Establishing a Microfluidic Tumor Slice Culture Platform to Study Drug Response
Zofia M. Komar, Zofia M. Komar, Dik C. van Gent, Dik C. van Gent, Sanjiban Chakrabarty, Sanjiban Chakrabarty
Abstract
Accurate models for tumor biology and prediction of drug responses of individual tumors require novel technology to grow tumor tissue ex vivo to maintain tumor growth characteristics in situ. Models containing only tumor cells, without the stromal components of the tumor, are suboptimal for many purposes and are generally problematic because the cells are passed through extensive culture and selection. Therefore, direct culture of (human) tumors is of considerable interest for basic tumor biology and diagnostic purposes. Microfluidic technologies have been proposed to accurately mimic physiological conditions for tissue growth. Most published systems build tissues from individual cell types in so-called Organ-on-Chip (OoC) cultures. We here describe a novel OoC device for growing tumor specimens. Thin tumor slices are grown in a microfluidic ‘chip’ that allows precisely controlled in vitro culture conditions. The performance of the OoC device was extensively validated for predicting therapeutic responses in human breast cancer patient-derived xenograft (PDX) tumor material. The system is amenable to primary tumor material from surgery or biopsies. In addition to using the model to predict and evaluate therapeutic responses, the model can also be used for mechanistic studies of human cancers, such as clonal evolution or immune responses, or to validate new or repurposed (cancer) drugs. The Bi/ond Cancer-on-Chip (CoC) device is designed to culture tumor slices and investigate aspects of tumor growth and drug responses. Here, we describe the step-by-step process of setting up tumor slice cultures using a Bi/ond CoC device and performing in vitro drug response evaluation. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Establishment of breast cancer tumor slice culture using a microfluidic cancer-on-chip platform for chemotherapy testing ex vivo
Basic Protocol 2 : Histology and immunohistochemistry-based analysis of tumor tissue architecture, cell proliferation, and cell death
INTRODUCTION
Organ-on-chip (OoC) platforms are microfluidic devices that provide a way to culture biological materials in a controlled environment that can mimic the in vivo functionalities of cells, tissues, and organs. Optimal OoC platforms combine 1) a microfluidic chip, 2) cells or a tissue specimen, 3) physiological levels of required stimuli for tissue maturation, and 4) sensors or optics for results readout. The capability of these models to effectively mimic the tumor microenvironment is expected to revolutionize oncology research and anticancer drug screening.
Most OoC systems are constructed as closed devices in which cells are inserted with the regular medium flow. This works well for cell lines or primary cells but cannot be used for larger pieces of tissue. Thin slices can be generated from tumor specimens and cultured for several days ex vivo in the presence or absence of chemotherapeutic drugs, antibodies, or radiation treatment (Ladan et al., 2022; Naipal et al., 2016). Such tissue slices can also be grown in OoC platforms, but only if the system can be closed after inserting the tissue specimen. Compared to standalone tumor slices, the slices on a chip more closely mimic the in vivo conditions with a continuous circulation of nutrients, oxygen gradients, and waste removal systems, making them attractive models to study chemosensitivity. For example, a reproducible microfluidic model was designed to culture precision-sliced thyroid tissue for up to 4 days to evaluate radioiodine sensitivity in real-time (Riley et al., 2019). Similarly, colorectal cancer tumor slices cultured in a microfluidic device demonstrated the potential utility of the platform for multiplexed drug testing, assays with various types of fluorescent cell death indicators, and real-time measurements (Rodriguez et al., 2020). A microfluidic device was developed for culturing tumor explants to investigate the responses of HNSCC tissue biopsies to chemotherapeutic drugs (Hattersley et al., 2012).
All these devices were custom-made and are not commercially available. Recently, we described a Cancer-on-Chip device with a 6-well plate design incorporating silicon-based microfluidics, which has application in the parallel culture of multiple tumor slices and diagnostic assays using primary tumor material (Chakrabarty et al., 2022). The CoC device is commercially available from BIOND Solutions B.V. (Bi/ond) (Delft, The Netherlands, www.gobiond.com), enabling other researchers to use the system for their own purposes. This device has a unique design that 1) facilitates culture of relatively large (tumor) tissue, 2) allows the tissue to receive media and nutrients through top and bottom flow, 3) combines polydimethylsiloxane (PDMS) and silicon-based components, and 4) has a relatively simple design that mimics 6-well culture plates, making it suitable for assessing drug response using patient biopsy material in a clinical setting. Earlier studies describing similar OoC devices had only a single flow (top or bottom) or a perfusion system for media flow (Astolfi et al., 2016; Horowitz et al., 2020; Kennedy et al., 2019; Rodriguez et al., 2020; Shirure and George, 2017). In the Bi/ond CoC device, the tumor biopsy material receives medium via top and bottom flow, which minimizes drug gradients throughout the culture period. The CoC device is connected to fluidics for constant replenishment with media and gas, minimizing culture-induced stress responses in the tumor material (Chakrabarty et al., 2022).
Earlier studies have revealed the limitations of PDMS-based OoC devices with hydrophobic drugs (Shirure and George, 2017). New materials have been proposed to modify or replace PDMS, such as polyethylene terephthalate (PET) and polystyrene (PS), thermoplastics that mitigate the inherent absorption limitations of PDMS materials (Campbell et al., 2021). However, these materials are less easily modified in a laboratory setting. One of the advantages of the Bi/ond CoC device is that a PDMS membrane is used to support the tumor tissue (enabling easy adaptation of the membrane properties). The rest of the microfluidic components are made of silicon, minimizing the risk of small molecule absorption. The device provides capacity for six simultaneous independent tumor specimens, making it amenable to a broad spectrum of experimental designs. Additionally, the silicon material in the microfluidic chip can be connected to sensors for real-time monitoring of gas concentrations, metabolites, and pH. A convenient thermoreversible hydrogel attaches the tumor tissue to the PDMS membrane, further reducing the effect of shear forces on the tissue as media flow above and below, sustaining optimal growth conditions.
We have demonstrated that the Bi/ond microfluidic CoC device can maintain breast tumor tissue slice cultures for up to 14 days with minimum alteration in cell viability, apoptosis, and tissue architecture. The application of the CoC device was validated for cisplatin response in a breast cancer PDX model and apalutamide (anti-androgen drug) response in prostate cancer PDX tumor slices (Chakrabarty et al., 2022). Here, we describe the Bi/ond CoC device, the first commercially available microfluidic platform for tumor slice culture, for its application in chemotherapeutic drug testing.
Basic Protocol 1 outlines the materials and methods for breast cancer PDX tumor slice preparation, establishing the CoC device setup for tumor slice culture, and cisplatin drug treatment.
Basic Protocol 2 outlines the materials and methods for preparing the breast cancer PDX tumor slices for histological and immunohistochemical analysis, evaluation of tumor tissue architecture, tumor cell proliferation, DNA damage response, and cell death in treatment-naive and cisplatin-treated samples.
NOTE : Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol 1: ESTABLISHMENT OF BREAST CANCER TUMOR SLICE CULTURE USING A MICROFLUIDIC CANCER-ON-CHIP PLATFORM FOR CHEMOTHERAPY TESTING EX VIVO
This protocol describes Breast Cancer (BrC) PDX tumor collection, preparation, and handling in detail. The method elaborates on the preparation of 300-µm tumor slices suitable for culture. Following the protocol allows the generation of tissue slices and culture conditions that preserve the viability of the original tissue. Next, the microfluidic device is described in more detail. The device is a closed version of the comPLATE (Bi/ond, Delft, The Netherlands). This protocol contains a step-by-step explanation of the procedure, including cleaning, assembly, control during the run, and disassembly of the device. This protocol allows breast cancer tissue slices to be cultured for up to 14 days with or without treatment. A maximum of six slices can be handled within a single setup. A video demonstration of how to assemble the comPLATE can be found at https://youtu.be/SKmaQeLZYP8.
Materials
-
Breast PDX tumors (or other tumor tissue)
-
Breast medium (see recipe; Naipal et al., 2016)
-
4% Low-gelling agarose (Sigma-Aldrich, cat. no. A9414) in PBS
-
Ethanol
-
0.1 g/ml Mebiol hydrogel (Cosmo Bio Co, cat. no. MGB-PMW20-1001-COS)
-
Store at 4°C. Dilute 1 g hydrogel in 10 ml breast medium
-
30 μM 5-Ethynyl deoxyuridine (EdU, 10 mM; Invitrogen, cat. no. A10044) in DMSO
-
Formalin
-
15- and 50-ml Falcon tubes (Greiner Bio-one, cat. no. 188271-N and 227261)
-
10-cm culture dishes (Cellstar, Greiner Bio-one, cat. no. 664160)
-
Disposable scalpel (e.g., Swann-Morton, cat. no. SM0501)
-
6-well plates (Starstedt, cat. no. 83.3920)
-
Leica VT 1200S Vibratome
-
60°C water bath
-
Incubator (37°C, 5% CO2)
-
Stuart SSM1 mini orbital shaker (Camlab Limited)
-
Blades (e.g., Super Chrome Blades, Shark)
-
Superglue (e.g., Bison)
-
10-ml plastic pipettes
-
Thermometer
-
Tubing, OD 1/16″ (Darwin Microfluidics, cat. no. BL-PTFE-1608-20)
-
Flangeless PEEK Fittings (IDEX, cat. no. CIL-XP-230X)
-
ETFE Ferrules ¼-28 to 1/16″ OD (IDEX, cat. no. CIL-XP-200NX)
-
comPLATE (Bi/ond, includes a bottom plate, top plates, rings, black rubber gasket, and chips)
-
FLPG Plus pumping system (Fluigent)
-
Fluigent MFCS-EZ (Fluigent)
-
Fluigent FLOW UNIT-S (Fluigent)
-
IO-Biofilm-Entferner (Umweltanalytik)
-
Paper tissues
-
Plastic precision tweezers (e.g., Cole-Parmer, cat. no. 15657418)
-
Metal precision tweezers (e.g., Fisher Scientific, cat. no. 16647753)
-
P20 pipette (Gilson)
-
P20 pipette tips, pre-cooled at −20°C
-
Scissors
1.Prepare a 50-ml Falcon tube with approximately 30 ml breast medium; tumor samples can be collected by placing tissue in this tube and transporting it on ice.
2.Place the tissue in a 10-cm dish filled with medium and prepare the tissue for slicing using a scalpel.
3.Clean the Leica VT 1200S vibratome and its surroundings with 70% ethanol and operate only while wearing gloves.
4.Pour the 4% agarose/PBS (kept in a 60°C water bath) into one well of a 6-well plate to fill it entirely.
5.Monitor the temperature until the agarose cools to 37°C.
6.Place the tissue in the 4% agarose and wait until the agarose solidifies (Fig. 1A).
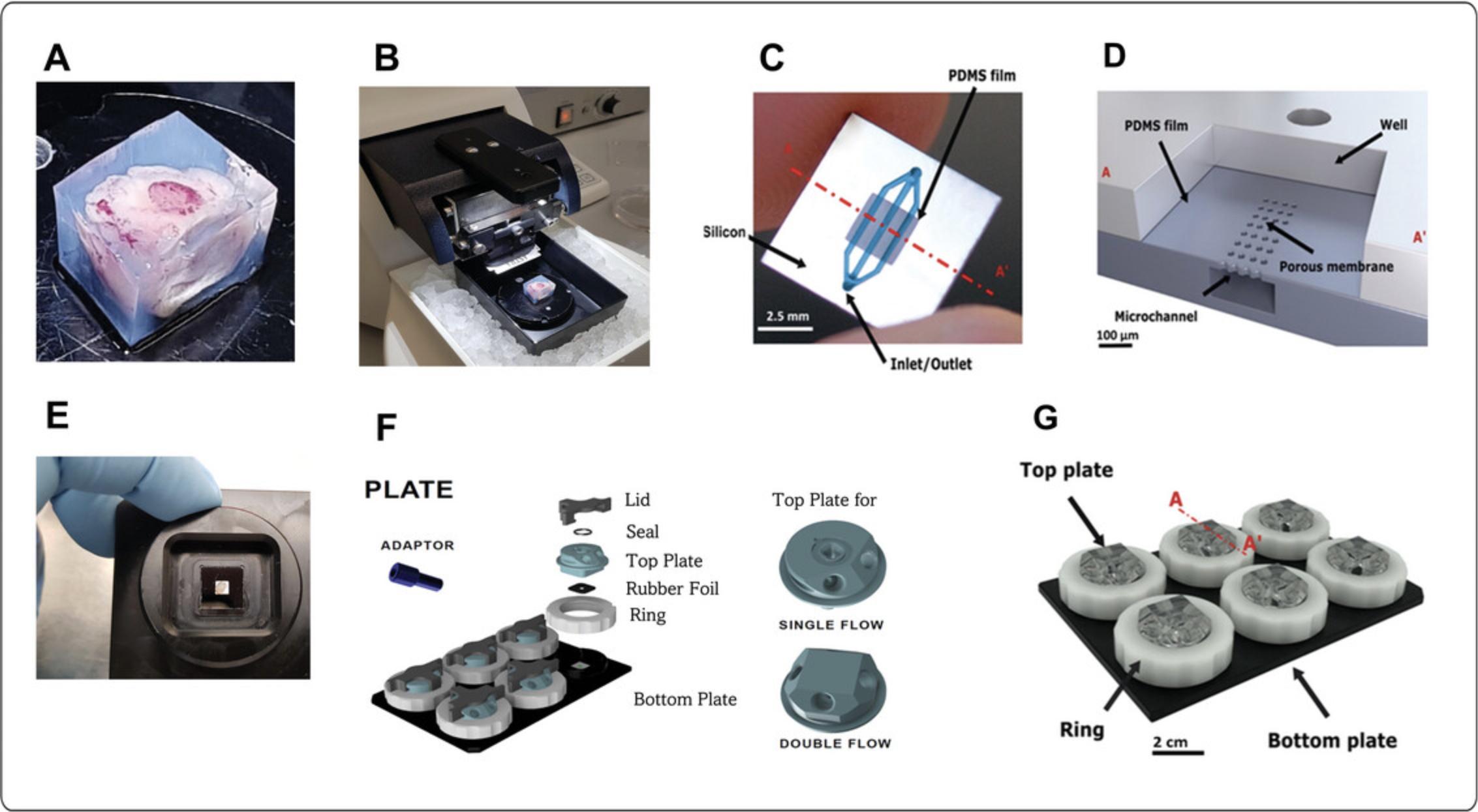
7.Remove the agarose block from the 6-well plate using a scalpel.
8.Trim the agarose block to ensure it fits in the Leica VT 1200S vibratome.
9.Glue the agarose block to the removable magnetic bottom of the Leica VT 1200S Vibratome using Bison superglue.
10.Place the magnetic bottom with the agarose block into the vibratome and fill the well with PBS until it covers the agarose block (Fig. 1B).
11.Begin slicing the tissue at a speed of 0.6 to 1 mm/s, amplitude 2.00 mm, and thickness 300 µm.
12.Use sterile metal tweezers to transfer the slices to a 10-cm dish filled with breast medium.
13.Place each slice in a separate well of a 6-well plate and place it on an orbital shaker at 60 rpm in a 37°C, 5% CO2 incubator.
14.Cut the tubing to appropriate lengths, clean it, and connect it to the perfusion system. Prepare the medium reservoir (15-ml Falcon tube) and collection tubes (15-ml Falcon tubes) and fix them in the appropriate tube holders connected to the shelf inside the CO2 incubator (Fig. 2A). Make sure the ends of the tubing are submerged in the medium in both reservoir and collection tubes to avoid air bubbles.
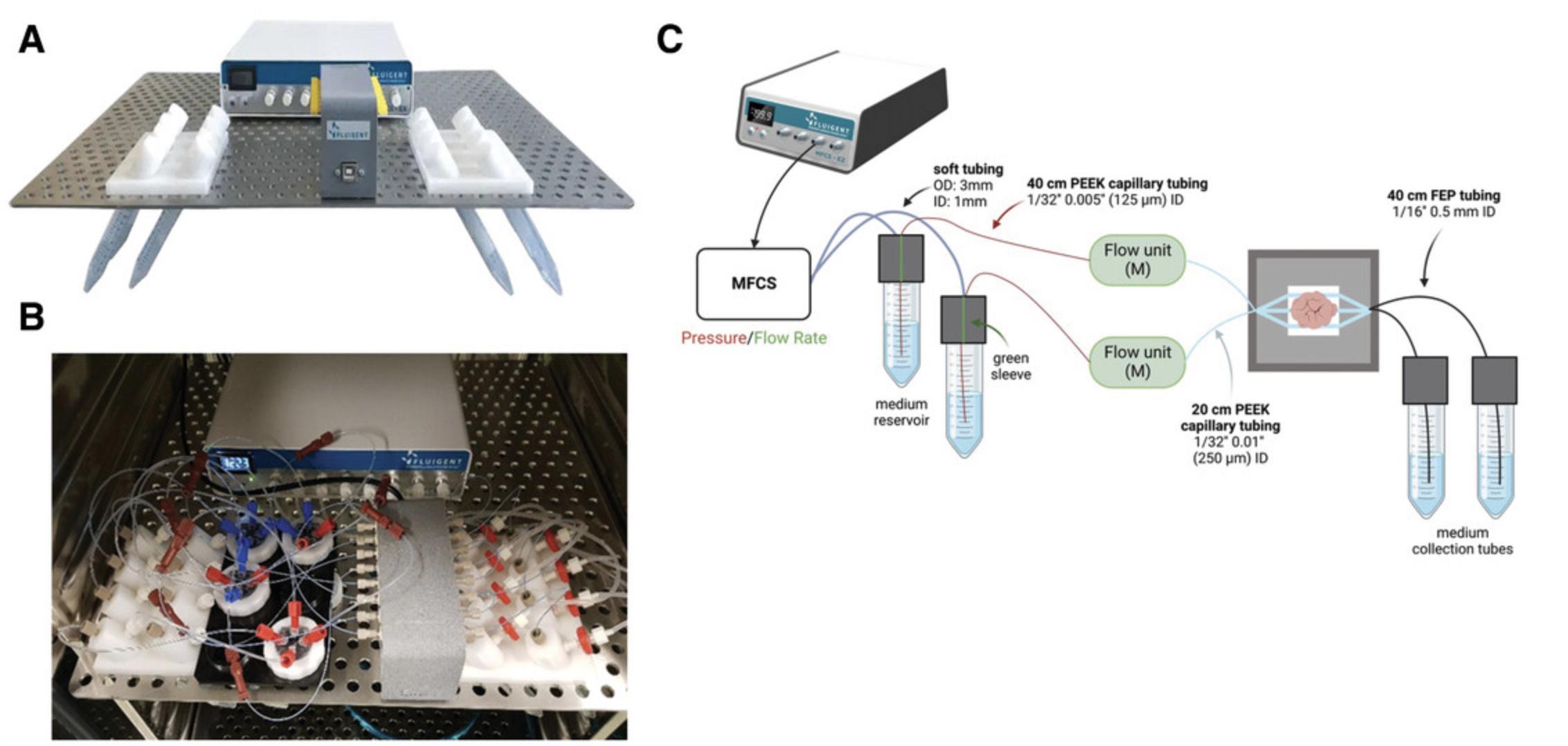
15.Connect the tubing with adaptors and ferrules to the plate (Fig. 2B).
16.Prepare the medium with or without additional (chemotherapeutic) compounds.
17.Prepare the comPLATE parts and connect the black rubber element with the top plate.
18.Prepare the tissue slices generated in step 13 and the pre-cooled hydrogel (keep on ice).
19.Place the tissue on the chip using the Mebiol hydrogel (Fig. 1C and 1D).
20.Close the system with a top plate and tighten with the white ring (Fig. 1E and 1F). Fill and connect the medium reservoir to the holder attached to the incubator shelf (Fig. 2A). Connect the collection reservoir to the holder opposite the medium reservoir. Ensure the tubing is fully submerged in both reservoirs.
21.Connect the CoC device to the FLPG Plus pumping system with an MFCS-EZ flow control system for continuous perfusion according to manufacturer directions.
22.Connect the microfluidic flow control system (Fluigent MFCS-EZ) and flow rate sensors (Fluigent FLOW UNIT-S) to the comPLATE containing tumor slices in each well using tubing and adaptors. Ensure the MFCS-EZ is connected to the MAESFLO software to check the flow rate during the run.
23.Make sure that the flow rate control system is connected to the Fluigent Microfluidic Automation Tool (MAT) with the predesigned program set for an inlet flow rate of 5 µl/min through the top and bottom channels.
24.Run the CoC device for the number of days required for the experiment while checking the system for leaks or disturbances daily. Ensure there is sufficient medium in the reservoir for at least 1 day.
25.After incubation, stop the system and disassemble the connectors from the Bi/ond plate.
26.Place the comPLATE on ice and incubate for 5-10 min to allow the thermoreversible Mebiol hydrogel to liquefy.
27.While incubating the comPLATE on ice, prepare a 6-well plate with 3 ml breast medium per well.
28.Disassemble the comPLATE and place tissue slices in the medium-filled wells.
29.Add EdU (3 µg/ml) to each well and incubate for 2 hr while placing the tissue on an orbital shaker at 60 rpm in a 37°C and 5% CO2 incubator.
30.After the run, thoroughly clean all tubing, adaptors, connectors along, and flow sensors with water, biofilm-removing detergent, and isopropanol. The detergent should be diluted with ultra-pure MQ water (20% IO-Biofilm-Entferner and 80% MQ-water). The comPLATE is cleaned with isopropanol followed by ultra-pure MQ water.
Basic Protocol 2: HISTOLOGY AND IMMUNOFLUORESCENCE ANALYSIS OF TUMOR TISSUE ARCHITECTURE, CELL PROLIFERATION, AND CELL DEATH
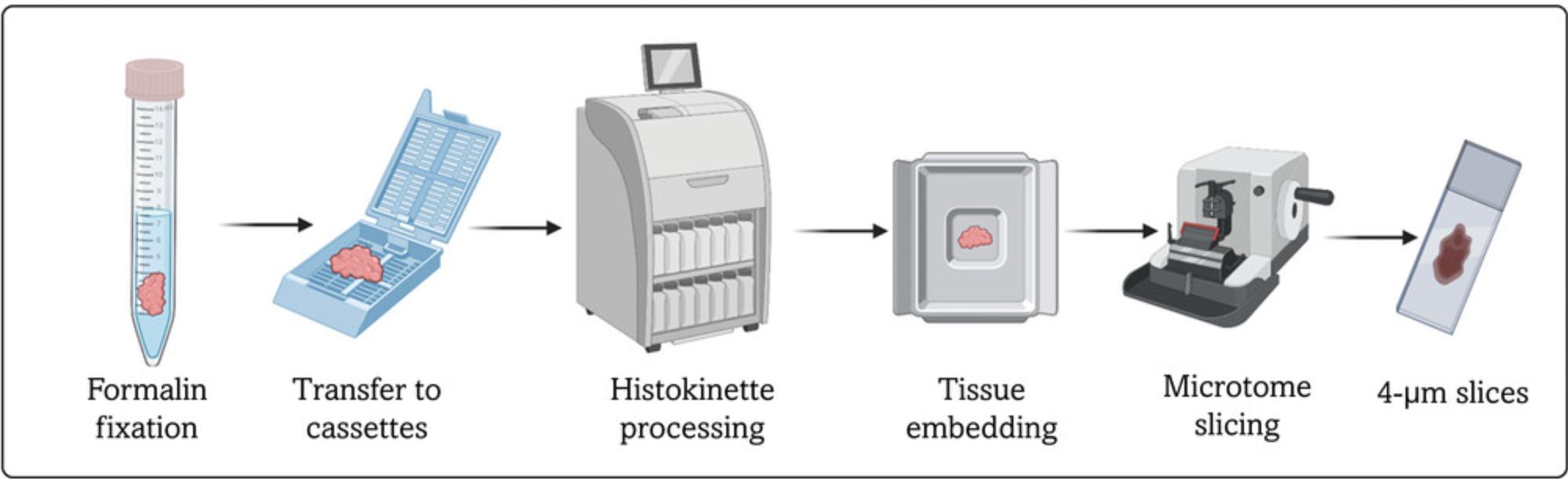
This protocol describes sample fixation and analysis. Tissue slices are fixed using the formalin-fixation paraffin embedding (FFPE) technique. Following this protocol allows the generation of 4-µm tissue sections that can be used for immunostaining or immunohistochemistry. Analyses described in this protocol allow assessment of tissue proliferation (EdU), apoptosis (TUNEL), DNA damage induction (53BP1), and histological tumor architecture (H&E) (Fig. 3).
Materials
-
Formalin
-
Ethanol
-
Xylene
-
Target Retrieval Solution (Dako, cat. no. S1699)
-
0.1% Tween-20/2% BSA (see recipe)
-
Click-iT cocktail (see recipe)
-
0.1% Tween-20 (see recipe)
-
TUNEL kit (Roche Diagnostics, cat. no. 11684795910)
-
Vectashield with DAPI (Vector Laboratories, cat. no. H-1200)
- Alternatively, Vectashield without DAPI (Vector Laboratories, cat. no. H-1000) can be used with a separate DAPI incubation (Sigma Aldrich, cat. no. 10236276001). Prepare a 1 mg/ml stock in MQ water and dilute to 1 µg/ml in PBS.
-
53BP1 primary antibody (Novus Biologicals, cat. no. NB100-305)
-
Alexa-Fluor-488 goat-anti-rabbit secondary antibody (ThermoFisher Scientific)
-
15-ml Falcon tube
-
Embedding Cassettes (Simport Scientific, cat. no. M490-4)
-
Biopsy foam pads (Simport Scientific, cat. no. M476-1)
-
Biopsy paper (Imeb, cat. no. BP-P)
-
EFTP Fast Tissue Processor (Adamas Instrumenten)
-
HM 335E Microtome (Microm International)
-
Superfrost Plus Adhesion Microscope Slides (VWR, cat. no. J1800AMNZ)
-
Microwave oven
-
Immunofluorescence Microscope (e.g., Automated Upright Microscope Leica DM4000 B)
-
Immedge Hydrophobic Barrier Pen (Vector Laboratories, cat. no. H-4000)
-
StainTray System with Black Cover (Simport Scientific, cat. no. 87000-136)
1.Place the tissue slice in a 15-ml Falcon tube filled with 10 ml formalin and incubate for 1-3 days at room temperature.
2.Move the tissue slices to 70% ethanol while placing them into cassettes.
3.Handle the material with the tissue processor Histokinette and embed samples in paraffin.
4.Prepare 4-µm tissue sections on microscope slides using a microtome.
5.Proceed to an appropriate staining protocol.
6.Remove the paraffin from the slides by washing in xylene for 3 min. Repeat 3 times.
7.Rehydrate the sample by washing the slides in an ethanol series, 1 min per wash: twice at 100%, twice at 95%, once at 80%, once at 70%, and once at 50%).
8.Wash the samples twice in demineralized water for 5 min.
9.Boil the samples in pre-heated antigen retrieval buffer for 15 min.
10.Let the samples cool to room temperature while keeping them in the antigen retrieval buffer for approximately 30 min.
11.Perform a series of 5-min washes: twice in demineralized water, twice in PBS, and twice in 0.1% Tween-20.
12.Mark tissue areas on the slide using the Immedge Hydrophobic Barrier pen.
13.Block using 0.1% Tween-20/2% BSA solution for 60 min.
14.Add the Click-iT cocktail and incubate for 30 min.
15.Wash with 0.1% Tween-20.
16.Add primary antibody diluted in 0.1% Tween-20/2% BSA and incubate for 90 min.
17.Wash three times with 0.1% Tween-20/2% BSA for 5 min.
18.Add a secondary antibody diluted in 0.1% Tween-20/2% BSA and incubate for 90 min.
19.Wash three times with 0.1% Tween-20 for 5 min.
20.Wash with PBS.
21.Embed the samples in Vectashield containing DAPI and seal the slides using nail polish.
22.Image samples using a fluorescence microscope.
23.Analyze images using ImageJ software.
REAGENTS AND SOLUTIONS
Use sterile, distilled, deionized water for all preparations unless otherwise stated. Prepare all reagents and solutions under sterile conditions.
Breast medium
- For reference, see Naipal et al., 2016
- DMEM:HAMs F12 (2:1)
- 2% FCS
- 0.3 µg/ml hydrocortisone
- 4 µg/ml insulin
- 4 µg/ml transferrin
- 1 ng/ml 3,3′,5′-Triiodothyronine
- 8 ng/ml EGF
- 7 ng/ml cholera toxin
- 0.2 mg/ml adenine and antibiotics
- Prepare and use immediately. Do not store.
Click-iT cocktail
- 30 µm Atto 488 azide (Atto-tex)
- 4 mM CuSO4
- 40 mM Tris Buffer (pH 7.5)
- 10 mM l-ascorbic acid (freshly made)
- Prepare 15 minutes before use. Do not store.
Tween-20, 0.1%/BSA, 2%
- In a 50-ml volume, prepare 0.1% Tween-20 (Sigma Aldrich) and 2% BSA (VWR) in PBS.
- Volume is sufficient for 20 samples. Unused solution can be stored for up to 6 months at −20°C.
Tween-20, 0.1%
- In a 20-ml volume, prepare 0.1% Tween-20 in PBS.
- Volume is sufficient for 20 samples. Unused solution can be stored for up to 6 months at room temperature.
COMMENTARY
Background Information
OoC devices provide a way to maintain cells or tissues under constant flow to mimic the natural environment inside the body as much as possible. Most devices are closed systems in which cells can be inserted via the flow channel(s) (Dsouza, Kuthethur, Kabekkodu, and Chakrabarty, 2022; Ingber, 2022). This protocol describes a method for culturing tissue slices loaded into the OoC device before the system is closed. Several designs have been described elsewhere (Table 1), but most are custom systems that are not commercially available. We described an OoC system that is commercially available (Chakrabarty et al., 2022). We considered several important features in addition to the option to load tissue specimens before closing the system.
| Cancer model | Selected material | Sample type | Anticancer drugs/compounds | Application | Maximum culture period | Option to test multiple drugs in a single run | Option to test multiple tumors in a single run | Option for on-chip imaging analysis | Tested in more than one tumor type | Flexibility to add sensors to test additional parameters | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Head and neck squamous cell carcinoma (HNSCC) | PDMS | Patient-derived tumor slices | 5-Fluorouracil and Cisplatin | Viability; studying tumor microenvironment; personalized treatment | 7 days | No | No | No | No | No | (Hattersley et al., 2012) |
| Glioblastoma multiforme | PDMS | PDX tumor slices | Temozolomide (TMZ) | Investigate tumor cell death in response to drug treatment; guide therapeutic decision-making | 2 days | Yes | No | Yes | No | No | (Chang et al., 2014) |
| Ovarian cancer | PDMS | Patient-derived tumor slices | Carboplatin | Drug testing and personalized therapy | 8 days | Yes | Yes | Yes | No | No | (Astolfi et al., 2016) |
| Adenocarcinoma of lung | PDMS | PDX tumor slices | Staurosporine | Assess tumor cell death in response to drug treatment | 5 days | Yes | Yes | Yes | No | No | (Holton et al., 2017) |
| Thyroid cancer | Polyether ether ketone (PEEK) | Patient-derived tumor slices | Etoposide and SP600125 | Cellular apoptosis; drug response; assessment of radioiodine sensitivity in real-time | 4 days | No | No | No | No | No | (Riley et al., 2019) |
| Head and neck squamous cell carcinoma (HNSCC) | Polyether ether ketone (PEEK) | Patient-derived tumor slices | Concurrent Cisplatin and irradiation | Caspase-dependent apoptosis, DNA damage, and proliferation; personalized treatment | 68 hr | No | No | No | No | No | (Kennedy et al., 2019) |
| Glioblastoma | Poly (methyl methacrylate) (PMMA) | Patient-derived tumor slices | Cisplatin and Staurosporine | Preclinical platform for cancer drug testing and improving cancer precision medicine | 3 days | Yes | No | Yes | No | No | (Horowitz et al., 2020) |
| Colorectal carcinoma liver metastasis | Poly (methyl methacrylate) (PMMA) | Patient-derived tumor slices | Fluorouracil and Oxaliplatin (FOLFOX); Fluorouracil and Irinotecan (FOLFIRI) | Cancer drug testing and precision medicine | 2 days | Yes | No | Yes | No | No | (Rodriguez et al., 2020) |
| Breast cancer and prostate cancer | PDMS and Silicon | PDX tumor slices | Cisplatin and Apalutamide | Cancer drug testing and precision medicine | 14 days | Yes | Yes | Yes | Yes | Yes | Chakrabarty et al., 2022 |
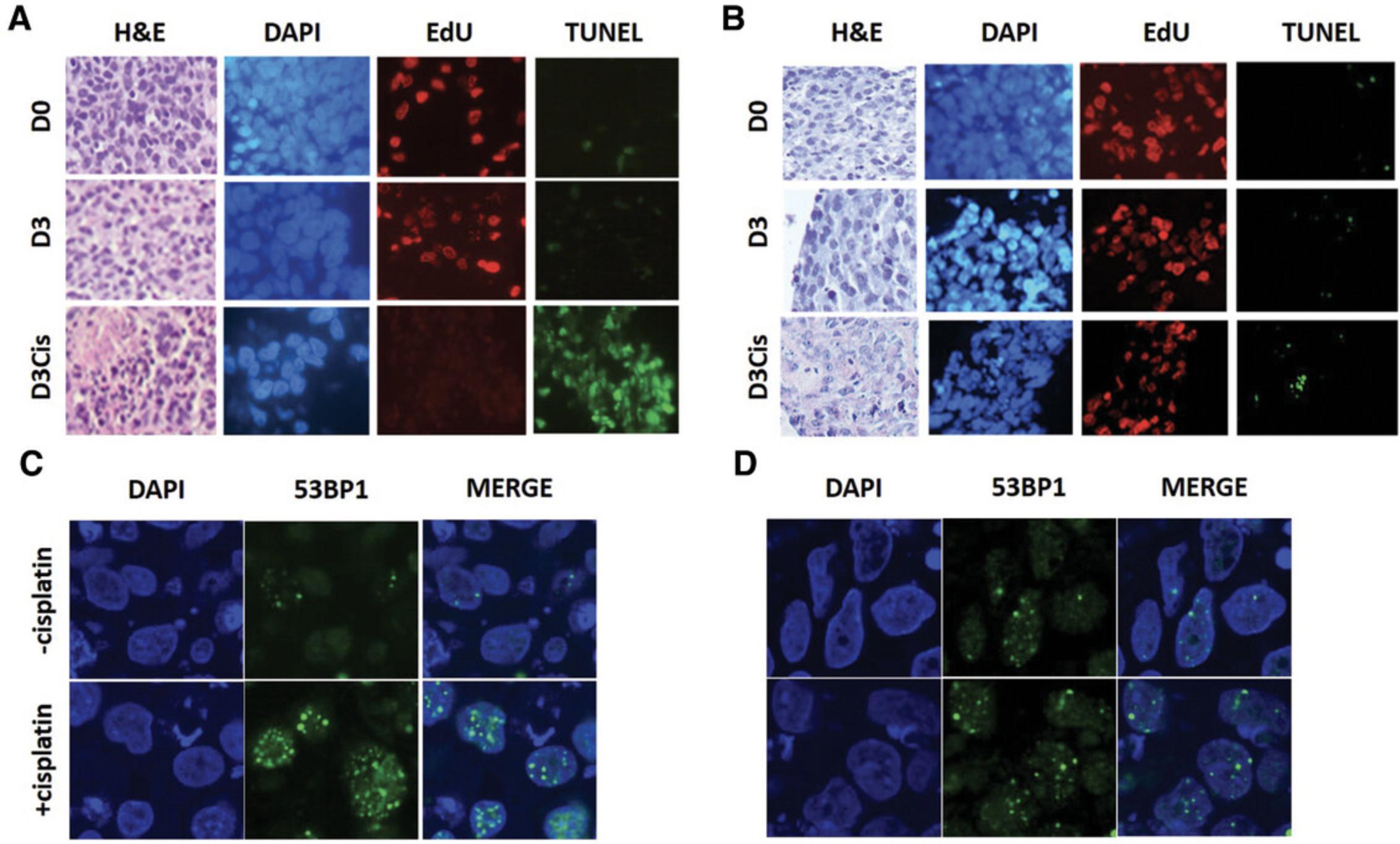
First, experiments must be reproducible. We investigated reproducibility in PDX tumors to demonstrate the ability to perform biological replicates in a controlled setting. Tumor slices from breast and prostate cancers showed good viability, proliferative capacity, and maintenance of tissue integrity for 7 to 14 days. We also confirmed that in vivo responses were mimicked in the OoC device: cisplatin sensitivity in the device was consistent the in vivo characteristics of breast cancer PDX models (Fig. 4), and androgen-dependent prostate tumors were sensitive to the anti-androgen apalutamide (Chakrabarty et al., 2022).
Second, the tumor microenvironment should be maintained for the duration of the experiment. We observed no significant differences in tumor architecture, including the condition of stromal fibroblasts. Gene expression analysis showed no significant changes between days 0 and 7, suggesting that the controlled environment was sufficient to maintain the integrity of all major cell types in the tissue. The primary limitation of these experiments is that PDX tumors are much less rich in stromal cells than primary or metastatic tumors directly derived from patients. Therefore, additional characterization of the culture system with these tumor samples provides a more complete understanding of its ability to faithfully maintain all stromal cell types (including immune cells) for the duration of the experiment.
Third, the OoC device should not influence therapeutic efficacy. We showed this is indeed the case for cisplatin and apalutamide, suggesting that absorption did not hamper drug efficacy. We hypothesize that the amount of PDMS in the device is sufficiently limited and absorption is minimal. This represents a substantial advantage of this device, which consists mainly of silicon in the membrane layer, leaving only some absorption by the PDMS. However, we cannot exclude the possibility that other compounds might behave differently. We are currently investigating paclitaxel and PARP inhibitors.
Fourth, multiple parallel experiments should be possible. The 6-well format of the device enables multiplexing. For example, duplicate experiments can be performed for untreated tumor slices and two drug concentrations to discriminate between sensitive and resistant tumors. Each culture well is large (3 × 3 mm), allowing relatively large pieces of tissue to be cultured, and the height of the chamber (several mm) is sufficient to allow growth. When the system is used for much smaller biopsies or parts of biopsies, it might be useful to redesign the chamber to optimize it for such specimens. The tissue slices are immobilized with a thermoreversible hydrogel to prevent movement after loading into the OoC device. This will also help to localize (very) small tissue pieces after the culture period, which is a major problem in standard 6- or 24-well ex vivo cultures.
Finally, the OoC device should be sufficiently flexible to adapt its configuration for membrane porosity, sensor insertion, and optical accessibility. The PDMS layer containing the membrane allows easy redesign of the membrane properties, for which only the chip must be reconfigured. Furthermore, sensors can be integrated into the chip, although this requires more extensive redesign. Optical access is possible from the bottom of the device through the transparent PDMS window. The main disadvantage is the relatively long working distance, as the tissue specimen is on top of the membrane, with the bottom channel between the microscope and the tissue.
We believe the Bi/ond OoC device is well suited for studies of tumor biology, treatment response, and tumor evolution because tumor characteristics can be maintained for a relatively long time. In addition to end-point measurements that can be done when the device is disassembled, it is also possible to measure tumor-derived components in the outflow (such as extracellular vesicles, cell-free DNA, and cytokines) and observe the tumor specimen directly using microscopic techniques via the transparent window. We have not yet explored these possibilities in detail, so additional analysis is needed to determine the level to which these analyses will be possible. Furthermore, the top and bottom flow design mimics the administration of therapeutic compounds via the bloodstream by coating the bottom channel with endothelial cells.
Critical Parameters
The microfluidic device used in this protocol has several critical parameters. A continuous flow of medium must be provided to the tissue to ensure its survival. Therefore, leaks should be avoided. If the system is set up properly, this issue should not occur, but leaks may occur if, for instance, bubbles form. Handling of the perfusion system should be done in a way that avoids bubble formation. A regular check of the system is required to ensure the experiment is carried out correctly. This should be done approximately once a day. For other issues with running the microfluidic setup, sample staining, and possible solutions, see Table 2.
| Problem | Possible cause | Solution |
|---|---|---|
| Leakage | Bubble formation | Inclusion of a bubble trap in the system |
| Chip running dry | Medium reservoir is empty | Daily monitoring of fluid levels in the medium reservoir. Alternatively, a syringe pump to infuse/withdraw medium can be used. |
| Overflow of medium in the incubator | Collection tubes are full | Replace the collection tube when it is full. |
| Tissue slicing difficulty | Necrosis within the tissue; tissue difficult to cut through | Decrease the speed and increase the amplitude of the Vibratome Leica VT |
| Inconsistent flow rate in the CoC | Blockage in the flow sensor or bubble in the tubing | Routine cleaning of flow sensors with biofilm remover diluted with ultrapure MQ water. Use a bubble trap to remove air bubbles from the tubes connecting the CoC and flow sensor |
| Gradient of DAPI channel signal within a tissue slice | DAPI does not fully penetrate the sample | Stain with DAPI before embedding using Vectashield without DAPI |
Understanding the Results
The Bi/ond microfluidic CoC device was developed to culture relatively large tumor slices inside a microfluidic chip connected to a system that circulates medium inside the chip and removes waste for an extended period. Precision-cut tumor slices are prepared using a Leica VT1200S vibratome and cultured inside the chip, which holds a silicon frame with microfluidic channels embedded in a PDMS film (Fig. 1A-D). The silicon frame has two openings (inlet and outlet) to the channels in the PDMS film (Fig. 1D). The design of comPLATE, which hosts the microfluidic chips, is based on a simple 6-well plate design that provides relative ease of handling for loading and disassembly (Fig. 1E-G). Each well can hold one tumor slice, enabling users to perform six independent experiments in a single run (Fig. 1E).
In each run, six chips can be located on the bottom and covered with a top (Fig. 1F-G). The optical window beneath the well of the microfluidic chip facilitates microscopy during culture (Fig. 1E). The design of the microfluidic chip facilitates culture of relatively large tumor slices for drug testing. This is important given that tumors are heterogenous, with varying proportions of tumor and stromal cells significantly impacting drug response. We used a thermoreversible hydrogel to attach the tumor slice inside the microfluidic chip to minimize the effect of shear forces during prolonged culture for up to 14 days.
The fluidic system consists of an FLPG pump connected to a microfluidic flow control system (Fluigent MFCS-EZ) and flow rate sensors (Fluigent FLOW UNIT-S) that maintain a flow rate of 5 µl/min. The entire system is placed on a shelf inside the CO2 incubator (Fig. 2A and 2B). Fluid pressure is monitored throughout the run in real-time using a Fluigent Microfluidic Automation Tool (MAT) (Fig. 2C). Previously, we showed that tumor slices could be cultured in our CoC device for up to 14 days without any gross change in tumor architecture, cell viability, proliferation, or death (Chakrabarty et al., 2022).
We cultured PDX-derived tumor slices for 3 days in the presence or absence of cisplatin. We compared cisplatin-sensitive and cisplatin-resistant breast PDX models to validate the system. Histology, cell proliferation (EdU staining), and cell death (TUNEL staining) analysis showed that proliferation decreased considerably in the cisplatin-sensitive model but not in the resistant model. Conversely, cell death increased in the sensitive model but not in the resistant model, demonstrating that the CoC device can accurately predict cisplatin sensitivity in these PDX models (Fig. 4A and 4B). Consistent with these results, DNA damage analysis showed increased DNA damage in the cisplatin-sensitive PDX (53BP1 foci formation). In contrast, cisplatin-resistant PDX showed minimal DNA damage upon cisplatin exposure, confirming that the sensitive model has impaired DNA damage repair (Fig. 4C and 4D).
Time Considerations
Basic Protocol 1 can be performed in one week. In case of the late arrival of the tumor tissue, overnight incubation at 4°C is possible. Alternatively, slicing can be performed immediately and the slices incubated at 37°C on a rotating platform overnight before assembling the CoC device the following day. Incubation time depends on the treatment chosen. In this setup, tissue slices were incubated with chemotherapy drugs for 3 days. Fixation requires 2 hr for EdU incubation and overnight incubation in formalin. Tissue processing following fixation requires 2-3 days. Immunostaining requires 4-5 hr. The entire procedure can be performed within 1 to 2 weeks from tumor sample retrieval.
Acknowledgments
This work was supported by grants from the Dutch Cancer Foundation (KWF 11011 and 13651); the Oncode Institute, which is partly financed by the Dutch Cancer Society; and the Netherlands Organization for Scientific Research (NWO Building Blocks of Life grant 737.016.011). The authors also thank William Quiros Solano and Nikolas Gaio for carefully checking our manuscript.
Author Contributions
Zofia M. Komar : Writing original draft; Dik C. van Gent : Writing review and editing; Sanjiban Chakrabarty : Writing original draft.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data available on request from the authors.
Literature Cited
- Astolfi, M., Péant, B., Lateef, M. A., Rousset, N., Kendall-Dupont, J., Carmona, E., … Gervais, T. (2016). Micro-dissected tumor tissues on chip: An ex vivo method for drug testing and personalized therapy. Lab on a Chip , 16, 312–325. doi: 10.1039/C5LC01108F
- Campbell, S. B., Wu, Q., Yazbeck, J., Liu, C., Okhovatian, S., & Radisic, M. (2021). Beyond polydimethylsiloxane: Alternative materials for fabrication of organ-on-a-chip devices and microphysiological systems. ACS Biomaterials Science & Engineering, 7, 2880–2899. doi: 10.1021/acsbiomaterials.0c00640
- Chakrabarty, S., Quiros-Solano, W. F., Kuijten, M. M. P., Haspels, B., Mallya, S., Lo, C. S. Y., … van Gent, D. C. (2022). A microfluidic cancer-on-chip platform predicts drug response using organotypic tumor slice culture. Cancer Research , 82, 510–520. doi: 10.1158/0008-5472.CAN-21-0799
- Chang, T. C., Mikheev, A. M., Huynh, W., Monnat, R. J., Rostomily, R. C., & Folch, A. (2014). Parallel microfluidic chemosensitivity testing on individual slice cultures. Lab on a Chip , 14, 4540–4551. doi: 10.1039/C4LC00642A
- Dsouza, V. L., Kuthethur, R., Kabekkodu, S. P., & Chakrabarty, S. (2022). Organ-on-Chip platforms to study tumor evolution and chemosensitivity. Biochimica et biophysica acta. Reviews on Cancer , 1877, 188717. doi: 10.1016/j.bbcan.2022.188717
- Hattersley, S. M., Sylvester, D. C., Dyer, C. E., Stafford, N. D., Haswell, S. J., & Greenman, J. (2012). A microfluidic system for testing the responses of head and neck squamous cell carcinoma tissue biopsies to treatment with chemotherapy drugs. Annals of Biomedical Engineering , 40, 1277–1288. doi: 10.1007/s10439-011-0428-9
- Holton, A. B., Sinatra, F. L., Kreahling, J., Conway, A. J., Landis, D. A., & Altiok, S. (2017). Microfluidic biopsy trapping device for the real-time monitoring of tumor microenvironment. PLoS ONE , 12, 1–21. doi: 10.1371/journal.pone.0169797
- Horowitz, L. F., Rodriguez, A. D., Dereli-Korkut, Z., Lin, R., Castro, K., Mikheev, A. M., … Rostomily, R. C. (2020). Multiplexed drug testing of tumor slices using a microfluidic platform. NPJ Precision Oncology , 4, 12. doi: 10.1038/s41698-020-0117-y
- Ingber, D. E. (2022). Human organs-on-chips for disease modelling, drug development and personalized medicine. Nature Reviews Genetics , 23, 467–491. doi: 10.1038/s41576-022-00466-9
- Kennedy, R., Kuvshinov, D., Sdrolia, A., Kuvshinova, E., Hilton, K., Crank, S., … Greenman, J. (2019). A patient tumour-on-a-chip system for personalised investigation of radiotherapy based treatment regimens. Scientific Reports , 9, 1–10. doi: 10.1038/s41598-019-42745-2
- Ladan, M. M., Meijer, T. G., Verkaik, N. S., Komar, Z. M., van Deurzen, C. H. M., den Bakker, M. A., … Jager, A. (2022). Functional ex vivo tissue-based chemotherapy sensitivity testing for breast cancer. Cancers , 14, 1252. doi: 10.3390/cancers14051252
- Naipal, K. A., Verkaik, N. S., Sanchez, H., van Deurzen, C. H., den Bakker, M. A., Hoeijmakers, J. H., … van Gent, D. C. (2016). Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer , 16, 78. doi: 10.1186/s12885-016-2119-2
- Riley, A., Green, V., Cheah, R., McKenzie, G., Karsai, L., England, J., & Greenman, J. (2019). A novel microfluidic device capable of maintaining functional thyroid carcinoma specimens ex vivo provides a new drug screening platform. BMC Cancer , 19, 259. doi: 10.1186/s12885-019-5465-z
- Rodriguez, A. D., Horowitz, L. F., Castro, K., Kenerson, H., Bhattacharjee, N., Gandhe, G., … Folcha, A. (2020). A microfluidic platform for functional testing of cancer drugs on intact tumor slices. Lab on a Chip , 20, 1658–1675. doi: 10.1039/C9LC00811J
- Shirure, V. S., & George, S. C. (2017). Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab on a Chip , 17, 681–690. doi: 10.1039/C6LC01401A
Internet Resources
A video demonstration of Complete assembly.
Citing Literature
Number of times cited according to CrossRef: 1
- Wenqi Hu, Ho-Pan Bei, Hongwei Jiang, Di Wu, Xiaorui Yu, Xintong Zhou, Qiuwan Sun, Qinrui Lu, Qijun Du, Liangwen Wang, Zhi Luo, Guohua Wu, Xin Zhao, Shuqi Wang, DLM–GelMA/tumor slice sandwich structured tumor on a chip for drug efficacy testing, Lab on a Chip, 10.1039/D4LC00278D, 24 , 15, (3718-3727), (2024).

