A Mouse Model for Eosinophilic Esophagitis (EoE)
Anish Dsilva, Anish Dsilva, Shmulik Avlas, Shmulik Avlas, Natalie Rhone, Natalie Rhone, Michal Itan, Michal Itan, Ariel Munitz, Ariel Munitz
atopic dermatitis (AD)
eosinophilic esophagitis (EoE)
eosinophils
esophagus
IL-13
IL-4
oxazolone (OXA)
Abstract
Eosinophilic esophagitis (EoE) is an emerging chronic T helper type 2 (Th2)-associated, allergic, and immune-mediated disease, characterized histologically by eosinophil-predominant mucosal inflammation and clinically by esophageal dysfunction. Over the past years, the prevalence of EoE has dramatically increased globally. Until recently, most studies of EoE focused on using human biopsies, which are also used for diagnostic purposes, or esophageal epithelial cell lines, which led to major advances in the understanding of EoE. Despite this, a robust mouse model that mimics human disease is still crucial for both understanding disease pathogenesis and as a preclinical model for testing future therapeutics. Herein, we describe a highly reproducible and robust model of EoE that can be performed using wild-type mice by ear sensitization with oxazolone (OXA) followed by intraesophageal challenges. Experimental EoE elicited by OXA mimics the main histopathological features of human EoE, including intraepithelial eosinophilia, epithelial and lamina propria thickening, basal cell hyperplasia, and fibrosis. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Induction of EoE in mice using oxazolone
Support Protocol 1 : Preparing the mouse esophagus for histological analysis
Support Protocol 2 : Assessment of epithelial and lamina propria thickness using H&E staining
Support Protocol 3 : Assessment of eosinophilic infiltration using anti-MBP and basal cell proliferation using anti-Ki-67 staining
Support Protocol 4 : Flow cytometry of mouse esophageal samples
Support Protocol 5 : ELISA on protein lysates of esophageal samples
INTRODUCTION
Eosinophilic esophagitis (EoE) is an emerging chronic T helper type 2 (Th2)-associated, allergic- and immune-mediated disease, characterized histologically by eosinophil-predominant mucosal inflammation and clinically by esophageal dysfunction (O'Shea et al., 2018). A significant number of people suffering from EoE have co-existent atopic diseases, such as atopic dermatitis, allergic rhinitis, and asthma. These patients likely have higher levels of peripheral blood eosinophils and circulating IgE levels (Mohammad et al., 2017). Since the original description of EoE (Attwood et al., 1993), its prevalence has dramatically increased (O'Shea et al., 2018) and is continuing to increase globally, including in the USA and Europe (Zifman et al., 2018). EoE has one of the lowest qualities of life, likely because of the restricted diet, chronic pain, relapsing nature, and need for recurrent endoscopies (O'Shea et al., 2018). Only one FDA-approved drug (Dupilumab) was approved for EoE in the USA (Al-Horani & Chiles, n.d.), whereas an orally dispersible steroid (budesonide) tablet is approved for use in Europe (Dohil et al., 2010).
Mouse models of human disease are an excellent tool for dissecting molecular and cellular mechanisms that govern disease pathogenesis. In addition, and considering that no mouse model can fully recapitulate human disease, they provide powerful tools to evaluate emerging therapies and molecular targets for disease. With the emergence of several biological therapeutic candidates in multiple allergic diseases including EoE (Salvati et al., 2022), establishing a reproducible and robust mouse model that can be used for identifying and testing new molecular targets in EoE is timely and important.
The experimental model of EoE described herein was developed using chronic administration of oxazolone (OXA), a common haptenizing agent that causes severe inflammation in mice characterized by the production of IL-4 and IL-13 (Boirivant et al., 1998). Recent studies have also shown OXA-like compounds to cause inflammation in the intestinal epithelial cells by modulating CD1d, iNKT cells, and Ahr pathways (Iyer et al., 2018). Using this chemically-induced model of EoE, we could recapitulate human EoE-like symptoms in the mouse esophagus including epithelial and lamina propria thickening, intraepithelial eosinophilic infiltration, basal cell hyper-proliferation, and fibrosis (Avlas et al., 2023). Furthermore, comparing the gene signature of experimental EoE to that of human disease revealed marked overlap of key type 2 cytokines/receptor and epithelial-related genes (Avlas et al., 2023). Thus, this experimental system serves as a unique and reliable model to study EoE and to evaluate future therapeutics that could ultimately be used in human trials.
This article describes an OXA-induced model of EoE using topical application of OXA followed by intraesophageal challenges of OXA to initiate EoE-like disease (Basic Protocol). Support Protocol 1 describes the preparation of the mouse esophagus for histological analysis and Support Protocol 2 describes how to perform and evaluate various pathologies, such as epithelial and lamina propria thickening using a simple H&E staining. Support Protocol 3 outlines immunohistochemical staining procedures for the identification of eosinophils and epithelial basal layer cell hyperplasia in the esophagus. Support Protocol 4 outlines the preparation of esophageal cells for flow cytometric analysis. Finally, Support Protocol 5 describes how to generate esophageal cell lysates, which enables the assessment of soluble factors in the tissue, such as cytokines and chemokines using ELISA.
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Institutional Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals.
Basic Protocol: INDUCTION OF EoE IN MICE USING OXAZOLONE
In this 35-day protocol, mice are skin-sensitized and challenged (5 challenges) with OXA, followed by intraesophageal challenges (8 challenges) using OXA to induce EoE.
Materials
-
Oxazolone (OXA) (Sigma, cat. no. E0753)
-
Acetone (Biolabs, Israel, cat. no. 000103052100)
-
Mice, C57BL/6J wild-type, 6 to 8 weeks old
-
Phosphate-buffered saline (PBS) (Sigma, cat. no. D8537)
-
IgE ELISA kit (BD, cat. no. 555248)
-
Olive oil, food grade, maximum acidity 0.5%
-
Ethanol, 96% (Yakev Hagalil, cat. no. 7290000484549)
-
2- to 20-µl pipette
-
Microcentrifuge tubes
-
Refrigerated centrifuge, 4°C
-
Plastic feeding tubes (Instech, cat. no. FTP-22-25)
-
25G needle
-
Vortex
-
1-ml syringe
Day 0 to 16: Allergic sensitization
1.Prepare 1% OXA solution in acetone.
Ear sensitization
2.On day 0, disperse 15 µl OXA on each side of each of the mouse ears using a 2- to 20-µl pipette (for a total of 60 µl per mouse). OXA can be administered on either one or two ears as shown in Video 1.
Support Protocol 1: PREPARING THE MOUSE ESOPHAGUS FOR HISTOLOGICAL ANALYSIS
There is no way to control the flow of OXA through the modified tube during the intraesophageal challenges. Thus, in initial experiments, it is recommended to divide the esophagus into four distinct segments and to assess the histopathology of each segment individually (see Fig. 1C).
Materials
-
Formaldehyde, 4% (Biolab, Israel, cat. no. 0010010445)
-
Paraplast plus (Leica, cat. no. 39602004)
-
Embedding cassette (BarNoar, Israel, cat. no. BN393YC)
-
Tissue processor (Leica, TP1020)
-
Embedding center (Leica, EG1160)
-
Microtome (Leica, RM2245)
-
Superfrost Plus adhesion microscope slides (Epredia, cat. no. J1800AMNZ)
1.Harvest the mouse esophagus and remove any connective tissue that is attached to it.
2.Place the esophagus in an embedding cassette and place the cassette in 4% formaldehyde for 24 to 48 hr.
3.Transfer the cassettes containing the fixed esophagus to a tissue processor for tissue dehydration and paraffin infusion.
4.Cut the esophagus into 0.5 cm pieces from the distal to the proximal end and embed each piece vertically into paraffin blocks as shown in Figure 1C.
5.Using a microtome, cut 5 µm sections of the esophageal blocks.
6.Float cut sections onto glass slides.
7.Dry the glass slides overnight at room temperature.
Support Protocol 2: ASSESSMENT OF EPITHELIAL AND LAMINA PROPRIA THICKNESS USING H&E STAINING
Experimental EoE recapitulates various histopathological features of human EoE including edema and thickening of the epithelial cell layer. These histopathological measurements can be determined by quantitative analysis of H&E-stained slides and graphically displayed as epithelial and lamina propria thickening.
Materials
-
Prepared mouse esophagus slides (see Support Protocol 1)
-
Harris hematoxylin (Sigma, cat. no. HH516)
-
Eosin Y (Leica, cat. no. 3801601E)
-
Microscope with 20× or 40× magnification
-
Aperio ImageScope (Leica)
-
Additional reagents and equipment for standard slide staining (Cardiff et al., 2014)
1.Stain the slides for H&E using a standard staining protocol.
2.Scan the slides using a microscope at 20× or 40× magnification.
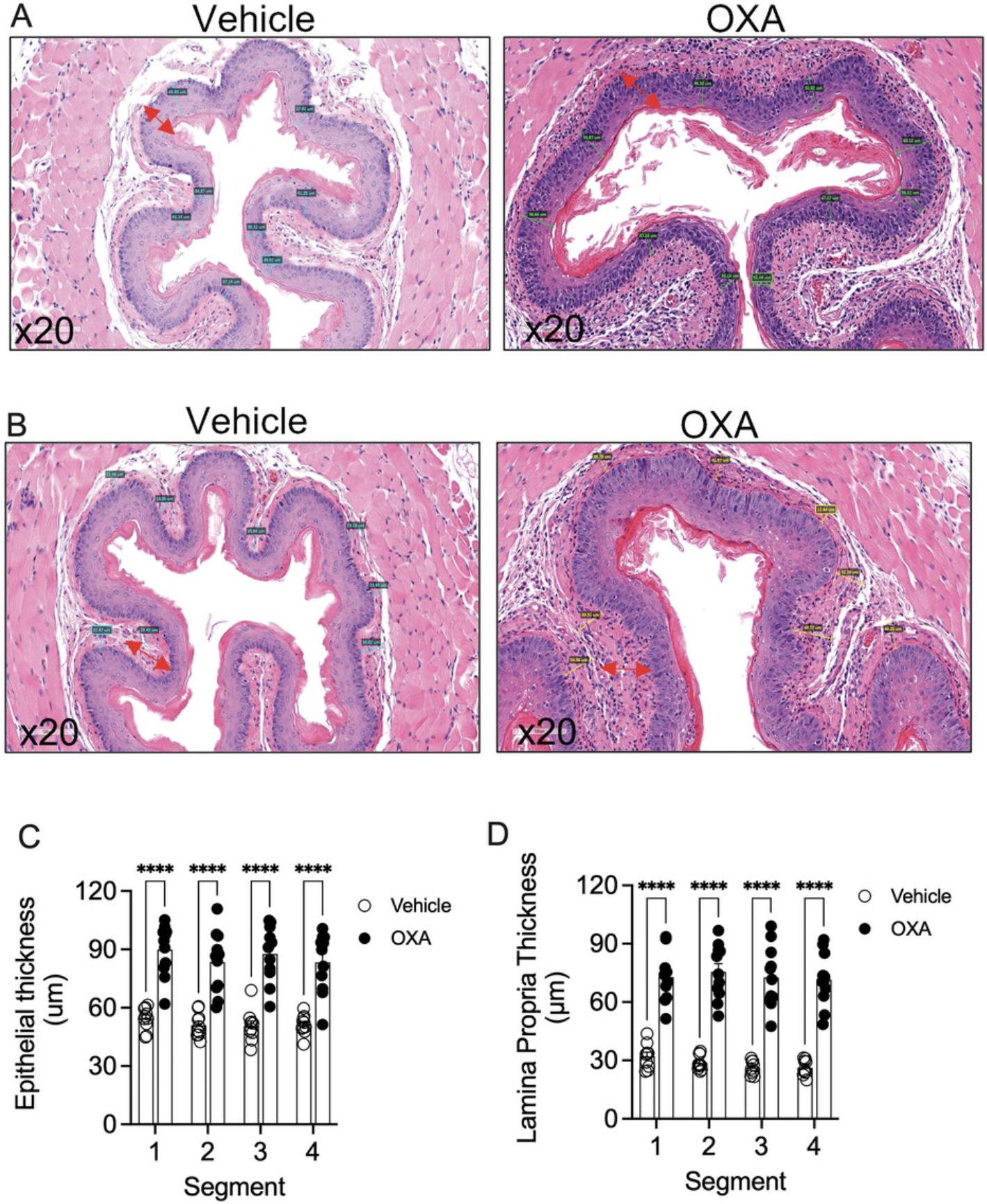
3.Measure epithelial thickness using the scale tool in the Aperio image scope software. Draw the scale from the keratin layer of the esophagus until the epithelial basal cell layer. Take an average of 8 measurements of epithelium for each segment. (Fig. 2A).
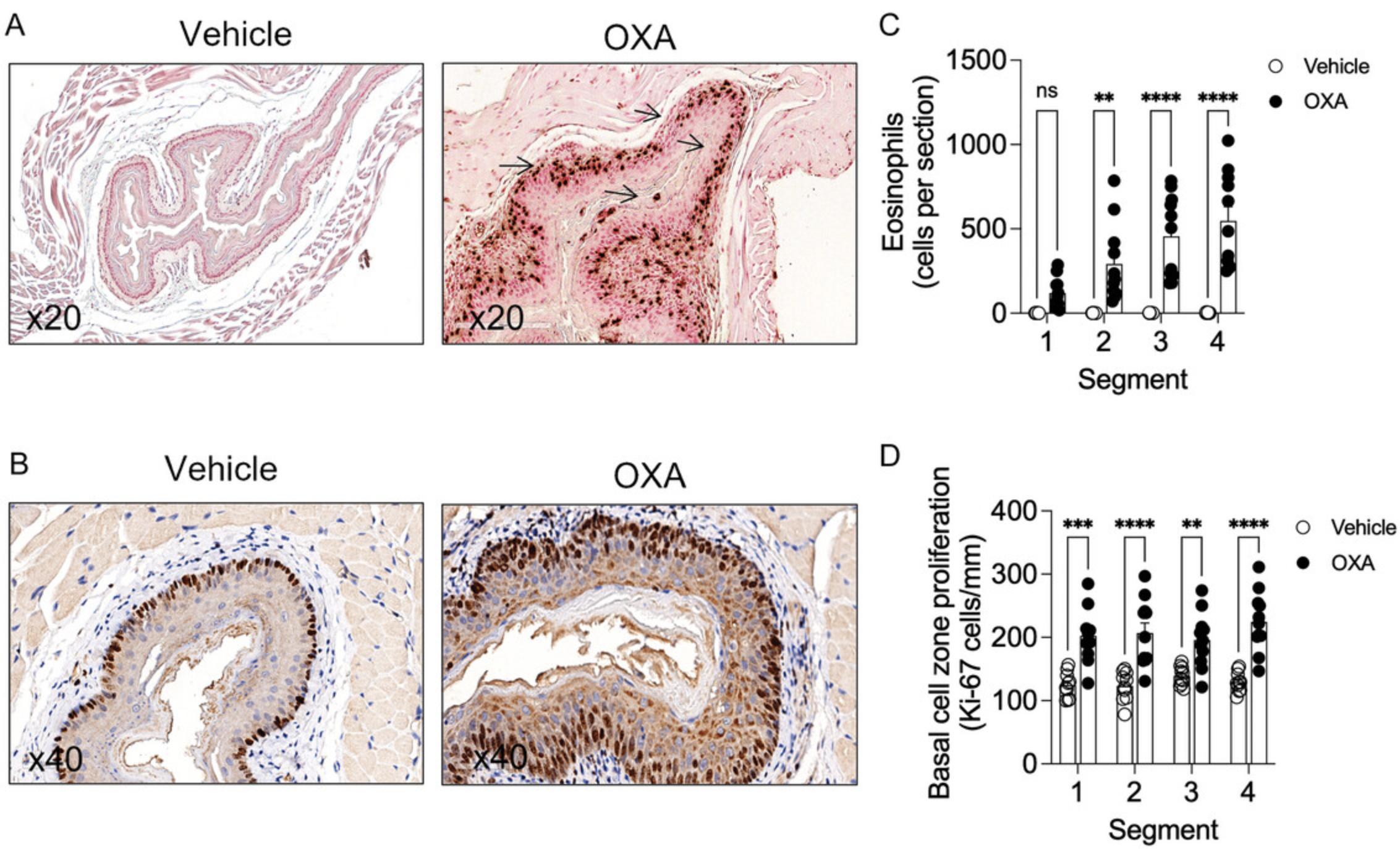
4.Measure lamina propria thickness using the scale tool in the Aperio image scope software. Draw the scale from the basal cell layer of the epithelium until the start of the muscle layer. Take an average of 8 different measurements for each segment (Fig. 2B)
5.Plot the data obtained from the averaged measurements. See Figure 2C and 2D.
Support Protocol 3: ASSESSMENT OF EOSINOPHILIC INFILTRATION USING ANTI-MBP AND BASAL CELL PROLIFERATION USING ANTI-Ki-67 STAINING
One of the striking features of experimental EoE is the ability to observe an influx of eosinophils into the epithelial layer (i.e., intraepithelial eosinophils). In addition, the proliferation of the basal cell layer of the esophageal epithelium is frequently observed. These characteristics are similar to findings in human EoE. Herein, a protocol for staining and quantitative analysis of eosinophil levels and epithelial cell proliferation is depicted.
Materials
-
Prepared mouse esophagus slides (see Support Protocol 1)
-
Xylene
-
Ethanol
-
PBS (Sigma, cat. no. D8537)
-
Quenching solution (see recipe)
-
Pepsin digest (Life technologies, cat. no. 003009)
-
Blocking solution: 3% normal goat serum (ThermoFisher, cat. no. 50062Z) in PBS
-
Rat anti-mouse MBP (provided by Dr. Elizabeth A. Jacobsen, Mayo Clinic, Scottsdale, AZ)
-
Rat anti-mouse Ki-67 (Novus, cat. no. NB500-170)
-
Biotinylated rabbit anti-rat (Vectastain, cat. no. BA-4000)
-
ABC reagent (Vectastain, cat. no. PK-4000)
-
DAB reagent (Vectastain, cat. no. SK-4100)
-
Harris hematoxylin (Sigma, cat. no. HH516)
-
H2O, distilled
-
Immunostaining tray
-
PAP pen (Daido, A-PAP PEN)
-
37°C incubator
-
4°C incubator
-
Clinical rotator
-
Coverslips
-
Microscope with 20× or 40× magnification
-
Aperio ImageScope (Leica)
-
Qpath (Queens University, Belfast)
1.Deparaffinize and rehydrate the tissue sections. Immerse the slides in each reagent specified below for 5 min in the following order:
-
Xylene.
-
Xylene.
-
Xylene.
-
100% ethanol.
-
100% ethanol.
-
95% ethanol.
-
95% ethanol.
-
70% ethanol.
2.Wash the slides in PBS for 5 min.
3.Incubate the slides in quenching solution for 20 min to remove endogenous peroxidase activity. Rinse the slides in PBS for 3 min.
4.Tap off excess liquid and place the slides in the immunostaining tray. Using a PAP pen creates a hydrophobic barrier around the tissue section.
5.Perform antigen retrieval by pipetting pepsin digest solution inside the tissue barrier. Incubate at 37°C for 10 min. Rinse the slides in PBS 3 times for 3 min.
6.For blocking, pipette blocking solution inside the tissue barrier, which was created by the PAP pen until it forms a liquid dome.
7.Incubate at room temperature for 2 hr.
8.Tap off the excess blocking solution from the slides.
9.Dilute primary antibody solution (rat anti-mouse MBP, 1:1000; rat anti-mouse Ki-67, 1:750) in blocking solution. Pipette the antibody solution into the tissue barrier. Incubate at 4°C overnight.
10.Rinse the slides in PBS 3 times for 3 min on a clinical rotator.
11.Dilute secondary antibody (biotinylated rabbit anti-rat, 1:250) in blocking solution. Pipette the antibody solution into the tissue barrier. Incubate at room temperature for 2 hr.
12.Rinse the slides in PBS 3 times for 3 min on a clinical rotator.
13.Apply ABC reagent to the tissue barrier. Incubate at room temperature for 45 min.
14.Rinse the slides in PBS 3 times for 3 min on a clinical rotator.
15.Prepare and add DAB solution into the tissue barrier. Incubate for 3 to 5 min until DAB develops. Rinse in PBS to stop the reaction.
16.Counterstain the slides in hematoxylin for 1 min.
17.Wash the tissues thoroughly in distilled water.
18.Add coverslips and observe under a microscope.
19.Scan the slides using a microscope at 20× or 40× magnification.
20.Count eosinophils and basal layer cells in the epithelium of the esophageal sections using Qpath cell detection software. Briefly, set vectors for DAB and hematoxylin colors in the Qpath software. Annotate the region of interest using the drawing tools, which are on the software.
21.Open the “analyze” bar and select for positive cell count option, which provides the number of cells in the annotated area.
22.Plot respective graphs using positive cell counts obtained from Qpath. See Figure 3.
Support Protocol 4: FLOW CYTOMETRY OF MOUSE ESOPHAGEAL SAMPLES
Induction of EoE in mice leads to the infiltration of several immune cells of myeloid and lymphoid origin into the esophagus. To determine and study these cell populations flow cytometry can be done on enzymatically digested and stained esophageal samples.
Materials
-
RPMI (Gibco, cat. no. 21875-034)
-
EDTA stripping buffer (see recipe)
-
PBS (Sigma, cat. no. D8537)
-
Collagenase A digestion buffer (see recipe)
-
HBA buffer (see recipe)
-
Antibodies:
- Anti-SiglecF PE (BD, cat. no. 552126)
- Anti-CD11b PerCP/Cy5.5 (eBioscience, cat. no. 45-0112-82)
- Anti-Ly6C PE/Cy7 (Biolegend, cat. no. 128018)
- Anti-CD45 APC (eBioscience, cat. no. 17-0451-82)
- Anti-Ly6G APC-Cy7 (Biogems, cat. no. 83112-87)
-
DAPI (Sigma, cat. no. 28718-90-3)
-
1.5-ml Eppendorf tube
-
Vortex
-
37°C incubator
-
Scissors (BarNoar, Israel, cat. no. BN11-440-09ASC)
-
50-ml tube
-
19G needle
-
70-μm cell strainer filter (Corning, cat. no. 431751)
-
Refrigerated centrifuge, 4°C
-
96-well cell culture plates, U-bottom (Greiner bio-one, cat. no. 650 180)
-
Refrigerated plate centrifuge, 4°C
-
5-ml polystyrene round-bottom tube (Corning, cat. no. 352008)
1.Excise the whole esophagus from the mouse (see Basic Protocol, step 11) and place it in a 1.5-ml Eppendorf tube containing RPMI for storage.
2.Carefully remove the RPMI from the tube using a pipette and add 1 ml pre-warmed (37°C) EDTA stripping buffer to the tube.
3.Incubate at 37°C for 10 min with vortexing every 2 to 3 min.
4.Remove the EDTA stripping buffer using a pipette and add 1 ml fresh EDTA stripping buffer to the tube.
5.Incubate the sample at 37°C for 5 min.
6.Vortex the sample for 10 s.
7.Remove the stripping buffer. Wash the tissue with 1 ml PBS by pipetting.
8.Remove the PBS solution and add 1 ml collagenase A buffer to the tube. Mince the tissue with a pair of scissors into small parts.
9.Incubate the sample at 37°C for 30 min, and vortex at high speed every 5 min.
10.Transfer the sample mixture into a 50-ml tube and dilute with 5 to 10 ml PBS.
11.Pass the sample through a 19G needle 3 to 4 times by sucking and releasing the sample.
12.Filter the sample using a 70-μm cell strainer.
13.Rinse the strainer once with PBS to allow the remaining tissue/cells to pass through.
14.Spin down the cells 5 min at 300 × g , 4°C, and carefully aspirate the supernatant.
15.Dilute the cells in an appropriate volume of HBA buffer.
16.Add 1 × 106 cells into the round-bottom 96-well plate.
17.Spin down the plate 2 min at 300 × g , 4°C.
18.Add 50 μl antibody mixture according to the manufacturer's instructions in HBA buffer to the wells and mix well by pipetting up and down.
19.Incubate in the dark for 30 min on ice.
20.Spin the plate 2 min at 300 × g , 4°C.
21.Resuspend the cells in an appropriate amount of HBA buffer with DAPI according to the manufacturer's instructions.
22.Transfer the cells to a 5-ml round-bottom polystyrene tube through a 70-μm cell strainer and proceed with flow analysis.
23.Refer to Figure 4 for the gating strategy of myeloid cell populations in the esophagus.
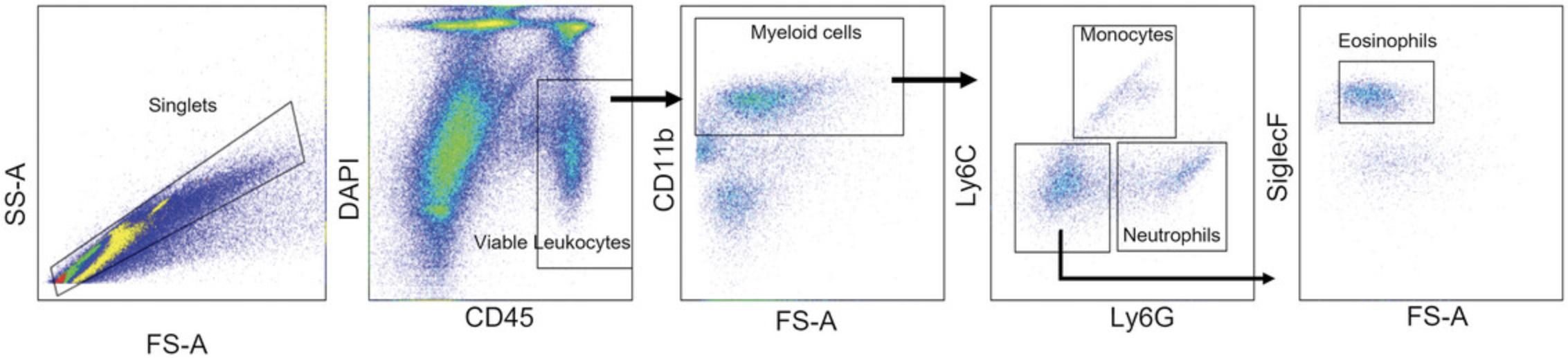
Support Protocol 5: ELISA ON PROTEIN LYSATES OF ESOPHAGEAL SAMPLES
EoE is associated with the expression of several chemokines and cytokines that play crucial roles in the progression of the disease. The protocol below can be utilized to obtain cell lysates from the esophagus tissue that could be used for ELISA.
Materials
-
PBS (Sigma, cat. no. D8537)
-
Protein lysis buffer (see recipe)
-
Kimwipes
-
Scale
-
5-ml polystyrene round-bottom tube (Corning, cat. no. 352008)
-
Tissue homogenizer (OMNI International, Tissue Master 125)
-
Ice
-
1.5-ml microcentrifuge tube
-
Refrigerated centrifuge, 4°C
-
Amicon 10K centrifugal filter (Millipore, cat. no. UFC501024)
1.Obtain the whole esophagus (see Basic Protocol, step 11) and rinse it with PBS, remove excess solution, and dry the esophagus using a Kimwipe.
2.Weigh the tissue.
3.Transfer the esophagus into a 5-ml round-bottom tube containing 750 μl protein lysis buffer.
4.Homogenize each esophagus using a tissue homogenizer starting at low speed and gradually increasing to high speed for 30 to 50 s.
5.Incubate the sample on ice for 5 min.
6.Transfer the homogenate into a new 1.5-ml tube and spin 10 min at 14,000 × g , 4°C.
7.Carefully pipette an equal volume of supernatant after the spin and transfer it into an Amicon 10K centrifugal filter.
8.Spin 25 min at 14,000 × g , 4°C, to concentrate the liquid.
9.Transfer the concentrated liquid into a new 1.5-ml tube.
10.Measure the volume of the concentrated liquid in each sample and normalize the volume of each sample using PBS.
11.Prepare aliquots of the protein lysate and store them at −20°C until use.
12.Perform ELISA using any basic kit according to the manufacturer's instructions. Normalize the results to the weight of each esophagus and multiply it by the concentration factor to quantify protein levels per ml/mg tissue.
13.Refer to Figure 5 to observe thymic stromal lymphopoietin (TSLP) quantification in the esophagus lysate by ELISA.
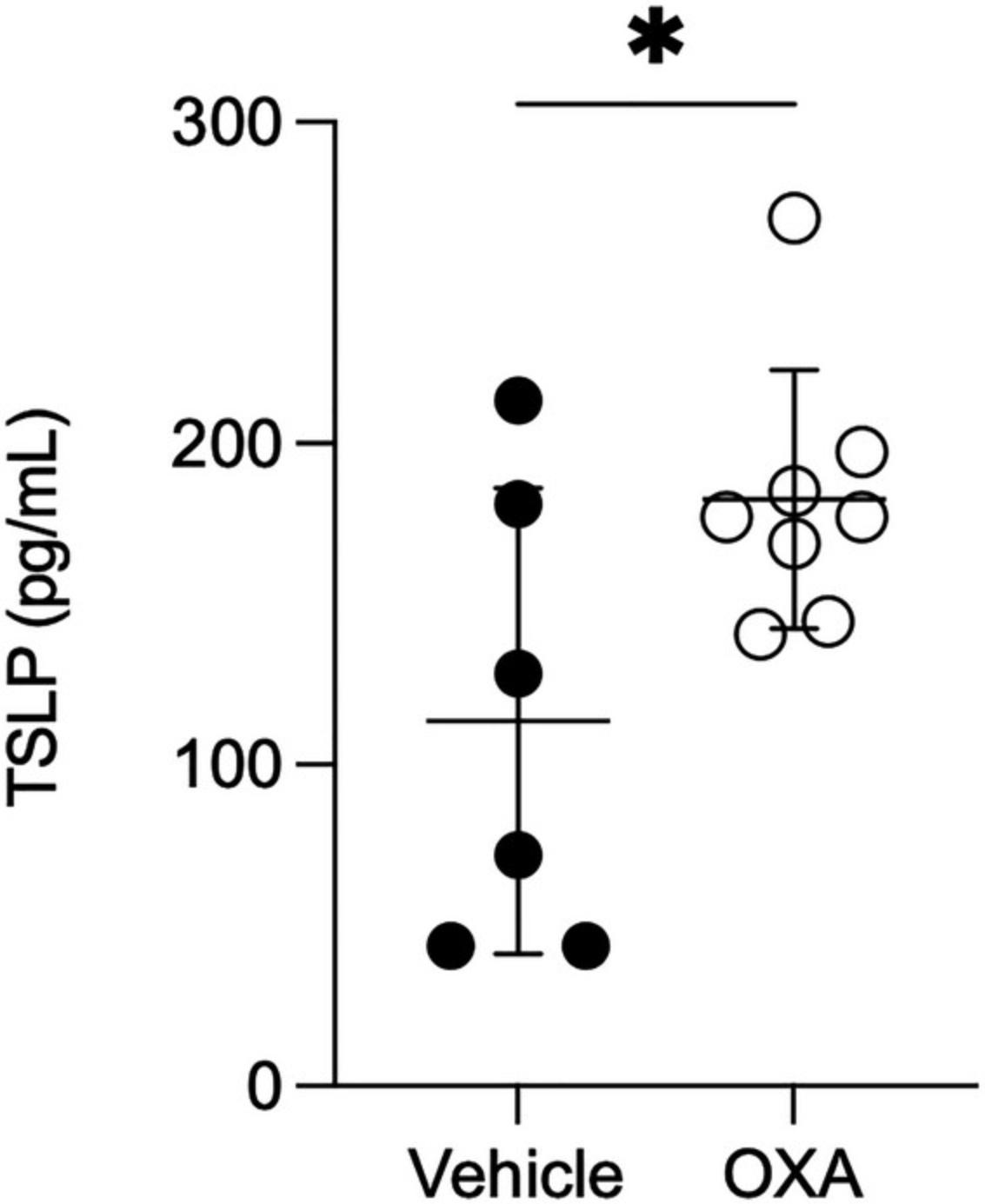
REAGENTS AND SOLUTIONS
Collagenase A digestion buffer
Dissolve 60 mg collagenase A (Roche, cat. no. 37170821) in 1.5 ml HBSS (Sartorius, cat. no. 02-018-1A) or PBS (Sigma, cat. no. D8537). Transfer 1.2 ml of collagenase solution into 18.8 ml RPMI (Gibco, cat. no. 21875-034). Prepare fresh and store at 4°C until use.
EDTA stripping buffer
- Mix in a sterile tube:
- 500 μl of 0.5 M EDTA (5 mM) (Biolab, Israel, cat. no. 009012230100)
- 1250 μl of 1 M HEPES (25 mM) (Sartorius, cat. no. 03-025-1B)
- 5 ml fetal calf serum (FCS) (10%)
- Bring up volume to 50 ml with HBSS (Sartorius, cat. no. 02-018-1A)
- Prepare fresh and store at 4°C until use
HBA buffer
- 500 mg bovine serum albumin (BSA) (0.1%) (Sigma-Aldrich, cat. no. A7030)
- 100 mg sodium azide (0.02%) (Sigma-Aldrich, cat. no. 71289)
- 500 ml HBSS (Sartorius, cat. no. 02-018-1A)
- Store up to 6 months at 4°C
Protein lysis buffer
Add 7.5 μl protease inhibitor (1:100) (Sigma, cat. no. P2714) per sample and 7.5 μl phosphatase inhibitor (Sigma, cat. no. S6508) to 750 μl Pierce protein lysis solution (Thermo Scientific, cat. no. 87787). Prepare fresh and store at 4°C until use.
Quenching solution
- 5 ml of 30% hydrogen peroxide (Bio Lab, cat. no. 000855032300)
- 70 ml of 99% methanol (Bio Lab, cat. no. 001368052100)
- Prepare fresh
COMMENTARY
Background Information
Animal models have been used in the study of various inflammatory diseases, such as asthma, inflammatory bowel disease, and colitis, and have helped in uncovering various pharmaceutical targets (Aun et al., 2017; Low et al., 2013). We have utilized the compound oxazolone (OXA) for inducing EoE. In this mouse model, OXA is a common haptenizing agent that causes severe acute inflammation in mice characterized by the production of IL-4 and IL-13 (Boirivant et al., 1998). Recent studies have also shown OXA-like compounds to cause inflammation in the intestinal epithelial cells by modulating CD1d, iNKT cells, and Ahr pathways (Iyer et al., 2018). This protocol has been developed as a robust model of EoE that can recapitulate human EoE-like conditions, such as epithelial and lamina propria layer thickening, basal cell hyperplasia, eosinophil infiltration, angiogenesis, and fibrosis.
Over the years various models of EoE have been developed: an aspergillus fumigatus-induced model, an ovalbumin-induced model, and a house dust mice-induced model (Akei et al., 2005; Brandt et al., 2006). IL-33 and TSLP-induced models of EoE are also under development using transgenic overexpression of these cytokines. The experimental EoE model described herein, provides a robust model that enables the study of EoE even in wild-type mice. Notably, this model closely recapitulates the symptoms seen in human EoE. Though the human and mouse esophagus differ, we have previously demonstrated that the transcriptome signature of experimental EoE overlaps with 25% of the genes upregulated in human EoE (Avlas et al., 2023). Using this model, we have also demonstrated a key role for the epithelial-expressed type II IL-4 receptor in EoE (Avlas et al., 2023)
Overall, using this model has the potential to uncover new cellular and molecular pathways that are involved in EoE pathogenesis. In addition, it could provide a potent tool to test and/or screen new molecular targets for EoE therapy.
Critical Parameters and Troubleshooting
Oxazolone dissolves easily in acetone but not in an ethanol and olive oil emulsion. To ensure proper dissolution, the emulsion needs to be vigorously mixed using a vortex for at least 10 to 25 min. It is essential to check for any undissolved OXA in the emulsion, as it could affect the experiment's accuracy.
During the intraesophageal challenge, precautions must be taken to avoid entering the airways of the mice. If the mice show any signs of discomfort, the feeding tube should be immediately withdrawn, and the challenge can be conducted again after allowing the mice some rest.
A syringe-fitted feeding tube may become difficult to use after 2 to 3 times. In such cases, the syringe can be changed every 2 to 3 uses. Occasionally, mice might bite the feeding tube, causing leaks that allow the emulsion to enter the air pipe. Therefore, the feeding tube should be checked before each challenge.
Following the 2nd intraesophageal challenge, the mice may appear weak due to the disease progression in the esophagus and gripping them might be challenging. In this situation, adjusting the angle of the feeding tube, preferably from the left side of the mouth, can help with a smoother passage. It is highly recommended to practice gavage using a flexible tube on the mice for at least a week before starting the experiment, as it requires more skill than using straightforward metal gavage tubes. After each intraesophageal challenge, the mice should be provided with wet food since swallowing hard food may be difficult due to the apparent disease in the esophagus.
Understanding Results
Application of OXA on the ear induces inflammation in the skin that is visible as early as after the 3rd skin challenge and becomes evident after the 5th challenge. Skin inflammation is noticeable in the form of skin lesions on the ears and scalp of mice; there is also visible hair loss around these regions. Following intraesophageal challenges, the mice become sick by the 2nd challenge. Indications, e.g., weight loss, shrunken pose, and lack of grooming, are quite visible. By the 3rd and 4th intraesophageal challenges, several mice (5% to 10%) die due to the disease; however, if the gavages are not performed properly the mortality could be 30% to 40%. By the 5th challenge, the mouse health stabilizes. Vehicle-treated mice show no signs of disease on intraesophageal challenges and any case of visible stress on the mice in this group can be attributed to errors in human handling procedures.
Time Considerations
This EoE model requires 35 days to complete, involving ear challenges until day 16 and intraesophageal challenges until day 35. The mice can be prepared to be euthanized on day 36. The process of preparing FFPE blocks and histological analysis takes ∼1 week to be completed, while esophageal cell lysate and cell suspensions are usually made on the same day.
In total, from the start of the model to the obtaining of slides for further analysis of the data, the process can take 42 to 45 days.
Acknowledgments
This work was supported by grants and fellowships to A.M. from the US-Israel Bi-national Science Foundation (grant no. 2015163), Israel Science Foundation (grants no. 542/20), Israel Cancer Research Fund (grant no. 1049171), Israel Cancer Association, Dotan Hemato Oncology Fund, Cancer Biology Research Center, Tel Aviv University, The Tel Aviv University Faculty of Medicine Recanati Fund, and Azrieli Foundation Canada-Israel.
Author Contributions
Dsilva Anish : Investigation; methodology; validation; visualization; writing original draft; writing review and editing. Avlas Shmulik : Conceptualization; formal analysis; investigation; methodology; validation. Rhone Natalie : Methodology. Itan Michal : Investigation; methodology. Munitz Ariel : Conceptualization; formal analysis; funding acquisition; investigation; methodology; project administration; supervision; validation; visualization; writing original draft; writing review and editing.
Conflict of Interest
Ariel Munitz serves on international advisory boards for GlaxoSmithKline, AstraZeneca, and Oravax, and received honorariums and grants from GlaxoSmithKline, AstraZeneca, Sanofi, and Sartorius. Other authors declare no conflict of interest.
Open Research
Data Availability Statement
All data will be shared upon request.
Literature Cited
- Akei, H. S., Mishra, A., Blanchard, C., & Rothenberg, M. E. (2005). Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology , 129(3), 985–994. https://doi.org/10.1053/J.GASTRO.2005.06.027
- Al-Horani, R. A., & Chiles, R. (n.d.). First Therapeutic Approval for Eosinophilic Esophagitis. https://doi.org/10.3390/gastroent13030024
- Attwood, S. E. A., Smyrk, T. C., Demeester, T. R., & Jones, J. B. (1993). Esophageal eosinophilia with dysphagia a distinct clinicopathologic syndrome. Digestive Diseases and Sciences , 38(1), 109–116. https://doi.org/10.1007/BF01296781
- Aun, M. V., Bonamichi-Santos, R., Arantes-Costa, F. M., Kalil, J., & Giavina-Bianchi, P. (2017). Animal models of asthma: Utility and limitations. Journal of Asthma and Allergy , 10, 293–301. https://doi.org/10.2147/JAA.S121092
- Avlas, S., Shani, G., Rhone, N., Itan, M., Dolitzky, A., Hazut, I., Grisaru-Tal, S., Gordon, Y., Shoda, T., Ballaban, A., Ben-Baruch, N. M., Rochman, M., Diesendruck, Y., Nahary, L., Bitton, A., Halpern, Z., Benhar, I., Varol, C., Rothenberg, M. E., & Munitz, A. (2023). Epithelial cell-expressed type II IL-4 receptor mediates eosinophilic esophagitis. Allergy , 78(2), 464–476. https://doi.org/10.1111/ALL.15510
- Boirivant, M., Fuss, I. J., Chu, A., & Strober, W. (1998). Oxazolone colitis: A murine model of T helper cell type 2 colitis treatable with antibodies to interleukin 4. The Journal of Experimental Medicine , 188(10), 1929. https://doi.org/10.1084/JEM.188.10.1929
- Brandt, E. B., Zimmermann, N., Muntel, E. E., Yamada, Y., Pope, S. M., Mishra, A., Hogan, S. P., & Rothenberg, M. E. (2006). The alpha4bbeta7-integrin is dynamically expressed on murine eosinophils and involved in eosinophil trafficking to the intestine. Clinical and Experimental Allergy : Journal of the British Society for Allergy and Clinical Immunology , 36(4), 543–553. https://doi.org/10.1111/J.1365-2222.2006.02456.X
- Cardiff, R. D., Miller, C. H., & Munn, R. J. (2014). Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harbor Protocols , 2014(6), 655–658. https://doi.org/10.1101/pdb.prot073411
- Dohil, R., Newbury, R., Fox, L., Bastian, J., & Aceves, S. (2010). Oral viscous budesonide is effective in children with eosinophilic esophagitis in a randomized, placebo-controlled trial. Gastroenterology , 139(2), 418–429.e1. https://doi.org/10.1053/J.GASTRO.2010.05.001
- Iyer, S. S., Gensollen, T., Gandhi, A., Oh, S. F., Neves, J. F., Collin, F., Lavin, R., Serra, C., Glickman, J., de Silva, P. S. A., Sartor, R. B., Besra, G., Hauser, R., Maxwell, A., Llebaria, A., & Blumberg, R. S. (2018). Dietary and microbial oxazoles induce intestinal inflammation by modulating aryl hydrocarbon receptor responses. Cell , 173(5), 1123. https://doi.org/10.1016/J.CELL.2018.04.037
- Low, D., Nguyen, D. D., & Mizoguchi, E. (2013). Animal models of ulcerative colitis and their application in drug research. Drug Design, Development and Therapy , 7, 1341–1356. https://doi.org/10.2147/DDDT.S40107
- Mohammad, A. A., Wu, S. Z., Ibrahim, O., Bena, J. M., Rizk, M., Piliang, M., & Bergfeld, W. F. (2017). Prevalence of atopic comorbidities in eosinophilic esophagitis: A case-control study of 449 patients. Journal of American Dermatology , 76, 559–560. https://doi.org/10.1016/j.jaad.2016.08.068
- O'Shea, K. M., Aceves, S. S., Dellon, E. S., Gupta, S. K., Spergel, J. M., Furuta, G. T., & Rothenberg, M. E. (2018). Pathophysiology of eosinophilic esophagitis. Gastroenterology , 154(2), 333–345. https://doi.org/10.1053/J.GASTRO.2017.06.065
- Salvati, L., Liotta, F., Annunziato, F., & Cosmi, L. (2022). Therapeutical Targets in Allergic Inflammation. In Biomedicines (Vol. 10, Issue 11). MDPI. https://doi.org/10.3390/biomedicines10112874
- Zifman, E., Banai, H., Shamir, R., Ringel-Kulka, T., & Zevit, N. (2018). Practice differences in the diagnosis and management of eosinophilic esophagitis among adult and pediatric gastroenterologists in Israel. Journal of Pediatric Gastroenterology and Nutrition , 67(1), 34–39. https://doi.org/10.1097/MPG.0000000000001909

