Gene Delivery and Analysis of Optogenetic Induction of Lytic Cell Death
Kai Zhang, Kai Zhang, Teak-Jung Oh, Teak-Jung Oh, Bryan Gworek, Bryan Gworek, Amna Mehfooz, Amna Mehfooz
Abstract
Necroptosis is a form of inflammatory lytic cell death involving active cytokine production and plasma membrane rupture. Progression of necroptosis is tightly regulated in time and space, and its signaling outcomes can shape the local inflammatory environment of cells and tissues. Pharmacological induction of necroptosis is well established, but the diffusive nature of chemical death inducers makes it challenging to study cell-cell communication precisely during necroptosis. Receptor-interacting protein kinase 3, or RIPK3, is a crucial signaling component of necroptosis, acting as a crucial signaling node for both canonical and non-canonical necroptosis. RIPK3 oligomerization is crucial to the formation of the necrosome, which regulates plasma membrane rupture and cytokine production. Commonly used necroptosis inducers can activate multiple downstream signaling pathways, confounding the signaling outcomes of RIPK3-mediated necroptosis. Opsin-free optogenetic techniques may provide an alternative strategy to address this issue. Optogenetics uses light-sensitive protein-protein interaction to modulate cell signaling. Compared to chemical-based approaches, optogenetic strategies allow for spatiotemporal modulation of signal transduction in live cells and animals. We developed an optogenetic system that allows for ligand-free optical control of RIPK3 oligomerization and necroptosis. This article describes the sample preparation, experimental setup, and optimization required to achieve robust optogenetic induction of RIPK3-mediated necroptosis in colorectal HT-29 cells. We expect that this optogenetic system could provide valuable insights into the dynamic nature of lytic cell death. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Production of lentivirus encoding the optogenetic RIPK3 system
Support Protocol : Quantification of the titer of lentivirus
Basic Protocol 2 : Culturing, chemical transfection, and lentivirus transduction of HT-29 cells
Basic Protocol 3 : Optimization of optogenetic stimulation conditions
Basic Protocol 4 : Time-stamped live-cell imaging of HT-29 lytic cell death
Basic Protocol 5 : Quantification of HT-29 lytic cell death
INTRODUCTION
Cell death is an essential mechanism to maintain the homeostasis of multicellular organisms. Apoptosis is a form of programmed cell death that clears apoptotic corpses in a “quiet” way, in that minimal inflammatory factors are released. Necrosis, which is typically not programmed, proceeds with membrane rupture and release of cellular contents into the extracellular space, resulting in inflammation. Necroptosis bears features of both apoptosis and necrosis in that cell rupture proceeds with concomitant upregulation of proinflammatory cytokine production. Defective necroptosis has been reported in multiple inflammatory diseases, such as inflammatory bowel diseases (IBD) and neurodegenerative diseases. Therefore, understanding the precise mechanism of necroptosis should benefit the development of future therapeutics.
A well-established canonical necroptosis pathway is initiated by tumor necrosis factor alpha (TNFα). TNFα binding to its receptor, TNF receptor 1 (TNFR1), recruits receptor-interacting protein kinase 1 (RIPK1) and other proteins to form a receptor complex called complex-I at the plasma membrane, which further promotes activation of the nuclear factor kappa B (NF-κB) pathway (Zhu et al., 2018). When apoptosis is blocked by the broad-spectrum caspase inhibitor zVAD-FMK, a membrane-less protein complex called a necrosome is formed, containing RIPK1 and RIPK3 (Cho et al., 2009). RIPK3 phosphorylates mixed lineage kinase domain-like (MLKL) and causes membrane rupture (Dondelinger et al., 2014; Sun et al., 2012). Non-canonical necroptosis also exists, for example, in the case of viral induction, and proceeds through RIPK3-dependent signaling processes. Highly inflammatory, necroptosis is regulated tightly in time and space.
The commonly used pharmacological approaches to stimulate necroptosis may face challenges in delineating the spatial and temporal regulation of necroptosis, in part due to the diffusive nature of chemical death inducers. Additionally, the potential off-target effects of small molecules make it difficult to define the function of specific signaling components such as RIPK3. One alternative strategy to address this issue is optogenetic technology, which engineers cells to harbor light-sensitive signaling molecules. Exposure to visible light induces conformational changes in the light-sensitive signaling molecules and activates downstream signaling pathways. Because light can be controlled precisely in time and space, signaling modulation can be achieved at the single-cell or even subcellular level (Fischer et al., 2022; Liu et al., 2022; Mazraeh & Di Ventura, 2022; Oh et al., 2021).
Here, we demonstrate the optical control of necroptosis in live cells. The system uses cryptochrome protein, a photoactivable protein that can be oligomerized by blue light, to induce RIPK3 activation in live cells. In this article, we introduce the procedure of preparing lentivirus encoding the optogenetic RIPK3 system (Basic Protocol 1 and Support Protocol), lentiviral transduction (Basic Protocol 2), optimization of optogenetic stimulation conditions (Basic Protocol 3), long-term time-stamped live-cell imaging and optogenetic induction (Basic Protocol 4), and data analysis (Basic Protocol 5).
STRATEGIC PLANNING
Successful achievement of optogenetic induction of necroptosis requires high-quality lentivirus, optimized optogenetic stimulation conditions, and familiarity with data analysis algorithms. Several steps can be performed in parallel, for example, Basic Protocols 1 and 3.
CAUTION : All procedures regarding lentivirus production, human cell line culturing, chemical transfection, and lentivirus transduction should be done in a certified biosafety level 2 (BSL2) biosafety cabinet. Please handle all materials with great care throughout the procedures to minimize potential microbe contamination. The imaging room for handling live human cell line imaging should meet the BSL2 requirement as well.
NOTE : All solutions and equipment coming into contact with cells must be sterile, and proper sterile technique should be used accordingly, including using a certified biosafety class II cabinet (Thermo Fisher 1300 Series A2).
NOTE : All culture incubations are performed in a 37°C, 5% CO2, 95% air incubator unless otherwise specified.
Basic Protocol 1: PRODUCTION OF LENTIVIRUS ENCODING THE OPTOGENETIC RIPK3 SYSTEM
This protocol outlines the steps to produce lentivirus encoding the optogenetic RIPK3 system. Lentivirus transduction is essential to achieve effective gene delivery into HT-29 cells (Basic Protocol 2).
Materials
-
Low-passage (<P15) HEK293T cells (ATCC, cat. no. CRL-3216)
-
Complete DMEM [supplemented with fetal bovine serum (FBS); see recipe], 37°C
-
Plasmids:
- Transfer plasmid (for optogenetic RIPK3)
- Packaging plasmid (psPAX2)
- Envelope plasmid (pMD2.G)
-
TurboFect Transfection Reagent (Thermo Fisher, cat. no. R0532)
-
Dulbecco's Modified Eagle's Medium (DMEM; Corning, cat. no. 10-013-CV) with antibiotics (1× penicillin/streptomycin, from 10,000 U/ml stock; Corning, cat. no. 30-002-CI) and without FBS
-
Lenti-X Concentrator (Takara, cat. no. 631231, lot no. 1710388A)
-
Complete medium of user's choice or Dulbecco's phosphate-buffered saline (DPBS; Thermo Fisher, cat. no. 14040141)
-
100-mm culture plates (Fisherbrand, cat. no. FB012924)
-
1.5-ml Eppendorf tubes (Thermo Fisher)
-
15- or 50-ml Falcon tubes (Thermo Fisher)
-
Standard tabletop centrifuge (Sorvall ST 8 Centrifuge, Thermo Fisher, cat. no. 75007200), room temperature and 4°C
-
0.4-μm syringe filters, sterile (Milltex PVDF 0.45 μm Sterile Syringe Filter, Millipore/Sigma-Aldrich, cat. no. SLHVR33RS)
-
10-ml syringes, sterile (Fisherbrand, cat. no. 14-955-459)
-
Additional reagents and equipment for measuring lentivirus concentration (see Support Protocol)
1.Grow low-passage HEK293T cells in a 100-mm culture plate in 10 ml complete DMEM (supplemented with FBS) at a 10% initial seeding density until 85% to 90% confluent to maximize the virus yield.
2.Prepare 800 μl transfection mix in a 1.5-ml Eppendorf tube, with a 3:2:1 molar ratio of transfer/packaging/envelope plasmid and a ratio of DNA/TurboFect Transfection Reagent of 1 μg to 3 μl.
3.Once the transfection mix is made, gently remove the old culture medium and replenish with 7.2 ml DMEM with antibiotics and without FBS (8 ml total volume).
4.Wait for 20 min for the lipid particle to generate complex at room temperature.
5.Add 800 μl of the transfection mix evenly to the plate dropwise.
6.Incubate the transfected cells in a 37°C, 5% CO2 incubator for 20 hr.
7.After 20 hr, replace the medium with 8 ml complete DMEM.
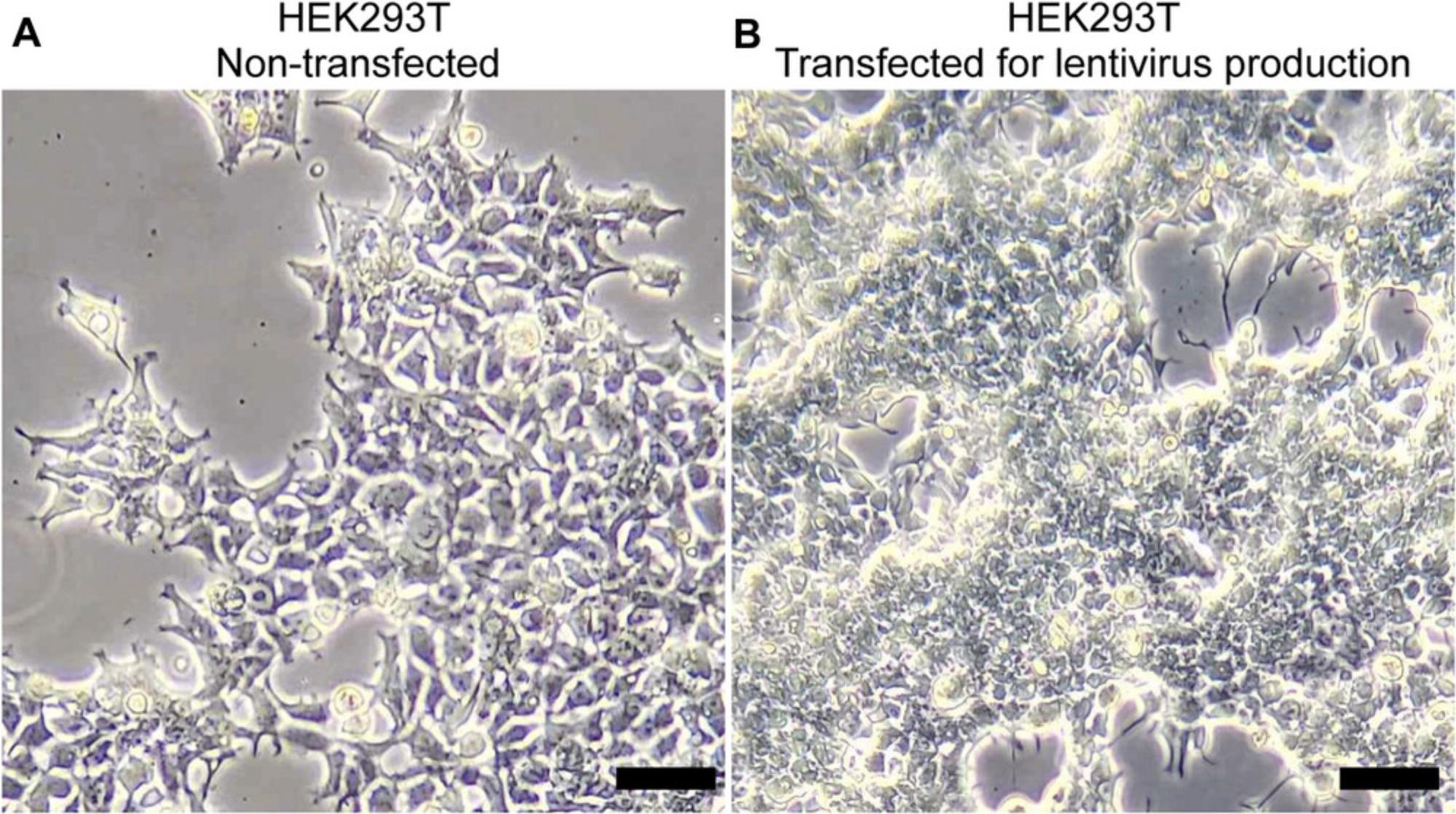
8.Incubate the plate again at 37°C and 5% CO2 for an additional 48 hr.
9.To harvest viral particles, proceed by gently pipetting virus-containing medium into a 15- or 50-ml Falcon tube.
10.Centrifuge virus-containing medium for 7 min at 500 × g to acquire pellet.
11.Gently draw the supernatant into a pipet without disturbing the cell pellet and filter through a 0.45-μm syringe filter using a 10-ml syringe into another 15- or 50-ml Falcon tube. Discard the pellet.
12.Measure virus concentration using Lenti-X GoStix Plus lentiviral titer kit (see Support Protocol).
13.Mix the filtered supernatant with Lenti-X concentrator at a 3:1 ratio of filtered virus medium/concentrator.
14.Incubate the sample on ice for ≥1 hr.
15.Centrifuge concentrated medium for 45 min at 1500 × g , 4°C.
16.Remove supernatant without disturbing the pellet.
17.Add complete medium of the user's choice or DPBS to the pellet at 1/10 or 1/100 of the original supernatant volume.
18.Gently resuspend the viral pellet by pipetting up and down 50 times without forming bubbles.
19.Once the viral pellet is resuspended, measure the concentration of the virus using the Lenti-X GoStix Plus lentiviral titer kit (see Support Protocol).
20.Store product for 2 to 4 months at −80°C. Thaw when needed.
Support Protocol: QUANTIFICATION OF THE TITER OF LENTIVIRUS
This support protocol lists steps to quantify the titer of lentivirus from Basic Protocol 1.Follow the guide on Takara's website for the Lenti-X GoStix Plus lentiviral titer kit.
Materials
-
Lenti-X GoStix Plus lentiviral titer kit (Takara, cat. no. 631280), containing cassette and chase buffer
-
Diluted virus sample (from concentrated virus from Basic Protocol 1)
-
Smartphone
1.Download the GoStix Plus app (available on Google Play or in the Apple App Store by searching “GoStix Plus” in the search bar) onto a smartphone.
2.Optional: Add an email address to have the result sent to that address.
3.Enter the lot number by scanning the QR code on the Lenti-X GoStix Plus lentiviral titer kit foil pouch. Press the QR code icon to activate the scanner.
4.Enter the number of tests to be scanned (1 to 8 tests).
5.Enter the sample names and the appropriate dilutions and then click “start.”
6.Following the prompts within the app, add 20 μl diluted virus sample to the cassette from the kit.
7.Add 80 μl chase buffer to the sample well and wait for 10 min using the timer in the app.
8.Ensure that the control band appears on the left and the test band appears on the right.
9.Using the app, align the cassette using the alignment tool.
10.Once all samples are scanned, view the results in the “Result detail” window. Press “done” if all samples have been scanned.
11.Interpret the results.
Basic Protocol 2: CULTURING, CHEMICAL TRANSFECTION, AND LENTIVIRUS TRANSDUCTION OF HT-29 CELLS
This protocol outlines the steps to culture the colorectal HT-29 cell line and perform chemical transfection or lentivirus transduction. A sufficient gene delivery rate can be achieved in HEK293T cells by either chemical transfection or lentiviral transduction. However, HT-29 cells require lentiviral transduction for effective gene delivery (Fig. 2).
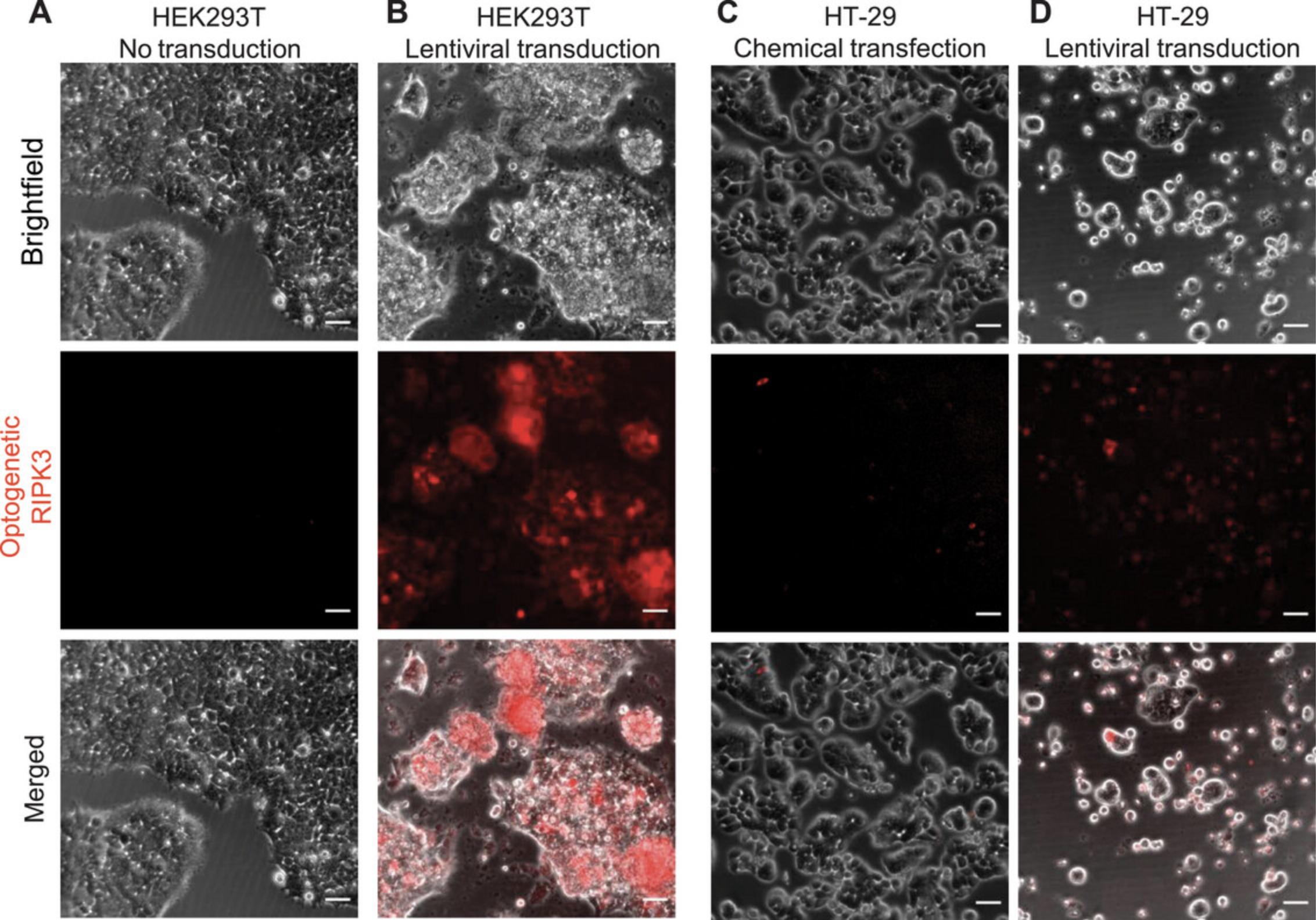
Materials
-
HT-29 cells (ATCC, cat. no. HTB-38)
-
Complete HT-29 cell culture medium (see recipe), 37°C
-
Optogenetic RIPK3 (fused with mCherry) plasmid
-
Opti-MEM medium (Thermo Fisher, cat. no. 31985062)
-
PEI MAX (Polysciences, cat. no. 24765-1)
-
Concentrated and quantified La-RIPK3 virus (see Basic Protocol 1 and Support Protocol)
-
Polybrene transfection reagent
-
70% (v/v) isopropyl alcohol
-
60-mm culture plates (Fisher Scientific, cat. no. FB012921)
-
24-well culture plates (Fisherbrand, cat. no. FB012929)
-
Standard tabletop centrifuge (Sorvall Legend XTR) with swinging-bucket rotor (Thermo Scientific, TX-1000) and plate adapter
-
Parafilm
-
Large Kimwipes
Cell culture
1.Culture HT-29 cells in 4 ml complete HT-29 cell culture medium in a 60-mm culture plate at an initial seeding density of 2 × 106 cells, changing the medium every 3 days and splitting the cells when they are 80% confluent.
Chemical transfection
2a. Plate 1.6 × 105 cells/ml in a 24-well culture plate for chemical transfection.
3a. Incubate the cells for 2 days. Supplement them with 500 µl fresh complete HT-29 cell culture medium 24 hr after cell plating.
4a. Gently remove the medium and add 450 µl fresh complete HT-29 cell culture medium before transfection.
5a. Prepare transfection mix using 500 ng of the optogenetic RIPK3 (fused with mCherry) plasmid 1.5 µl PEI MAX, and Opti-MEM medium to 50 µl.
6a. Incubate the transfection mix for 15 min at room temperature.
7a. Add 50 µl transfection mix to each well dropwise and incubate the cells for 4 hr.
8a. After 4 hr, remove the transfection medium and add 500 µl fresh complete HT-29 cell culture medium to the cells.
Lentivirus transduction
2b. Plate 1.6 × 105 cells/ml in a 24-well culture plate for lentivirus transduction.
3b. Pre-warm the centrifuge with a swinging-bucket rotor to 30°C.
4b. Make a virus cocktail to be added to the target cells.
1.Concentrated and quantified La-RIPK3 virus amount: 1501 GV.
GV units are custom units from Takara and are ng/ml of the p24 protein of the lentivirus capsid.
This procedure is for 24-well plates. Ideally, the amount of virus should be increased or decreased based on the area of the plate.
- Polybrene transfection reagent: final concentration 8 µg/ml.
- Make up the rest of the volume with the complete medium of the target cells.
5b. Add the virus cocktail to the HT-29 cells in the 24-well plate (see step 2b).
6b. Incubate the cells in the 37°C incubator for 10 min.
7b. Proceed to spin infection:
- Sterilize the centrifuge plate adapter with 70% isopropyl alcohol.
- Parafilm the entire junction of the cell plate and its lid and then wrap it with a large Kimwipe (Fig. 3).
This is done so the CO2 and humidity can be maintained better during the 1-hr centrifugation (see step 7b, iii). Wrapping the plate using a Kimwipe minimizes the chance of contamination by bacteria likely to be present in the centrifuge.
- Place the Kimwipe-wrapped plate into the centrifuge adapter and balance the other side of the rotor with a plate of similar weight (within 1 g of difference).
- Proceed to centrifugation for 1 hr at 3000 rpm in a TX-1000 swinging-bucket rotor, 30°C.
Spin infection can improve the viral transduction of certain cells (this is mandatory for HT-29 cells).

8b. Once centrifugation is done, remove the Parafilm and return the plate to the incubator.
9b. Replace medium with 500 µl fresh complete medium 24 hr post-transduction.
Basic Protocol 3: OPTIMIZATION OF OPTOGENETIC STIMULATION CONDITIONS
This protocol lists steps to optimize the optogenetic stimulation conditions for minimal phototoxicity.
Materials
-
Stable HT-29 cells expressing the optogenetic RIPK3 system (see Basic Protocol 2)
-
SYTOX Green (Thermo Fisher, cat. no. S7020; see Basic Protocol 4, step 3)
-
Environmental control enclosure system (In Vivo Scientific)
-
Inverted microscope (Leica DMI8)
-
Imaging software (Leica LAS-X, Micromanager 1.4)
1.Treat the stable HT-29 cells expressing the optogenetic RIPK3 system with 100 µl SYTOX Green per well for 10 min in a sterile biosafety cabinet.
2.Turn on the environmental control enclosure system (5% CO2 and 37°C; Fig. 4A) 1 hr before imaging. Adjust the flow rate to 300 cm3/min (ccm).
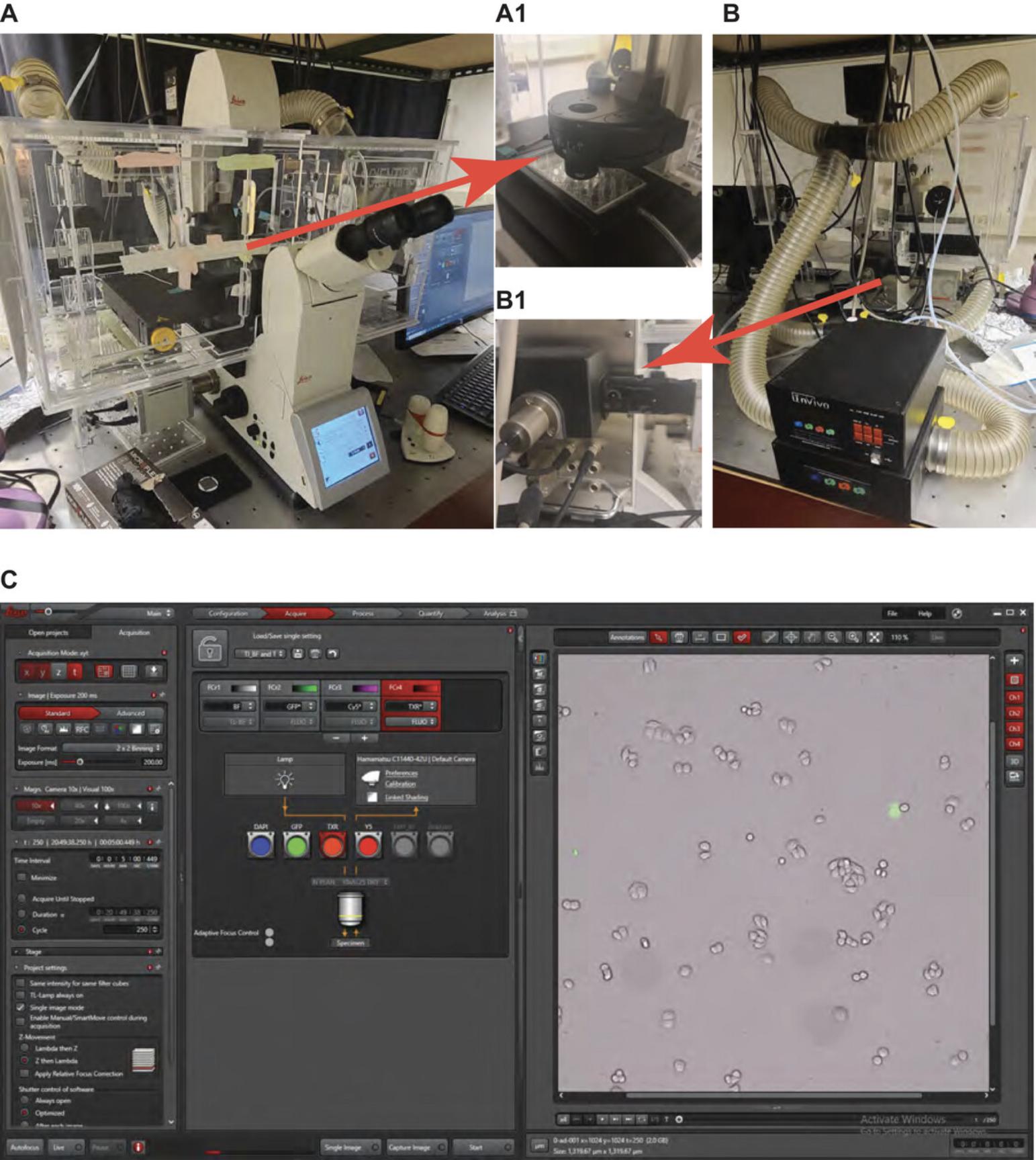
3.Turn on the inverted microscope shortly before imaging (see Fig. 4 for setup of hardware and software).
4.Set the fluorescence intensity manager (Fig. 4B) at the second position.
5.Use a 10× objective and set the plate of stable HT-29 cells expressing the optogenetic RIPK3 system under the microscope.
6.Set up imaging parameters in imaging software as follows:
- Texas red (TXR): 100 ms.
Purpose: To identify cells expressing the optogenetic RIPK3 system with mCherry.
- GFP: 15 ms.
Purpose: To capture SytoxGreen-stained lytic cells and provide optogenetic stimulation.
- GFP: 80 ms.
Purpose: To provide an extra dose of light for optogenetic stimulation. Two-step GFP stimulation is used so that signals for SytoxGreen-stained cells will be saturated and sufficient blue light is supplied for optogenetic stimulation.
- Brightfield: 100 ms.
Purpose: To capture cell morphology.
- Dark interval: 5 min.
Purpose: To minimize phototoxicity.
-
Total imaging acquisition time: 20 hr.
7.Examine cell membrane rupture.
Basic Protocol 4: TIME-STAMPED LIVE-CELL IMAGING OF HT-29 LYTIC CELL DEATH
This protocol outlines the steps to achieve time-stamped live-cell imaging that captures HT-29 lytic cell death.
Materials
-
Stable HT-29 cells expressing the optogenetic RIPK3 system (see Basic Protocol 2)
-
Wild-type HT-29 cells (ATCC, cat. no. HTB-38)
-
Complete HT-29 cell culture medium (see recipe), 37°C
-
SYTOX Green (Thermo Fisher, cat. no. S7020)
-
12-well culture plates (Fisherbrand)
-
Inverted microscope (Leica DMI8) with optimized optogenetic stimulation conditions (see Basic Protocol 3)
-
Imaging software (Leica LAS-X, Micromanager 1.4.)
-
ImageJ software
1.Plate two wells of stable HT-29 cells expressing the optogenetic RIPK3 system and two wells of wild-type HT-29 cells (Gene-/Dark, Gene-/Light, Gene+/Dark, and Gene+/Light) in complete HT-29 cell culture medium in a 12-well culture plate at a seeding density of 0.25 × 106 cells per well and incubate for 24 hr in a 37°C and 5% CO2 incubator.
2.Remove old medium from each well and wash away dead cells three times using fresh medium. Then, add 900 µl fresh medium to the cells and incubate for 30 min at 37°C and 5% CO2.
3.Stain the cells with 100 µl SYTOX Green per well for 10 min at room temperature in a sterile biosafety cabinet.
4.Start the imaging software and create a new project to be saved as an LIF file.
5.Proceed with live-cell imaging under the inverted microscope with optimized optogenetic stimulation conditions (see Basic Protocol 3), including the following imaging parameters:
-
TXR: 100 ms.
-
GFP: 15 ms.
-
GFP: 80 ms.
-
Brightfield: 100 ms.
-
Dark interval: 5 min.
-
Fluorescence intensity attenuator: position 2.
-
Total data acquisition time: 20 hr.
6.Remove the plate after 20 hr of imaging, save the project LIF file (or TIFF stack of images), and analyze data using ImageJ software.
Basic Protocol 5: QUANTIFICATION OF HT-29 LYTIC CELL DEATH
This protocol outlines the data analysis steps to quantify HT-29 lytic cell death.
Materials
- FIJI (https://imagej.net/software/fiji/)
- CellProfiler (https://cellprofiler.org)
- Desktop computer (computer with ≥16 GB RAM and fast CPU; preferred, Intel i7 or AMD Ryzen 7 equivalent)
- TIFF stack acquired from microscope software (see Basic Protocol 4)
- Microsoft Excel or equivalent quantification software (Matlab, Origin)
1.Open up FIJI and CellProfiler on a desktop computer.
2.Using FIJI, open up the TIFF stack acquired from microscope software by selecting File > Open > Browse.
3.If the option is available, check the box “Open Virtual Stack” to reduce the amount of RAM used. Proceed by clicking “Okay”.
4.Select Image > Color > Split Channels to split the individual color channels into Bright Field (Grey), SytoxGreen (Green), and mCherry-labeled Lentivirus (Texas Red).
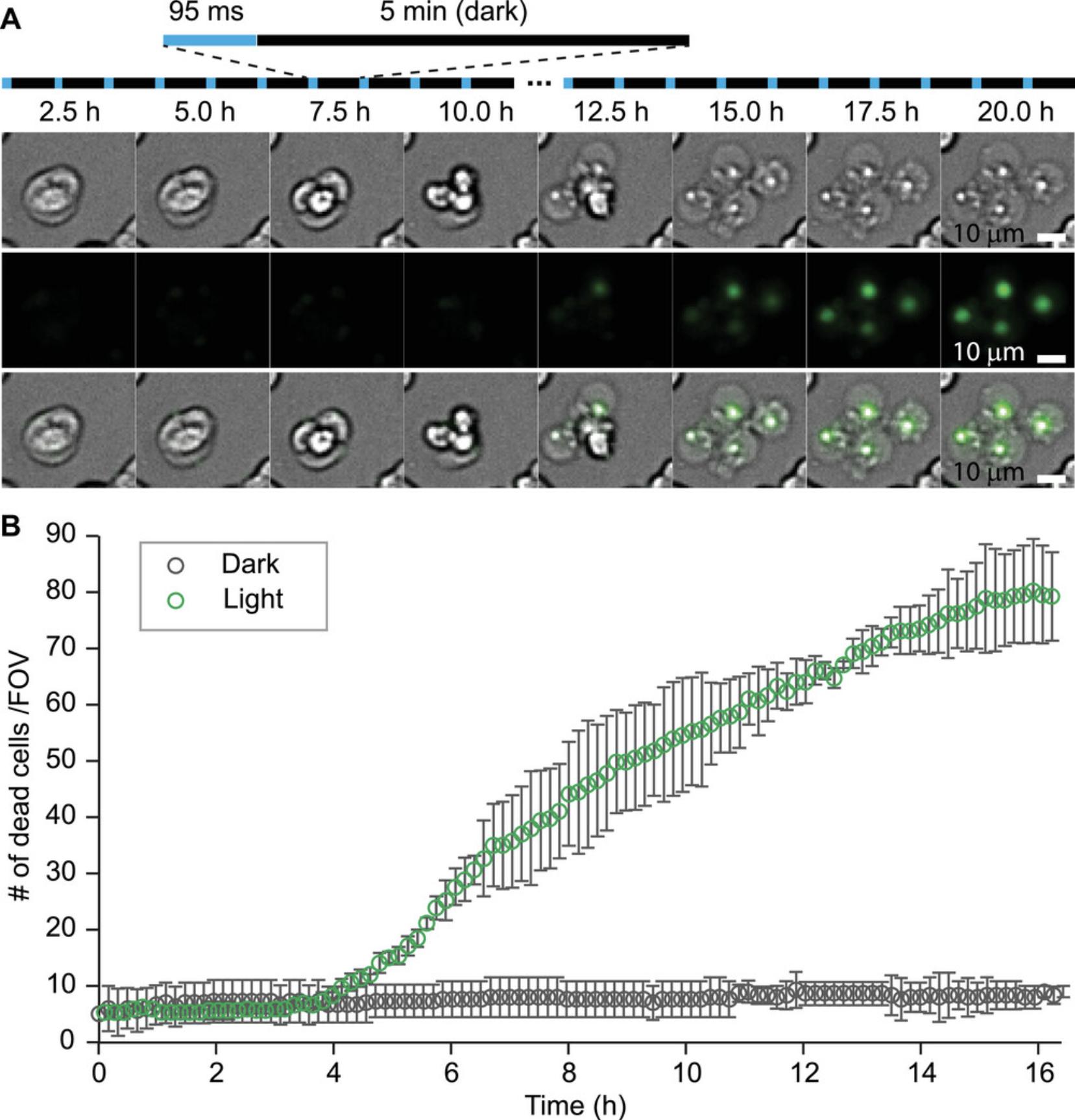
5.Save each color channel separately in Image Sequence format by clicking File > Save as > Image Sequence (for easy documentation, these should be saved to a separate folder). Save representative cell death images by cropping the stack images (Fig. 5A).
6.For all constructs needing to be analyzed, open and organize all files in the same manner as in steps 2 to 5 for easy identification in CellProfiler.
7.Using CellProfiler, open an Image Sequence file previously created in step 5 by dragging the folder into CellProfiler.
8.In the Adjust Modules panel, click the “+” icon and search for IdentifyPrimaryObjects. Double-click to add a module.
9.In the same panel, search for ExportToSpreadsheet. Double-click to add a module.
10.In IdentifyPrimaryObjects, adjust the following parameters:
-
Use Advanced Settings by clicking “Yes”.
-
Select the input image stack previously created in step 7.
-
Adjust the typical diameter size of the cells to be analyzed.
For HT29 cells, this value could range between 10 (min) to 20 (max) μm.
-
Use the “Otsu” thresholding method.
-
Adjust thresholding correction value to between 0.5 and 1.
See Troubleshooting for why this is an important value.
-
Allow all other settings to be the default.
11.In ExportToSpreadsheet, use the following settings:
-
Direct the output file to the desired folder.
-
Double-check that “Comma” is selected for column delimiter.
-
Allow all other settings to be the default unless changes are desired.
12.Once all settings are completed, click “Analyze Images” to start the analysis.
13.Repeat steps 7 to 12 for all other sets of images in three biological replicates.
14.Open the CSV file generated by CellProfiler into Microsoft Excel or equivalent quantification software to further analyze the data into a time series or to perform other quantification methods.
15.Plot the time-dependent cell death from the compiled results (Fig. 5B).
REAGENTS AND SOLUTIONS
Complete DMEM
- DMEM (Corning, cat. no. 10-013-CV)
- 10% (v/v) FBS (Sigma-Aldrich)
- 1× penicillin/streptomycin (10,000 U/ml stock; Corning, cat. no. 30-002-CI)
- Store ≤1 week at 4°C
Complete HT-29 cell culture medium
- McCoy's 5A medium (ATCC, cat. no. 30-2007)
- 10% (v/v) FBS (Sigma-Aldrich)
- 1× penicillin/streptomycin (10,000 U/ml stock; Corning, cat. no. 30-002-CI)
- Store ≤1 week at 4°C
COMMENTARY
Background Information
Well-established genetic and cell biological strategies have played a pivotal role in the study of cell death pathways. These previous studies provide the foundation for the design of optogenetic systems. Optogenetic stimulation provides high resolution in spatial and temporal control of cell signaling and therefore could provide new insights into the dynamic nature of cell death pathways. However, careful design of optogenetic experiments is warranted to verify the effect of the optogenetic response.
Critical Parameters
Gene delivery methods: Chemical versus lentivirus
Two commonly used gene delivery methods are chemical transfection and lentiviral transduction (see Basic Protocol 2). Chemical transfection uses cationic molecules that can partially neutralize the negative charge of DNA plasmids, which facilitates transport through the plasma membrane. Lentivirus packages the encoding genes in the transfer plasmid and forms non-propagated viral particles that infect the target cells. Chemical transfection can be done within 2 to 4 hr. Gene expression starts at ∼6 hr post-transfection. Lentivirus requires initial transfection in host cells (typically HEK293T cells), collection of the conditioned medium in 48 to 72 hr, and purification and concentration if needed. Upon transduction, gene expression takes another 3 days to 1 week.
The choice of gene delivery method depends on the cell model and the purpose of the application. Chemical transfection, despite its convenience and fast timeline, cannot provide sufficient transfection efficiency for all cell lines. In extremely challenging cell lines like HT-29 cells or primary culture, the transfection efficiency could be as low as 1%. Additionally, the gene expression profile is extremely heterogeneous. Lentiviral transduction provides a titer-dependent transduction rate. In our experience, the HT-29 transduction rate can reach 40% to 50% with lentiviral transduction. The expression profile from lentiviral transduction is more homogeneous. Whereas chemical transfection might be sufficient if the purpose is to observe single-cell behavior (e.g., through imaging), biochemical and cell biological analysis of cells (e.g., phosphorylation of target proteins or metabolic responses) may benefit from transduction from lentivirus or other viral vectors.
Choice of optogenetic systems and optimization of stimulation conditions
When working with a new cell or tissue model, optogenetic stimulations should be optimized to minimize phototoxicity (Basic Protocol 3). Commonly used optogenetic systems use visible light as a stimulating source. Light with shorter wavelengths introduces greater phototoxicity than that with longer wavelengths. Additionally, longer-wavelength light has a slightly deeper penetration depth than shorter-wavelength light and therefore is preferred for in vivo studies. However, different optogenetic systems also show different light sensitivity, e.g., different light-to-dark dynamic ranges or Kd ranges. Thus, the efficacy needs to be individually tested. Once a working system is confirmed, optical stimulation conditions should be optimized. Continuous exposure will introduce greater phototoxicity. Thus, an intermittent stimulation pattern could be used. The bottom line is to identify a safe range of experimental parameters, including wavelength, instantaneous light power, and total dosage, that do not introduce phototoxicity to wild-type cells.
Necessary controls
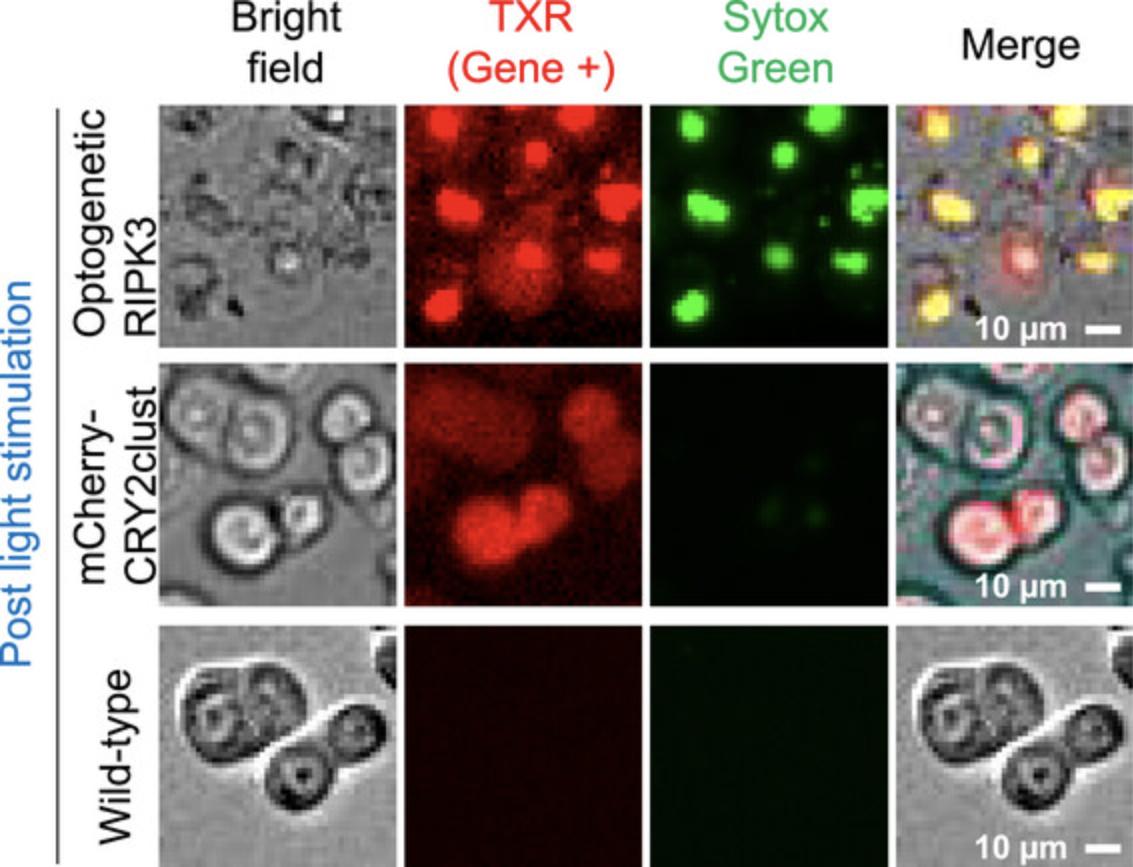
Whenever possible, optogenetic experiments (Basic Protocol 4) should be carried out with quadrant conditions in mind. Three controls are needed to delineate the optogenetic response, including Gene-/Dark, Gene-/Light, and Gene+/Dark. Only when all three controls show distinct cell responses from Gene+/Light could one safely interpret the observed phenotype as an optogenetic response. Figure 6 shows the distinct plasma membrane rupture phenotype in wild-type (non-transfection), mCherry-CRY2clust-transfected (without RIPK3), and optogenetic RIPK3–transfected cells after blue light stimulation. As expected, wild-type and mCherry-CRY2clust-transfected HT29 cells were not stained by SYTOX Green, indicating intact membranes, whereas optogenetic RIPK3–transfected cells were clearly stained by SYTOX Green.
Real-time monitoring of cell response and healthiness
For long-term illumination, one needs an environmental chamber (Basic Protocols 3 and 4) that provides a sustained temperature (37°C) and acidity (5% CO2) to ensure the healthiness of cells. Light sources from the optical microscope can be directly used to stimulate the optogenetic systems. Different channels of emission can be programmed together to monitor the cellular response to optogenetic stimulation in real time.
Troubleshooting
Common problems with the protocols, their causes, and potential solutions are shown in Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| Cells are over-confluent in the culture plate after adding the transfection mix | DMEM with FBS was used | Remember to use DMEM without FBS for a reduction in cell growth during the transfection period. |
| Poor transfection efficiency | There are many possible reasons for this: low-confluent cells; unhealthy cells; wrong amounts of transfer, packaging, and envelope plasmids; poor centrifugation; and/or inadequate cell conditions | Follow the proper molar ratio of plasmids and adhere to incubation times. Take pictures of any viral pellet found after concentrating for further verification that the transfection was successful. |
| Loss of viral product after concentrating | Incubation without ice or not incubated long enough | Incubate for >1 hr. Use ice during incubation. |
| No pellet seen after concentrating | Inadequate incubation time after mixing the virus with concentrator | Incubate the virus and concentrator for a longer time. |
| No cells were identified in CellProfiler; fewer green objects were identified, and more yellow/magenta objects were identified | The thresholding correction factor will help CellProfiler differentiate a cell from the background. If the thresholding factor is too low, CellProfiler cannot differentiate. However, the thresholding factor cannot be too high because CellProfiler will misidentify objects in the frame that are not cells. | Adjust the thresholding correction factor according to each image stack. This will require some trial and error. |
Understanding Results
Interpretation of optogenetic induction of lytic cell death requires data analysis of multi-color fluorescence images (Basic Protocol 5). Using CellProfiler, the number of dead cells versus time is shown (Fig. 5B). With optogenetic blue light simulation, the amount of cell deaths increases over time. When compared to control experiments undergoing no stimulation, the number of dead cells remains constant. One can compare the amount of SytoxGreen versus mCherry signal to account for the percentage of cell death by necroptosis.
Lentiviral transduction (Basic Protocol 2) is crucial for effective gene delivery. When producing lentivirus (Basic Protocol 1), a good benchmark for sufficient yield is the morphology of host cells (HEK293T) after transfection with the mix of transfer, packaging, and envelope plasmids. Clustering and shrinkage of cells are signs of effective transfection and production of lentivirus particles (Fig. 1). Because different cell lines show significant variation in the transduction rate, lentivirus transduction becomes mandatory for cell lines like HT-29 cells (Fig. 2). Fluorescence imaging provides a quick benchmark for transgene expression. Biochemical analysis, such as western blot or immunofluorescence staining, can be used to validate the expression level. Practically, we found that spinning down the cells together with lentivirus increased the transduction rate, but this procedure needs to be optimized for different cell lines.
Long-term imaging (Basic Protocol 4) tracks the cell response in real time and is selected as the primary way to quantify cell death in this article. A full environmental control enclosure system (Basic Protocols 3 and 4) is used to maintain the temperature, humidity, and CO2 levels during data acquisition (Fig. 4A). Regular maintenance and checkups are warranted to ensure the appropriate functioning of the unit (e.g., the gas tank, CO2 regulator, water batch, and temperature measurement). For data analysis (Basic Protocol 5), automated algorithms minimize human bias and are used in practice. Cells from different fluorescence channels are selected, and their intensity changes are recorded to calibrate cell death (Fig. 5). Cells under control conditions (e.g., wild-type, mCherry-CRY2clust) show a distinct cell death phenotype compared to optogenetic RIPK3–transfected cells (Fig. 6).
Time Considerations
Basic Protocol 1: Production of lentivirus encoding the optogenetic RIPK3 system: 3 days.
Basic Protocol 2: Culturing, chemical transfection, and lentivirus transduction of HT-29 cells: 3 days.
Basic Protocol 3: Optimization of optogenetic stimulation conditions: 24 hr.
Basic Protocol 4: Time-stamped live-cell imaging of HT-29 lytic cell death: 24 hr.
Basic Protocol 5: Quantification of HT-29 lytic cell death: 2 hr.
Acknowledgments
K. Z. is supported by the National Institute of General Medical Sciences (R01GM132438) and National Institute of Mental Health (R01MH124827) of the National Institutes of Health, the National Science Foundation (award no. 2121003), NSF grant 2243257, the National Science Foundation Science and Technology Center for Quantitative Cell Biology, and the Cancer Center at Illinois (CCIL) at UIUC.
Author Contributions
Teak-Jung Oh : Data curation; formal analysis; methodology; validation; writing—original draft. Bryan Gworek : Data curation; formal analysis; software; writing—original draft. Amna Mehfooz : Data curation; formal analysis; writing—original draft. Kai Zhang : Conceptualization; funding acquisition; project administration; resources; software; supervision; writing—original draft.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The complete raw data of time-stamped images of RIPK3-mediated lytic cell death are available from the corresponding author upon request.
Literature Cited
- Cho, Y. S., Challa, S., Moquin, D., Genga, R., Ray, T. D., Guildford, M., & Chan, F. K. (2009). Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell , 137(6), 1112–1123. https://doi.org/10.1016/j.cell.2009.05.037
- Dondelinger, Y., Declercq, W., Montessuit, S., Roelandt, R., Goncalves, A., Bruggeman, I., Hulpiau, P., Weber, K., Sehon, C. A., Marquis, R. W., Bertin, J., Gough, P. J., Savvides, S., Martinou, J. C., Bertrand, M. J., & Vandenabeele, P. (2014). MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Reports , 7(4), 971–981. https://doi.org/10.1016/j.celrep.2014.04.026
- Fischer, A. A. M., Kramer, M. M., Radziwill, G., & Weber, W. (2022). Shedding light on current trends in molecular optogenetics. Current Opinion in Chemical Biology , 70, 102196. https://doi.org/10.1016/j.cbpa.2022.102196
- Liu, R., Fang, M., Chen, X., & Yang, Y. (2022). The status and challenges of optogenetic tools for precise spatiotemporal control of RNA metabolism and function. Clinical and translational medicine , 12(10), e1078. https://doi.org/10.1002/ctm2.1078
- Mazraeh, D., & Di Ventura, B. (2022). Synthetic microbiology applications powered by light. Current Opinion in Microbiology , 68, 102158. https://doi.org/10.1016/j.mib.2022.102158
- Oh, T. J., Fan, H., Skeeters, S. S., & Zhang, K. (2021). Steering molecular activity with optogenetics: recent advances and perspectives. Advanced Biology , 5(5), e2000180. https://doi.org/10.1002/adbi.202000180
- Sun, L., Wang, H., Wang, Z., He, S., Chen, S., Liao, D., Wang, L., Yan, J., Liu, W., Lei, X., & Wang, X. (2012). Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell , 148(1-2), 213–227. https://doi.org/10.1016/j.cell.2011.11.031
- Zhu, K., Liang, W., Ma, Z., Xu, D., Cao, S., Lu, X., Liu, N., Shan, B., Qian, L., & Yuan, J. (2018). Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression. Cell Death & Disease, 9(5), 500. https://doi.org/10.1038/s41419-018-0524-y

