Critical Conditions for Studying Interleukin-11 Signaling In Vitro and Avoiding Experimental Artefacts
Sivakumar Viswanathan, Sivakumar Viswanathan, Benjamin Ng, Benjamin Ng, Anissa A. Widjaja, Anissa A. Widjaja, Chee Jian Pua, Chee Jian Pua, Nevin Tham, Nevin Tham, Jessie Tan, Jessie Tan, Stuart A. Cook, Stuart A. Cook, Sebastian Schafer, Sebastian Schafer
Abstract
Interleukin (IL) 11 is a member of the IL6 family of cytokines which require the ubiquitous gp130 receptor to activate canonical (JAK/STAT) and non-canonical (e.g., ERK) signaling pathways. The IL11 cytokine is upregulated in a number of fibro-inflammatory diseases and cancer, where it binds the cognate IL11 receptor alpha subunit (IL11RA) to form a hexameric IL11:IL11RA:gp130 signaling complex. The specific IL11RA receptor is highly expressed on cells of the stromal and parenchymal niche but expressed at low levels on immune cells, highly passaged cells, or transformed cell lines. Consequently, primary cells such as hepatic stellate cells, fibroblasts, and hepatocytes are ideal experimental systems to study IL11 signaling in vitro. In contrast to immortalized cell lines, primary cells better display relevant cellular physiology and pathobiology. This collection of protocols details experimental and culturing conditions for primary cells that preserve meaningful cellular states and physiological responses ex vivo in conventional 2D cell culture systems. Readouts of cellular activity are chosen carefully to capture the non-canonical, post-transcriptional activity of IL11 signaling. Our data suggest that cell type, cell culture conditions, passage number, concentrations of stimuli, timing, and other factors have major implications for studies of IL11 signaling. In vitro experiments with primary cell material need to be planned and executed with great caution. Otherwise, physiologically relevant mechanisms may become dysfunctional and reproducible experimental artefacts can obscure our view of true cytokine biology. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Expansion of primary human hepatic stellate cells (HSCs) and human renal proximal tubular epithelial cells (HRPTEpiCs)
Basic Protocol 2 : Expansion of primary human lung fibroblasts (HLFs)
Alternate Protocol 1 : Isolation and expansion of primary mouse lung fibroblasts
Support Protocol 1 : Freezing and thawing of primary cells
Support Protocol 2 : Operetta high-content imaging-based phenotyping
Support Protocol 3 : Colorimetric assay of solubilized collagen
Support Protocol 4 : Quantification of fibrosis marker secretion
Support Protocol 5 : Western blotting studies of IL11 signaling in HSCs, HLFs, and HRPTEpiCs
Basic Protocol 3 : IL11 stimulation of primary human hepatocytes
Alternate Protocol 2 : IL11 stimulation of primary mouse hepatocytes
Support Protocol 6 : Alanine transaminase (ALT) secretion by human and mouse hepatocytes
INTRODUCTION
The interleukin (IL) 6 family of cytokines consists of IL6, IL11, oncostatin M, leukemia inhibitory factor (LIF), and others which all share the ubiquitously expressed gp130 receptor to initiate signaling. Specificity within the family is introduced by individual receptor subunits that are present in distinct subsets of cells. The IL11-specific receptor alpha subunit (IL11RA) is predominantly expressed on stromal and parenchymal cells. In response to IL11, cardiac fibroblasts, hepatic stellate cells (HSCs), and human lung fibroblasts (HLFs) transform into myofibroblasts (Ng et al., 2019; Schafer et al., 2017; Widjaja et al., 2019b). Interestingly, this transition can occur within 24 hr without any apparent changes in the transcriptome. This suggests that the IL11-dependent fibrotic response is not driven by the activation of the canonical JAK/STAT signaling cascade which primarily regulates gene transcription. In contrast to its effect on stromal cells, IL11 induces cell death of hepatocytes, a parenchymal cell type that expresses IL11RA.
In vitro gain-of-function experiments show that both transforming growth factor-beta 1 (TGFβ1) and IL11 effectively activate HSCs and HLFs at low passages. However, we found cells that have undergone multiple rounds of cell doubling are not activated to the same extent. The fibrogenic effect of both cytokines is either completely lost or severely reduced in later passage primary cells (Fig. 1). This is a major challenge for the field because growing cells in flat dishes on coated and uncoated surfaces is the most common format of in vitro cell expansion and experiments. Beyond passaging, other factors such as excessive centrifugation, harsh pipetting, unsuitable seeding density, insufficient medium exchange, freeze-thaw cycles, or the source and/or quality of the primary cells can also have a detrimental impact on in vitro studies of IL11 signaling. The phenotypic integrity of primary cells must be preserved in order to obtain meaningful results ex vivo. Cytokine levels used in culture are also critical, as high concentrations can lead to nonspecific interactions with receptor subunits and trigger unphysiological signaling events, which may be reproducible but are likely unrelated to true biology. If the cytokine and cell type are not species-matched, we observe differences in signaling pathway activation and effects that arise from binding competition between species-foreign IL11 and endogenous species-matched IL11.
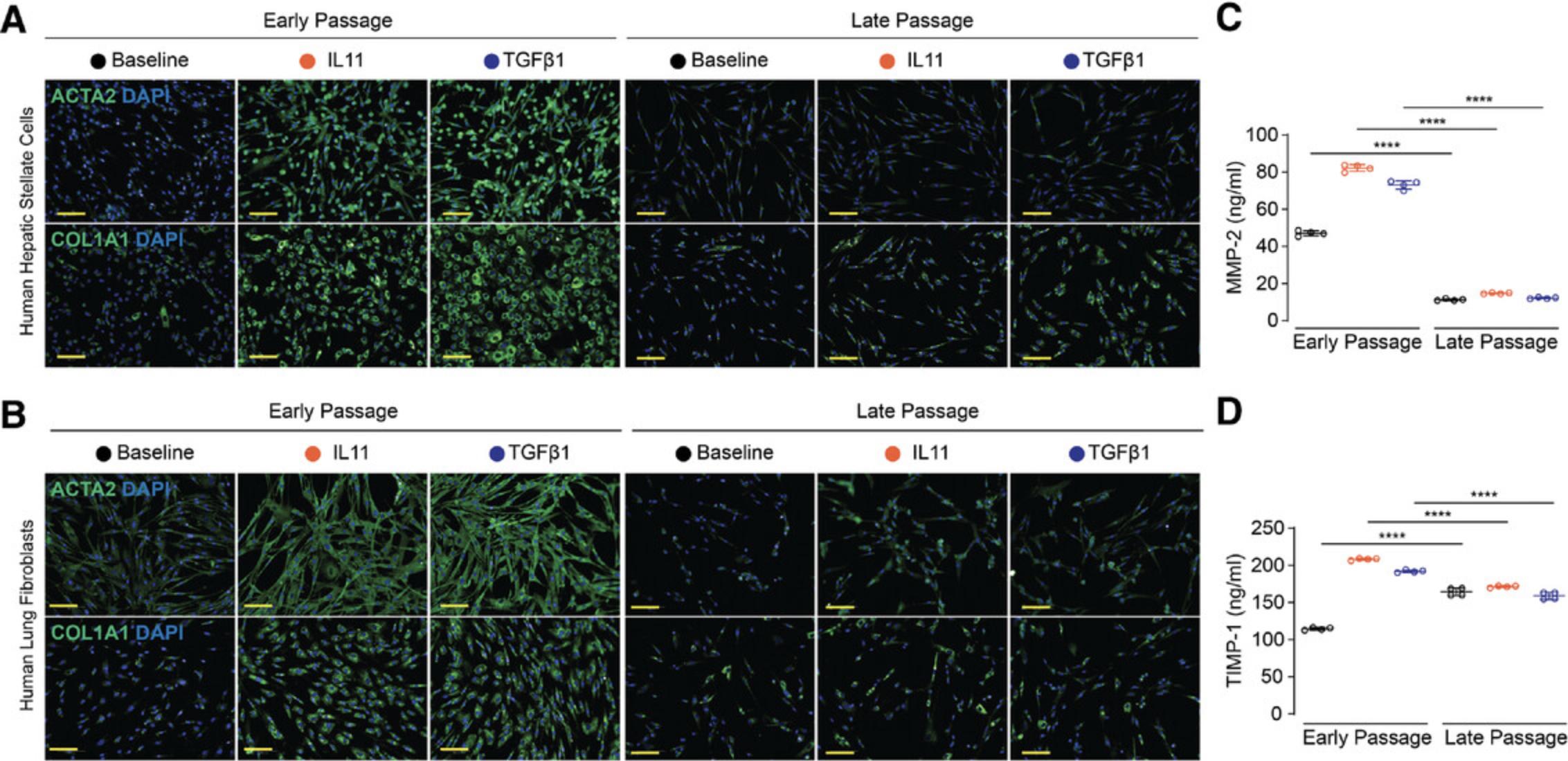
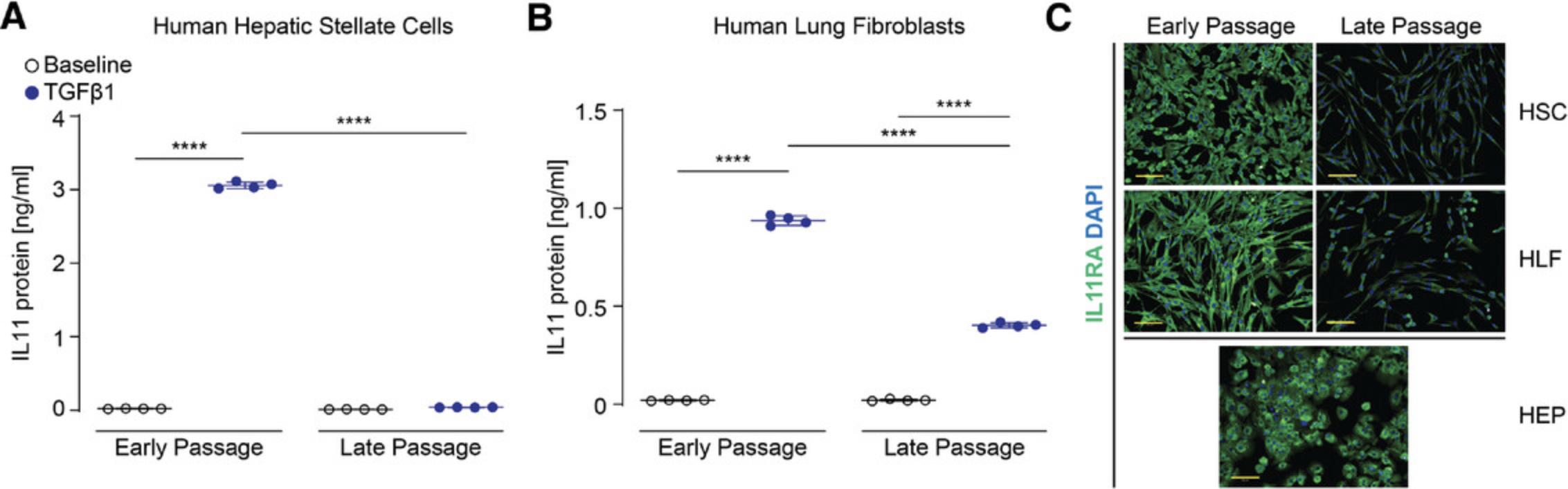
To better understand how suboptimal in vitro conditions can affect the IL11 pathway, we compared responsive HSCs and HLFs (early passage) to non-responsive cells (late passage). First, we measured IL11 secretion after stimulation with TGFβ1, a powerful inducer of IL11 secretion from stromal cells. In contrast to early passage HSCs, late passage cells did not upregulate IL11. Similarly, IL11 upregulation was greatly reduced in late passage HLFs. Immunohistochemical staining for the IL11 receptor confirmed that IL11RA is expressed on HSCs, HLFs, and hepatocytes (Fig. 2). However, when stromal cells were expanded beyond three passages, HSCs and HLFs had greatly reduced expression of IL11RA on their surface. Western blotting confirmed that primary cells at later passages express significantly less IL11RA protein (Fig. 3). Interestingly, IL11RA RNA transcripts were not downregulated in late passaged HLFs and only slightly downregulated in HSCs. This suggests that post-transcriptional mechanisms affecting protein turnover could be responsible for the loss of IL11RA. The IL11-specific receptor could also be shed from the surface of the cells, which can be triggered by proteases such as ADAM10 (Lokau et al., 2016). We have previously shown that the TGFβ1 effect is dependent on IL11 (Ng et al., 2019; Schafer et al., 2017; Widjaja et al., 2019b). The data on the IL11 cytokine and its specific receptor subunit shown here suggest that this dependency is gradually lost when primary cells are maintained ex vivo.
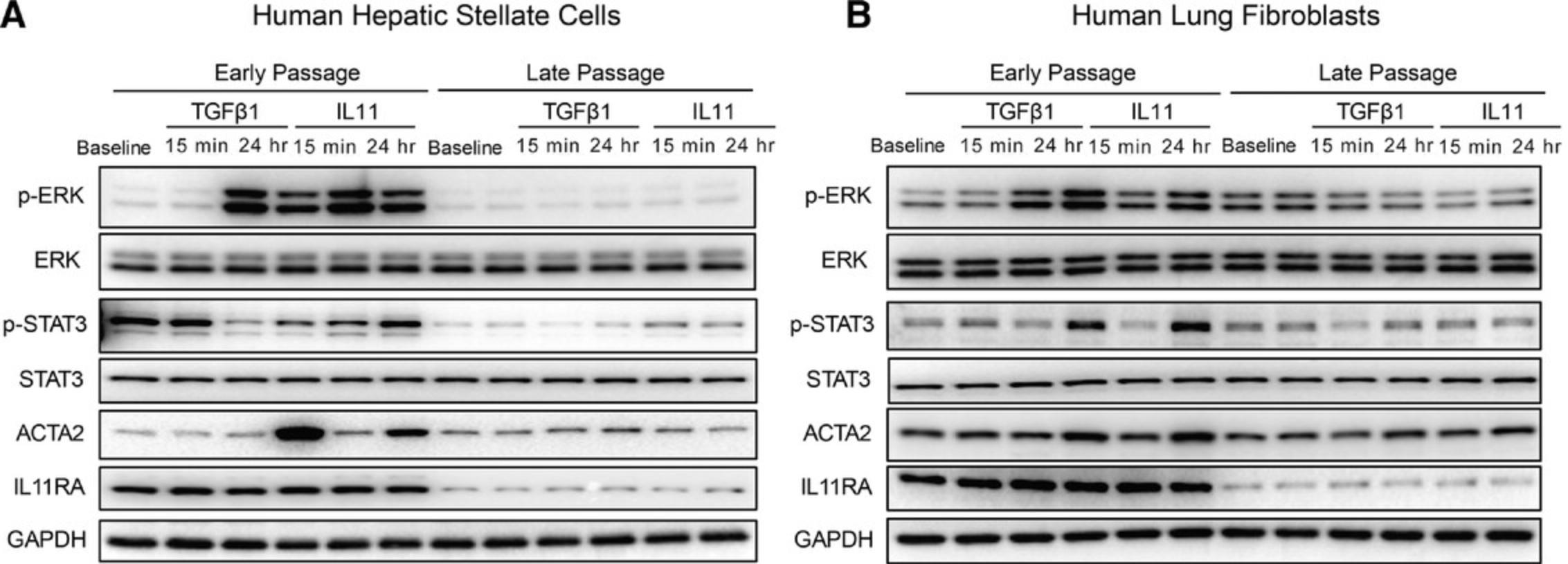
Non-canonical signaling is of central importance for the IL11-driven fibrotic response. As expected, we observed early (15 min) and sustained (24 hr) phosphorylation of ERK in both HSCs and HLFs at early passages, which coincided with ACTA2 upregulation at 24 hr. The ERK pathway was not activated in late passage cells (Fig. 3). The canonical STAT pathway was strongly activated in early passage HLFs at 24 hr after stimulation. This effect might be driven by a secondary factor that is upregulated by TGFβ1 or IL11. We also observed weak activation of STAT in late passage HSCs after IL11 stimulation. However, this did not result in the activation of HSCs (Fig. 1). These results highlight that signaling pathway activation is highly dependent on the time points and the cell type chosen. Non-canonical pathway activation is only observed in cells that are also effectively activated and transformed by IL11 stimulation.
The cellular phenotype of primary cells is likely to change on multiple levels during extended culturing ex vivo. To better understand this cellular transition at a genome-wide level, we performed RNA-sequencing (RNA-seq), as previously described (Widjaja et al., 2019b), with both unstimulated HSCs and HLFs at early and late passage. We found hundreds of differentially expressed genes in late passage HSCs and HLFs (Fig. 4A and 4C), which implies that the primary cells undergo extensive changes on the molecular level when cultured in vitro. Gene set enrichment analysis of both datasets revealed that target genes of several signaling pathways are affected: In HSCs, target genes of MYC, MTORC1, TNFα, TGFβ, and JAK/STAT were found to be upregulated in late passage cells (Fig. 4B). In late passage HLFs, target genes of the IFNα, MTORC1, JAK/STAT, and P53 pathways, among others, were upregulated (Fig. 4D). It is possible that external triggers, such as mechanical stress, initiate multiple autocrine signaling loops. This eventually renders the cellular material unusable for experiments that are supposed to model true cell biology ex vivo. Cells undergoing extensive culturing may not only lose the TGFβ or IL11 effect, as shown above. The RNA-seq analysis suggests that signaling cascades, in particular, might be very sensitive to changes in the cellular phenotype. This likely has detrimental effects on loss- and gain-of-function experiments with cytokines in general.
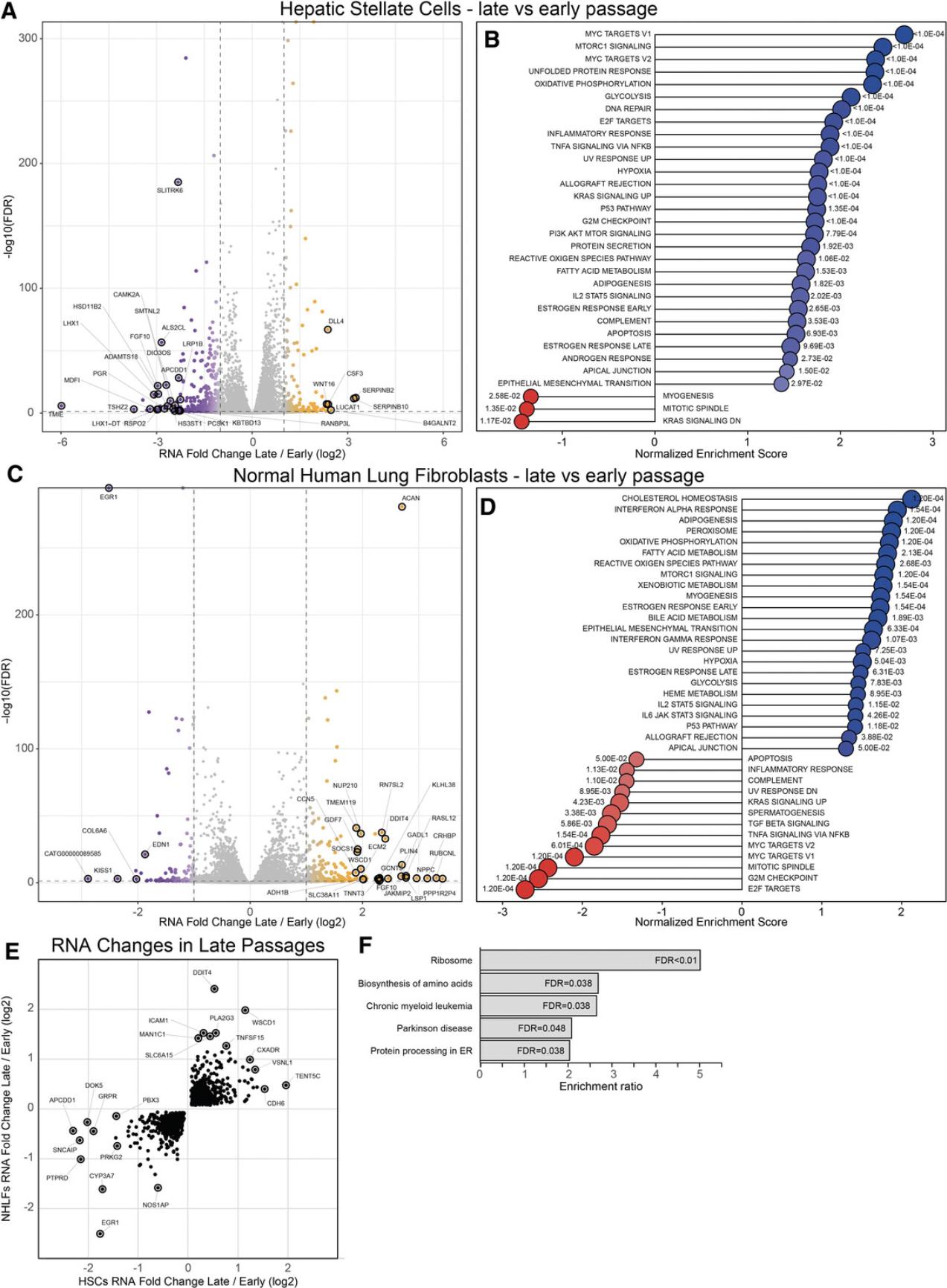
To identify commonalities of the HSC and HLF phenotype switch in vitro , we next looked only at genes that significantly change in the same direction during prolonged culturing of both cell types. Upregulated genes include targets of TNF and IL-1 signaling such as TNFSF15 (Migone et al., 2002) or PLA2G3, associated with oxidative stress (Martínez-García et al., 2010). Expression of cancer suppressor genes such as the transcription factor EGR1 (Baron, Adamson, Calogero, Ragona, & Mercola, 2006) is lost. Interestingly, KEGG pathway analysis of these genes revealed that components of the ribosomal complex and genes associated with the biosynthesis of amino acids and protein processing in the endoplasmic reticulum are differentially expressed in late passage cells. This suggests that protein translation and processing is altered in these cells. These intracellular mechanisms may be an additional reason, beyond the loss of IL11RA, why extensively passaged cells lose the IL11 mechanism.
The protocols in this collection detail IL11 signaling studies in 2D cell cultures, the most common form of ex vivo experimental systems. A major focus is on maintaining cellular phenotypes of relevant primary cells in culture to keep the IL11 mechanism intact. To test whether cells are in a suitable state for experiments, we recommend measuring the IL11RA protein, not RNA, levels. IL11RA expression can be lost in culture with detrimental effects on the IL11 pathway. Other marker genes such as those shown in (Fig. 4E) might be suitable to assess the integrity of primary cells. Readouts of cellular activity must capture post-transcriptional regulation and include western blotting, immunofluorescence staining, and ELISA-based assays.
Basic Protocols 1 and 2 detail the expansion of commercially available human HSCs, HLFs, and human renal proximal tubular epithelial cells (HRPTEpiCs) in preparation for studies of IL11 activity. Alternate Protocol 1 describes the isolation and culture of primary mouse lung fibroblasts. Support Protocol 1 includes instructions on freezing, thawing, and recovery of primary cells. Extracellular matrix production and α-SMA (ACTA2) expression can be monitored via immunocytochemistry as shown in Support Protocol 2. The secretion of fibrosis markers can be measured by colorimetric methods such as Sirius Red staining (Support Protocol 3), ELISA (Support Protocol 4), or western blotting (Support Protocol 5). Basic Protocol 3 and Alternate Protocol 2 outline the primary culture of human and mouse hepatocytes, respectively, for the study of IL11 activity on this parenchymal cell type. Support Protocol 6 explains how alanine transaminase (ALT) levels in the culture supernatants of IL11-treated human and mouse hepatocytes can be measured.
STRATEGIC PLANNING
When planning in vitro experiments with primary cells, it is important to develop a workflow that preserves cellular integrity while being time and cost effective. For cells that can be expanded before the experiment, we recommend using cells without a freeze/thaw cycle for the best results. It perhaps goes without saying but becoming an expert in culturing a cell type can take many months and truly reproducible and excellent culture techniques take years to master. Overall, an experiment can last up to 2 weeks from the start of the initial HSCs, HLFs, and HRPTEpiCs culture to the signaling and phenotyping assays (Fig. 5).
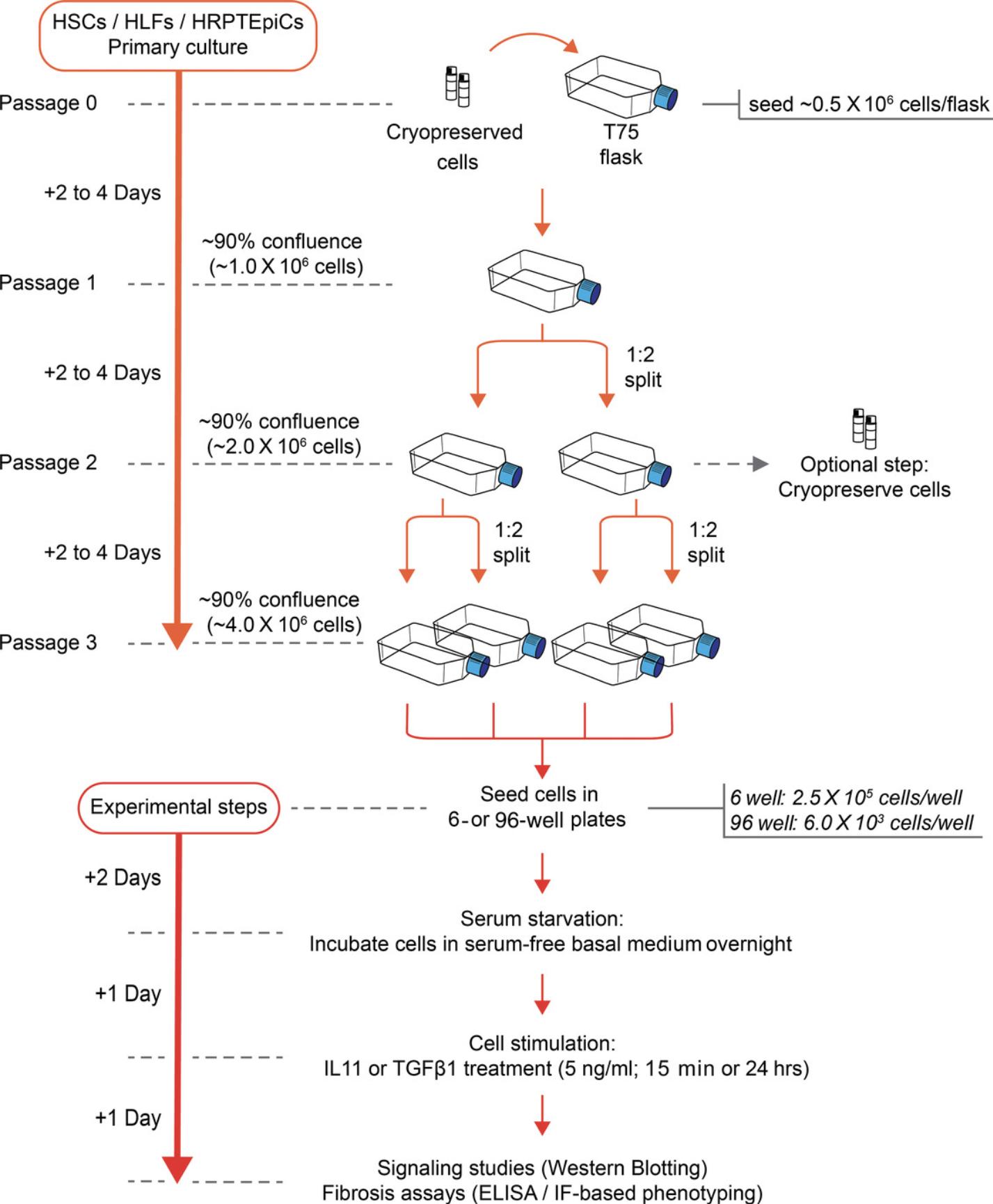
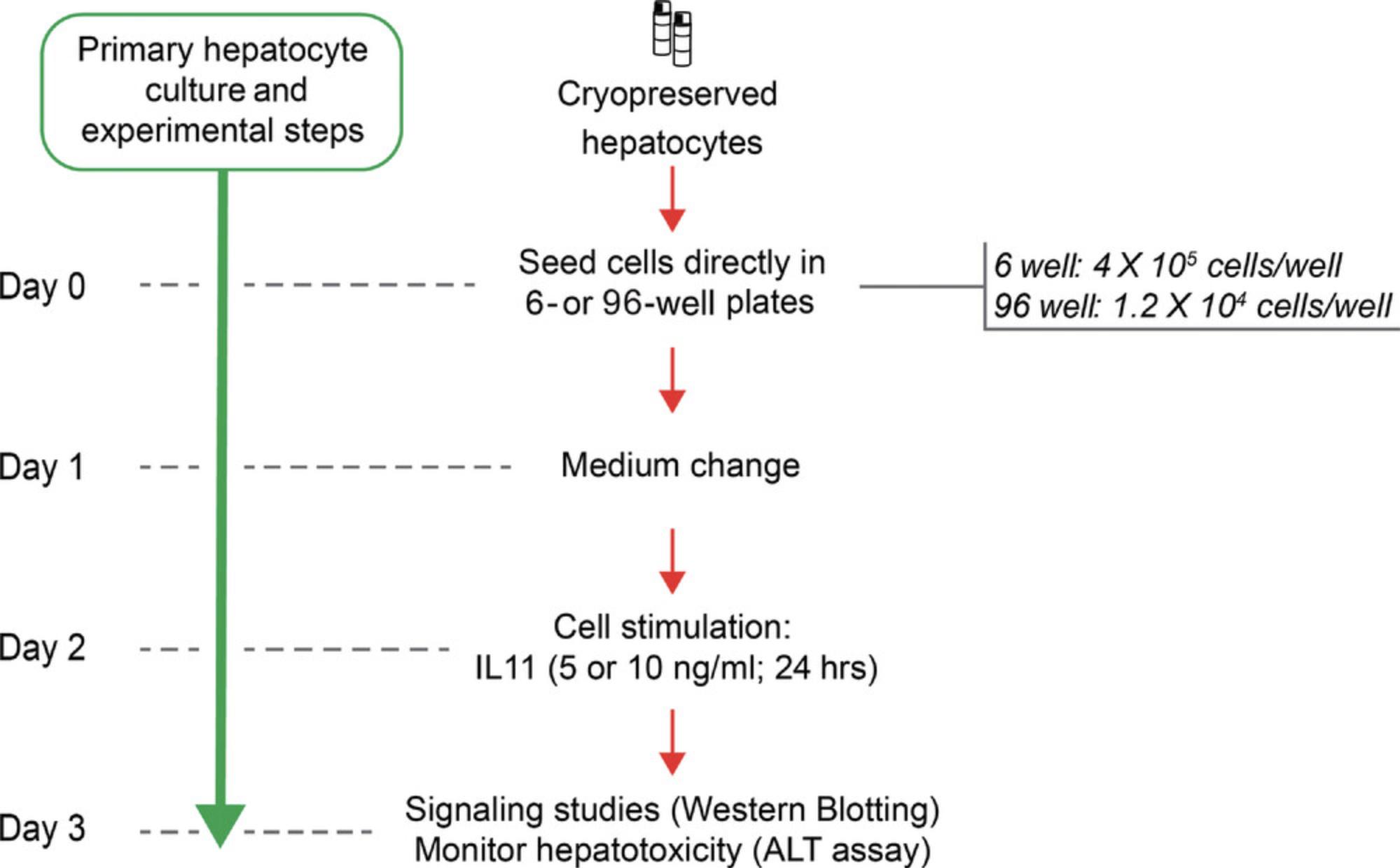
Primary hepatocytes should be used for downstream experiments within 2 days after they have been seeded. This severely limits the cellular starting material available for downstream analyses, which should be taken into account when planning the experiments (Fig. 6).
NOTE : All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals.
Basic Protocol 1: EXPANSION OF PRIMARY HUMAN HEPATIC STELLATE CELLS (HSCs) AND HUMAN RENAL PROXIMAL TUBULAR EPITHELIAL CELLS (HRPTEpiCs)
HSCs are the precursor cells of fibrogenic myofibroblasts in the liver. Isolated cells are grown in a monolayer and should only be subjected to limited cell expansion. After passage two, cells can be frozen and stored for future experiments. Controlled population doublings are achieved by plating HSCs at the cell densities specified in this protocol, which also ensures that the cells are maintained at an exponential growth phase throughout the primary culture. Depending on the donor source and handling by commercial vendors, the passage number where cells are still suitable for IL11 experiments can vary. We recommend measuring IL11RA protein expression to monitor primary cell integrity (see Introduction).
Epithelial cells form a thin protective layer around the outer layers of body organs and vessels and express IL11RA. Similar to fibroblasts, they are frequently used in an in vitro model system to study tissue repair and remodeling. In disease, epithelial cells can be converted into stromal-like cells through a process called epithelial-mesenchymal transition (EMT). This disrupts secretion, sensing, selective absorption of the epithelial layer, and eventually impairs organ function. Here we provide details for the culturing of HRPTEpiCs from ScienCell Research Laboratories. These cells are isolated from the human kidney, cryopreserved at passage one and supplied frozen. This protocol is optimized to reduce the loss of IL11RA expression in vitro and we recommend measuring IL11RA protein expression to monitor primary cell integrity (see Introduction). The EMT can be monitored by assessing ACTA2 and COL1A1 expression (see Support Protocol 2) as well as intracellular signaling (see Support Protocol 5).
Materials
-
Human Hepatic Stellate Cells (HSCs; ScienCell Research Laboratories, cat. no. 5300)
-
SteCM complete medium (see recipe)
-
Human Renal Proximal Tubular Epithelial Cells (HRPTEpiCs; ScienCell, cat. no. 4100)
-
EpiCM complete medium (see recipe)
-
TrypLE Express with Phenol Red (Gibco brand, Thermo Fisher Scientific, cat. no. 12605010)
-
Dulbecco's Phosphate Buffered Saline (DPBS)/Modified (Hyclone, cat. no. SH30028.02)
-
Trypan Blue Stain (0.4%; Thermo Fisher Scientific, cat. no. T10282)
-
Poly-L-lysine (MilliporeSigma, cat. no. P4707)
-
Water, deionized, distilled (Hyclone, cat. no. SH30538.02)
-
Ethanol, 70%
-
T-75 culture flask (BD Falcon, cat.no. 353136)
-
15-ml Falcon tubes (BD Falcon, cat. no. 352096)
-
50-ml Falcon tubes (BD Falcon, cat. no. 352070)
-
1.7-ml microtubes (Axygen, cat. no. MCT-175-C)
-
Disposable Serological Pipet, 5 ml (Costar, cat. no. 4487)
-
Disposable Serological Pipet, 10 ml (Costar, cat. no. 4488)
-
S1 Pipet Fillers (Thermo Fisher Scientific, cat. no. 9521)
-
CountessTM Cell Counting Chamber Slides (Thermo Fisher Scientific, cat. no. C10314)
-
Countess® II Automated Cell Counter (Thermo Fisher Scientific, cat. no. AMQAX1000)
-
Tissue Culture Centrifuge (Thermo Sorvall Legend XIR)
-
Water bath, SUB Aqua 12 Plus (Grant Scientific)
-
Nikon Eclipse TS100 Inverted Microscope
-
CO2 incubator
-
Biosafety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
NOTE : An alternative to traditional trypsin/EDTA is TrypLE Express with phenol red from Thermo Fisher Scientific. It is stable at room temperature, does not require inactivation by an inhibitor, and is gentle on cells which results in high viability rates.
Initiating the culture
1.Prepare SteCM and EpiCM complete media (see Reagents and Solutions).
2.Prepare poly-L-lysine-coated T-75 flask (see Reagents and Solutions).
3.Place the vial of frozen human HSCs or HRPTEpiCs in a 37°C water bath. Hold vial with forceps and rotate vial gently until the contents completely thaw.
4.In a biosafety cabinet, carefully remove the cap of the human HSCs or HRPTEpiCs vial without touching the interior threads. Gently resuspend the cells with a 1-ml pipet and dispense the contents of the entire vial into a poly-L-lysine-coated T-75 culture flask containing 10 ml pre-warmed SteCM complete medium.
5.Incubate cells in a 37°C, CO2 incubator.
Maintaining the culture
6.After at least 16 hr of incubation, perform a medium change by removing and discarding spent medium and replacing it with 10 ml pre-warmed SteCM or EpiCM complete medium.
7.Change medium every 2 days until the cells reach ∼90% confluence.
Subculturing
8.Once the cells have reached ∼90% confluence, remove and discard spent cell culture medium from the T-75 culture flask.
9.Wash cells twice with 5 ml DPBS and discard the last wash.
10.Add 4 ml of dissociation reagent (TrypLE™ Express) to the cells and gently rock container to get complete coverage of the cell layer. Incubate cells at 37°C for 3-5 min.
11.After 3-5 min of incubation, observe flask under the microscope to monitor for cell detachment.
12.When ≥90% of the cells have detached, tilt the flask at an ∼45° angle to allow the detached cells to collect at the bottom corner of the flask.
13.Add 8 ml (or twice the volume of dissociation reagent used) of pre-warmed SteCM or EpiCM complete medium and disperse cells by pipetting the medium over the cell layer surface several times.
14.Transfer cell suspension to a 50-ml conical tube and centrifuge at 500 × g for 5 min at room temperature (25°C).
15.Discard supernatant and gently resuspend cell pellet in 1 ml pre-warmed SteCM or EpiCM complete medium.
16.Remove a 10-μl aliquot of cells and mix with 10 μl trypan blue and perform a cell count.
17.The cells are then split at a 1:2 ratio by first diluting the cell suspension with SteCM or EpiCM complete medium and then seeding cells into two new poly-L-lysine coated T-75 flasks.
18.Incubate cells in a 37°C, CO2 incubator.
19.On the next day, change medium with 10 ml pre-warmed SteCM or EpiCM complete medium to remove unattached/dead cells. Subsequently, perform a fresh medium change every other day, until culture reaches ∼90% confluent.
20.To acquire more cells for experiments, cells are subcultured an additional time by repeating steps 8-19.Cells obtained from the step above are split in a 1:2 ratio by seeding cells from two T-75 flasks into four new T-75 flasks.
21.The cells are cultured until they are ∼90% confluent, before being seeded in 6-well or 96-well plates for further downstream experiments as described in Support Protocols 2-5.
Basic Protocol 2: EXPANSION OF PRIMARY HUMAN LUNG FIBROBLASTS (HLFs)
Pulmonary fibroblasts are a frequently used in vitro model system to study lung tissue repair and remodeling. HLFs can be isolated from healthy human lung tissue. These cells are adherent and adopt a spindle-like morphology when grown on plastic tissue culture dishes. Lung fibroblasts must be grown in monolayers before being stimulated with profibrotic cytokines. After passage two, cells can also be frozen and stored for future experiments. Cells should not be plated at lower densities as stated below, which limit total cell numbers available for downstream experiments. We recommend measuring IL11RA protein expression to monitor primary cell integrity (see Introduction).
Materials
-
HLF FGMTM-2 Cryo Amp (LONZA, cat. no. CC-2512)
-
FGM-2 fibroblast complete medium (see recipe)
-
Trypan Blue Stain (0.4%; Thermo Fisher Scientific, cat. no. T10282)
-
TrypLE Express with Phenol Red (Gibco brand, Thermo Fisher Scientific, cat. no. 12605010)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
Ethanol, 70%
-
T-75 culture flask (BD Falcon, cat. no. 353136)
-
15-ml Falcon tubes (BD Falcon, cat. no. 352096)
-
50-ml Falcon tubes (BD Falcon, cat. no. 352070)
-
1.7-ml microtubes (Axygen, cat. no. MCT-175-C)
-
Disposable Serological Pipet, 5 ml (Costar, cat. no. 4487)
-
Disposable Serological Pipet, 10 ml (Costar, cat. no. 4488)
-
S1 Pipet Fillers (Thermo Fisher Scientific, cat. no. 9521)
-
CountessTM Cell Counting Chamber Slides (Thermo Fisher Scientific, cat. no. C10314)
-
Countess® II Automated Cell Counter (Thermo Fisher Scientific, cat. no. AMQAX1000)
-
Tissue Culture Centrifuge (Thermo Sorvall Legend XIR)
-
Water bath, SUB Aqua 12 Plus (Grant Scientific)
-
Nikon Eclipse TS100 Inverted Microscope
-
CO2 incubator
-
Biosafety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
NOTE : An alternative to traditional trypsin/EDTA is TrypLE Express with phenol red from Thermo Fisher Scientific. It is stable at room temperature, does not require inactivation by an inhibitor, and is gentle on cells which results in high viability rates.
Initiating the culture
1.Prepare FGM-2 fibroblast complete medium for growing and maintenance of HLFs (see recipe).
2.Pre-warm FGM-2 fibroblast complete medium in a 37°C water bath prior to use.
3.Add 10 ml pre-warmed FGM-2 fibroblast complete medium to a T-75 flask.
4.Thaw the vial of frozen HLFs in a 37°C water bath. Hold vial with forceps and rotate vial gently until the contents are completely thawed.
5.Wipe the outside of the vial with 70% ethanol.
6.In a biosafety cabinet, carefully remove the cap without touching the interior threads. Gently resuspend cell suspension and dispense contents of the entire vial into a T-75 flask containing pre-warmed FGM-2 fibroblast complete medium.
7.Incubate cells in a 37°C, CO2 incubator overnight.
Maintaining the culture
8.The next day, remove medium from the T-75 flask and replace with 10 ml pre-warmed FGM-2 complete medium to remove residual DMSO and unattached or dead cells.
9.Change medium every 2 days, until cells reach ∼90% confluent (2-4 days).
Subculturing
10.Once cells have reached ∼90% confluence, the cells are ready to be subcultured at a 1:2 split ratio.
11.Remove and discard spent cell culture medium from the culture flask.
12.Wash cells twice with 5 ml DPBS and discard the last wash.
13.Add 4 ml dissociation reagent (TrypLE™ Express) to the side of the flask.
14.Incubate cells for 3-5 min in a 37°C, CO2 incubator.
15.When ≥90% of the cells have detached, tilt the flask at a ∼45° angle to allow the detached cells to collect at the bottom corners of the flask.
16.Add 8 ml (or twice the volume of dissociation reagent used) of pre-warmed FGM-2 fibroblast complete medium and disperse cells by pipetting medium over the cell layer surface several times.
17.Transfer cells to a 50-ml conical tube and centrifuge at 500 × g for 5 min at room temperature (25°C).
18.Discard supernatant and resuspend cell pellet in 1 ml pre-warmed FGM-2 fibroblast complete medium.
19.Remove a10-μl aliquot of cells and mix with 10 μl trypan blue and perform a cell count.
20.Dilute cell suspension with FGM-2 fibroblast complete medium and seed cells into two new T-75 flasks (1:2 split ratio).
21.Incubate cells in a 37°C, CO2 incubator.
22.On the next day, remove medium from the cells and then add 10 ml pre-warmed FGM-2 fibroblast complete medium to remove unattached or dead cells. Subsequently, perform a fresh medium change every other day, until the culture is ∼90% confluent.
23.To obtain more cells for downstream experiments, repeat steps 10-22 to further expand cells into new T-75 flasks (1:2 split ratio).
24.The cells are cultured until they are ∼90% confluent, at which point they can be seeded in 6-well or 96-well plates for further downstream experiments as described in Support Protocols 2-5.
Alternate Protocol 1: ISOLATION AND EXPANSION OF PRIMARY MOUSE LUNG FIBROBLASTS
In this alternate protocol, we describe a method of fibroblast isolation from wild type C57BL/6J mice by explanting digested lung tissue, followed by cell purification and culture maintenance of mouse primary lung fibroblasts. It is important to highlight that due to the highly proliferative nature of lung fibroblasts in culture, these primary cells enter replicative senescence beyond Passage 5 and hence should be used for experiments only at early passages. We have shown previously that mouse lung fibroblasts isolated using this protocol retain many of their in vivo effector functions including myofibroblast differentiation, cell migration and invasion, and extracellular matrix secretion when stimulated with various profibrotic stimuli (Ng et al., 2019). We recommend measuring IL11RA protein expression to monitor primary cell integrity (see Introduction).
NOTE : All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals.
Materials
-
C57BL/6J mice, 8-10 weeks of age (The Jackson Laboratory)
-
DMEM complete medium (see recipe)
-
Digest medium (see recipe)
-
Ice-cold cell sorting buffer (see recipe)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
TrypLE Express with Phenol Red (Gibco brand, Thermo Fisher Scientific, cat. no. 12605010)
-
Trypan Blue Stain (0.4%; Thermo Fisher Scientific, cat. no. T10282)
-
LiberaseTM (Roche, cat. no. 5401119001)
-
CD45 MicroBeads, mouse (Miltenyi Biotec, cat. no. 130-052-301)
-
CD31 MicroBeads, mouse (Miltenyi Biotec, cat. no. 130-097-418)
-
CD326 (EpCAM) MicroBeads, mouse (Miltenyi Biotec, cat. no. 130-105-958)
-
Ethanol, 70%
-
10-cm culture dish (BD Falcon, cat. no. 430167)
-
T-75 culture flask (BD Falcon, cat. no. 353136)
-
15-ml Falcon tubes (BD Falcon, cat. no. 352096)
-
50-ml Falcon tubes (BD Falcon, cat. no. 352070)
-
40-µm cell strainer (BD Falcon, cat. no. 352340)
-
1.7-ml microtubes (Axygen, cat. no. MCT-175-C)
-
Disposable Serological Pipet, 5 ml (Costar, cat. no. 4487)
-
Disposable Serological Pipet, 10 ml (Costar, cat. no. 4488)
-
S1 Pipet Fillers (Thermo Fisher Scientific, cat. no. 9521)
-
CountessTM Cell Counting Chamber Slides (Thermo Fisher Scientific, cat. no. C10314)
-
Sterile Scalpel blades (B. Braun, cat. no. BB510)
-
LD columns (Miltenyi Biotec, cat. no. 130-042-901)
-
QuadroMACS™ Separator (Miltenyi Biotec, cat. no. 130-091-051)
-
Countess® II Automated Cell Counter (Thermo Fisher Scientific, cat. no. AMQAX1000)
-
Tissue Culture Centrifuge (Thermo Sorvall Legend XIR)
-
Water bath, SUB Aqua 12 Plus (Grant Scientific)
-
Nikon Eclipse TS100 Inverted Microscope
-
CO2 incubator
-
Biosafety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
NOTE : An alternative to traditional trypsin/EDTA is TrypLE Express with phenol red from Thermo Fisher Scientific. It is stable at room temperature, does not require inactivation by an inhibitor, and is gentle on cells which results in high viability rates.
Isolation of mouse lung fibroblast
1.Anaesthetize mouse and dissect out the lungs and separate the lobes.
2.Place lung tissue into a 1.7-ml microtube containing 1.5 ml DMEM complete medium and transfer microtube onto wet ice.
3.Wipe the outside of the microtube with 70% ethanol and place samples into the biosafety cabinet.
4.Use aseptic techniques from this point forward. Use sterile scalpel blades or scissors to dissect out and discard the trachea. Next, mince the rest of the lungs into ∼1-2 mm size pieces.
5.Transfer minced lung pieces (up to 80 mg) into a 15-ml Falcon tube containing 4 ml digest medium.
6.Incubate for 30 min at 37˚C.
7.After digestion, add 10 ml DMEM complete medium into the Falcon tube and break up the digested tissue pieces by pipetting the tissue up and down vigorously to neutralize the Liberase enzymes.
8.Centrifuge digested tissue at 500 × g at room temperature (25˚C) for 5 min.
9.Discard supernatant and resuspend tissue pellet in 10 ml DMEM complete medium and thoroughly wash the pellet as described in step 7.
10.Centrifuge digested tissue at 500 × g at room temperature (25˚C) for 5 min.
11.Remove supernatant and add 10 ml DMEM complete medium to the tube and resuspend tissue pellet.
12.Pipet digested tissue into a 10-cm tissue culture dish and ensure that the tissue pieces are evenly distributed around the 10-cm dish by gently rocking the dish backwards and forwards.
13.Incubate digested tissue in a 37˚C, CO2 tissue culture incubator.
14.After 3 days, adherent cells should start to migrate out from the tissue pieces and other adherent cells should also start to proliferate at this point.
15.On day 4, replace half the volume of medium in the dish by gently removing 5 ml medium from the cells and add 5 ml fresh DMEM complete medium to the cells.
16.Replace half the volume of medium every 2 days until the cells reach ∼80%-90% confluence.
17.Upon reaching 80%-90% confluence on the 10-cm dish, the cells can now be passaged.
18.Remove medium in the dish and wash cells twice with 5 ml/wash of DPBS.
19.Add 3 ml TrypLE Express to the cells and incubate for 5 min at 37˚C.
20.Add 6 ml DMEM complete medium to the cells and resuspend cells and any remaining tissue pieces.
21.Filter the mixture of cells and tissue pieces through a 40-µm cell strainer fitted into a 50-ml Falcon tube to remove any debris or dislodged tissue pieces.
22.Pass an additional 5 ml DMEM complete medium through the cell strainer to dislodge any retained cells on the filter and then discard the filter.
Magnetic cell sorting procedure for purifying mouse lung fibroblasts
23.Centrifuge cells at 500 × g for 5 min at room temperature (25˚C).
24.Remove supernatant and gently resuspend cell pellet in 1 ml ice-cold cell sorting buffer.
25.Transfer cells into a 1.7-ml microtube.
26.Remove a 10-μl aliquot of cells and mix with 10 μl trypan blue and perform a cell count.
27.Centrifuge remaining cell suspension at 300 × g for 10 min at room temperature (25˚C).
28.Remove supernatant and gently resuspend cell pellet in 70 μl ice-cold cell sorting buffer.
29.To the cell suspension, directly add 10 μl CD45 microbeads, 10 µl CD31 microbeads, and 10 μl CD326 microbeads and resuspend antibody and cell mixture by gently tapping the sides of the tube.
30.Incubate cells for 15 min at 2° to 8°C.
31.After incubation, wash cells with 1 ml ice-cold cell sorting buffer.
32.Centrifuge cells at 300 × g for 10 min at room temperature (25˚C).
33.Prepare the LD column and cell collection tube in a QuadroMACS™ Separator. To each LD column add 2 ml ice-cold cell sorting buffer to wash the column. Discard wash from the collection tube and place the empty collection tube back beneath the LD column.
34.Remove supernatant and gently resuspend cell pellet in 500 μl ice-cold cell sorting buffer.
35.Load 500 μl resuspended cells onto the LD column and wait 1-2 min until cells have been taken up by the column.
36.Wash column two times with 1 ml ice-cold cell sorting buffer.
37.After the column has been cleared of the buffer, centrifuge the eluted cells in the collection tube at 300 × g for 10 min at room temperature (25˚C).
38.Remove supernatant and gently resuspend cells in 1 ml DMEM complete medium.
39.Remove a 10-μl aliquot of cells and mix with 10 μl trypan blue and perform cell counts.
40.Cell concentrations are adjusted to 2.0 × 105 cells/ml in DMEM complete medium and 10 ml of cells are seeded into a T-75 flask (i.e., seed 2 × 106 cells per T-75 flask).
41.Incubate cells in a 37˚C, CO2 incubator.
42.On the next day, remove medium from the cells and add 10 ml/flask of DMEM complete medium to remove any dead cells.
43.The cells are cultured until they are ∼90% confluent, at which point they can be seeded in 6-well or 96-well plates for downstream experiments as described in Support Protocols 2-5.
Support Protocol 1: FREEZING AND THAWING OF PRIMARY CELLS
Cryopreservation of cells in culture is a routine process. Cryoprotectants used to maintain cellular integrity during storage include glycerol, ethylene glycol, and dimethyl sulfoxide.
Materials
-
Primary HSCs or HLFs or HRPTEpiCs (see Basic Protocols 1 and 2)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
TrypLE Express with phenol red (Gibco brand, Thermo Fisher Scientific, cat. no. 12605010)
-
SteCM complete medium (see recipe)
-
FGMTM-2 fibroblast complete medium (see recipe)
-
EpiCM complete medium (see recipe)
-
Trypan Blue Stain (0.4%; Thermo Fisher Scientific, cat. no. T10282)
-
Freezing medium (see recipe)
-
15-ml Falcon tubes (BD Falcon, cat. no. 352096)
-
50-ml Falcon tubes (BD Falcon, cat. no. 352070)
-
Tissue Culture Centrifuge (Thermo Sorvall Legend XIR)
-
Countess Cell Counting Chamber Slides (Thermo Fisher Scientific, cat. no. C10228)
-
Countess® II Automated Cell Counter (Thermo Fisher Scientific, cat. no. AMQAX 1000)
-
Cryovials (Thermo Fisher Scientific, cat. no. 368632)
-
Mr. Frosty™ Freezing Container (Thermo Fisher Scientific, cat. no. 5100-0001)
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
NOTE : An alternative to traditional trypsin/EDTA is TrypLE Express with phenol red from Thermo Fisher Scientific. It is stable at room temperature, does not require inactivation by an inhibitor, and is gentle on cells which results in high viability rates.
Freezing of primary cells
1.Freezing is performed when the Passage 2 (P2) culture reaches ∼90% confluence.
2.Remove and discard spent cell culture medium from the T-75 culture flask.
3.Wash cells twice with 5 ml/wash of DPBS.
4.Add 4 ml dissociation reagent (TrypLE™ Express) to the side of the flask and gently rock the container to get complete coverage of the cell layer.
5.Incubate cells in a 37°C, CO2 incubator for 3-5 min.
6.When ≥90% of the cells have detached, add 8 ml specific complete medium and disperse cells by pipetting the medium over the cell layer surface several times.
7.Transfer cells into a 50-ml conical tube and centrifuge at 500 × g for 5 min at room temperature (25°C).
8.Discard supernatant and gently resuspend cell pellet in 1 ml pre-warmed specific complete medium.
9.Remove a 10-μl aliquot of cells and mix with 10 µl trypan blue and perform cell counts.
10.Centrifuge remaining cell suspension (described in step 8) at 500 × g for 5 min at room temperature (25°C).
11.Discard supernatant and resuspend cell pellet in 2-4 ml freezing medium, depending upon the cell number.
12.Make aliquots of ∼0.5 × 106 cells per cryotube.
13.Store cryotubes in a Mr. Frosty freezing container at -80°C for up to 24 hr.
14.The next day, transfer frozen cryotubes into a liquid nitrogen tank for long term storage.
Thawing and initiating the culture
15.Thaw and initiate cell culture as described in Basic Protocols 1 and 2.
Support Protocol 2: OPERETTA HIGH-CONTENT IMAGING-BASED PHENOTYPING
This support protocol details how the Operetta platform, a high-content imaging tool, can be used for phenotyping of primary cells in 96-well plates. Immunohistochemical staining of markers such as smooth muscle actin or collagen allows for the monitoring of fibroblast activation and extracellular matrix production. If IL11RA protein is still detected at high levels, the primary cells have retained their original cellular phenotype.
Materials
-
Primary HSCs, HLFs, or HRPTEpiCs (see Basic Protocols 1 and 2)
-
SteCM complete medium (see recipe)
-
Stellate Cell Medium-basal (SteCM-b; ScienCell Research Laboratories, cat. no. 5301-b)
-
FGMTM-2 fibroblast complete medium (see recipe)
-
FBMTM fibroblast basal medium (LONZA, cat. no. CC-3131)
-
EpiCM complete medium (see recipe)
-
Epithelial cell medium-basal (EpiCM-b; ScienCell Research Laboratories, cat. no. 4101-b)
-
Human Recombinant TGFβ1 (Bio-Rad, cat. no. PHP143B)
-
Human Recombinant IL11 (Genscript, cat. no. UniProtKB; accession no. P20809.1; protein used for this work) or IL11 Recombinant Human Protein (Thermo Fisher Scientific, cat. no. PHC0115)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
4% paraformaldehyde (PFA; see recipe)
-
Permeabilization buffer (see recipe)
-
Blocking solution (see recipe)
-
Wash buffer (see recipe)
-
α-Smooth muscle actin/ACTA2 antibody (abcam, cat. no. ab7817)
-
Anti-collagen I antibody (abcam, cat. no. ab34710)
-
IL11RA antibody (abcam, cat. no. ab125015)
-
Anti-mouse Alexa Fluor 488 antibody (abcam, cat. no. ab150113)
-
Anti-rabbit Alexa Fluor 488 antibody (abcam, cat. no. ab150077)
-
DAPI (Thermo Fisher Scientific, cat. no. 1306)
-
Disposable Serological Pipet, 5 ml (Costar, cat. no. 4487)
-
Disposable Serological Pipet, 10 ml (Costar, cat. no. 4488)
-
S1 Pipet Fillers (Thermo Fisher Scientific, cat. no. 9521)
-
Reagent Reservoir (Costar, cat. no. 4870)
-
Multi-Channel Pipet (Eppendorf, cat. no. ES-12-300)
-
Operetta 96-well black CellCarrier plates (PerkinElmer, cat. no. 6005550)
-
Operetta high-content imaging system (PerkinElmer, model no. 1483)
-
Tissue Culture Centrifuge (Thermo Sorvall Legend XIR)
-
CO2 incubator
-
Biosafety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
Culturing of primary cells for Operetta high-content phenotyping
1.HSCs, HLFs, or HRPTEpiCs cultured as described in Basic Protocols 1 and 2 are seeded in 96-well black cell carrier plates with SteCM complete medium, FGM-2 fibroblast complete medium, or EpiCM complete medium, respectively, at a density of 6 × 103 cells/well and 100 μl/well and incubated in a 37°C, CO2 incubator for 24 hr.
2.After 24 hr of incubation, completely remove medium from the cells and wash cells twice with 100 μl/well of either SteCM-b (for HSCs) or FBM fibroblast basal medium (for HLFs) or epithelial cell basal medium (for HRPTEpiCs).
3.To serum-starve the HSCs, HLFs, or HRPTEpiCs, add 100 μl of cell-type specific serum-free basal medium (SteCM-b, FBM fibroblast basal medium, or EpiCM-b, respectively) and incubate cells in a 37°C, CO2 incubator overnight (∼16 hr).
4.After overnight incubation, prepare medium for cell stimulation. To cell-type specific serum-free basal medium, add recombinant cytokines (i.e., IL11 or TGFβ1) in 1.7-ml microtubes to a final working concentration of 5 ng/ml.
5.Completely remove and discard medium and add 100 μl/well of cell-type specific serum-free basal medium containing recombinant cytokines.
6.Incubate plate in a 37°C, CO2 incubator for 24 hr.
Immunostaining of primary cells for Operetta high-content phenotyping
7.After 24 hr of cell stimulation, remove medium from the cells.
8.Wash cells twice with 100 μl/well of DPBS and discard the last wash.
9.Fix cells with 100 μl/well of 4% paraformaldehyde for 20 min at room temperature (25°C).
10.Remove 4% paraformaldehyde from cells and wash cells three times with 100 μl/well of DPBS; discard the last wash.
11.Add 100 μl/well of permeabilization buffer and incubate 15 min at room temperature to permeabilize the cells.
12.Remove permeabilization buffer from cells and wash cells twice with 100 μl/well of DPBS; discard the last wash.
13.Add 100 μl/well of blocking solution and incubate 1 hr at room temperature (25°C).
14.While cells are in blocking solution, prepare sufficient quantities of diluted primary antibodies in blocking solution in separate 1.7-ml microtubes and store at 4°C:
-
anti-ACTA2 (1:500 dilution);
-
anti-COL1A1 (1:500 dilution);
-
anti-IL11RA (1:100 dilution).
15.After 1 hr of incubation, remove blocking solution from cells and incubate cells with 100 μl/well of diluted primary antibody solution and incubate plate overnight at 4°C.
16.The next day, prepare the following secondary antibody dilutions in blocking solution in separate 1.7-ml microtubes. Protect diluted secondary antibodies from light.
-
for ACTA2, use anti-mouse Alexa Fluor 488-conjugated secondary antibody (1:1,000 dilution);
-
for COL1A1, use anti-rabbit Alexa Fluor 488-conjugated secondary antibody (1:1,000 dilution);
-
for IL11RA, use anti-rabbit Alexa Fluor 488-conjugated secondary antibody (1:200 dilution).
17.Remove diluted primary antibodies from cells and wash cells once with 100 μl/well of wash buffer.
18.Add 100 μl/well of diluted secondary antibodies to the cells and incubate plate for 1 hr at room temperature (25°C), protected from light.
19.After 1 hr of incubation, counterstain cells with 100 µl/well of DAPI (1 µg/ml) in blocking solution for 15 min at room temperature.
20.Wash cells three times with 100 μl/well of DPBS and after the last wash, fill wells with 100 μl/well of DPBS (to avoid cells drying out).
21.Immunofluorescence images are acquired using the Operetta high-content imaging system (PerkinElmer).
Support Protocol 3: COLORIMETRIC ASSAY OF SOLUBILIZED COLLAGEN
IL11 and TGFβ1 treatment induces collagen secretion from human HSCs and HLFs in vitro (Ng et al., 2019; Widjaja et al., 2019b). We here describe a simple quantitative assay to quantify the total amount of solubilized collagen in the supernatant of IL11-treated cells. Sirius red is a unique dye that specifically binds to the [Gly-X-Y]n helical structure on fibrillar collagen (type I to V) and thus does not discriminate between collagen species and types. Due to the low level of collagen in cell culture medium, an additional concentration step is necessary to detect collagen in culture medium samples within the standard range of the Sirius Red Collagen Detection kit.
Materials
-
Primary HSCs or HLFs (see Basic Protocols 1 and 2)
-
SteCM complete medium (see recipe)
-
Stellate cell medium-basal (SteCM-b; ScienCell, cat. no. 5301-b)
-
FGMTM-2 fibroblast complete medium (see recipe)
-
FBMTM fibroblast basal medium (LONZA, cat. no. CC-3131)
-
Human Recombinant TGFβ1 (Bio-Rad, cat. no. PHP143B)
-
Human Recombinant IL11 (Genscript, cat. no. UniProtKB; P20809)
-
Sirius Red Collagen Detection Kit (Chondrex, cat. no. 9062)
-
Concentrating Solution (Chondrex, cat. no. 90626)
-
Microtubes, 2 ml (Axygen, cat. no. MCT-200-C)
-
6-well culture plates (BD Falcon, cat. no. 353046)
-
Disposable Serological Pipet, 5 ml (Costar, cat. no. 4487)
-
Disposable Serological Pipet, 10 ml (Costar, cat. no. 4488)
-
Vortex-Genie 2 (Scientific Industries)
-
Centrifuge (Sorvall, Legend Micro 21R)
-
ELISA plate reader (Tecan Infinite® M200PRO)
Stimulation of primary cells and sample preparation
1.HSCs or HLFs (cultured as described in Basic Protocols 1 and 2) are seeded in 6-well plates at a density of 2 × 105 cells per well.
2.After reaching ∼80% confluence, remove complete medium from cells and wash cells twice with 2 ml/wash of either SteCM-b (for HSCs) or FBM fibroblast basal medium (for HLFs).
3.To serum-starve HSCs or HLFs, add 100 μl/well of either SteCM-b or FBM fibroblast basal medium and incubate cells in a 37°C, CO2 incubator overnight (∼16 hr).
4.After overnight incubation, prepare medium for cell stimulation. To SteCM-b or FBM fibroblast basal medium, add recombinant cytokines (i.e., IL11 or TGFβ1) to a final working concentration of 5 ng/ml.
5.For cell simulation of HSCs or HLFs, completely remove and discard medium from cells and add 1 ml/well of specific serum-free basal medium containing recombinant cytokines (i.e., IL11 or TGFβ1).
6.Incubate cells in a 37°C, CO2 incubator for 24 hr.
7.After 24 hr of cell stimulation, the supernatants are collected in 2-ml microtubes for Sirius red collagen assay.
8.Due to the low level of collagen in cell culture medium, the supernatant must first be concentrated.
9.To concentrate the supernatant, add 250 μl of Concentrating Solution (Chondrex) to 1 ml of supernatant.
10.Vortex sample briefly.
11.Incubate at 4°C for 16-24 hr.
12.Centrifuge at 10,000 rpm (∼13,000 × g) for 3 min.
13.Remove supernatant by pipetting carefully without disturbing the pellet.
14.Add 100 μl of 0.05 M acetic acid (diluted from 0.5 M acetic acid stock) to dissolve the pellet.
Quantification of solubilized collagen in the culture supernatant
15.Proceed to quantify the quantity of solubilized collagen (µg/ml) in the supernatant according to the manufacturer's protocol (Sirius Red Collagen Detection Kit; Chondrex).
Support Protocol 4: QUANTIFICATION OF FIBROSIS MARKER SECRETION
Liver and lung fibrosis are associated with elevated expression of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs). Activated HSCs and HLFs upregulate MMP-2 and TIMP-1, respectively. Both cell types also secrete IL11 when stimulated with TGFβ1.This protocol details how HSC or HLF activation can be studied by monitoring the concentration of fibrosis markers in the supernatant.
Materials
-
Primary HSCs or HLFs (see Basic Protocols 1 and 2)
-
SteCM complete medium (see recipe)
-
Stellate cell medium-basal (SteCM-b; ScienCell, cat. no. 5301-b)
-
FGM-2 fibroblast complete medium (see recipe)
-
FBMTM fibroblast basal medium (LONZA, cat. no. CC-3131)
-
Human Recombinant TGFβ1 (Bio-Rad, cat. no. PHP143B)
-
Human Recombinant IL11 (Genscript, cat. no. UniProtKB; P20809)
-
Human IL11 Quantikine ELISA Kit (R&D Systems, cat. no. D1100)
-
Total MMP-2 Quantikine ELISA Kit (R&D Systems, cat. no. MMP200)
-
Human TIMP-1 Quantikine ELISA Kit (R&D Systems, cat. no. DTM100)
-
Disposable Serological Pipet, 5 ml (Costar, cat. no. 4487)
-
Disposable Serological Pipet, 10 ml (Costar, cat. no. 4488)
-
S1 Pipet Fillers (Thermo Fisher Scientific, cat. no. 9521)
-
6-well culture plates (BD Falcon, cat. no. 353046)
-
1.7-ml microtubes (Costar, cat. no. MCT-175-C)
-
Reagent Reservoir (Costar, cat. no. 4870)
-
Multi-Channel Pipet (Eppendorf, cat. no. ES-12-300)
-
ELISA Plate Reader (Tecan Infinite® M200PRO)
Measurement of fibrosis markers in the culture supernatant
1.HSCs or HLFs are seeded in 6-well culture plates, with 2 ml SteCM complete medium or FGM-2 fibroblast complete medium, respectively, at a density of 2.5 × 105 cells/well and incubated in a 37°C tissue culture incubator for 24 hr.
2.After reaching ∼80% confluence, remove complete medium from cells and wash cells twice with 2 ml/wash of either SteCM-b (for HSCs) or FBM fibroblast basal medium (for HLFs).
3.To serum-starve the HSCs or HLFs, add 100 μl/well of either SteCM-b (for HSCs) or FBM fibroblast basal medium (for HLFs) and incubate cells in a 37°C, CO2 incubator overnight (∼16 hr).
4.After overnight incubation, prepare medium for cell stimulation. To SteCM-b (for HSCs) or FBM fibroblast basal medium (for HLFs), add recombinant cytokines (i.e., IL11 or TGFβ1) to a final working concentration of 5 ng/ml.
5.For cell stimulation of HSCs or HLFs, completely remove and discard medium from cells and add 1 ml/well of specific serum-free basal medium containing recombinant cytokines (i.e., IL11 or TGFβ1).
6.Incubate cells in a 37°C, CO2 incubator for 24 hr.
7.After 24 hr of stimulation, the culture supernatant is collected in 1.7-ml microtubes.
8.For sample preparation prior to running the ELISA, the culture supernatant is first diluted accordingly:
-
Use 100 μl of culture supernatant directly for each well of the IL11 ELISA.
-
Dilute culture supernatant 1:4 (12.5 μl of culture supernatant + 37.5 μl of 1:5 diluted RD5P; supplied in the R&D Systems ELISA kit) for each well of the MMP-2 ELISA.
-
Dilute culture supernatant 1:15 (3.5 μl of culture supernatant + 46.5 μl of 1:5 diluted RD5P) for each well of the TIMP-1 ELISA.
9.Perform ELISA according to the manufacturer's protocol.
Support Protocol 5: WESTERN BLOTTING STUDIES OF IL11 SIGNALING IN HSCs, HLFs, and HRPTEpiCs
IL11 binds to its specific alpha receptor (IL11RA), which then signals via the ubiquitously expressed gp130 receptor to trigger a range of signaling pathways including JAK/STAT, ERK, and AKT (Cook & Schafer, 2020). To investigate the IL11-induced signaling pathways in HSCs, HLFs, or HRPTEpiCs, the cells cultured from Basic Protocols 1 and 2 are seeded in 6-well plates and subsequently grown to ∼70% to 80% confluence. To eliminate the influence of growth factors present in complete culture medium on the activation of multiple downstream signaling pathways, the cells are first cultured overnight (∼16 hr) in serum-free basal medium before stimulation with the profibrotic cytokines.
Materials
-
Primary HSCs, HLFs, or HRPTEpiCs (see Basic Protocols 1 and 2)
-
SteCM complete medium (see recipe)
-
Stellate cell medium-basal (SteCM-b; ScienCell, cat. no. 5301-b)
-
FGM-2 fibroblast complete medium (see recipe)
-
FBMTM fibroblast basal medium (LONZA, cat. no. CC-3131)
-
EpiCM complete medium (see recipe)
-
Epithelial cell medium-basal (EpiCM-b; ScienCell, cat. no. 4101-b)
-
Human Recombinant TGFβ1 (Bio-Rad, cat. no. PHP143B)
-
Human Recombinant IL11 (Genscript, cat. no. UniProtKB; accession no. P20809.1; protein used for this work) or IL11 Recombinant Human Protein (Thermo Fisher Scientific, cat. no. PHC0115)
-
Tris-buffered saline (TBS) buffer (4 L; Axil Scientific, cat. no. BUF-3030-20X)
-
Tris-buffered saline and Tween 20 (TBST) wash buffer (see recipe)
-
Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, cat. no. 23225)
-
RIPA lysis and extraction buffer (see recipe)
-
Tris-Glycine Buffer, 10× (Bio-Rad, cat. no. 161-0771)
-
Laemmli Sample Buffer, 4× (Bio-Rad, cat. no. 161-0747)
-
Spectra multicolor Broad Range Ladder (Thermo Fisher Scientific, cat. no. 26634)
-
Trans-Blot Turbo mini PVDF (Bio-Rad, cat. no. 170-4156)
-
Western blot blocking buffer (see recipe)
-
Antibody incubation buffer (see recipe)
-
Water, deionized, distilled (Hyclone, cat. no. SH30538.02)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
IL11RA antibody (Santa Cruz, cat. no. 130920)
-
ACTA2 antibody (CST, cat. no. 19245)
-
p-ERK1/2 antibody (CST, cat. no. 4370)
-
ERK1/2 antibody (CST, cat. no. 4695)
-
p-STAT3 antibody (CST, cat. no. 4113)
-
STAT3 antibody (CST, cat. no. 4904)
-
GAPDH antibody (CST, cat. no. 2118)
-
Mouse HRP antibody (CST, cat.no. 7076)
-
Rabbit HRP antibody (CST, cat. no. 7074)
-
ECL Plus Western Blotting Substrate (Thermo Fisher Scientific, cat. no. 32132)
-
SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, cat. no. 34096)
-
6-well culture plates (BD Falcon, cat. no. 353046)
-
Cell Scraper, 30 cm (SPL, cat. no. 90030)
-
Disposable Serological Pipet, 5 ml (Costar, cat. no. 4487)
-
Disposable Serological Pipet ,10 ml (Costar, cat. no. 4488)
-
Microtubes, 1.7 ml (Axygen, cat. no. MCT-175-C)
-
Microtubes, 0.6 ml (Axygen, cat. no. MCT-060-C-S)
-
Centrifuge (Sorvall, Legend Micro 21R)
-
Trans-Blot Turbo Transfer System (Bio-Rad, cat. no. 1704150EDU)
-
ELISA plate reader (Tecan Infinite® M200PRO)
-
ChemiDoc Imaging System (Bio-Rad)
-
CO2 incubator
-
Biological safety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
Cell stimulation and sample preparation
1.HSCs, HLFs, or HRPTEpiCs are seeded in 6-well plates at a density of 2.5 × 105 cells/well in 2 ml/well of specific complete medium (SteCM complete medium, FGM-2 fibroblast complete medium, or EpiCM complete medium, respectively) and incubated in a 37°C, CO2 incubator.
2.Once cells have grown to ∼80% confluence (usually within 24 hr), remove culture medium and wash cells twice with cell-type specific serum-free basal medium (SteCM-b, FBM fibroblast basal medium, or EpiCM-b, respectively).
3.Serum-starve the cells by adding 1 ml/well of cell-type specific serum-free basal medium and incubate cells in a 37°C, CO2 incubator overnight.
4.For cell stimulation of HSCs, HLFs, or HRPTEpiCs, remove medium and replace with 1 ml/well of cell-type specific serum-free basal medium containing the appropriate concentrations of recombinant cytokine (IL11 or TGFβ1).
5.Incubate cells for 24 hr in a 37°C, CO2 incubator.
6.Following 24 hr of cell stimulation, plates containing cells are placed on ice.
7.Remove medium from cells and gently wash cell monolayer with ice-cold DPBS.
8.Add 150 μl of ice-cold RIPA lysis and extraction buffer.
9.Scrape cells using a cell scraper and transfer the lysate to 1.7-ml microtubes.
10.Incubate tubes containing cell lysates on ice for 10 min.
11.Centrifuge tubes at 18,000 × g for 20 min at 4°C. Collect supernatant in a new 1.7-ml microtube and place it on ice. Discard pellet.
12.The protein concentration of each sample can be measured using the BCA Protein Assay Kit following manufacturer's protocol (Pierce).
13.The samples are aliquoted and stored at -30°C or directly used for western blotting. Proteins are boiled (in Laemmli sample buffer) at 95°C for 5 min before proceeding to the electrophoresis step in the subsequent section.
Western blotting
14.Load equal amounts of boiled protein from step 13 (e.g., 10 µg in 10 μl) into the wells of the SDS-PAGE gel, along with a lane for the molecular weight markers such as the Spectra multicolor broad range ladder from Thermo Fisher Scientific.
15.Run gel for 30 min at 80 V in 1× tris-glycine buffer (diluted from 10× stock) and then change the voltage to 120 V for 1-2 hr.
16.Transfer proteins from the gel to a PVDF membrane using a transfer pack according to the manufacturer's protocol.
17.Block membrane with a western blot blocking buffer for 1 hr at room temperature.
18.Incubate membrane with appropriate dilutions of primary antibodies (normally 1:1,000) in the incubation buffer overnight at 4°C.
19.Wash membrane three times with TBST, 5 min for each wash.
20.Incubate membrane with the recommended dilution of species-specific HRP-conjugated secondary antibodies (1:2,000) in the incubation buffer at room temperature for 1-2 hr.
21.Wash membrane three times with TBST, 10 min for each wash.
22.Wash membrane once for 10 min with TBS and proceed to develop the membrane.
23.Proteins are visualized using either ECL Plus Western Blotting Substrate or Femto Maximum Sensitivity Substrate and the membrane is imaged using a chemiluminescent detection system such as the ChemiDoc Imaging System (Bio-Rad).
Basic Protocol 3: IL11 STIMULATION OF PRIMARY HUMAN HEPATOCYTES
Studies in our laboratory have shown that IL11 causes hepatocyte dysfunction which underlies non-alcoholic steatohepatitis (NASH) disease pathology (Widjaja et al., 2019b). In primary hepatocyte cultures, IL11-treatment induces hepatocyte dysfunction by activating ERK, JNK, and STAT3 signaling (Dong, Viswanathan, Adami, & Chothani, 2021; Widjaja, Dong, Adami, Viswanathan, & Ng, 2019a). This protocol describes optimized culture and experimentation conditions for the study of IL11 signaling in hepatocytes. Once they have been recovered from the initial thaw cycle, primary human hepatocytes are used directly for downstream experiments. When seeded at low densities (<4.0 × 105 cells per well of a 6-well plate), hepatocytes will de-differentiate and fail to replicate in vivo cellular physiology. Therefore, it is important to follow the counting and seeding guidelines described below to ensure that hepatocyte functions are preserved ex vivo.
Materials
-
Cryo Human Hepatocytes Plateable, Passage 0 (LONZA, cat. no. HUCPI)
-
Cryo Human Hepatocyte thawing medium (LONZA, cat. no. MCHT50)
-
Human Hepatocyte plating medium (see recipe)
-
Human Hepatocyte complete medium (see recipe)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
Recombinant human IL11 (Genscript, cat. no. UniProtKB; P20809)
-
Trypan Blue Stain (0.4%; Thermo Fisher Scientific, cat. no. T10282)
-
Collagen I, Coated Plate, 6 well (Thermo Fisher Scientific, cat. no. A1142801)
-
15-ml Falcon tubes (BD Falcon, cat. no. 352096)
-
50-ml Falcon tubes (BD Falcon, cat. no. 352070)
-
Tissue Culture Centrifuge (Thermo Sorvall Legend XIR)
-
Water bath, SUB Aqua 12 Plus (Grant Scientific)
-
Nikon Eclipse TS100 Inverted Microscope
-
CO2 incubator
-
Biosafety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
Initiating the culture
1.Prepare human hepatocyte plating and complete medium (see Reagents and Solutions).
2.Warm human hepatocyte thawing medium, plating medium, and complete medium in a 37°C water bath before use.
3.Remove a vial of cryopreserved hepatocytes from liquid nitrogen storage and quickly thaw cells in a 37°C water bath. Remove vial from the water bath when a small frozen cell pellet remains.
4.Wipe the outside of the vial with 70% ethanol.
5.In a biosafety cabinet, carefully transfer cell pellet and cell suspension from the vial to a 50-ml conical tube containing 49 ml of pre-warmed human hepatocyte thawing medium.
6.Rinse vial with 1 ml of human hepatocyte thawing medium and add dropwise back to the cells. Cap tube and gently invert the tube to resuspend cells.
7.Centrifuge cell suspension at 100 × g for 8 min at room temperature (25°C).
8.Carefully remove supernatant with a pipet, leaving a small amount of medium to ensure that the cell pellet is not disturbed.
9.Add 1 ml plating medium to the cells and gently resuspend cells with a pipet.
10.For cell counting, remove a 10-µl aliquot of cells, mix with 10 µl of trypan blue, and perform cell counts.
Plating and culturing human hepatocytes
11.Dilute cell suspension with human hepatocyte plating medium to a concentration of 2 × 105 cells/ml and seed 4 × 105 cell/well in a collagen I-coated 6-well plate. Gently swirl plate to distribute the cell suspension evenly.
12.Incubate cells in a 37°C, CO2 incubator.
13.Without removing the plate from the incubator, swirl the 6-well plates 15, 30, and 45 min post-seeding (except 96-well plates).
14.After 60 min post-seeding, carefully remove human hepatocyte plating medium to remove any non-attached and/or dead cells.
15.Gently add 2 ml/well of fresh human hepatocyte plating medium to the cells.
16.Incubate cells for an additional 4-6 hr.
17.Remove plating medium from the cells and add 2 ml/well of warm human hepatocyte complete medium to the cells.
18.Incubate cells in a 37°C, CO2 incubator overnight.
19.On the next day (Day 1), remove any unattached and/or dead cells by removing medium from the cells and add 1.5 ml/well of pre-warmed human hepatocyte complete medium to the cells.
20.Incubate cells in a 37°C, CO2 incubator for an additional day before cell stimulation.
Cell stimulation
21.On the next day (Day 2), prepare medium containing recombinant human IL11 (5 or 10 ng/ml in human hepatocyte complete medium).
22.Remove medium from cells.
23.Add 1 ml/well of human hepatocyte complete medium containing recombinant human IL11.
24.Incubate cells in a 37˚C, CO2 incubator for 24 hr.
25.The cells and culture supernatants are now ready for use in downstream studies including:
-
Cell signaling studies by western blot (see Support Protocol5).
-
ALT assay (see Support Protocol6).
Alternate Protocol 2: IL11 STIMULATION OF PRIMARY MOUSE HEPATOCYTES
Mouse hepatocytes are isolated from CD-1 mouse livers from 8 to 12 week old mice after liver perfusion and low-speed differential centrifugation. The hepatocytes are cryopreserved immediately after the isolation process. Similar to primary human hepatocytes, primary mouse hepatocytes should be seeded directly at suitable densities for downstream experiments.
Materials
-
Mouse Hepatocytes (Accegen Biotech, cat. no. ABC-TC3928)
-
Mouse Hepatocyte Thawing medium (Accegen Biotech, cat. ABS-MT3928)
-
Mouse Hepatocyte Plating medium (Accegen Biotech, cat. no. ABS-TP3928)
-
Mouse Hepatocyte complete medium (see recipe)
-
Recombinant mouse IL11 (Genscript, cat. no. UniProtKB; P47873)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
Trypan Blue Stain (0.4%; Thermo Fisher Scientific, cat. no. T10282)
-
Collagen I, Coated Plate, 6 well (Thermo Fisher Scientific, cat.no. A1142801)
-
15-ml Falcon tubes (BD Falcon, cat. no. 352096)
-
Tissue Culture Centrifuge (Thermo Sorvall Legend XIR)
-
Water bath, SUB Aqua 12 Plus (Grant Scientific)
-
Nikon Eclipse TS100 Inverted Microscope
-
CO2 incubator
-
Biosafety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
Thawing of frozen mouse hepatocyte cells
1.Prepare mouse hepatocyte complete medium (see Reagents and Solutions).
2.Warm up mouse hepatocyte thawing medium and plating medium in a 37°C water bath before use.
3.Take out the vial of mouse hepatocytes from the liquid nitrogen storage unit and quickly thaw the vial in a 37°C water bath by gently shaking the vial. Remove vial from the water bath when a small frozen cell pellet remains.
4.Wipe the outside of the vial with 70% ethanol.
5.In a biosafety cabinet, transfer cell pellet and cell suspension from the vial into a 50-ml conical tube containing 49 ml mouse hepatocyte thawing medium. Cap tube and gently invert the tube to fully thaw and resuspend cells in thawing medium.
6.Centrifuge cell suspension at 100 × g for 5 min at room temperature (25°C).
7.Carefully remove supernatant, leaving a small amount of medium to ensure that the cell pellet is not disturbed.
8.Gently add 1 ml mouse hepatocyte plating medium to the tube and gently resuspend cells using a pipet.
9.For cell counting, remove a 10-µl aliquot of cells and mix cells with 10 µl of trypan blue and perform a cell count.
Plating and culturing mouse hepatocytes
10.Dilute cell suspension with mouse hepatocyte plating medium to a concentration of 2 × 105 cells/ml and seed 4 × 105 cell/well in a collagen I-coated 6-well plate. Gently swirl plate to distribute the cell suspension evenly.
11.Incubate cells in a 37°C, CO2 incubator for 2-4 hr to allow cells to attach to the culture plates.
12.After the cells have attached, gently swirl culture plate and aspirate medium to remove any non-attached and/or dead cells.
13.In the 6-well plates, add 1.7 ml/well of mouse hepatocyte complete growth medium to the cells.
14.Incubate cells in a 37°C, CO2 incubator overnight.
15.On the next day (Day 1), remove medium from the cells.
16.Gently add 1.7 ml/well of mouse hepatocyte complete growth medium to the cells.
17.Incubate cells in a 37°C, CO2 incubator for an additional day before cell stimulation.
Cell stimulation
18.On the next day (Day 2), prepare medium containing recombinant mouse IL11 (10 ng/ml in mouse hepatocyte complete medium).
19.Remove medium from the cells.
20.Add 1 ml/well of mouse hepatocyte complete medium containing recombinant mouse IL11.
21.Incubate cells in a 37˚C, CO2 incubator for 24 hr.
22.The cells and culture supernatants are now ready for use in downstream studies including:
-
Cell signaling studies by western blot (see Support Protocol5).
-
ALT assay (see Support Protocol6).
Support Protocol 6: ALANINE TRANSAMINASE (ALT) SECRETION BY HUMAN AND MOUSE HEPATOCYTES
ALT is the most widely used clinical biomarker for liver injury. ALT catalyzes the transamination of alanine to α-ketoglutarate and it is released from hepatocytes following hepatocellular injury. The hepatotoxic effects of IL11 can be monitored by quantifying ALT activity in the culture supernatant of IL11-treated human or mouse hepatocytes using a commercially available ALT activity assay kit.
Materials
-
Cryo Human and Mouse Hepatocytes (see Basic Protocol 3 and Alternate Protocol 2)
-
Human Hepatocyte complete medium (see recipe)
-
Mouse Hepatocyte complete medium (see recipe)
-
DPBS/Modified (Hyclone, cat. no. SH30028.02)
-
Recombinant human IL11 (Genscript, cat. no. UniProtKB; P20809)
-
Recombinant mouse IL11 (Genscript, cat. no. UniProtKB; P47873)
-
Alanine Transaminase Activity Assay kit (abcam, cat. no. ab105134)
-
Collagen I, Coated Plate, 6 well (Thermo Fisher Scientific, cat. no. A1142801)
-
15-ml Falcon tubes (BD Falcon, cat. no. 352096)
-
ELISA plate reader (TECAN Infinite® 200 PRO)
-
Water bath, SUB Aqua 12 Plus (Grant Scientific)
-
CO2 incubator
-
Biosafety cabinet
NOTE : DPBS/Modified: For primary culture, use a buffered salt solution that is Ca2+ and Mg2+ free to wash cells. Ca2+ and Mg2+ in the salt solution can cause cells to stick together.
1.Human or mouse hepatocytes are cultured as described in Basic Protocol 3 and Alternate Protocol 2 and seeded in 6-well plates at a density of 4 × 105 cells/well.
2.Incubate cells in a 37˚C, CO2 incubator for 24 hr.
3.After 24 hr (Day 1), remove medium from cells and replace with 2 ml pre-warmed species-specific hepatocyte complete medium to remove the unattached or dead cells.
4.Incubate cells in a 37˚C, CO2 incubator for an additional 24 hr.
5.The following day (Day 2), stimulate cells with recombinant IL11 (5 or 10 ng/ml) in species-specific hepatocyte complete medium (1 ml/well) for 24 hr.
6.After 24 hr of cell stimulation (Day 3), collect culture supernatants in 1.7-ml microtubes for ALT assay.
7.ALT measurements for each sample are performed by first diluting 5 µl supernatant with 15 µl assay buffer (abcam). The subsequent steps are performed according to the manufacturer's protocol.
REAGENTS AND SOLUTIONS
Ensure that all reagents and solutions used for cell culture are prepared and handled using aseptic techniques to prevent potential contamination of cell culture. Use deionized, distilled water (Hyclone, cat. no. SH30538.02).
Antibody incubation buffer
- 1× TBST (see recipe) with 1% (w/v) BSA (MilliporeSigma, cat. no. A7906).
- Prepare fresh for each experiment.
For western blotting.
Blocking buffer
- 1× TBST (see recipe) with 5% (w/v) Blotting Grade Blocker (Bio-Rad, cat. no. 170-6404).
- Prepare fresh for each experiment.
Store up to 1 week at 4°C (for immunofluorescence staining). Prepare fresh for each experiment (for western blotting).
For western blotting.
Blocking solution
- 0.5% (w/v) BSA (MilliporeSigma, cat. no. A7906) and 0.1% (v/v) Tween-20 (Bio-Rad, cat. no. 170-6531) in Dulbecco's phosphate-buffered saline (DPBS; Hyclone, cat. no. SH30028.02).
- Store up to 1 week at 4°C (for immunofluorescence staining). Prepare fresh for each experiment (for western blotting).
Cell sorting buffer
- 1× PBS containing 0.5% (w/v) BSA (MilliporeSigma, cat. no. A7906) and 2 nM EDTA (MilliporeSigma, cat. no. E6758).
- Store at 4°C for up to 3 months.
DMEM complete medium
- Dulbecco's modified Eagle medium (DMEM; Gibco brand, Thermo Fisher Scientific, cat. no. 11995-065) containing 10% (v/v) FBS (heat-inactivated, Brazil; Hyclone, cat. no. 10500064) and 1% (v/v) penicillin/streptomycin (Gibco brand, Thermo Fisher Scientific, cat. no. 15140-122).
- Filter sterilize and store ≤1 month at 4°C.
Digest medium
- DMEM (Gibco brand, Thermo Fisher Scientific, cat. no. 11995-065) containing 1% (v/v) penicillin/streptomycin (Gibco brand, Thermo Fisher Scientific, cat. no. 15140-122) and 0.14 Wunsch U/ml Liberase™ (Roche, cat. no. 5401119001).
- Prepare fresh and store at room temperature (20° to 25°C). Use within 1 hr of preparation to prevent loss of enzyme activity.
FGM-2 fibroblast complete medium
- Decontaminate external surfaces of all vials and medium bottle with 70% ethanol.
- Then transfer the contents of the FGM™-2 SingleQuots™ supplements (LONZA, cat. no. CC-4126), containing the following growth supplements: hFGF-B (0.5 ml), insulin (0.5 ml), FBS (10 ml), and GA-1000 (0.5 ml), into 500 ml FBM™ Fibroblast Basal Medium (LONZA, cat. no. CC-3131) with a pipet and rinse each vial with medium. Filter sterilize and store ≤1 month at 4°C.
When preparing these BulletKit™ Media (LONZA), it may not be possible to recover the entire volume listed for each vial. Small losses (up to 10%) should not affect the cell growth characteristics of the supplemented medium.
If there is concern that sterility was compromised during the supplementation process, the entire newly prepared culture medium may be re-filtered with a 0.2-µm filter to assure sterility. Routine re-filtration is not recommended.
Freezing medium
- Mix FBS and dimethyl sulfoxide (MilliporeSigma, cat. no. D2660) in a ratio of 9:1.
- Store for up to 1 month at 4°C.
EpiCM complete medium
- Thaw epithelial cell growth supplements (ScienCell, cat. no. 4152), FBS (ScienCell, cat. no. 0010), and antibiotic solution (ScienCell, cat. no. 0503) at 37°C.
- Once the reagents are thawed, gently tilt the tubes several times to ensure the contents are completely mixed.
- Spray the medium bottle and tubes with 70% ethanol, and wipe to remove excess liquid.
- Mix all the reagents above mentioned with 500 ml of epithelial cell medium (EpiCM; ScienCell, cat. no. 4101).
- Because several components are light-sensitive, the medium should not be exposed to the light for extended periods.
- When stored in the dark at 4°C, the reconstituted complete medium is stable for 1 month.
- Warm epithelial cell complete medium in a 37°C water bath prior to use.
Human hepatocyte complete medium
- Decontaminate external surfaces of all vials and the medium bottle with 70% ethanol.
- To formulate hepatocyte culture medium (HCM™ Medium), transfer the contents of the HCM™ SingleQuots™ Kit [LONZA, cat. no. CC-4182 containing ascorbic acid, bovine serum albumin – fatty acid free (BSA-FAF), hydrocortisone, human epidermal growth factor (hEGF), transferrin, insulin, and gentamicin/amphotericin-B (GA)] to HBM™ Basal Medium (LONZA, cat. no. CC3199) with a pipet, and rinse each vial with medium. Filter sterilize and store ≤1 month at 4°C.
When preparing these BulletKit™ Media, it may not be possible to recover the entire volume listed for each vial. Small losses (up to 10%) should not affect the cell growth characteristics of the supplemented medium.
- If there is concern that sterility was compromised during the supplementation process, the entire newly prepared culture medium may be re-filtered with a 0.2-µm filter to assure sterility. Routine re-filtration is not recommended.
Human hepatocyte plating medium
- Decontaminate external surfaces of all vials and the medium bottle with 70% ethanol.
- To complete the hepatocyte plating medium (LONZA, cat. no. MP100 and MP250), pour the entire contents of the vial labeled “Plating Supplement” into the medium.
- Filter sterilize and store ≤1 month at 4°C.
Incubation buffer
- 1× TBST (see recipe) with 1% w/v BSA.
- Buffer can be prepared and stored at 4°C for up to 1 month.
For western blotting.
Mouse hepatocyte complete medium
- Thaw penicillin/streptomycin (P/S) solution at 37°C.
- After thawing, gently tilt the tube several times to ensure the contents are completely mixed.
- Mix P/S with mouse hepatocyte medium (Accegen Biotech, cat. no. ABC-TM3928) and warm in a 37°C water bath prior to use.
- Filter sterilize and store ≤1 month at 4°C.
RIPA lysis and extraction buffer
- 50 ml RIPA buffer (Thermo Fisher Scientific, cat. no. 89900) with one tablet of protease inhibitor (EDTA-free; Thermo Fisher Scientific, cat. no. A32965) and five tablets of mini phosphatase inhibitors (Thermo Fisher Scientific, cat. no. A32957).
- Make aliquots and store for up to 1 month at −30°C.
Paraformaldehyde, 4%
- Dilute 16% paraformaldehyde (PFA) stock solution (Thermo Fisher Scientific, cat. no. 28906) in DPBS.
- Make fresh before use.
Permeabilization buffer
- 0.1% (v/v) Triton X-100 (MilliporeSigma, cat. no. T8787) in DPBS.
- Prepare fresh and use immediately.
For immunofluorescence staining.
Preparation of poly-L-lysine-coated T-75 flask
- Add 10 ml of sterile water to a T-75 flask and then add 15 µl of poly-L-lysine stock solution (0.01%; MilliporeSigma, cat. no. P4707) for a final concentration of 2 µg/cm2 (T-75 flask is recommended) poly-L-lysine in the mixture.
- Leave the flask in a 37°C incubator overnight or for a minimum of 1 hr.
- After overnight incubation, aspirate the poly-L-lysine mixture from the T-75 flask and rinse the poly-L-lysine-coated T-75 flask twice with sterile water then add 15 ml of human hepatic stellate cell complete medium.
- Leave the flask in the sterile biosafety cabinet or CO2 incubator and proceed to seed the cells.
SteCM complete medium
- Thaw stellate cell growth supplements (ScienCell, cat. no. 5352), FBS (ScienCell, cat. no. 0010), and penicillin/streptomycin solution (ScienCell, cat. no. 0503) at 37°C.
- Once the reagents are thawed, gently tilt the tubes several times to ensure the contents are completely mixed.
- Spray the medium bottle and tubes with 70% ethanol, and wipe to remove excess liquid.
- Mix all the reagents above mentioned with 500 ml of stellate cell medium (SteCM; ScienCell, cat. no. 5301).
- Because several components are light-sensitive, the medium should not be exposed to the light for extended periods.
- When stored in the dark at 4°C, the reconstituted complete medium is stable for 1 month.
- Warm stellate cell complete medium in a 37°C water bath prior to use.
Tris-buffered saline and Tween 20 (TBST) wash buffer
- Tris-buffered saline (TBS) buffer (Axil Scientific, cat. no. BUF-3030-20X-4L)
- with 0.1% (v/v) Tween-20 (Bio-Rad, cat. no. 170-6531).
- Store at room temperature (20° to 25°C) for up to 1 month.
For western blotting.
Wash buffer
- 0.25% (w/v) BSA (MilliporeSigma, cat. no. A7906) and 0.1% (v/v) Tween-20 (Bio-Rad, cat. no. 170-6531) in DPBS.
- Store for up to 1 month at 4°C (for immunofluorescence staining).
Store at room temperature (20° to 25°C) for up to 1 month (for western blotting).
COMMENTARY
Background Information
2D cell cultures are the most common form of in vitro experimental systems as they are typically the easiest to handle and scale. This makes them suitable for a wide range of applications including high-throughout target discovery screenings or in-depth studies of cytokine activity. However, growing cells on hard surfaces can induce mechanical stress, which has adverse effects on primary cells. 3D culture systems, and co-culture systems with several cell types, are viable alternatives. However, each approach comes with its own advantages and disadvantages. This series of protocols describes how to perform experiments with primary cells in 2D culture with minimal impact on cell physiology.
Critical Parameters
Several factors were found to influence the cellular response to IL11, including passage number and cell seeding density. IL11 was also found to have a different effect when cell and cytokine species were not matched. Human IL11 can activate mouse cells at higher concentrations (Schafer et al., 2017) but the underlying signaling pathway activation is different and likely non-physiological (Widjaja et al., 2019a). Furthermore, the cell type should be chosen based on the native expression levels of IL11 receptor alpha (IL11RA). IL11RA is highly expressed by primary parenchymal and stromal cells in the lung and liver but is not expressed by cells of the immune system, which predominantly express the IL6 receptor (IL6R). Immortalized cells usually do not express the IL11 receptor, or at very low levels, and are therefore not suitable for IL11 experiments.
Troubleshooting
When an experiment is not showing the expected results, we recommend that the cellular phenotype of the primary cells is carefully assessed. The expression levels of IL11RA protein on the cell surface should be confirmed. Genes that are up or downregulated in both highly passaged HSCs or HLFs (Fig. 4E) may be suitable RNA-based markers for cell dedifferentiation ex vivo. In addition, the activity of recombinant IL11 must be confirmed and species-matched cytokines should be used in all experiments.
Understanding Results
For anticipated results, refer to Figures 1, 2, and 3 (early passage).
Time Considerations
Stromal cells can be expanded before experiments, which results in an overall duration of ∼14 days from the initial thawing of cells to the last day of cell culture (Fig. 5). Hepatocytes are used in vitro without prior passaging, which shortens the duration of experiments but severely limits the amount of cells available for downstream experiments (Fig. 6).
Acknowledgments
This work was funded and supported by the Open Fund - Young Individual Research Grant (OFYIRG18nov-0014), Duke-NUS-GCR/2018/0017A Grant and the Academic Clinical Programme Charles Toh Cardiovascular Fellowship Award to S. Schafer, NMRC/OFYIRG/0053/2017 to A. Widjaja, and the National Medical Research Council (NMRC) Singapore STaR awards (NMRC/STaR/0011/2012 and NMRC/STaR/0029/2017), the NMRC Centre Grant to the NHCS, Goh Foundation, Tanoto Foundation, and a grant from the Fondation Leducq to S. A. Cook.
Author Contributions
Sivakumar Viswanathan : Conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing original draft, writing review and editing; Benjamin Ng : Conceptualization, data curation, methodology, visualization, writing original draft, writing review and editing; Anissa Widjaja : Conceptualization, data curation, formal analysis, investigation, methodology, writing original draft, writing review and editing; Chee Jian Pua : Investigation; Nevin Tham : Investigation; Jessie Tan : Investigation; Stuart Cook : Conceptualization, resources, supervision, writing review and editing; Sebastian Schaefer : Conceptualization, formal analysis, investigation, project administration, supervision, visualization, writing original draft, writing review and editing.
Conflict of Interest
SAC, SS, AAW, BN are co-inventors on a number of patent applications relating to the role of IL11 in human diseases: WO2017103108, WO2017103108 A2, WO 2018/109174 A2, WO 2018/109170 A2. SAC and SS are co-founders and shareholders of Enleofen Bio PTE LTD, a company that develops anti-IL11 therapeutics, which was acquired for further development by Boehringer Ingelheim.
Open Research
Data Availability Statement
All data generated and analyzed in the current study are presented in the article or available from the corresponding author upon request.
Literature Cited
- Baron, V., Adamson, E. D., Calogero, A., Ragona, G., & Mercola, D. (2006). The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53, and fibronectin. Cancer Gene Therapy , 13, 115–124. doi: 10.1038/sj.cgt.7700896.
- Blancher, C., & Jones, A. (2001). SDS-PAGE and Western blotting techniques. In S. A. Brooks & U. Schumacher (Eds), Metastasis Research Protocols. Methods in Molecular Medicine (Vol. 57, pp. 145–162). Totowa, NJ: Humana Press. doi: 10.1385/1-59259-136-1:145.
- Cook, S. A., & Schafer, S. (2020). Hiding in plain sight: Interleukin-11 emerges as a master regulator of fibrosis, tissue integrity, and stromal inflammation. Annual Review of Medicine , 71, 263–276. doi: 10.1146/annurev-med-041818-011649.
- Dong, J., Viswanathan, S., Adami, E., Singh, B. K., Chothani, S. P., Ng, B., … Widjaja, A. A. (2021). Hepatocyte-specific IL11 cis-signaling drives lipotoxicity and underlies the transition from NAFLD to NASH. Nature Communications , 12, 66. doi: 10.1038/s41467-020-20303-z.
- Korotkevich, G., Sukhov, V., & Sergushichev, A. (2021). Fast gene set enrichment analysis. bioRxiv. 060012. doi: 10.1101/060012.
- Lokau, J., Nitz, R., Agthe, M., Monhasery, N., Aparicio-Siegmund, S., Schumacher, N., … Garbers, C. (2016). Proteolytic cleavage governs Interleukin-11 Trans-signaling. Cell Reports , 14, 1761–1773. doi: 10.1016/j.celrep.2016.01.053.
- Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology , 15, 550. doi: 10.1186/s13059-014-0550-8.
- Martínez-García, A., Sastre, I., Recuero, M., Aldudo, J., Vilella, E., Mateo, I., … Bullido, M. J. (2010). PLA2G3, a gene involved in oxidative stress induced death, is associated with Alzheimer's disease. Journal of Alzheimer's Disease , 22, 1181–1187. doi: 10.3233/JAD-2010-101348.
- Migone, T. S., Zhang, J., Luo, X., Zhuang, L., Chen, C., Hu, B., … Wei, P. (2002). TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity , 16, 479–492. doi: 10.1016/S1074-7613(02)00283-2.
- Ng, B., Dong, J., D'Agostino, G., Viswanathan, S., Widjaja, A. A., Lim, W.-W., … Schafer, S. (2019). Interleukin-11 is a therapeutic target in idiopathic pulmonary fibrosis. Science Translational Medicine , 11, eaaw1237. doi: 10.1126/scitranslmed.aaw1237.
- Schafer, S., Viswanathan, S., Widjaja, A. A., Lim, W.-W., Moreno-Moral, A., DeLaughter, D. M., … Cook, S. A. (2017). IL-11 is a crucial determinant of cardiovascular fibrosis. Nature , 552, 110–115. doi: 10.1038/nature24676.
- Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., … Mesirov, J. P. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America , 102, 15545–15550. doi: 10.1073/pnas.0506580102.
- Su, G. H. (2005). Pancreatic cancer: Methods and protocols. Heidelberg, Germany: Springer Science & Business Media.
- Widjaja, A. A., Dong, J., Adami, E., Viswanathan, S., & Ng, B. (2019a). Redefining Interleukin 11 as a regeneration-limiting hepatotoxin. bioRxiv. doi: 10.1101/830018.
- Widjaja, A. A., Singh, B. K., Adami, E., Viswanathan, S., Dong, J., D'Agostino, G. A., … Cook, S. A. (2019b). Inhibiting interleukin 11 signaling reduces hepatocyte death and liver fibrosis, inflammation, and steatosis in mouse models of nonalcoholic steatohepatitis. Gastroenterology , 157, 777–792.e14. doi: 10.1053/j.gastro.2019.05.002.
Citing Literature
Number of times cited according to CrossRef: 5
- Puyan Rafii, Christiane Seibel, Hendrik T. Weitz, Julia Ettich, Anna Rita Minafra, Patrick Petzsch, Alexander Lang, Doreen M. Floss, Kristina Behnke, Karl Köhrer, Jens M. Moll, Jürgen Scheller, Cytokimera GIL-11 rescued IL-6R deficient mice from partial hepatectomy-induced death by signaling via non-natural gp130:LIFR:IL-11R complexes, Communications Biology, 10.1038/s42003-023-04768-4, 6 , 1, (2023).
- Tongtong Song, Yiwen Gu, Wenting Hui, Xiaoyu Yang, Yanqing Liu, Xia Chen, Oxygen–Glucose Deprivation Promoted Fibroblast Senescence and Collagen Expression via IL11, International Journal of Molecular Sciences, 10.3390/ijms232012090, 23 , 20, (12090), (2022).
- Benjamin Ng, Sivakumar Viswanathan, Anissa A. Widjaja, Wei-Wen Lim, Shamini G. Shekeran, Joyce Wei Ting Goh, Jessie Tan, Fathima Kuthubudeen, Sze Yun Lim, Chen Xie, Sebastian Schafer, Eleonora Adami, Stuart A. Cook, IL11 Activates Pancreatic Stellate Cells and Causes Pancreatic Inflammation, Fibrosis and Atrophy in a Mouse Model of Pancreatitis, International Journal of Molecular Sciences, 10.3390/ijms23073549, 23 , 7, (3549), (2022).
- Pengsen Wu, Bingying Lin, Siyu Huang, Jie Meng, Fan Zhang, Min Zhou, Xiangqing Hei, Yu Ke, Huasheng Yang, Danping Huang, IL-11 Is Elevated and Drives the Profibrotic Phenotype Transition of Orbital Fibroblasts in Thyroid-Associated Ophthalmopathy, Frontiers in Endocrinology, 10.3389/fendo.2022.846106, 13 , (2022).
- Anissa A. Widjaja, Sivakumar Viswanathan, Joyce Goh Wei Ting, Jessie Tan, Shamini G. Shekeran, David Carling, Wei-Wen Lim, Stuart A. Cook, IL11 stimulates ERK/P90RSK to inhibit LKB1/AMPK and activate mTOR initiating a mesenchymal program in stromal, epithelial, and cancer cells, iScience, 10.1016/j.isci.2022.104806, 25 , 8, (104806), (2022).

