC-3'
-
Heating block
-
Thermal cycler
-
1.5-ml microcentrifuge tubes
-
PCR tubes
-
Vortex mixer
-
Microcentrifuge (e.g., Eppendorf Centrifuge 5424)
-
Additional reagents and equipment for agarose gel electrophoresis (see Current Protocols article: Armstrong & Schulz, 2015) and Sanger sequencing (see Current Protocols article: Shendure et al., 2011)
1.Preheat heating block to 95°C.
2.Transfer 10 µl fresh bacterial culture to a 1.5-ml microcentrifuge tube or PCR tube.
3.Add 10 µl buffer 1 and vortex briefly.
4.Spin down mixture briefly to collect the liquid.
5.Incubate mixture at 95°C for 30 min.
6.Add 10 µl buffer 2 and vortex.
7.Spin down mixture briefly to collect the liquid.
8.Set up the following PCR reaction to amplify the 16S rRNA region (25 μl final volume):
- 2.5 μl of 10× Taq incomplete buffer
- 0.5 μl of 100 mM MgCl2
- 0.5 μl of 10 mM dNTPs
- 0.75 μl of 10 µM forward primer
- 0.75 μl of 10 µM reverse primer
- 0.4 μl of 5 U/μl Taq polymerase
- 1.0 to 3.0 μl template DNA
- Bring to 25 µl with water.
We use the BIORON DFS-Taq system as an example. The PCR reaction mixture can be changed according to the manufacturer's suggestion.
The primer pair will amplify the V5V7 hypervariable region of the 16S rRNA gene.
9.Carry out PCR in a thermal cycler using the following amplification cycles:
| Initial step: | 2 min | 94°C |
| 34 cycles: | 30 s | 94°C |
| 30 s | 55°C | |
| 60 s | 72°C | |
| Final step: | 5 min | 72°C |
| Hold: | indefinitely | 4°C. |
10.Confirm presence of PCR product on a 1.5% (w/v) agarose gel, and then confirm sequences by Sanger sequencing.
Basic Protocol 2: COINOCULATION OF PLANTS GROWN ON AGAR WITH ELICITOR AND MICROBIOTA
Plants grown in natural soil host a complex microbiota. Studying plant‒microbiota associations, however, remains a challenge because plant‒microbe and microbe‒microbe interactions occur simultaneously. Germ-free plants are commonly grown on a sterile agar-based system to exclude interference from microbes in the environment. Selected bacterial root commensals can then be supplemented either as a drop inoculum or homogeneously embedded into the agar, as illustrated in this protocol. Such a gnotobiotic system provides a convenient means to grow plants axenically or in cocultivation with a specific microbial inoculum (SynCom) and harvest different plant parts (e.g., roots and shoots), which are compatible with downstream applications including transcriptomics and community profiling through genomic marker sequencing.
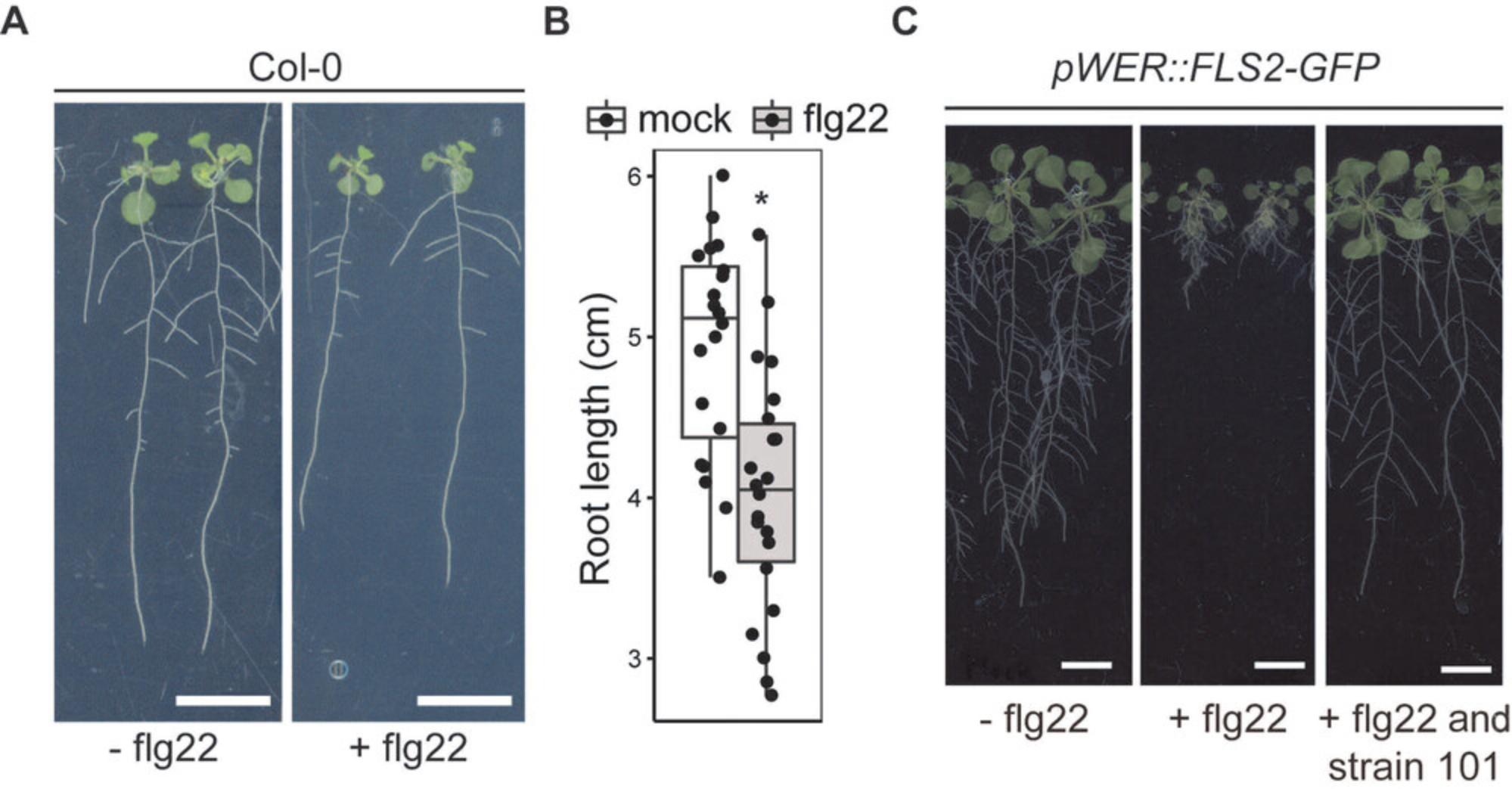
Due to the solid agar matrix with aqueous pores, different elicitors, including the immunogenic peptide elicitor flg22, can be coinoculated with the SynCom to stimulate chronic immune responses in roots. We previously showed the use of the agar-based system together with a flg22-hypersensitive A. thaliana line (i.e., pWER::FLS2-GFP ; Wyrsch, Dominguez-Ferreras, Geldner, & Boller, 2015) significantly increases the signal-to-noise ratio of flg22-mediated root growth inhibition. However, plant roots grown with the agar-based system are continuously exposed to light without extensive physical damage normally experienced when plants are grown in soil or peat matrix. Readers may refer to the Alternate Protocol or other published gnotobiotic systems (Kremer et al., 2021) as possible alternatives. Up to 30 individual bacterial strains can be assayed per day routinely.
Materials
-
Bacterial strains (see Basic Protocol 1)
-
Murashige and Skoog (MS) medium, half strength (see recipe)
-
Chemical elicitors
-
10 mM MgSO4, sterile (e.g., Sigma-Aldrich, cat. no. M7506)
-
A. thaliana seeds, sterile (see Support Protocol 2)
-
Optional: 50°C water bath
-
50-ml conical tube
-
Optional: magnetic stir bar and stir plate with heating function
-
Spectrophotometer (e.g., Implen) and compatible cuvettes (e.g., Ratiolab, cat. no. 2712120)
-
Square plates, 12 cm × 12 cm (e.g., Greiner Bio-One, cat. no. 688102)
-
3M Micropore Surgical Tape, 1.25 cm width (e.g., VWR, cat. no. 115-8172)
-
Phytochamber (e.g., Panasonic Chamber MLR-352)
1.After obtaining resuspended bacterial culture (Basic Protocol 1), calculate volume needed for the inoculation experiment.
2.Prepare and autoclave half-strength MS medium containing agar. Allow medium to cool to ∼50°C by leaving medium at room temperature or in a preset water bath.
3.Pour 50 ml warm half-strength MS medium into a 50-ml conical tube. Add chemical elicitors when necessary.
4.Add the corresponding volume of bacterial stock culture as calculated from Basic Protocol 1, step 10.Add extra MgSO4 to bring to a total volume of 150 µl (30 μM MgSO4 final concentration).
5.If a SynCom is inoculated, add each strain to a final concentration corresponding to an OD600 of 0.0005.
6.Pour medium into a 12 cm × 12 cm square plate. Mix medium well by inverting the conical tube gently upside-down twice to minimize the trapping of air bubbles.
7.Dry plates in a laminar flow hood by leaving plates open for ∼10 min to minimize excessive condensation during incubation.
8.Allow plates to stand for 1 hr to overnight before adding A. thaliana seeds on top of the solidified agar plate.
9.Depending on experimental requirements, add five to 15 seeds (two rows for more than eight seeds) on each plate.
10.Seal stacked petri dish with 3M micropore surgical tape, and allow plants to grow vertically in phytochamber for 2 to 3 weeks. Randomize position of the plates once a week.
Expected outcome
A. thaliana plants can be grown under the indicated conditions for 2 to 3 weeks. It is typical for wild-type plants (i.e., Col-0) to develop a primary root up to 5 cm in 2 weeks. Addition of immunogenic elicitor such as flg22 generally results in about 20% reduction in primary root length (Fig. 1A,B). The use of a hypersensitive line (i.e., pWER::FLS2-GFP) can boost flg22-mediated root growth inhibition from 20% to 80% (Fig. 1C).
Alternate Protocol: INOCULATION OF PLANTS CULTIVATED IN A PERLITE-BASED GROWTH SYSTEM
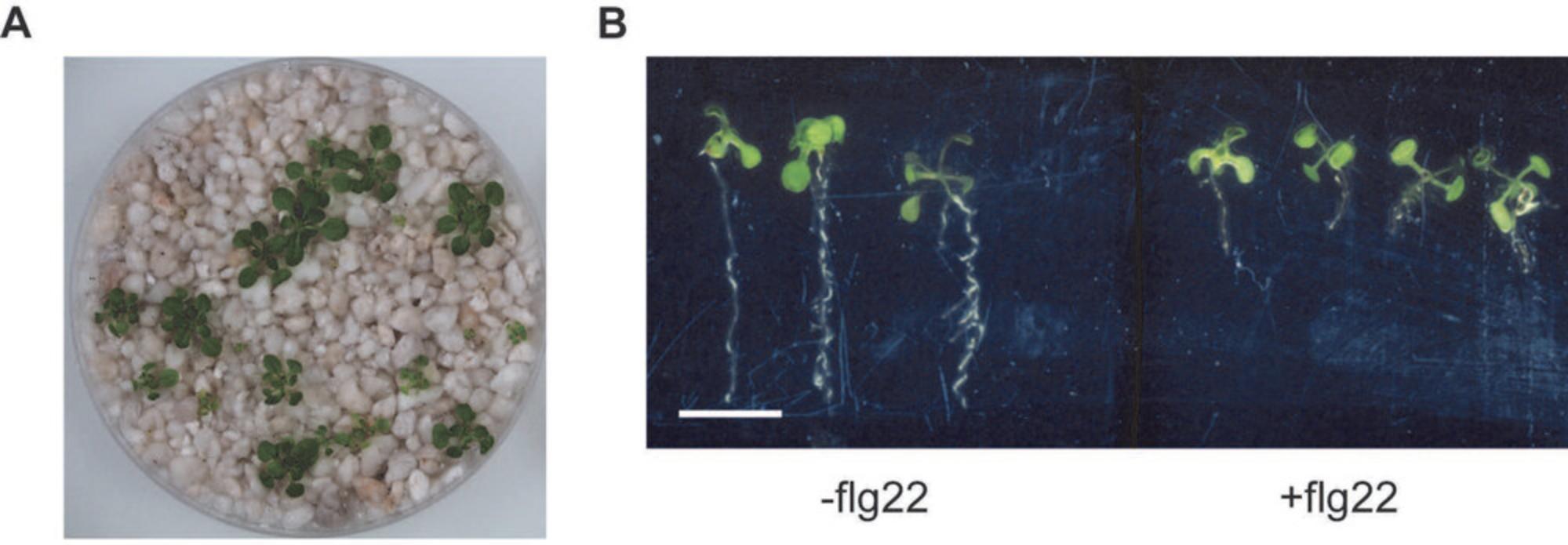
Growing plants in an agar-based system provides multiple advantages from easy visual assessment of the root architecture to compatibility of harvesting for downstream assays. However, extrapolating results gained from plants grown on agar plates is complicated by their artificial environment: root exposure to light and lack of physical damage for surface-grown roots. Additionally, the choice of gelling agent (i.e., agar) or even batches can impose significant variation on nutrients (Gruber, Giehl, Friedel, & von Wirén, 2013). In contrast, plants grown in soil-like peat matrix are closer to natural cultivation conditions. Kremer and colleagues have recently developed a peat-based gnotobiotic system known as flowpot. However, the setup of flowpot requires additional equipment and is more time-consuming. The jiffy pot‒based gnotopot (Kremer et al., 2021) is easier to set up but is nevertheless incompatible with the exogenous application of certain microbial elicitors, possibly due to absorption by peat matrix and/or chemical modification by this growth substrate (Sasse et al., 2020). Furthermore, plants grown in peat are difficult to harvest and can complicate downstream assays that rely on accurate and clean tissue amounts for normalization or quick harvesting for transcriptomic analyses. Lastly, these peat-based systems are incompatible with mineral malnutrition experiments owing to nutrient leaching. To address these concerns, we have developed an alternative gnotobiotic system that takes advantage of the perlite material as expanded volcanic glass matrix that is commonly used in hydroponics. The perlite system is advantageous due to its compatibility with some nutritional stress (e.g., phosphate starvation, peptide elicitor treatment) and the feasibility to harvest relatively intact and clean roots.
Materials
-
Perlite (e.g., Knauf, PERLIGRAN)
-
MS medium, half strength (see recipe)
-
Elicitor (see Basic Protocol 2)
-
Bacteria (see Basic Protocol 2)
-
A. thaliana seeds, sterile (see Support Protocol 2)
-
Autoclave-resistant container
-
92 mm × 16 mm petri dish (e.g., Sarstedt, cat. no. 82.1473.001)
-
3M Micropore Surgical Tape, 1.25 cm width (e.g., VWR, cat. no. 115-8172)
1.Wash perlite three times with tap water. Then wash two additional times with Milli-Q water. Transfer washed perlite into a deep autoclave-resistant container.
2.Autoclave perlite once at 121°C for 30 min. Let autoclaved material cool overnight, and repeat autoclave process.
3.Fully fill a regular 92 mm × 16 mm petri dish with sterile perlite. Pour half-strength MS medium supplemented with elicitor or bacteria according to the procedures described in Basic Protocol 2.
4.Germinate up to 10 seeds in each petri dish.
5.Use lower half of another petri dish as a cap. Seal stacked petri dish with 3M tape.
6.Grow plants in this system up to 3 weeks before harvesting.
Expected outcome
It is feasible to grow plants in a regular 92 mm × 16 mm petri dish for 3 weeks (Fig. 2A). Additional nutrient solution or water may be added to keep the plants hydrated. In addition, immunogenic elicitors (i.e., flg22) can be applied to induce plant immune responses, as evidenced by flg22-mediated root growth inhibition (Fig. 2B). Relatively intact plant roots can be harvested after 2 to 3 weeks of growth.
Support Protocol 2: SURFACE STERILIZATION OF Arabidopsis thaliana SEEDS
In contrast to some plant species, most of the seed-associated microbiota of A. thaliana are found only on the surface. A typical surface sterilization protocol using ethanol or bleach is sufficient to remove the seed-associated microbiota and prepare the germ-free plants for subsequent experiments. For these experiments, we recommend using seeds collected from plants propagated in greenhouses without any infestation.
Materials
-
A. thaliana seeds
-
70% and 96% ethanol
-
Optional: 50% bleach
-
1.5-ml microcentrifuge tubes
-
Optional: rocking platform
1.Aliquot an appropriate amount of seeds into a 1.5-ml microcentrifuge tube.
2.Wash seeds with 70% ethanol for 5 min.
3.Remove 70% ethanol by pipetting, avoiding taking seeds in the process. Repeat step 2 twice.
4.Remove 70% ethanol, and wash with 96% ethanol for 2 min.
5.Remove 96% ethanol, and add sterile water.
6.Remove water by pipetting. Repeat washing step for a total of three times, ∼1 min each.
7.Leave a small amount of water that barely covers the seeds. Keep seeds in the dark at 4°C for at least 2 days.
Basic Protocol 3: INOCULATION USING Pseudomonas OPPORTUNISTIC PATHOGEN
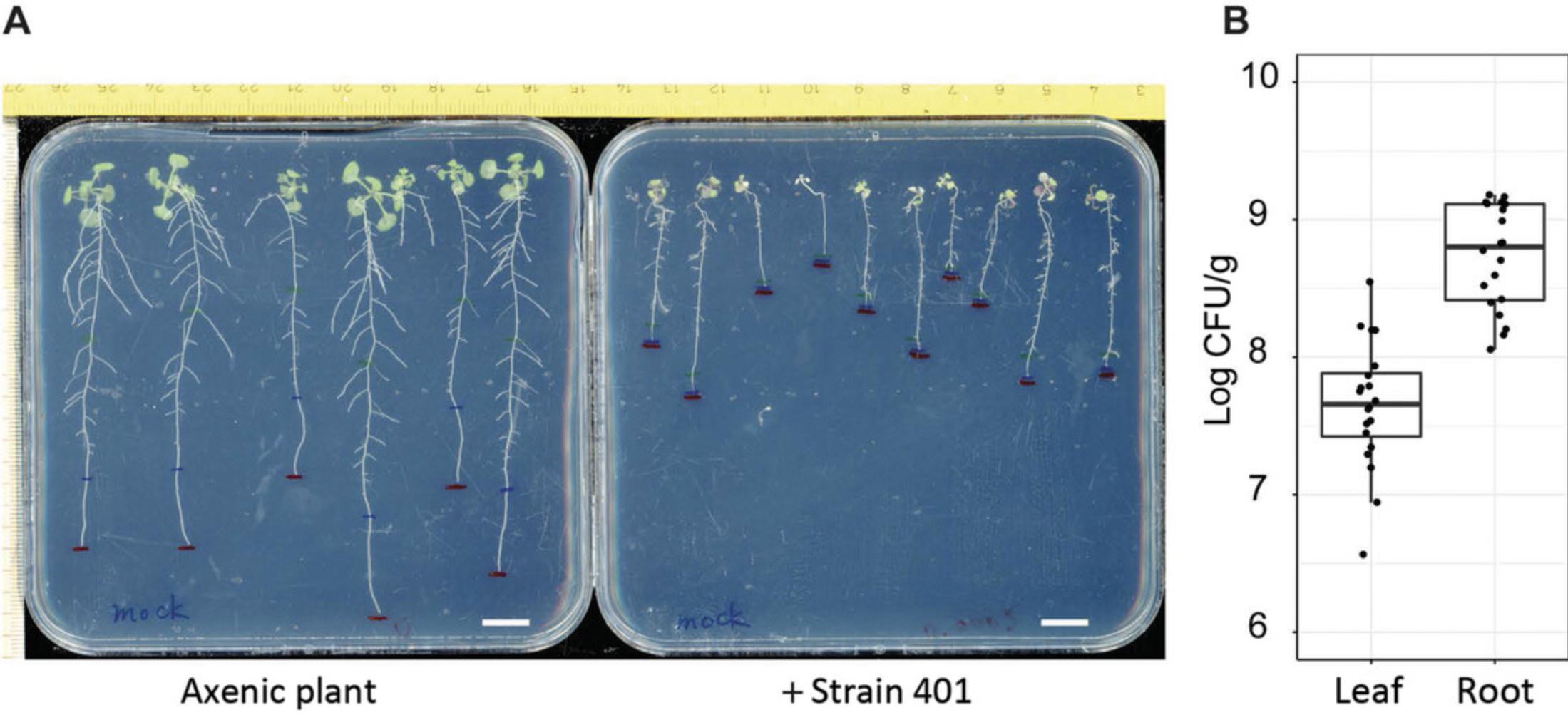
Pseudomonas is a highly diversified genus within the Gammaproteobacteria that can associate with multiple eukaryotic hosts. While some Pseudomonas spp. are beneficial microbes, others are infectious agents, and together they form one of the core lineages of the A. thaliana root microbiota. There are numerous Pseudomonas pathovars that can successfully establish and infect A. thaliana leaves including the commonly used P. syringae pv. tomato DC3000. However, only a limited number of Pseudomonas root pathogens have been described so far. Previously, we characterized an opportunistic Pseudomonas pathogen strain R401 from the At -R-SPHERE collection. Inoculation of nonimmunosuppressive or immunosuppressive SynComs, designed based on the immunosuppressive activity of individual strains in mono-association, affect plant susceptibility to the opportunistic pathogen R401 (Ma et al., 2021). Therefore, following the proliferation of R401 can serve as an output of root immunity. Here, we provide a detailed infection protocol of A. thaliana by R401.
Materials
-
MS agar plate (see recipe)
-
Elicitor, bacterium, or SynCom (see Basic Protocol 2)
-
A. thaliana seeds, sterile (see Support Protocol 2)
-
Bacterial suspension of pathogens (e.g., Pseudomonas strain R401)
-
10 mM MgSO4, sterile (e.g., Sigma-Aldrich, cat. no. M7506)
-
Silwet l-77 (e.g., Lehle Seeds, cat. no. VIS-30)
-
TSB plate (see recipe)
-
Spectrophotometer
-
Scalpel, sterile (e.g., VWR, cat. no. 233-5315)
-
Filter paper, sterile (e.g., Whatman)
-
2-ml tubes, sterile
-
Metal beads, sterile (e.g., Biospec, cat. no. 11079113c)
-
Scale
-
Tissue homogenizer (e.g., Bertin Technologies Precellys 24)
-
96-well plate or equivalent
-
12-channel pipette
-
25°C incubator
1.Prepare half-strength MS agar plate without sugar so that the inoculated bacteria depend on plants for organic carbon source. Supplement plate with elicitor, single bacterium, or SynCom as described in Basic Protocol 2.Pregerminate surface-sterilized seeds on half-strength MS agar plate for 14 days.
2.Flood inoculate plants with a bacterial suspension of pathogens (e.g., Pseudomonas strain R401) at a final OD600 of 0.0001 (in 10 mM MgSO4 supplemented with 0.005% Silwet).
3.Incubate plants with bacterial culture for 5 min. Afterward, remove excessive liquid.
4.After 1 to 2 days, separate shoots and roots using a sterile scalpel, and pool roots and shoots from five and three individual plants, respectively. Wash samples briefly in sterile water, and blot dry with sterile filter paper.
5.Prepare a sterile tube, and add sterile metal beads to about 1 cm depth. Weigh tubes with metal beads.
6.Transfer washed and dried samples from step 4 to tubes with beads. Weigh tubes with plant samples, and calculate the difference as the weight of your samples.
7.Homogenize samples with metal beads once. Homogenize sample again in 500 µl sterile 10 mM MgSO4 using a tissue homogenizer (6200 rpm, 2 × 30 s, 15 s pauses in between).
8.Perform serial dilutions in a sterile 96-well plate using the format shown in Figure 3A.
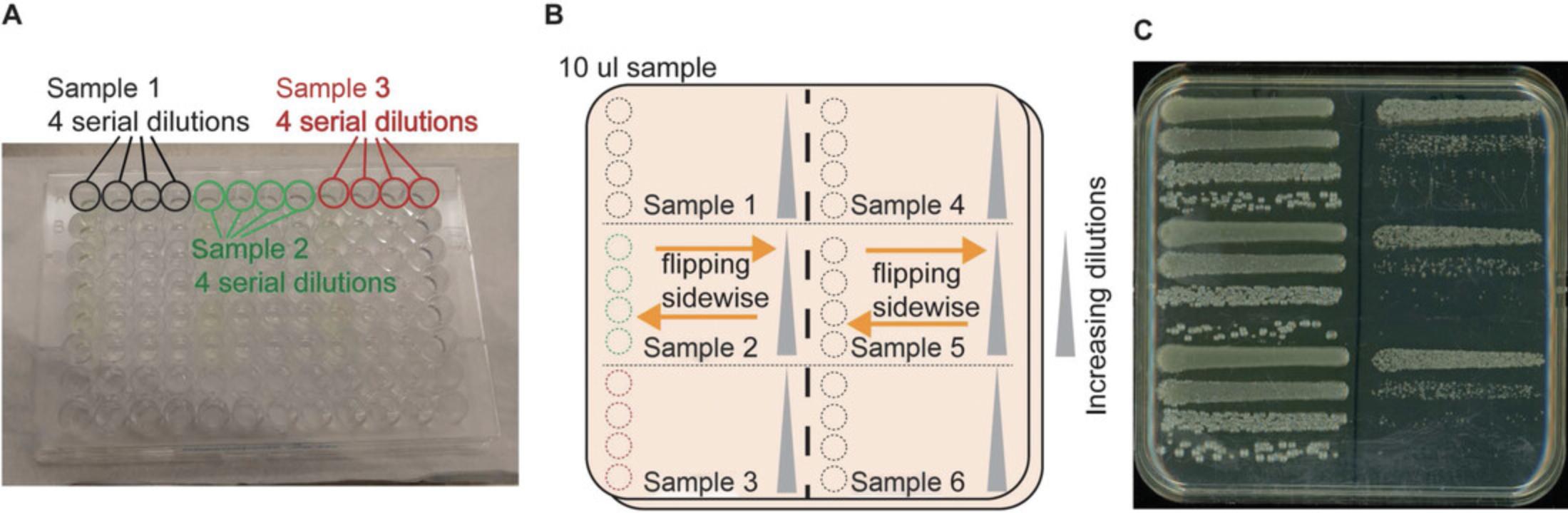
9.After preparing serial dilutions, use a 12-channel pipette to aliquot 10 µl samples on a half-strength TSB plate, and flip plate sidewise to let the liquid flow evenly, moving the plate back and forth a few times (Fig. 3B).
10.Incubate plates at 25°C for 1 to 4 days until single colonies appear (Fig. 3C).
Expected outcome
Depending on the starting bacterial inoculum and the developmental stage of the plants upon inoculation, plants infected with R401 will be inhibited in growth and will develop leaf chlorosis symptoms (Fig. 4A). Typically, axenic 14-day-old plants inoculated with R401 at a starting inoculum of OD600 = 0.0001 will have a titer of 108 to 109 colony forming units (CFUs) per gram of root tissue 2 days after inoculation (Fig. 4B).
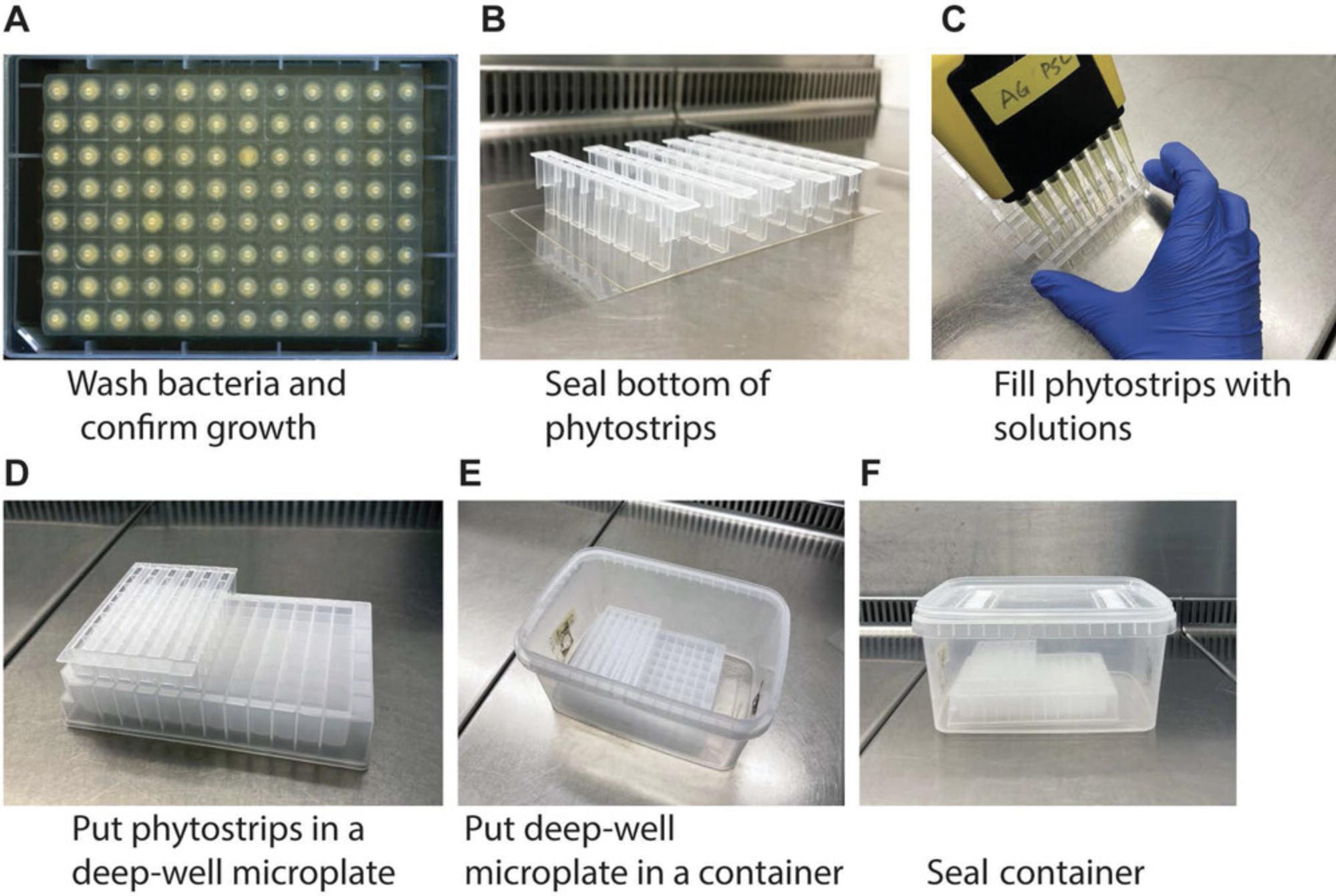
Basic Protocol 4: ASSESSMENT OF COMMENSAL-MEDIATED ROOT PHENOTYPES USING PHYTOSTRIPS
In mono-association studies of bacteria with plant roots, roots are inoculated with individual bacterial strains. The system described in Basic Protocol 2 is limited in terms of the capacity to perform high-throughput screening of bacteria resulting in altered plant phenotypes. We adopt and modify the previously published phytostrip system (Burrell et al., 2017) based on a 96-well microplate format to allow parallel A. thaliana mono-association experiments with one bacterial strain or SynCom per well in the presence or absence of microbial elicitors. Routinely, 380 bacterial strains in 96-well formats can be tested per day. Here, we demonstrate the feasibility to screen for bacterial root-derived commensal mutants of a transposon library to modulate flg22-mediated root growth inhibition by using the A. thaliana flg22-hypersensitive transgenic line pWER::FLS2-GFP (Wyrsch et al., 2015).
Materials
-
Bacterial stocks
-
TSB medium (see recipe)
-
10 mM MgSO4, sterile (e.g., Sigma-Aldrich, cat. no. M7506)
-
MS medium (see recipe)
-
Phytagel (e.g., Sigma-Aldrich, cat. no. P8169)
-
A. thaliana seeds, sterile (see Support Protocol 2)
-
96-deep-well plates, 2 ml (e.g., Eppendorf, cat. no. 0030506308)
-
Micropore foil (e.g., AeraSeal; Sigma-Aldrich, cat. no. A9224)
-
Thermoshaker
-
Microcentrifuge with swinging bucket rotor and plate adapter
-
Multichannel pipettes
-
Vortex mixer
-
Glass petri dish, minimum diameter: length of phytostrips (e.g., Sigma-Aldrich, cat. no. BR455701-10EA)
-
50°C incubator
-
Phytostrips (e.g., Brooks Life Sciences, cat. no. 4ti-0310)
-
Adhesive PCR foil (e.g., Bio-Budget, cat. no. 30-SP-0027)
-
96-deep-well square microplates, 1.2 ml, U bottom (e.g., Brooks Life Sciences, cat. no. 4ti-0126)
-
Rectangular container (e.g., Sac O2, TP1600+TPD1200)
-
3M Micropore Surgical Tape, 1.25 cm width (e.g., VWR, cat. no. 115-8172)
-
Growth cabinet
-
Scanner
Prepare bacterial culture
1.Fill 96-deep-well plates with 1.2 ml half-strength TSB medium, and inoculate with bacteria.
2.Seal 96-deep-well plate with micropore foil.
3.Incubate inoculated plates in a thermoshaker until the bacterial culture reaches prestationary phase titers.
4.Pellet bacteria by centrifuging 96-deep-well plates 10 to 20 min at 3200 × g , room temperature, using a compatible plate adapter.
5.Discard supernatant by pipetting.
6.Use a 12-channel pipette to add 1 ml of 10 mM MgSO4.
7.Resuspend bacteria using a vortex mixer or by pipetting.
8.Repeat steps 4 to 7.
9.Pellet bacteria by centrifuging 96-deep-well plates 10 to 20 min at 3200 × g , room temperature, using a compatible plate adapter.
10.Discard supernatant.
11.Add half-strength MS medium to get a bacterial suspension for inoculation.
Prepare phytostrips
12.Transfer a sterile glass petri dish to a laminar flow hood, and prewarm plate to about 50°C.
13.Hold a single 8-well phytostrip in position by taping to adhesive PCR foil (Fig. 5B).
14.Fill 100 µl resuspended bacteria culture into sealed phytostrips using a multichannel pipette (Fig. 5C).
15.Prepare warm half-strength MS medium supplemented with 1.2% (w/v) Phytagel.
16.Pour supplemented half-strength MS medium into a prewarmed glass petri dish.
17.Add 230 µl supplemented half-strength MS medium into individual wells of the phytostrips using an 8-channel pipette, and mix well with bacterial culture by pipetting up and down once.
18.Wait for medium to solidify (∼15 min).
19.Fill 96-deep-well square microplates with 1 ml half-strength MS medium.
20.Place prepared phytostrips in the storage microplates (Fig. 5D).
21.Add two to five A. thaliana seeds on top of the agar-supplemented medium in each well.
22.Place 96-well plate holding the phytostrips in a rectangular container (Fig. 5E).
23.Close container with the lid, and seal with 3M micropore surgical tape if necessary (Fig. 5F).
24.Transfer phytostrips to a growth cabinet, and grow plants for up to 3 weeks.
25.Scan individual 8-well phytostrips on a scanner with a black background to phenotype root architecture.
Expected outcome
At the end of a 2- to 3-week cultivation period, primary roots of untreated wild-type (Col-0) or hypersensitive (pWER::FLS2-GFP) plants (no flg22) should reach the half-strength MS medium reservoir. Application of the immunogenic elicitor flg22 in combination with the hypersensitive line (pWER::FLS2-GFP) is expected to induce strong root growth inhibition, in which roots are barely visible to the naked eye. Depending on the ability of up to 90 individual mutant bacterial strains to suppress flg22-induced root growth inhibition, intermediate phenotypes in terms of root length will be observed. Due to the fact that each phytostrip (12 per plate) can be moved individually, the effect of single bacteria can be quantified not only for the shoot (Fig. 6A) but also for the root organ by a side view over time (Fig. 6B). Note that roots can also be directly harvested from individual wells for downstream assays.

REAGENTS AND SOLUTIONS
Buffer 1, pH 12
- 25 mM NaOH
- 0.2 mM EDTA
- Store at room temperature for up to 1 year
Buffer 2, pH 7.4
- 40 mM Tris·HCl
- Store at room temperature for up to 1 year
MS medium, half strength, pH 5.6 to 5.7
- 2.2 g/L MS (e.g., Duchefa Biochemie, cat. no. M0222; Sigma-Aldrich, cat. no. M5519)
- 0.1 g/L MES monohydrate (e.g., Duchefa Biochemie, cat. no. M1503)
- 10 g/L Bacto agar or 12 g/L Phytagel
- Store autoclaved medium at room temperature for up to 3 months
To prepare MS plates, dissolve MS and MES powder in Milli-Q water. Adjust pH with 0.5 M NaOH. Add agar before autoclaving, and then pour into plates.
Microwave the medium before use.
TSB medium
- 15 g/L TSB (e.g., Sigma-Aldrich, cat. no. T8907)
- 15 g/L Bacto agar
- Store autoclaved medium at room temperature for up to 3 months
To prepare TSB plates, dissolve TSB powder in Milli-Q water. There is no need to adjust pH. Add agar before autoclaving, and then pour into plates.
Microwave the medium before use.
COMMENTARY
Background Information
Advances in short-read NGS of microbial markers have uncovered the composition and variation of the root microbiota associated with different plant species grown in diverse natural soils (Bulgarelli et al., 2012; Fitzpatrick et al., 2018; Lundberg et al., 2012; Thiergart et al., 2020; Yeoh et al., 2017). Bulgarelli and colleagues proposed a model in which edaphic factors, rhizodeposition, cell wall features, and host genotypes account for the gradual selection of microbiota members from the soil biome and proliferation on and inside plant roots with an enrichment of Proteobacteria, Actinobacteria, and Bacteroidetes as phyla of the core root microbiota (Bulgarelli et al., 2013; Hacquard et al., 2015). Surveys with field-collected samples are limited in their ability to draw conclusions about the biological function(s) of the root microbiota. This is due to the numerous interactions between and within the different microbial classes of the root microbiota, interactions with the plant host, and the influence of edaphic and other environmental parameters on microbial community composition and activity (Bulgarelli et al., 2013; Duran et al., 2018; He et al., 2021; Thiergart et al., 2020). This makes it almost impossible to disentangle these interactions and assign particular plant phenotypes to microbial services. Genomic marker‒based amplicon sequencing together with reconstitution experiments of defined SynComs using gnotobiotic plant systems offers opportunities to monitor community dynamics with strain-specific resolution and study causal relationships between particular microbiota activities and plant phenotypes. However, any simplified system for reconstituting the plant microbiota in the laboratory must be critically evaluated to determine whether it reflects, at least in part, the patterns of root microbial communities found in field biology experiments in natural environments.
For example, use of A. thaliana root-derived bacterial and fungal culture collections (Bai et al., 2015; Duran et al., 2018), isolated from the same soil, revealed in a peat-based flowpot system (Kremer et al., 2021) the essential role of the bacterial microbiota in protecting the host against detrimental root-associated fungal communities (Duran et al., 2018). This laboratory finding recapitulates mainly antagonistic interactions between root-associated bacterial and fungal communities as determined by microbial interkingdom network analyses in root samples collected from natural A. thaliana populations (Duran et al., 2018). Similarly, competition experiments with mixtures of bacterial SynComs derived from two culture collections of the root microbiota of Lotus japonicus and A. thaliana showed that native SynComs outcompeted the non-native communities when two plant species were cultivated in the peat-based flowpot system (Wippel et al., 2021). This laboratory finding recapitulated host species‒specific signatures of the root microbiota when these plant species were grown in the same natural soil. Subsequent sequential inoculation experiments with the same native and non-native SynComs demonstrated that host preference of the bacterial commensals resulted from the invasiveness and persistence of the native bacterial communities on either host (Wippel et al., 2021). An agar-based gnotobiotic plant growth system was used together with At -R-SPHERE to reveal a role of individual bacterial commensals and a SynCom in alleviating A. thaliana iron starvation. This beneficial interaction is elicited by root-secreted coumarins and is independent of their known antimicrobial activity (Harbort, Hashimoto, Inoue, & Schulze-Lefert, 2020). Root commensal-mediated iron mobilization for plant growth in the agar-based system is thought to partly explain why A. thaliana coumarin biosynthesis mutants grow poorly in natural iron-limiting soil and exhibit shifts in the bacterial root microbiota compared with wild-type species (Harbort, Hashimoto, Inoue, & Schulze-Lefert, 2020). Collectively, these examples demonstrate the need to establish plant species-specific microbial culture collections (Zhang et al., 2021) from contrasting natural environments and gnotobiotic systems for reconstitution experiments. Ultimately, this might provide a toolbox to test whether beneficial functions of the plant microbiota are the result of adaptation or coadaption of the host with its microbiota to the respective environment.
There are multiple choices for gnotobiotic plant cultivation. The first system described here involves growth of plants according to their natural gravitropism on vertical agar plates. In Basic Protocol 2, bacteria are homogenously distributed in the agar matrix at a low titer (OD600 = 0.0005) in the absence of an external organic carbon source such as sucrose. In this system root colonization and subsequent proliferation of the bacteria is mainly driven by photoassimilate-derived root exudates. This contrasts with drop inoculation experiments in which a high initial bacterial titer is often directly applied to the roots. In our hands, we never observe bacterial overgrowth on or inside the agar matrix, indicating that the cultured root commensals from At -R-SPHERE cannot efficiently metabolize agar carbohydrates as energy source. In comparison to natural soil substrate, individual parameters including mineral nutrient bioavailability, gel strength, and pH can be manipulated according to experimental needs. All plant organs, including roots can be collected with ease for subsequent analyses. This versatility makes agar plates one of the most popular gnotobiotic plant system to study root-microbiota interactions. However, roots of agar-grown plants are exposed to light, and this triggers light-stimulated responses in the root organ, for which consequences on root-microbe associations are unknown (Silva-Navas et al., 2016). Damage of the root epidermis occurs naturally through lateral root emergence, and these sites are known as hotspots for bacterial root colonization, which can be detected on agar plate‒grown plants (Lundberg et al., 2012). However, surface growth of roots on vertical plates prevents extensive damage to the root system seen in natural soil (Bulgarelli et al., 2012). In mono-associations, coincidence of root damage and perception of microbial elicitors is needed for localized immune responses to a tested pathogenic and commensal bacterium in the root differentiation zone (Zhou et al., 2020). We describe in Basic Protocol 2 how to inoculate a single bacterial strain or SynComs in agar matrix. As a potential application, we describe in Basic Protocol 3 how to quantify plant susceptibility to the opportunistic Pseudomonas pathogen strain R401 using a viable plate counting method. We consider the viable plate counting protocol to be advantageous compared with qPCR-based bacterial quantification because viable plate counting assesses absolute bacterial abundance and discriminates between live and dead microbes.
Kremer and colleagues developed two gnotobiotic systems using sterile peat-based matrix, called flowpot and gnotopot (Kremer et al., 2021). Plant growth in these systems more closely mimics growth in natural soil because it provides soil organic matter to both soilborne microbes and the host, permits root growth in the dark, and likely exposes roots to physical damage. However, peat matrix is not suitable to mimic variation in edaphic factors found in different soil types. In addition, defined mineral malnutrition is almost impossible to apply in these two gnotobiotic systems due to nutrient leaching from peat. Application of defined concentrations of exogenously applied chemicals or microbial elicitors such as immunogenic flg22 peptide also remains challenging, possibly due to their absorption by peat. Finally, collecting intact A. thaliana roots grown in peat matrix is impossible due to intimate association between fine roots and peat fibers, limiting the interpretation of data collected from downstream analysis such as root transcriptome and microscopic studies. Here, we developed the Alternate Protocol to grow A. thaliana plants on perlite matrix. The perlite system is easy to set up without the need for specialized equipment as required for the assembly of the flowpot system (Kremer et al., 2021). The perlite system is also advantageous in terms of the relative ease in harvesting intact roots. Roots are grown in the dark and likely experience physical damage during gravitropic growth. We tested whether the perlite system was suitable to apply defined mineral nutrient stresses, including phosphate starvation, and an immunogenic peptide elicitor such as flg22. Recently, a tailor-made gnotobiotic system, designated EcoFAB, has been reported (Zengler et al., 2019). Users can use 3D printing to design their own physical chambers to accommodate growth of different plant species, each with different space requirements or growth periods. However, some experience in engineering is required. Whatever systems are used, these simplified systems inevitably fall short in recapitulating the complexity of natural soil and fluctuating environment. However, this simplification is essential to tease apart the contribution of the plant microbiota to indirect protection against pathogens, mineral nutrition of the host, abiotic stress tolerance, and phytohormone-mediated plant growth promotion. In summary, studies on different beneficial functions of the microbiota require different gnotobiotic plant systems.
Due to an increasing demand for mechanistic studies underlying root associations with commensal microorganisms, we provide Basic Protocol 4, enabling high-throughput screening of bacterial mutant libraries, microbiota culture collections in mono-associations, or SynCom contexts. The protocol described here was successful to identify bacterial Tn5 transposon insertion mutants of a Rhodanobacter root commensal, present in At -R-SPHERE, with altered immunosuppressive activities using root growth inhibition as an output (Ma et al., 2021). This 96-well-based system enables nondestructive visual inspection of root growth within the agar matrix from a side view over time. We expect a wider application of this setup to screen for other commensal-mediated beneficial traits such as rescue of plant growth under defined mineral malnutrition or altered root architecture.
Critical Parameters
For commensal microorganisms, selectable markers such as antibiotic resistance are often unknown. This necessitates stringent quality control by DNA sequencing of the 16S rRNA marker gene to validate absence of contaminating strains. We experienced that a change in pH or agar supplier can exert a profound impact on microbe-mediated plant phenotypes, which is probably partly explained by batch-to-batch variation of mineral nutrients in the agar preparations (Gruber et al., 2013). For the agar plate protocol without sucrose, a fresh seed batch will increase germination rate. Materials including seeds and culture medium should be kept sterile by means of surface sterilization or autoclaving, respectively.
Concerning the high-throughput screening based on Basic Protocol 4, attention should be paid to avoid cross-contamination of neighboring strains in the 96-well format. Variation in the microenvironment between individual wells can confound the identification of commensal bacteria‒mediated plant root phenotypes (e.g., due to uneven distribution of microbes or inclusion of air bubbles into solidified agar). The combination of high-throughput screening using phytostrips (Basic Protocol 4) and subsequent validation with vertical square plates (Basic Protocol 2) reduces the number of false-positive candidates.
Troubleshooting
See Table 1 for common problems encountered during these protocols, their potential causes, and suggested solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Bacteria show different morphologies | Contamination of original bacterial stock | Restreak cells to obtain single colonies; validate by sequencing using Support Protocol 1 |
| Bacterial culture is too old | Prepare fresh bacterial cultures from glycerol stocks | |
| Bacteria do not colonize plant roots to degree expected | Bacteria are dead | Prepare fresh bacterial culture |
| Ensure bacteria are inoculated in agar medium cooled to about 50°C | ||
| Some bacteria form aggregates, resulting in uneven distribution of bacteria on agar plate | Use fresh bacterial culture, and mix agar medium well after inoculation with bacteria | |
| Infection assay shows too much variation | Inoculated plants are of different ages | Ensure plants inoculated are of similar size and age |
| Bacteria are not inoculated evenly | Ensure plants are completely flooded with bacterial culture; sometimes gentle mixing is needed to remove air bubbles trapped on plant surfaces, which prevents direct contact of bacteria with plants | |
| Humidity is too high | Add Silwet to inoculation medium in Basic Protocol 3 step 2 | |
| Ensure excessive liquid is removed from plate before placing back into phytochamber |
Understanding Results
With Basic Protocol 2 or Support Protocol 2, primary plant roots will grow to be ∼5 cm long after 2 weeks. Addition of flg22 will lead to about 20% (wild-type Col-0) to 80% (pWER::FLS2-GFP) reduction in plant root growth depending on the plant genotype. Supplementation of individual bacteria or SynCom can exaggerate or rescue growth-reduction phenotypes. Plants grown in both systems could be harvested with ease for downstream application including quantification of bacterial titer, DNA/RNA extraction for determination of marker gene expression, or community profiling. If viable plate counting will be performed using Basic Protocol 3, plants should be cleaned to remove agar or perlite still attached to plant roots and blotted dry before weighing. Plants inoculated with strain R401 are expected to accommodate up to 108 to 109 CFU per gram of tissue 2 days after inoculation. With Basic Protocol 4, plants can be coinoculated with chemicals or bacteria for up to 3 weeks. Plants can be phenotyped continuously until the end of the experiment, and chemicals can be applied to the roots continuously from the liquid reservoir.
Time Considerations
In general, plants can be inoculated with bacteria before germination or any time afterward. For the infection assay, 2-week-old plants are ideal (Basic Protocol 3). Screening using phytostrip (Basic Protocol 4) usually takes 2 to 3 weeks after plants are inoculated. Plants can be grown on the agar plate (Basic Protocol 2) or perlite system (Alternate Protocol) for up to 3 weeks.
Acknowledgments
Funding was provided by the Max Planck Society and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy, EXC-Nummer 2048/1 project 390686111, and SPP 2125 DECRyPT to P. S.-L.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Ka-Wai Ma and Jana Ordon : methodology, investigation, writing of original draft, reviewing, and editing; Paul Schulze-Lefert : conceptualization, supervision, manuscript review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Armstrong, J. A., & Schulz, J. R. (2015). Agarose gel electrophoresis. Current Protocols Essential Laboratory Techniques , 10, 7.2.1‒7.2.22. doi: 10.1002/9780470089941.et0702s10.
- Bai, Y., Muller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., … Schulze-Lefert, P. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature , 528(7582), 364–369. doi: 10.1038/nature16192.
- Berendsen, R. L., Vismans, G., Yu, K., Song, Y., de Jonge, R., Burgman, W. P., … Pieterse, C. M. J. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. The ISME Journal , 12(6), 1496–1507. doi: 10.1038/s41396-018-0093-1.
- Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., … Schulze-Lefert, P. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature , 488(7409), 91–95. doi: 10.1038/nature11336.
- Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren van Themaat, E., & Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annual Review of Plant Biology , 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106.
- Burrell, T., Fozard, S., Holroyd, G. H., French, A. P., Pound, M. P., Bigley, C. J., … Forde, B. G. (2017). The Microphenotron: A robotic miniaturized plant phenotyping platform with diverse applications in chemical biology. Plant Methods , 13, 10. doi: 10.1186/s13007-017-0158-6.
- Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., & Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods , 13(7), 581–583. doi: 10.1038/nmeth.3869.
- Carrion, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., de Hollander, M., Ruiz-Buck, D., … Raaijmakers, J. M. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science , 366(6465), 606–612. doi: 10.1126/science.aaw9285.
- Castrillo, G., Teixeira, P. J., Paredes, S. H., Law, T. F., de Lorenzo, L., Feltcher, M. E., … Dangl, J. L. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature , 543(7646), 513–518. doi: 10.1038/nature21417.
- Duran, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., & Hacquard, S. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell , 175(4), 973–983.e914. doi: 10.1016/j.cell.2018.10.020.
- Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., & Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proceedings of the National Academy of Sciences of the United States of America , 115(6), E1157–E1165. doi: 10.1073/pnas.1717617115.
- Garrido-Oter, R., Nakano, R. T., Dombrowski, N., Ma, K. W., AgBiome, T., McHardy, A. C., & Schulze-Lefert, P. (2018). Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host & Microbe, 24(1), 155–167.e155. doi: 10.1016/j.chom.2018.06.006.
- Gruber, B. D., Giehl, R. F. H., Friedel, S., & von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiology , 163(1), 161–179. doi: 10.1104/pp.113.218453.
- Hacquard, S., Garrido-Oter, R., Gonzalez, A., Spaepen, S., Ackermann, G., Lebeis, S., … Schulze-Lefert, P. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host & Microbe, 17(5), 603–616. doi: 10.1016/j.chom.2015.04.009.
- Harbort, C. J., Hashimoto, M., Inoue, H., Niu, Y., Guan, R., Rombola, A. D., … Schulze-Lefert, P. (2020). Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host & Microbe, 28(6), 825–837.e826. doi: 10.1016/j.chom.2020.09.006.
- Harbort, C. J., Hashimoto, M., Inoue, H., & Schulze-Lefert, P. (2020). A gnotobiotic growth assay for Arabidopsis root microbiota reconstitution under iron limitation. STAR Protocols , 1(3), 100226. doi: 10.1016/j.xpro.2020.100226.
- He, X. Q., Zhang, Q., Jin, Y., Jiang, L. B., Wu, R. L., & Li, B. (2021). Network mapping of root-microbe interactions in Arabidopsis thaliana. npj Biofilms and Microbiomes , 7(1), 72. doi: 10.1038/s41522-021-00241-4.
- Hou, S., Thiergart, T., Vannier, N., Mesny, F., Ziegler, J., Pickel, B., & Hacquard, S. (2021). A microbiota-root-shoot circuit favours Arabidopsis growth over defence under suboptimal light. Nature Plants , 7(8), 1078–1092. doi: 10.1038/s41477-021-00956-4.
- Kearl, J., McNary, C., Lowman, J. S., Mei, C., Aanderud, Z. T., Smith, S. T., … Nielsen, B. L. (2019). Salt-tolerant halophyte rhizosphere bacteria stimulate growth of alfalfa in salty soil. Frontiers in Microbiology , 10, 1849. doi: 10.3389/fmicb.2019.01849.
- Kremer, J. M., Sohrabi, R., Paasch, B. C., Rhodes, D., Thireault, C., Schulze-Lefert, P., … He, S. Y. (2021). Peat-based gnotobiotic plant growth systems for Arabidopsis microbiome research. Nature Protocols , 16(5), 2450–2470. doi: 10.1038/s41596-021-00504-6.
- Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., … Kim, J. F. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nature Biotechnology , 36, 1100‒1109. doi: 10.1038/nbt.4232.
- Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., … Dangl, J. L. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature , 488(7409), 86–90. doi: 10.1038/nature11237.
- Ma, K. W., Niu, Y., Jia, Y., Ordon, J., Copeland, C., Emonet, A., … Schulze-Lefert, P. (2021). Coordination of microbe-host homeostasis by crosstalk with plant innate immunity. Nature Plants , 7(6), 814–825. doi: 10.1038/s41477-021-00920-2.
- Maier, B. A., Kiefer, P., Field, C. M., Hemmerle, L., Bortfeld-Miller, M., Emmenegger, B., … Vorholt, J. A. (2021). A general non-self response as part of plant immunity. Nature Plants , 7(5), 696–705. doi: 10.1038/s41477-021-00913-1.
- Santos-Medellin, C., Liechty, Z., Edwards, J., Nguyen, B., Huang, B., Weimer, B. C., & Sundaresan, V. (2021). Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nature Plants , 7(8), 1065–1077. doi: 10.1038/s41477-021-00967-1.
- Sasse, J., Kosina, S. M., de Raad, M., Jordan, J. S., Whiting, K., Zhalnina, K., & Northen, T. R. (2020). Root morphology and exudate availability are shaped by particle size and chemistry in Brachypodium distachyon. Plant Direct , 4(7), e00207. doi: 10.1002/pld3.207.
- Schlaeppi, K., Dombrowski, N., Oter, R. G., Ver Loren van Themaat, E., & Schulze-Lefert, P. (2014). Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proceedings of the National Academy of Sciences of the United States of America , 111(2), 585–592. doi: 10.1073/pnas.1321597111.
- Shendure, J. A., Porreca, G. J., Church, G. M., Gardner, A. F., Hendrickson, C. L., Kieleczawa, J., & Slatko, B. E. (2011). Overview of DNA sequencing strategies. Current Protocols in Molecular Biology , 96, 7.1.1‒7.1.23. doi: 10.1002/0471142727.mb0701s96.
- Silva-Navas, J., Moreno-Risueno, M. A., Manzano, C., Tellez-Robledo, B., Navarro-Neila, S., Carrasco, V., … del Pozo, J. C. (2016). Flavonols mediate root phototropism and growth through regulation of proliferation-to-differentiation transition. Plant Cell , 28(6), 1372–1387. doi: 10.1105/tpc.15.00857.
- Staley, J. T., & Konopka, A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annual Review of Microbiology , 39, 321–346. doi: 10.1146/annurev.mi.39.100185.001541.
- Stringlis, I. A., Proietti, S., Hickman, R., Van Verk, M. C., Zamioudis, C., & Pieterse, C. M. J. (2018). Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant Journal , 93(1), 166–180. doi: 10.1111/tpj.13741.
- Teixeira, P., Colaianni, N. R., Law, T. F., Conway, J. M., Gilbert, S., Li, H., … Dangl, J. L. (2021). Specific modulation of the root immune system by a community of commensal bacteria. Proceedings of the National Academy of Sciences of the United States of America , 118(16), e2100678118. doi: 10.1073/pnas.2100678118.
- Thiergart, T., Duran, P., Ellis, T., Vannier, N., Garrido-Oter, R., Kemen, E., … Hacquard, S. (2020). Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nature Ecology & Evolution, 4(1), 122–131. doi: 10.1038/s41559-019-1063-3.
- Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., & Singh, B. K. (2020). Plant-microbiome interactions: From community assembly to plant health. Nature Reviews Microbiology , 18(11), 607–621. doi: 10.1038/s41579-020-0412-1.
- Vogel, C., Bodenhausen, N., Gruissem, W., & Vorholt, J. A. (2016). The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytologist , 212(1), 192–207. doi: 10.1111/nph.14036.
- Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nature Reviews Microbiology , 10(12), 828–840. doi: 10.1038/nrmicro2910.
- Wippel, K., Tao, K., Niu, Y., Zgadzaj, R., Kiel, N., Guan, R., … Garrido-Oter, R. (2021). Host preference and invasiveness of commensal bacteria in the Lotus and Arabidopsis root microbiota. Nature Microbiology , 6(9), 1150–1162. doi: 10.1038/s41564-021-00941-9.
- Wyrsch, I., Dominguez-Ferreras, A., Geldner, N., & Boller, T. (2015). Tissue-specific FLAGELLIN-SENSING 2 (FLS2) expression in roots restores immune responses in Arabidopsis fls2 mutants. New Phytologist , 206(2), 774–784. doi: 10.1111/nph.13280.
- Yeoh, Y. K., Dennis, P. G., Paungfoo-Lonhienne, C., Weber, L., Brackin, R., Ragan, M. A., … Hugenholtz, P. (2017). Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nature Communication , 8(1), 215. doi: 10.1038/s41467-017-00262-8.
- Yu, P., He, X., Baer, M., Beirinckx, S., Tian, T., Moya, Y. A. T., … Hochholdinger, F. (2021). Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nature Plants , 7(4), 481–499. doi: 10.1038/s41477-021-00897-y.
- Zengler, K., Hofmockel, K., Baliga, N. S., Behie, S. W., Bernstein, H. C., Brown, J. B., … Northen, T. R. (2019). EcoFABs: Advancing microbiome science through standardized fabricated ecosystems. Nature Methods , 16(7), 567–571. doi: 10.1038/s41592-019-0465-0.
- Zhang, J., Liu, Y. X., Guo, X., Qin, Y., Garrido-Oter, R., Schulze-Lefert, P., & Bai, Y. (2021). High-throughput cultivation and identification of bacteria from the plant root microbiota. Nature Protocols , 16(2), 988–1012. doi: 10.1038/s41596-020-00444-7.
- Zhou, F., Emonet, A., Tendon, V. D., Marhavy, P., Wu, D. S., Lahaye, T., & Geldner, N. (2020). Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell , 180(3), 440–453.e18. doi: 10.1016/j.cell.2020.01.013.
Internet Resources
Curated database that provides detailed information on the A. thaliana culture collection as described previously (Bai et al., 2015).
Citing Literature
Number of times cited according to CrossRef: 6
- Ryohei Thomas Nakano, Tomohisa Shimasaki, Long-Term Consequences of PTI Activation and Its Manipulation by Root-Associated Microbiota, Plant And Cell Physiology, 10.1093/pcp/pcae033, 65 , 5, (681-693), (2024).
- Jana Ordon, Julien Thouin, Ryohei Thomas Nakano, Ka-Wai Ma, Pengfan Zhang, Bruno Huettel, Ruben Garrido-Oter, Paul Schulze-Lefert, Chromosomal barcodes for simultaneous tracking of near-isogenic bacterial strains in plant microbiota, Nature Microbiology, 10.1038/s41564-024-01619-8, 9 , 4, (1117-1129), (2024).
- Qian Zhou, Jilian Wang, Tian Zhang, Mingyuan Li, Special Fungal Community Structure Formed by Typical Halophytes in the Rhizosphere Soil Under the Synergistic Action of Different Saline and Alkaline Environments, Journal of Plant Growth Regulation, 10.1007/s00344-024-11422-8, (2024).
- M. J. Poupin, T. Ledger, R. Roselló-Móra, B. González, The Arabidopsis holobiont: a (re)source of insights to understand the amazing world of plant–microbe interactions, Environmental Microbiome, 10.1186/s40793-023-00466-0, 18 , 1, (2023).
- Reza Sohrabi, Bradley C. Paasch, Julian A. Liber, Sheng Yang He, Phyllosphere Microbiome, Annual Review of Plant Biology, 10.1146/annurev-arplant-102820-032704, 74 , 1, (539-568), (2023).
- Ankita Bhattacharyya, Olga Mavrodi, Niladri Bhowmik, David Weller, Linda Thomashow, Dmitri Mavrodi, Bacterial biofilms as an essential component of rhizosphere plant-microbe interactions, Biofilms, 10.1016/bs.mim.2023.05.006, (3-48), (2023).

