Cultivation and Genomic DNA Extraction of Klebsiella pneumoniae
Drew A. Pariseau, Drew A. Pariseau, Brooke E. Ring, Brooke E. Ring, Saroj Khadka, Saroj Khadka, Laura A. Mike, Laura A. Mike
Abstract
Klebsiella pneumoniae is a Gram-negative, rod-shaped bacterium of medical significance. It typically exists as part of the normal flora of the human intestine but can cause severe infections in the healthcare setting due to its rapid acquisition of antibiotic resistance. Cultivating and extracting genomic DNA from this bacterium is crucial for downstream characterization and comparative analyses. To provide a standardized approach for growing K. pneumoniae in the laboratory setting, this collection of protocols provides step-by-step procedures for routine culturing, generating growth curves, storing bacteria, and extracting genomic DNA. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Reviving K. pneumoniae from frozen stocks
Basic Protocol 2 : Cultivating K. pneumoniae in rich growth medium
Alternate Protocol : Cultivating in minimal liquid growth medium
Basic Protocol 3 : Enumerating K. pneumoniae colony forming units
Basic Protocol 4 : Growth curves
Basic Protocol 5 : Genomic DNA extraction
Basic Protocol 6 : Characterizing K. pneumoniae strains based on genomic sequence
Basic Protocol 7 : Storage of K. pneumoniae frozen stocks in glycerol
Basic Protocol 8 : Storage of K. pneumoniae in agar stabs
INTRODUCTION
Klebsiella pneumoniae is a member of the Enterobacteriaceae family of bacteria and is associated with hospital-acquired urinary tract infections, septicemia, and pneumonia (Magill et al., 2014). This species causes approximately 10% of all nosocomial infections and is frequently found to be resistant to empirical antimicrobial treatments, such as carbapenem (Magill et al., 2014). Like other members of the Enterobacteriaceae family, members of Klebsiella do not require special growth medium and will grow on common growth media such as LB and MacConkey media.
K. pneumoniae has similar growth conditions to other Enterobacteriaceae. The bacterium has an optimal growth temperature of between 35°C to 37°C and a pH of 7.2 (Ristuccia & Cunha, 1984). In some instances, temperatures of 30°C may be preferable to prevent overgrowth in assays visualizing colonies, such as counting colony forming units (CFUs). K. pneumoniae are also facultative anaerobes and can survive solely on citrate and glucose as carbon sources and ammonia as a nitrogen source (Ristuccia & Cunha, 1984).
This article describes general methods for growing colonies on LB agar, cultivating in LB-based and M9-based liquid media, generating growth curves, extracting genomic DNA extraction, performing genomic analyses, and long-term storage. Using the outlines provided, the reader will be able to cultivate and purify DNA from K. pneumoniae for the use in studying growth kinetics, the effects of specific nutrients on growth, down-stream genetic analyses and manipulation, and other biochemical assays such as quantifying capsule production (Khadka et al., 2023a; Ring et al., 2023).
CAUTION : K. pneumoniae is a Biosafety Level 2 [BSL-2] pathogen. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms.
STRATEGIC PLANNING
Strain Selection
K. pneumoniae has several strains commonly used in the laboratory. Prior to choosing a K. pneumoniae strain, identify the experimental purpose and downstream applications. A list of commonly used strains can be found in Table 1.
| Strain (alternate names) | Notable information | Reference/source |
|---|---|---|
| 342 | ST146 K. variicola isolated from interior of nitrogen-efficient maize plants; pKP187; previously described as K. pneumoniae | Chelius & Triplett, 2000; Fouts et al., 2008 |
| 1084 | ST23 K1, non-mucoid | Lin et al., 2012 |
| AJ218 | Nosocomial urinary tract infection isolate | Jenney et al., 2006 |
| ATCC 43816 (KPPR1/KPPR1S/MKP220) | ST493 clinical pneumonia isolate; O1:K2 | Bakker-Woudenberg et al., 1985; Roosendaal et al., 1985 |
| KPPR1: Rifampin-resistant derivative; used to generate an arrayed transposon library | Lawlor et al., 2005; Broberg et al., 2014; Mike et al., 2021 | |
| KPPR1S: Streptomycin- and rifampin-resistant derivative | Walker et al., 2019 | |
| MKP220: Spectinomycin-resistant derivative; used to generate an arrayed transposon library | Paczosa et al., 2020 | |
| BJ1 | K2; encodes no phages and few defense systems. | Blin et al., 2017 |
| CG43 | ST86 hypermucoid bacteremia isolate with K2 and pLVPK; highly virulent in mice | Peng et al., 1991 |
| hvKP1 | hvKp isolated from blood and liver abscess in Buffalo, NY, USA | Pomakova et al., 2012; Russo & Gill, 2013 |
| KP-1 | CG10193 environmental isolate | Lee et al., 2013 |
| Kp52.145 (B5055/52145) | ST66 hvKp strain with O1:K2, Kp52.145pI and Kp52.145pII (pKP100) | Goslings et al. 1936; Lery et al., 2014 |
| KPNIH1 (MKP103) | ST258 nosocomial outbreak strain isolated from the groin of an organ transplant patient; pAAC154-a50, pKPN-498, pKpQIL-6e6 (blaKPC) | Snitkin et al., 2012 |
| MKP103 is a derivative cured of pKpQIL-6e6 (blaKPC) used to generate an arrayed transposon library | Ramage et al., 2017 | |
| LM21 | Nosocomial isolate | Favre-Bonte et al., 1998 |
| MGH78578 (ATCC 700721) | ST52 sputum isolate with K52 pKPN3, pKPN4, pKPN5, pKPN6, pKPN7 | McClelland et al., 2001; Ogawa et al., 2005 |
| SGH10 | ST23 (CG23-I) hvKp isolate from human liver abscess with K1, pK2044-like and typical CG23-I virulence loci intact | Lam et al., 2018 |
| TOP52 1721 (TOP 52) | Acute cystitis isolate | Rosen et al., 2008; Johnson et al., 2014 |
| NTUH-K2044 | ST23 hvKp isolate from a patient with liver abscesses and meningitis; O1:K1; pK2044 | Wu et al., 2009 |
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals. Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol 1: REVIVING K. pneumoniae FROM FROZEN STOCKS
K. pneumoniae can be stored long-term at −80°C when diluted in sterile glycerol (at a final concentration of 25% vol./vol. glycerol). As frozen strain stocks are frequently accessed, contamination is a threat to stock integrity. Therefore, rigorous aseptic technique is necessary. For example, the use of a biosafety cabinet will greatly reduce the chance of contamination when reviving a frozen stock of bacteria.
Materials
-
K. pneumoniae frozen stock
-
Inoculating loop or wooden applicator sticks, sterile (Fisher Scientific, cat. no. 22-029-491)
-
Luria broth (LB) agar plates (see recipe)
-
Incubator, static, set to 30°C
1.Using a sterile wooden applicator stick, remove a small chunk of frozen bacteria from the stock vial.
2.Streak the bacteria onto the LB agar plate.
3.Incubate the plate overnight for 14 to 18 hr at 30°C or 37°C.
Basic Protocol 2: CULTIVATING K. pneumoniae IN RICH GROWTH MEDIUM
K. pneumoniae can be cultured in a variety of liquid media, each with its own unique composition and applications. Lysogeny broth (LB) is the primary medium used for the cultivation of the bacterium, and the medium can be prepared in three primary formulations: Miller, Lennox, and Luria (Sezonov et al., 2007). The Luria formulation is the preferred formulation due to its lower sodium chloride content, which enhances K. pneumoniae mucoidy (Fig. 1). See Reagents and Solutions for media recipes.
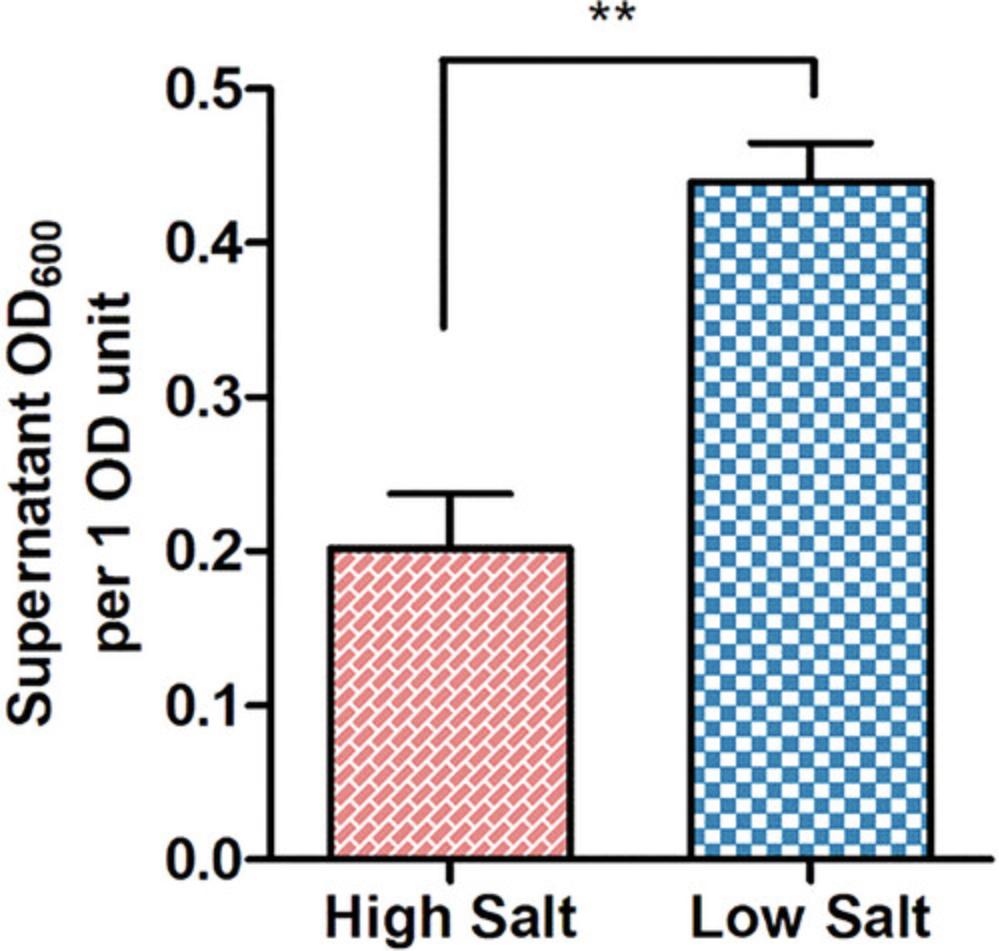
Each medium can be adapted to the precise requirements for individual experimental conditions. Modifications can be made by incorporating supplements, such as antibiotics at desired concentrations (Table 2), to tailor the different media to suit the specific requirements of the experiment.
| Antibiotic | Solvent | Stock concentration (mg/ml) | Final concentration (μg/ml) |
|---|---|---|---|
| Kanamycin | Water | 25 | 25 |
| Chloramphenicol | Ethanol | 80 | 80 |
| Spectinomycin | Water | 50 | 50 |
| Gentamicin | Water | 100 | 10 |
| Tetracycline | Water | 6 | 6 |
| Apramycin | Water | 50 | 50 |
| Hygromycin | Water | 100 | 100 |
| Rifampin | Methanol | 30 | 30 |
| Streptomycin | Water | 500 | 500 |
- 1
Note that K. pneumoniae isolates are ampicillin-resistant.
Materials
-
Individual colonies of K. pneumoniae on LB agar plate
-
Luria broth (LB), sterile (see recipe)
-
Aeration culture tube, sterile, 16-ml, 17 mm diameter (Genesee Scientific, cat. no. 21-131 or equivalent)
-
Inoculating loop or wooden applicator sticks, sterile (Fisher Scientific, cat. no. 22-029-491 or equivalent)
-
Shaking incubator, set to 37°C, 200 rpm
1.Using aseptic technique, transfer 3 ml of LB broth to a sterile 16 ml aeration culture tube.
2.Use a sterile inoculating loop or wooden applicator stick to collect a single colony from the LB agar plate.
3.Inoculate the LB broth in the culture tube by agitating the colony on the stick in the LB broth.
4.Replace the cap onto the culture tube to the aerated position.
5.Incubate the tube in a 37°C shaking incubator at 200 rpm for 16 to 18 hr, overnight.
Alternate Protocol: CULTIVATING IN MINIMAL LIQUID GROWTH MEDIUM
This protocol is a modified version of Jeffery H. Miller's original work on M9 minimal growth medium (Miller, 1972). It includes updated information on the best practices for cultivating K. pneumoniae. In contrast to Basic Protocol 2, the composition of this medium includes a mixture of salts, calcium, magnesium, and can be supplemented with glucose, amino acids, and vitamins, as needed. This variation enables enhanced control over the provided nutrients. The procedure entails preparing a 10× mixture of salts and supplements, performing chelation treatment, sterilizing through filtration, and subsequently introducing metals. By following this protocol, minimal medium enables precise control, reproducibility, and flexibility in the study of organism growth, metabolism, and genetic manipulation. As a result, it serves as a valuable tool in various scientific contexts.
Materials
-
M9 minimal salts, 10×, sterile (see recipe)
-
Calcium chloride (CaCl2), 100 mM, sterile (see recipe)
-
Magnesium sulfate (MgSO4), 1 M, sterile (see recipe)
-
Glucose, 20% (w/v), sterile (see recipe)
-
Thiamine, 10 mg/ml, sterile (see recipe)
-
Casamino acids (CAA), 10% (w/v), sterile (see recipe)
-
Chelex 100 chelating resin (BioRad, cat. no. 1421253)
-
Ferrous sulfate (FeSO4), 36 mM in 0.25 N HCl, 36 mM, sterile (see recipe)
-
1000-ml plastic beaker
-
Stir bar
-
Stir plate
-
Vacuum filter, 0.22 µm (Fisher Scientific, cat. no. S2GPU02RE or equivalent)
Preparing 10× M9 Minimal Salts
1.Make a 1 L volume of 10× M9 minimal salts (see recipe).
2.Autoclave the 10× M9 minimal salts solution to sterilize.
Preparing Stock Solutions
3.Prepare a 100 mM stock solution of CaCl2, 1 M stock solution of MgSO4, and a 20% (w/v) stock solution of glucose (see recipes).
4.Filter sterilize all stock solutions using a sterile vacuum filtration system with 0.22 µm pore size.
Assembling the Medium
5.Add 25 ml of 10× M9 minimal salts and 5 ml of 20% (v/v) glucose, bring the volume up to 250 ml with ultrapure water in a plastic beaker.
6.Add 0.25 g of Chelex 100 resin (1 g/L) to the solution.
7.Cover the medium with plastic wrap and stir with a stir bar for 3 hr to facilitate metal chelation.
8.Sterilize the solution and remove the Chelex by filtering the medium through a vacuum filter unit with a 0.22 µm membrane under aseptic conditions.
9.Add 0.25 ml of 100 mM CaCl2 stock solution and 0.25 ml of 1 M MgSO4 stock solution to the sterile-filtered medium.
10.Culture K. pneumoniae according to Basic Protocol 2 using the prepared M9 minimal medium.
Basic Protocol 3: ENUMERATING K. pneumoniae COLONY FORMING UNITS
The following protocol outlines a method for enumerating K. pneumoniae colony forming units (CFUs). Accurate quantification of bacterial CFUs is essential for assessing microbial load and understanding pathogenic potential. This protocol encompasses step-by-step instructions for sample preparation, serial dilution, plating techniques, and incubation conditions to enable the precise enumeration of K. pneumoniae colonies.
Materials
-
K. pneumoniae liquid suspensions
-
LB agar plates (see recipe)
-
Phosphate-buffered saline (PBS), sterile (see recipe)
-
Vortex mixer
-
96-well, flat-bottom microplates, sterile
-
Multichannel pipette, 10 to 100 μl
-
Incubator, static, set to 37°C
Preparing Serial Dilutions
1.Vortex a liquid suspension of K. pneumoniae and transfer 100 μl of each sample into row A of a 96-well microplate.
2.Fill rows B to H with 90 μl of sterile PBS.
3.Transfer 10 μl from row A into row B. Mix well by pipetting 10 to 20 times.
4.Transfer 10 μl from row B into row C. Mix well by pipetting.
5.Continue this serial dilution series until the final dilution is made in row H, mixing well by pipetting up and down 10 to 20 times and changing pipette tips between each dilution step.
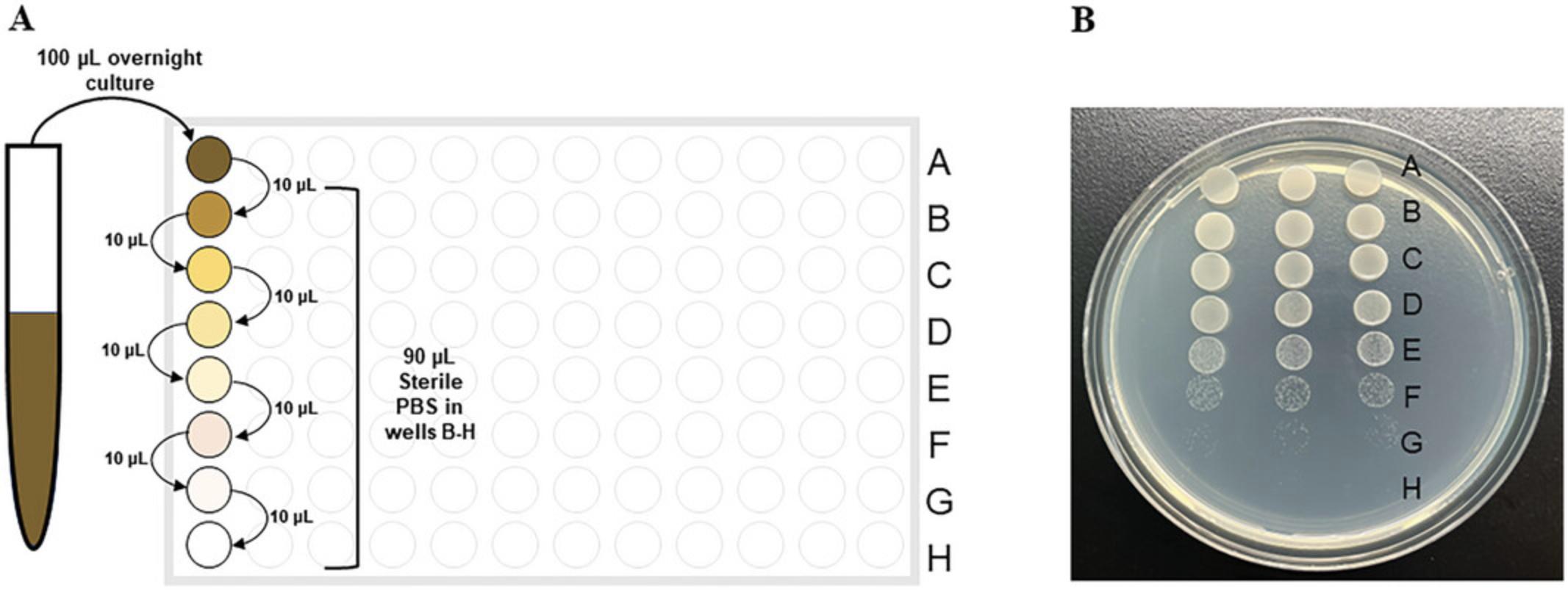
6.Using a multichannel pipette, spot 10 μl of each dilution from one column to an individual LB agar plate. Three columns usually fit on a standard LB agar plate.
7.Incubate the LB agar plate with serial dilution spots in a 37°C incubator for 1 to 2 hr and then leave the plate overnight at room temperature.
Enumerating Colonies
8.Count the colonies at the most diluted spot that can be seen.
9.If possible, count the colonies at the second most diluted spot that can be seen.
10.If two countable spots are present, average the number of CFUs counted in each spot.
Basic Protocol 4: GROWTH CURVES
Growth curve analyses provide insight into the growth kinetics and population dynamics of bacterial cultures. Like evaluating growth on solid media, it is important to maintain rigorous aseptic techniques throughout the entire protocol to ensure reliable results. Uninoculated growth medium represents a critical negative control to evaluate aseptic technique. See Understanding Results for sample data and calculations.
Materials
-
Individual colonies of K. pneumoniae on a LB agar plate
-
LB broth (see recipe)
-
1× PBS, sterile (see recipe)
-
Aeration culture tube, sterile, 16-ml, 17 mm diameter (Genesee Scientific, cat. no. 21-131 or equivalent)
-
1.7-ml disposable cuvettes (Fisher Scientific, cat. no. 14-955-127 or equivalent)
-
1.7-ml microcentrifuge tubes, sterile
-
96-well, flat-bottom plates, sterile (Genesee Scientific, #25-104)
-
Parafilm
-
Shaking incubator, set to 37°C, 200 rpm
-
Visible spectrophotometers (for 1.7-ml cuvettes and microplates)
Subculturing
1.Prepare overnight cultures of single K. pneumoniae colonies in 3 ml of LB broth as described in Basic Protocol 2.
2.Transfer 3 μl of each overnight culture into three additional 16 ml aeration culture tubes, each filled with 3 ml of LB broth.
3.Incubate in a 37°C, 200 rpm for 5 to 7 hr.
Standardization of Inoculum
4.Pipette 900 μl of PBS into one disposable cuvette per sample.
5.Transfer 100 μl of a K. pneumoniae sub-culture to the disposable cuvette containing 900 μl PBS. Mix well by pipetting.
6.Measure the optical density of the sub-culture at a wavelength of 600 nm (OD600) using a visible spectrophotometer.
7.Calculate the amount of bacterial suspension needed to achieve an initial OD600 of 0.0001 in 1 ml of LB broth.
Preparing the Growth Curve Plate
8.Fill 1.7-ml centrifuge tubes with 1 ml of LB broth.
9.Pipette the calculated volume of diluted sub-culture to 0.0001 OD600 in each tube.
10.Pipette 100 μl from each tube containing 0.0001 OD600 of K. pneumoniae culture into a well in a sterile 96-well, flat-bottom microplate.
11.Wrap the edge of the 96-well microplate in laboratory Parafilm.
12.Load the 96-well microplate into a microplate spectrophotometer.
13.Measure the OD600 for the baseline 0 hr timepoint.
14.Run a program that incubates the microplate at 37 °C with orbital shaking (282 cpm) and read the OD600 every 30 min for 16 to 18 hr.
Basic Protocol 5: GENOMIC DNA EXTRACTION
This protocol describes a method to extract K. pneumoniae genomic DNA using the Wizard® Genomic DNA kit (Promega). The method involves treating cell pellets with cell lysis solution, RNase solution, and proteinase solution to remove contaminants. The extracted DNA is purified and precipitated using isopropanol and ethanol. This protocol provides high-quality and high-yield genomic DNA.
Materials
-
Individual colonies of K. pneumoniae on a LB agar plate
-
LB broth (see recipe)
-
Wizard® Genomic DNA purification Kit (Promega, #A1125)
-
Isopropanol (room temperature)
-
70% ethanol (room temperature)
-
Aeration culture tube, sterile, 16-ml, 17 mm diameter (Genesee Scientific, cat. no. 21-131 or equivalent)
-
1.7-ml microcentrifuge tubes, sterile
-
Shaking incubator, set to 37°C, 200 rpm
-
Vortex mixer
-
80°C heat block or water bath
-
Microcentrifuge, minimum speed 17,000 × g
-
Clean paper towel
Overnight Culture
1.Inoculate a single, isolated colony of K. pneumoniae into 3 ml of LB in a 16-ml aeration culture tube. Incubate at 37°C for 16 to 18 hr with shaking at 200 rpm as described in Basic Protocol 2.
Cell Lysis and RNA Removal
2.Add 1 ml of gently vortexed overnight culture to a 1.7 ml centrifuge tube.
3.Pellet the cells: centrifuge 2 min at 16,000 × g , room temperature. Carefully remove the supernatant.
4.Completely resuspend the cell pellet in 600 µl Nuclei Lysis Solution (provided in the kit). Ensure that no visible portions of pellet remain.
5.Incubate the mixture at 80°C for 5 min. Then, cool down the cell lysate to room temperature.
6.Add 3 µl of RNase solution (provided in the kit) to the cell lysate and mix well by inverting the tube 5 times.
7.Incubate RNase-treated cell lysate for 60 min at 37°C. Then, cool down the RNase-treated cell to room temperature.
Protein Precipitation
8.Add 200 µl of Protein Precipitation Solution (provided in the kit) to the tube and mix by vortexing vigorously for 20 sec.
9.Incubate the mixture on ice for 5 min.
10.Centrifuge 3 min at 16,000 × g , room temperature.
DNA Precipitation and Rehydration
11.Following centrifugation, transfer the supernatant (contains DNA) to the tube containing 600 µl of isopropanol.
12.Mix the isopropanol-supernatant solution by gently inverting the tube 6 to 10 times, or until a visible mass of DNA strands is formed.
13.Centrifuge for 2 min at 16,000 × g , room temperature.
14.Slowly pour off the supernatant and drain the remaining solution by inverting the tube on a clean paper towel for 10 to 20 sec.
15.Add 600 µl of 70% ethanol (room temperature) to the DNA pellet and invert the tubes 10 to 15 times.
16.Centrifuge the ethanol-washed DNA for 2 min at 16,000 × g , room temperature.
17.Gently remove the supernatant using a pipette. Drain the remaining ethanol by inverting the tube on a clean paper towel for 10 to 20 sec.
18.To ensure complete removal of ethanol, allow the tube to air dry for 15 min.
19.Add 100 µl of DNA Rehydration Solution (provided in the kit) to the tube. Resuspend precipitated DNA by incubating at 65°C for 1 hr with periodic mixing of the solution.
20.Store rehydrated genomic DNA at −20°C or 2 to 8°C.
Basic Protocol 6: CHARACTERIZING K. pneumoniae STRAINS BASED ON GENOMIC SEQUENCE
Basic Protocol 6 illustrates a guide to characterizing K. pneumoniae species complex strains based on genomic sequence. The genomic sequence of a strain of interest is run through several databases and tools, including BIGSdb and Kleborate by Pathogenwatch (Argimón et al., 2021; Lam et al., 2018; Olson et al., 2023; Wyres et al., 2016). Through these established bioinformatic pipelines, a report is generated highlighting key characteristics of the strain based on the genomic sequence. The report generated using this method can be used to characterize K. pneumoniae with information such as capsule locus type and lipopolysaccharide (LPS) locus type.
Materials
- Desktop or laptop computer with a minimum screen resolution of 1366 × 768 and Chrome, Safari, or Firefox browser
- Bacterial and Viral Bioinformatics Resource Centre software, version 3.30.19 (https://www.bv-brc.org)
- Pathogenwatch software, version 20.5.5 (https://pathogen.watch)
- Short-reads or assembled whole-genome sequence of Klebsiella pneumoniae
Assembly of K. pneumoniae Short-Read Sequence
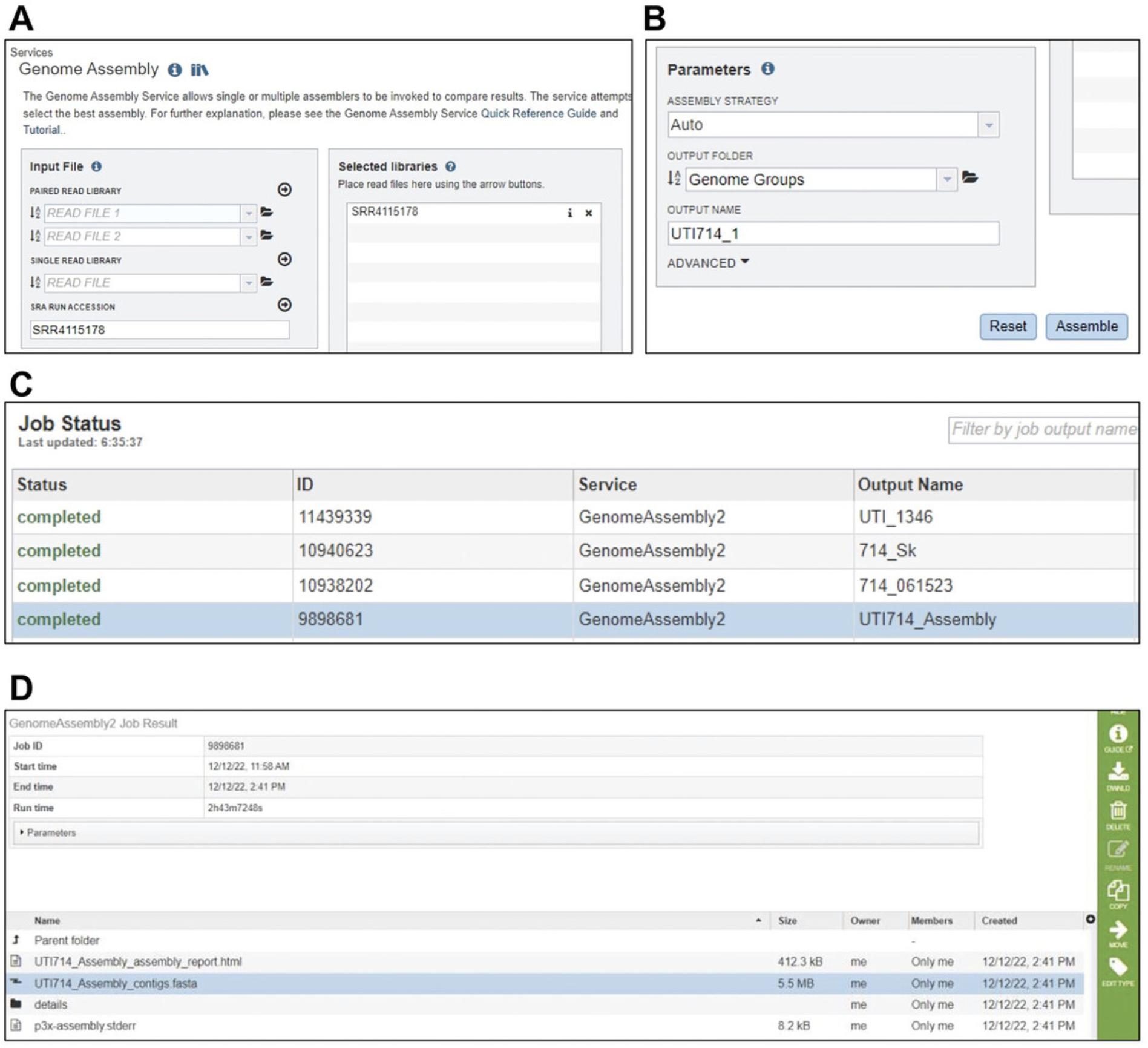
1.Sign up and log in to BV-BRC (https://www.bv-brc.org).
2.At the top of the webpage, click on “Genome Assembly (B)” within the “TOOLS & SERVICES” tab. A new Assembly page will be loaded.
3.If the whole genome sequence of the strain is uploaded in the NCBI Sequence Read Archive (SRA) database, enter the SRA Run Accession number in ‘SRA RUN ACCESSION’ textbox (Fig. 3A).
4.Click on the adjacent arrow button to upload read files to the “Selected libraries” box. If the input SRA number or read file is valid, the library name will appear in the “Selected libraries” list (Fig. 3B).
5.In the “Parameters” box, select output folder to choose the location for storing analyzed data. Also, type the name of the output file in the “OUTPUT NAME” textbox. Then, click “Assemble” to submit the genome assembly job.
6.In the lower-right corner of the home page, click on “Jobs” to locate the completed assembly. A new page will load.
7.Double click on the completed job. A new window will open containing a list of several files associated with the assembly. Select the file with extension “.fasta” and click on “DWNLD” to download the assembled sequence (Fig. 3C–D).
Characterization of K. pneumoniae using Assembled Whole Genome Sequence
8.Load Pathogenwatch (https://pathogen.watch) in a browser and click on “UPLOAD” from the bar at the top. A new page will load.
9.Sign in using one of the available options. Upon signing in, a page to upload sequence files will load.
10.Click on “Single Genome FASTAs”. A new page will load.
11.Click on the “+” button at the lower-right corner of the page. Then upload the assembled sequence downloaded from BV-BRC.
12.Once completed, directly click on “View Genomes” or go to “My Genomes” from the top left menu.
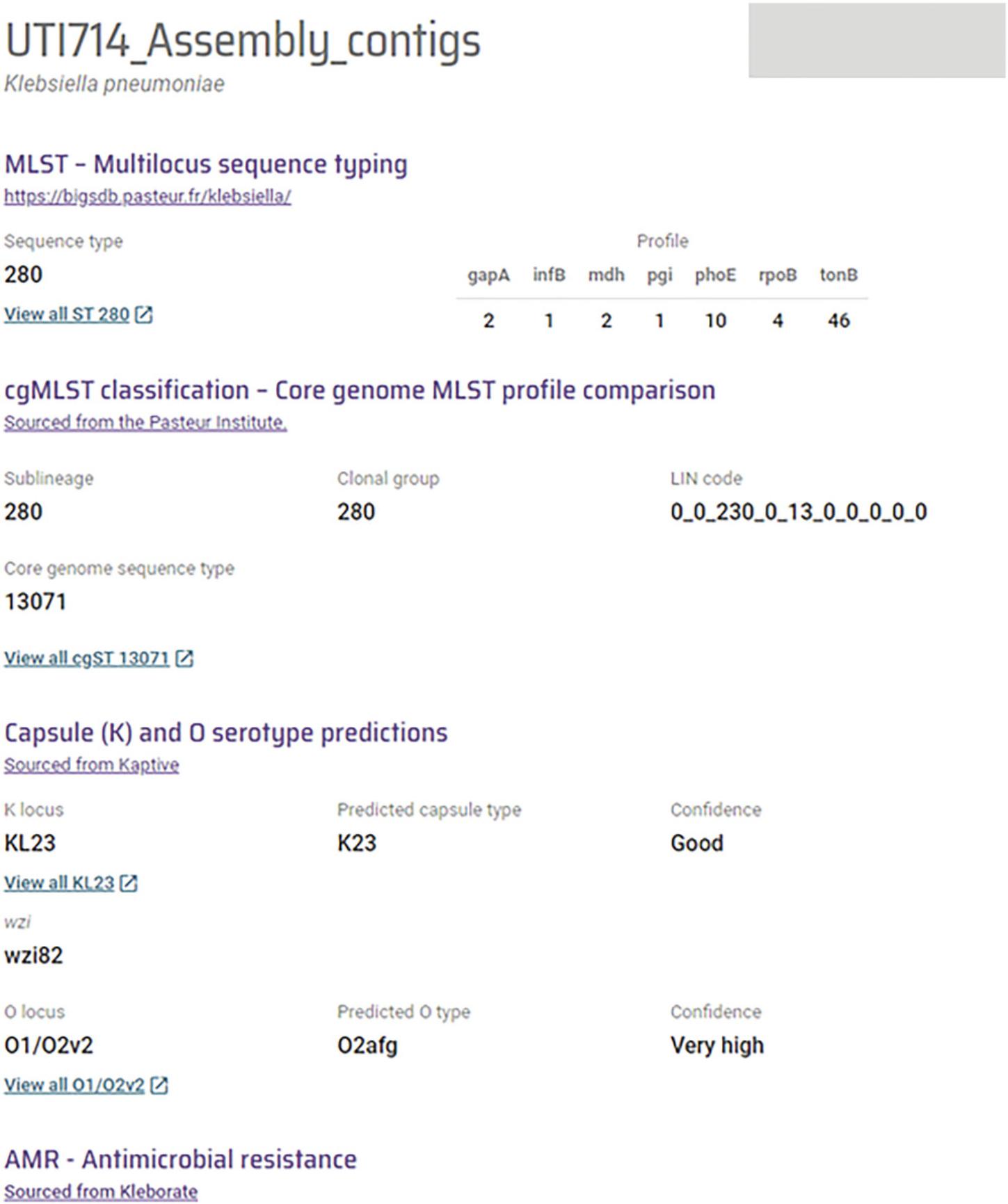
13.From the completed jobs list, open the job with the same file name as the uploaded fasta file. A final report highlighting the species and subspecies of Klebsiella , capsule and LPS locus types, virulence-associated genes, and antimicrobial resistance associated genes will load (Fig. 4).
Basic Protocol 7: STORAGE OF K. pneumoniae FROZEN STOCKS IN GLYCEROL
Freezing bacterial cultures as glycerol stocks is a common method for long-term storage and allows preservation of cell viability and genetic integrity. Glycerol serves as a cryoprotectant, minimizing the damage caused by ice crystal formation during the freeze-thaw cycle.
Materials
-
K. pneumoniae overnight liquid culture
-
50% glycerol in ultrapure water, sterile
-
1.8-ml cryovials (Genesee Scientific, #24-203P or equivalent)
-
Vortex mixer
-
−80°C freezer
1.Transfer 0.5 ml of an overnight culture into a cryovial containing 0.5 ml of 50% glycerol, vortex well.
2.Label the vial with the strain information and other pertinent information.
3.Transfer to an ultra-low temperature freezer for long-term storage at −80°C.
Basic Protocol 8: STORAGE OF K. pneumoniae IN AGAR STABS
Agar stabs are sometimes used for short-term storage of strains at temperatures above freezing, such as when shipping strains. When shipping strains, it is important to adhere to all federal, state, and institutional guidelines. Please consult with your institutional Environmental Health and Safety Office for a full list of federal, state, and institutional guidelines.
Materials
-
Individual colonies of K. pneumoniae on a LB agar plate or in liquid suspension
-
Luria broth (LB) agar stabs (see recipe)
-
Toothpicks, sterile
-
Incubator, static, set to 37°C
1.Touch a sterile toothpick to an individual colony on a solid media plate, or into an overnight liquid culture of K. pneumoniae.
2.Stab almost to the bottom of the vial and then pull the toothpick straight out.
3.Slightly loosen the cap of the agar stab vial and incubate at 37°C for approximately 12 to 18 hr.
4.Screw the cap on tightly and store the vial at 4°C for up to 3 weeks, or room temperature for shipping.
REAGENTS AND SOLUTIONS
Use ultrapure water to prepare all recipes.
Calcium chloride (CaCl2), 100 mM
- 1.47 g calcium chloride
- 100 ml chelated water
Dissolve 1.47 g calcium chloride in 100 ml chelated water. Sterilize with a 0.22 μm filter. Store at room temperature for up to one year.
Casamino acids (CAA), 10 % (w/v)
- 10 g casamino acids
- 100 ml ultrapure water
Dissolve 10g casamino acids in 100 ml ultrapure water. Sterilize with a 0.22 μm filter. Store at 4˚C for up to 6 months.
Chelated water
- 1 g Chelex 100 chelating resin (BioRad, cat. no. 1421253)
- 1000 ml ultrapure water
Dissolve 1 g Chelex 100 chelating resin in 1000 ml ultrapure water with stirring for 3 hours. Remove Chelex 100 by sterile-filtering with a 0.22 μm filter. Store indefinitely at room temperature.
Ferrous sulfate in 0.25 N hydrochloric acid, 36 mM
- 1 g ferrous sulfate (FeSO4)
- 100 ml 0.25 N HCl
Dissolve 1g ferrous sulfate in 100 ml of 0.25 N HCl. Sterilize with a 0.22 μm filter. Store at room temperature.
Glucose, 20% (w/v)
- 20 g glucose
- 100 ml ultrapure water
Dissolve 20 g glucose in 100 ml ultrapure water. Sterilize with a 0.22 μm filter. Store at 4˚C for up to 6 months.
Luria broth (LB), low-salt
- 5 g yeast extract
- 10 g tryptone
- 0.5 g sodium chloride (NaCl)
Add ingredients and ultrapure water to a final volume of 1000 ml in a 4-L beaker. Stir until all solids are dissolved, then aliquot into clean glass bottles. Autoclave the media bottles with the lids slightly loosened.
Store at room temperature up to 6 months.
Luria broth (LB) agar
- 5 g yeast extract
- 10 g tryptone
- 0.5 g sodium chloride (NaCl)
- 20 g agar
Add ingredients and ultrapure water to a final volume of 1000 ml in a 2-L flask. Stir while heating until all solids are dissolved. Autoclave to sterilize. Allow the flask to cool enough to handle without heat-protective gloves and then add antibiotics, as necessary.
To pour agar plates: pour ∼25 ml into sterile 100 mm × 15 mm Petri dishes. Allow to solidify at room temperature. Dry at room temperature for 1 to 2 days. Store in sealed bags at 4°C for up to 3 months.
To make agar stabs: Aliquot 1 ml warm LB agar into a 1.8 ml cryotube. Close the lid loosely and allow the agar to solidify at an angle. Store at 4°C for up to 3 months.
If antibiotics are added, it may be necessary to store the plates protected from light.
M9 minimal salts, 10×
- 60 g sodium dihydrogen phosphate (Na2HPO4)
- 30 g monopotassium phosphate (KH2PO4)
- 5 g sodium chloride (NaCl)
- 10 g ammonium chloride (NH4Cl)
- 1000 ml ultrapure water
Dissolve salts in ultrapure water and bring to 1000 ml. Autoclave and store indefinitely at room temperature.
Magnesium sulfate (MgSO4), 1 M
- 12.03 g magnesium sulfate
- 100 ml chelated water
Dissolve 12.03 g magnesium sulfate in 100 ml chelated water. Sterilize with a 0.22 μm filter. Store at room temperature for up to one year.
Phosphate-buffered saline (PBS), 10×
- 80 g sodium chloride (NaCl)
- 2 g potassium chloride (KCl)
- 14.4 g sodium dihydrogen phosphate (Na2HPO4)
- 2.4 g monopotassium phosphate (KH2PO4)
Dissolve salts in 800 ml ultrapure water. Adjust pH to 7.4 with sodium hydroxide (NaOH). Add ultrapure water to 1 L total. Autoclave and store indefinitely at room temperature.
Thiamine stock solution, 10 mg/ml
- 100 mg thiamine
- 10 ml ultrapure water
Dissolve 100 mg thiamine in 10 ml of ultrapure water. Sterilize with a 0.22 μm filter. Store at 4˚C for up to 2 weeks.
COMMENTARY
Background Information
K. pneumoniae has emerged as a significant global health concern in recent decades. The bacterium can infect a wide range of tissues and rapidly develop resistance to antibiotics, making it considerably potent in the hospital setting. Traditionally, the classical K. pneumoniae (cKp) strains are commonly known to be opportunistic pathogens, affecting those with comorbidities and the immunocompromised. However, the rise of hypervirulent strains of K. pneumoniae (hvKp) has allowed the bacterium to cause infections in young and healthy individuals without traditional risk factors. These strains are frequently associated with serious infections such as liver abscesses, meningitis, and endophthalmitis (Russo & Marr, 2019).
Although there is a rising threat and prevalence of the hvKp strains, there are still knowledge gaps in understanding host-pathogen interactions, virulence determinants, and pathogenesis within different tissue types. The cultivation of laboratory strains and extraction of genomic DNA for study are essential first steps to developing a working understanding of K. pneumoniae pathogenesis mechanisms.
Critical Parameters and Troubleshooting
The quality of the final report generated by Basic Protocol 6 depends on the input genomic sequence. It is critical to ensure that the correct genomic sequence (short-reads or SRR) is submitted to BV-BRC for assembly. Based on the sequence read quality, the platform may fail to generate coherent contigs, ultimately affecting analysis by Pathogenwatch. Pathogenwatch may not be able to identify capsule or lipopolysaccharide type in the final report if it is a novel K- or O-type yet to be updated in the database. Further, it is important to ensure that the most recent versions of both programs are used for analysis. Troubleshooting guides for reviving K. pneumoniae from frozen stocks and growth curve analysis can be found in Tables 4 and 5.
Statistical Analysis
Basic Protocol 4 describes the generation of growth curves. From the data obtained in this experiment, the doubling time can be calculated by identifying an early (t1 and OD1) and a late (t2 and OD2) time point within the most linear portion of the logarithmic growth phase and then applying the following equation.
If utilizing GraphPad Prism (version 8.3.0 or later) for data analysis, the area under the curve can also be calculated to quantify the overall growth of the strain (Mike et al., 2021).
Understanding Results
Basic Protocol 1 describes the revival of K. pneumoniae from a frozen stock culture. After streaking and overnight incubation in 37°C, single, isolated colonies should be visible away from the initial streaking area and should be large, round, and white in color. Basic Protocol 2 should yield a turbid liquid culture with no significant change in medium color. Basic Protocol 3 describes counting CFUs on an LB agar plate. After 37°C incubation, single colonies should only be present where the plates were spotted. Optimal growth is achieved when two of the most dilute spots have distinct, individual colonies.
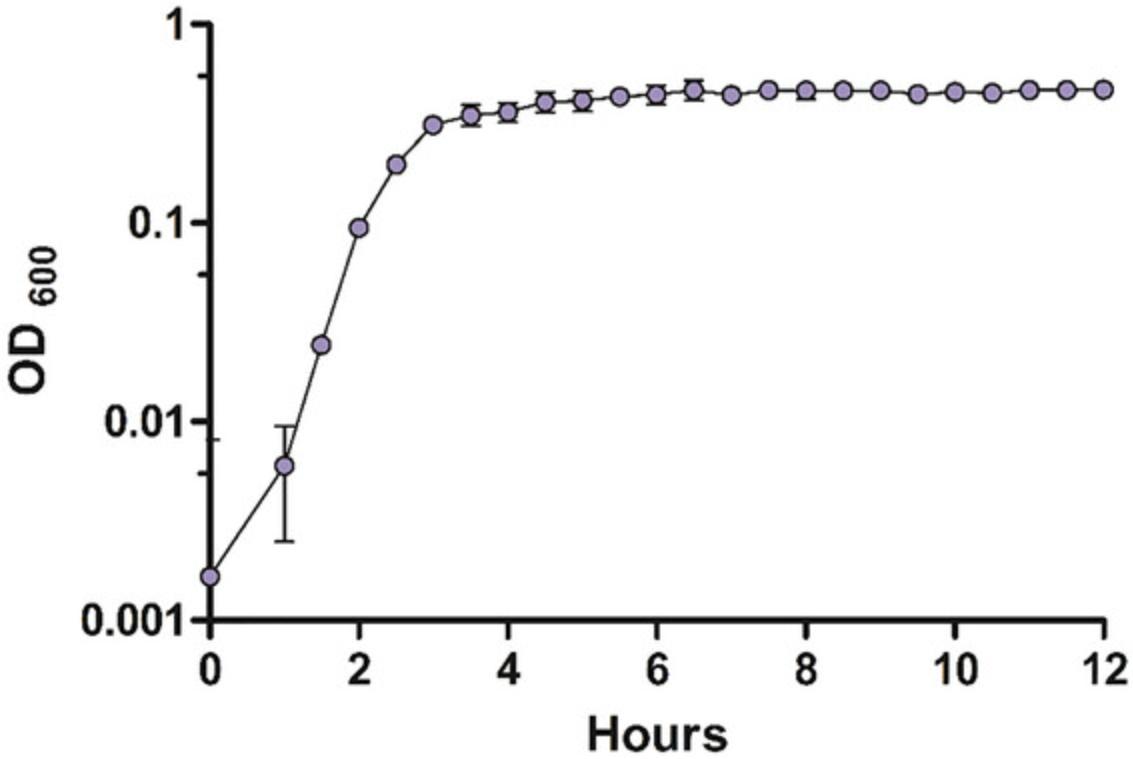
Basic Protocol 4 describes the generation of growth curves over a period of time. An expected OD600 data set may look like the data generated in Table 3. The values shown have the background value subtracted from the culture OD600. This data set illustrates three samples of the NTUH-K2044 laboratory strain over a period of 12 hr. 16 to 18 hrs is recommended when conducting growth curves for the first time. The time course for subsequent experiments may be adjusted for fast or slow-growing strains. A growth curve can then be plotted from this data. An example of a curve generated from the data in Table 3 is shown in Figure 5. Note that the decline in growth during the late stationary phase may not always be seen, such as in this experiment. Longer monitoring may be necessary to see every phase.
| Time (hr) | Replicate 1 (OD600)1 | Replicate 2 (OD600)1 | Replicate 3 (OD600)1 |
|---|---|---|---|
| 0 | −0.00833 | −0.00033 | 0.013667 |
| 0.5 | −0.007 | −0.004 | 0.003 |
| 1 | 0.003 | 0.002 | 0.013 |
| 1.5 | 0.023333 | 0.023333 | 0.026333 |
| 2 | 0.094 | 0.097 | 0.093 |
| 2.5 | 0.218333 | 0.179333 | 0.193333 |
| 3 | 0.356333 | 0.262333 | 0.314333 |
| 3.5 | 0.422333 | 0.273333 | 0.350333 |
| 4 | 0.423667 | 0.285667 | 0.375667 |
| 4.5 | 0.478 | 0.314 | 0.424 |
| 5 | 0.502 | 0.333 | 0.403 |
| 5.5 | 0.488333 | 0.353333 | 0.455333 |
| 6 | 0.535333 | 0.366333 | 0.427333 |
| 6.5 | 0.563333 | 0.379333 | 0.454333 |
| 7 | 0.503333 | 0.394333 | 0.425333 |
| 7.5 | 0.536333 | 0.402333 | 0.462333 |
| 8 | 0.555333 | 0.407333 | 0.432333 |
| 8.5 | 0.533333 | 0.415333 | 0.440333 |
| 9 | 0.523333 | 0.423333 | 0.445333 |
| 9.5 | 0.471333 | 0.423333 | 0.441333 |
| 10 | 0.486333 | 0.433333 | 0.449333 |
| 10.5 | 0.465333 | 0.436333 | 0.453333 |
| 11 | 0.486333 | 0.443333 | 0.468333 |
| 11.5 | 0.481333 | 0.445333 | 0.469333 |
| 12 | 0.48 | 0.449 | 0.475 |
- 1 OD600 = Sample OD600 – Average Background OD600.
| Problem | Possible cause | Solution |
|---|---|---|
| No visible growth | Insufficient bacteria transferred from stock vial | Retry streaking process with a larger portion of bacteria-containing ice from the frozen stock |
| Improper agar medium and/or supplements used | Ensure that a rich agar medium is used, such as LB, and all correct supplements, such as antibiotics, were introduced into the agar medium at the correct concentration | |
| Viability of the bacteria in the frozen stock has decreased | Plate the entire frozen stock and make another frozen stock vial | |
| Growth in the negative control | Growth medium is contaminated | Prepare a new batch of growth medium for use |
| Problem | Possible cause | Solution |
|---|---|---|
| Insufficient growth | Inoculum size is too small | Increasing the inoculum size to an OD600 above 0.0001, such as 0.001 or 0.01 |
| Temperature fluctuations within the microplate reader | Ensure that the microplate reader is maintaining a constant temperature of 37°C for the entire duration of data measurement | |
| Growth in the negative control | Growth medium is contaminated | Prepare a new growth curve with new growth medium |
Basic Protocol 5 describes the extraction of genomic DNA from a K. pneumoniae culture. The anticipated result is the production of a genomic DNA stock that can be used for downstream experiments. Basic Protocol 6 describes characterizing K. pneumoniae strains based on genomic sequence. A genome report (Fig. 4) with detailed information about the genus and species of the organism, sequency type, lineage, capsule and LPS type, antimicrobial resistance profile, genome clustering and plasmids is anticipated upon successful completion of Basic Protocol 1. Serotype predictions are provided with ‘confidence’ level. Generally, serotype reports with ‘good’ or higher confidence are desirable.
Basic Protocol 7 describes the storage of K. pneumoniae in glycerol as frozen stocks. The anticipated result is the production of frozen stocks for long-term use and extended maintenance of bacterial integrity. Basic Protocol 8 is a similar method that produces more short-term storage of K. pneumoniae agar stabs with the additional benefits of being stable at room temperature and thus easily shippable.
Time Considerations
Preparing input bacterial samples for a growth curve requires approximately 24 hr of mostly incubation time. Depending on the number of samples, subculturing for growth curves (Basic Protocol 4) can be completed in 5 to 15 min. Measuring and calculating the inoculation size to achieve an optical density of 0.0001 can be completed in 10 to 20 min. Preparing the 96-well microplate with 100 μl of each sample can be completed in 10 to 15 min. Not including culture of the input bacteria and the 16 to 18 hr data collection, the preparation of the bacteria for growth curves should take 25 to 50 min.
When extracting genomic DNA (Basic Protocol 5), allow 16 to 18 hr for the bacteria to grow overnight. Incubating the RNase-added cell lysate at 37°C will take 60 min and resuspending the precipitated DNA at 65°C will take an additional 60 min. Several shorter incubation periods total approximately 25 min, and several centrifugation steps total approximately 9 min. To remove the ethanol from the DNA pellet, it is recommended to air-dry the tube for 15 min. Basic Protocol 5 should take about 2.5 to 3 hr, not including the overnight culture.
In Basic Protocol 6, the genome assembly process within the BV-BRC online applet may take several hours depending on genome size and the availability of servers. The analysis of whole genome sequences within Pathogenwatch may take a few minutes to an hour depending on server availability.
The remaining Basic and Alternate protocols should take no more than few minutes each, not including the time required to grow the bacteria overnight (e.g., 16 to 18 hr). With larger sample sets, additional time should be allotted to handle multiple samples.
Acknowledgments
The authors thank Drs. Caitlyn Holmes, Olaya Rendueles Garcia, Yi Han Tan, and Jay Vornhagen for critical input. Support for the authors and development of these methods was provided by the University of Toledo College of Medicine and Life Sciences, American Heart Association 23CDA1056712 (L.A.M.), and K22 AI145849 (L.A.M.) and R35 GM150588 (L.A.M.) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the American Heart Association or the National Institutes of Health.
Author Contributions
Drew Pariseau : Conceptualization; data curation; methodology; supervision; writing—original draft; writing—review and editing. Brooke Ring : Writing—original draft; writing—review and editing. Saroj Khadka : Writing—original draft; writing—review and editing. Laura Mike : Conceptualization; methodology; project administration; supervision; writing—original draft; writing—review and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Literature Cited
- Argimón, S., David, S., Underwood, A., Abrudan, M., Wheeler, N. E., Kekre, M., Abudahab, K., Yeats, C. A., Goater, R., Taylor, B., Harste, H., Muddyman, D., Feil, E. J., Brisse, S., Holt, K., Donado-Godoy, P., Ravikumar, K. L., Okeke, I. N., Carlos, C., … Stelling, J. (2021). Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clinical Infectious Diseases , 73(Supplement_4), S325–S335. https://doi.org/10.1093/cid/ciab784
- Bakker-Woudenberg, I. A., van den Berg, J. C., Vree, T. B., Baars, A. M., & Michel, M. F. (1985). Relevance of serum protein binding of cefoxitin and cefazolin to their activities against Klebsiella pneumoniae pneumonia in rats. Antimicrobial Agents and Chemotherapy , 28(5), 654–659. https://doi.org/10.1128/AAC.28.5.654
- Blin, C., Passet, V., Touchon, M., Rocha, E. P. C., & Brisse, S. (2017). Metabolic diversity of the emerging pathogenic lineages of Klebsiella pneumoniae. Environmental Microbiology , 19(5), 1881–1898. https://doi.org/10.1111/1462-2920.13689
- Broberg, C. A., Wu, W., Cavalcoli, J. D., Miller, V. L., & Bachman, M. A. (2014). Complete genome sequence of Klebsiella pneumoniae strain ATCC 43816 KPPR1, a rifampin-resistant mutant commonly used in animal, genetic, and molecular biology studies. Genome Announcements , 2(5), e00924–e1014. https://doi.org/10.1128/genomeA.00924-14
- Chelius, M. K., & Triplett, E. W. (2000). Immunolocalization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays L. Applied and Environmental Microbiology , 66(2), 783–787. https://doi.org/10.1128/AEM.66.2.783-787.2000
- Favre-Bonté, S., Forestier, C., & Joly, B. (1998). Inhibitory effect of roxithromycin on adhesion of Klebsiella pneumoniae strains 3051, CF504 and LM21. The Journal of Antimicrobial Chemotherapy , 41 Suppl B, 51–55. https://doi.org/10.1093/jac/41.suppl_2.51
- Fouts, D. E., Tyler, H. L., DeBoy, R. T., Daugherty, S., Ren, Q., Badger, J. H., Durkin, A. S., Huot, H., Shrivastava, S., Kothari, S., Dodson, R. J., Mohamoud, Y., Khouri, H., Roesch, L. F., Krogfelt, K. A., Struve, C., Triplett, E. W., & Methé, B. A. (2008). Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genetics , 4(7), e1000141. https://doi.org/10.1371/journal.pgen.1000141
- Goslings, W. R. O., & Snijders, E. P. (1936). Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. Abt I Orig , 136, 1–24.
- Jenney, A. W., Clements, A., Farn, J. L., Wijburg, O. L., McGlinchey, A., Spelman, D. W., Pitt, T. L., Kaufmann, M. E., Liolios, L., Moloney, M. B., Wesselingh, S. L., & Strugnell, R. A. (2006). Seroepidemiology of Klebsiella pneumoniae in an Australian tertiary hospital and its implications for vaccine development. Journal of Clinical Microbiology , 44(1), 102–107. https://doi.org/10.1128/JCM.44.1.102-107.2006
- Johnson, J. G., Spurbeck, R. R., Sandhu, S. K., & Matson, J. S. (2014). Genome Sequence of Klebsiella pneumoniae urinary tract isolate Top52. Genome Announcements , 2(4), e00668–e00714. https://doi.org/10.1128/genomeA.00668-14
- Khadka, S., Ring, B. E., Pariseau, D. A., & Mike, L. A. (2023a). Characterization of Klebsiella pneumoniae extracellular polysaccharides. Current Protocols , 3(11), e937. https://doi.org/10.1002/cpz1.937
- Khadka, S., Ring, B. E., Walker, R. S., Krzeminski, L. R., Pariseau, D. A., Hathaway, M., Mobley, H. L. T., & Mike, L. A. (2023b). Urine-mediated suppression of Klebsiella pneumoniae mucoidy is counteracted by spontaneous Wzc variants altering capsule chain length. mSphere , 8(5), e0028823. 10.1128/msphere.00288-2
- Lam, M. M. C., Wyres, K. L., Duchêne, S., Wick, R. R., Judd, L. M., Gan, Y. H., Hoh, C. H., Archuleta, S., Molton, J. S., Kalimuddin, S., Koh, T. H., Passet, V., Brisse, S., & Holt, K. E. (2018). Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nature Communications , 9(1), 2703. https://doi.org/10.1038/s41467-018-05114-7
- Lawlor, M. S., Hsu, J., Rick, P. D., & Miller, V. L. (2005). Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Molecular Microbiology , 58(4), 1054–1073. https://doi.org/10.1111/j.1365-2958.2005.04918.x
- Lee, K. W., Arumugam, K., Purbojati, R. W., Tay, Q. X., Williams, R. B., Kjelleberg, S., & Rice, S. A. (2013). Draft genome sequence of Klebsiella pneumoniae strain KP-1. Genome Announcements , 1(6), e01082–e01113. https://doi.org/10.1128/genomeA.01082-13
- Lery, L. M., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., Barbe, V., Bengoechea, J. A., Sansonetti, P., Brisse, S., & Tournebize, R. (2014). Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biology , 12, 41. https://doi.org/10.1186/1741-7007-12-41
- Lin, A. C., Liao, T. L., Lin, Y. C., Lai, Y. C., Lu, M. C., & Chen, Y. T. (2012). Complete genome sequence of Klebsiella pneumoniae 1084, a hypermucoviscosity-negative K1 clinical strain. Journal of Bacteriology , 194(22), 6316. https://doi.org/10.1128/JB.01548-12
- Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., Lynfield, R., Maloney, M., McAllister-Hollod, L., Nadle, J., Ray, S. M., Thompson, D. L., Wilson, L. E., Fridkin, S. K., & Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. (2014). Multistate point-prevalence survey of health care-associated infections. The New England Journal of Medicine , 370(13), 1198–1208. https://doi.org/10.1056/NEJMoa1306801
- McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., Hou, S., Layman, D., Leonard, S., Nguyen, C., Scott, K., Holmes, A., Grewal, N., Mulvaney, E., Ryan, E., … Wilson, R. K. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature , 413(6858), 852–856. https://doi.org/10.1038/35101614
- Mike, L. A., Stark, A. J., Forsyth, V. S., Vornhagen, J., Smith, S. N., Bachman, M. A., & Mobley, H. L. T. (2021). A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathogens , 17(3), e1009376. https://doi.org/10.1371/journal.ppat.1009376
- Miller, J. H. (1972). Experiments in molecular genetics. Cold Spring Harbor Laboratory.
- Ogawa, W., Li, D. W., Yu, P., Begum, A., Mizushima, T., Kuroda, T., & Tsuchiya, T. (2005). Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biological & Pharmaceutical Bulletin, 28(8), 1505–1508. https://doi.org/10.1248/bpb.28.1505
- Olson, R. D., Assaf, R., Brettin, T., Conrad, N., Cucinell, C., Davis, J. J., Dempsey, D. M., Dickerman, A., Dietrich, E. M., Kenyon, R. W., Kuscuoglu, M., Lefkowitz, E. J., Lu, J., Machi, D., Macken, C., Mao, C., Niewiadomska, A., Nguyen, M., Olsen, G. J., … Stevens, R. L. (2023). Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Research , 51(D1), D678–D689. https://doi.org/10.1093/nar/gkac1003
- Paczosa, M. K., Silver, R. J., McCabe, A. L., Tai, A. K., McLeish, C. H., Lazinski, D. W., & Mecsas, J. (2020). Transposon mutagenesis screen of Klebsiella pneumoniae identifies multiple genes important for resisting antimicrobial activities of neutrophils in mice. Infection and Immunity , 88(4), e00034–e00120. https://doi.org/10.1128/IAI.00034-20
- Peng, H. L., Wang, P. Y., Wu, J. L., Chiu, C. T., & Chang, H. Y. (1991). Molecular epidemiology of Klebsiella pneumoniae. Chinese Journal of Microbiology and Immunology , 24(3), 264–271.
- Pomakova, D. K., Hsiao, C. B., Beanan, J. M., Olson, R., MacDonald, U., Keynan, Y., & Russo, T. A. (2012). Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia : An emerging and under-recognized pathogenic variant. European Journal of Clinical Microbiology & Infectious Diseases, 31(6), 981–989. https://doi.org/10.1007/s10096-011-1396-6
- Ramage, B., Erolin, R., Held, K., Gasper, J., Weiss, E., Brittnacher, M., Gallagher, L., & Manoil, C. (2017). Comprehensive arrayed transposon mutant library of Klebsiella pneumoniae outbreak strain KPNIH1. Journal of Bacteriology , 199(20), e00352–e00417. https://doi.org/10.1128/JB.00352-17
- Ring, B. E., Khadka, S., Pariseau, D. A., & Mike, L. A. (2023). Genetic manipulation of Klebsiella pneumoniae. Current Protocols , 3(10), e912. https://doi.org/10.1002/cpz1.912
- Ristuccia, P., & Cunha, B. (1984). Klebsiella. Infection Control & Hospital Epidemiology, 5(7), 343–347. https://doi.org/10.1017/S0195941700060549
- Roosendaal, R., Bakker-Woudenberg, I. A., van den Berg, J. C., & Michel, M. F. (1985). Therapeutic efficacy of continuous versus intermittent administration of ceftazidime in an experimental Klebsiella pneumoniae pneumonia in rats. The Journal of Infectious Diseases , 152(2), 373–378. https://doi.org/10.1093/infdis/152.2.373
- Rosen, D. A., Pinkner, J. S., Jones, J. M., Walker, J. N., Clegg, S., & Hultgren, S. J. (2008). Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infection and Immunity , 76(7), 3337–3345. https://doi.org/10.1128/IAI.00090-08
- Russo, T. A., & Gill, S. R. (2013). Draft genome sequence of the hypervirulent Klebsiella pneumoniae strain hvKP1, isolated in Buffalo, New York. Genome Announcements , 1(2), e0006513. https://doi.org/10.1128/genomeA.00065-13
- Russo, T. A., & Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clinical Microbiology Reviews , 32(3), e00001–19. https://doi.org/10.1128/CMR.00001-19
- Sezonov, G., Joseleau-Petit, D., & D'Ari, R. (2007). Escherichia coli physiology in Luria-Bertani broth. Journal of Bacteriology , 189(23), 8746–8749. https://doi.org/10.1128/JB.01368-07
- Snitkin, E. S., Zelazny, A. M., Thomas, P. J., Stock, F., NISC Comparative Sequencing Program Group, Henderson, D. K., Palmore, T. N., & Segre, J. A. (2012). Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Science Translational Medicine , 4(148), 148ra116. https://doi.org/10.1126/scitranslmed.3004129
- Walker, K. A., Miner, T. A., Palacios, M., Trzilova, D., Frederick, D. R., Broberg, C. A., Sepúlveda, V. E., Quinn, J. D., & Miller, V. L. (2019). A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio , 10(2), e00089–e00119. https://doi.org/10.1128/mBio.00089-19
- Wu, K. M., Li, L. H., Yan, J. J., Tsao, N., Liao, T. L., Tsai, H. C., Fung, C. P., Chen, H. J., Liu, Y. M., Wang, J. T., Fang, C. T., Chang, S. C., Shu, H. Y., Liu, T. T., Chen, Y. T., Shiau, Y. R., Lauderdale, T. L., Su, I. J., Kirby, R., & Tsai, S. F. (2009). Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. Journal of Bacteriology , 191(14), 4492–4501. https://doi.org/10.1128/JB.00315-09
- Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., & Holt, K. E. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microbial Genomics , 2(12), e000102. https://doi.org/10.1099/mgen.0.000102

