nanochromosome arrays combinatory assembly
Dariusz Abramczyk, Dariusz Abramczyk
Abstract
The yeast Pichia pastoris is used widely to biomanufacture high-value recombinant proteins. Its cells can secrete copious quantities of post-translationally modified proteins. Currently, however, heterologous genes must first be integrated into the genome in order to achieve expression.
We produced a synthetic 'chromosome-like' construct called here nanochromosome, an autonomously replicating and mitotically stable synthetic construct expressing heterologous genes. The nanochromosome contains essential scaffolding features as telomeres, centromere and yeast replication origins along with a versatile platform for genetic engineering. This platform could be used for the accurate and controlled insertion of multiple expression cassettes placed on 'landing pad', in which an array of genes of interest alternate with ~1kp non-coding DNA sequences (LHR) chosen to facilitate simultaneous double cross-overhomologous recombination and serve as spacers. The landing zone translates along the nanochromosome in an inchworming mode of sequential gene integrations that recycles a pair of antibiotic-resistance markers

Module with expression cassettes and LHR spacers as a pre-prepared in-vitro assembled DNA parts (insertion or integration arrays) is delivered to Pichia by transformation
Both the “integration” (receptor) and “insertion” (donor) arrays referred to herein consist of long, HR-ready, regions (LHRs) alternating with gene-expression cassettes. We constructed array parts containing either two or three LHRs e.g.LHRn-GoIn-AMRH/Z-LHRZ or LHRn-GoIn-LHRm-AMRH/Z-LHRZ, respectively, wherein: LHRn, LHRm and LHRZ are unique sequences of ~1000 bp from a library of LHRsA-Z, with LHRZ reserved as the last LHR in the array; GoIn is a cassette consisting of a gene-of-interest with promoter and terminator regions; and AMRZ/H is a zeocin-resistance gene or hygromycin-resistance gene also with promoter and terminator regions.
Preparation of ZeoR-LHRZ and HygR-LHRZ pair tandems in separate protocol https://www.protocols.io/view/preparation-of-parts-hygr-lhrz-and-zeor-lhrz-cqujvwun
The protocol for multi-DNA parts arrays preparation is based on combinatory assembly and molecular biology techniques customised to create a DNA parts (arrays) library in pUC19 plasmid.
Major 3 experimental steps:
Step 1 - DNA parts generated by PCR with a complementary overhangs designed to create a combined linear dsDNA suitable for insertion into BsmBI-sites of pUC19 (DNA arrays library)
-
a part generating by PCR, oligos deliver a complementary overhangs for digestion with BsmBI (Esp3I) or BsaI,
-
PCR product digestion with BsmBI or BsaI
-
checking digested PCR products on agarose gel
-
PCR clean up OR gel extraction of DNA and clean up (only in the case if a PCR product is not homogenous
-
estimation of DNA concentration (molar)
Note: (*) stands for AsiSI-FspI RE recognition site provides in Fd-oligo1 and RV-olig4

Step 2 - preparation of semi-products before the the final ligation with linearlized pUC adaptor plasmid
-
ligation part1+2 with T7 DNA ligase
-
ligation part 3+4 with T7 DNA ligase
-
loading ligation mixtures and separation on agarose gel,
-
gel extraction of estimated DNA band
-
agarose gel verification, DNA concentration (molar)

Step3 - the final ligation of pre-assembled parts with BsmBI-linearized pUC19
-
part1+2 and part 3+4 and linearized pUC19 ligation (DNA T7 ligase)
-
transformation into E.coli and selection on LB ampicillin plates
-
verification by bacterial colony PCR
-
plasmid isolation and Sanger (Plasmidsaurus) sequencing
-
positively verified clone storage in glycerol stock

The assembled part can be retained by:
-
PCR amplification using flanking oligonucleotides
-
Digested by AsiSI or FspI (need to be checked for the absence RE site in internal array) and digestion mixture can be used for Pichia transformation
Note: LHR part (e.g. LHRA) in flanking region (as a part 1) sometimes called 'OUT' while the LHRA in internal site (as a part 3) sometimes called 'IN'
Steps
Preparation of triple-LHR integration array LHRA-PAOXPDI-LHRE-HygR-LHRZ
DNA parts preparation by PCR reactions
List of DNA parts
| A | B | C | D | E |
|---|---|---|---|---|
| LHRA (OUT) BsmBI | F315-CCGTCTCCgggaGCGATCGCCTGTGTTAAACCGTCTTTAAGTCAACCC | R301-GCGTCTCCtgtcCAAACTAAGATTGGACTATTGCTATC | 883 bp | eDA8 |
| PAOX1PDIhis(BsmBI) | F302-CCGTCTCCgacaAACATCCAAAGACGAAAGGTTG | R303-GCGTCTCCctgaTCTCACTTAATCTTCTGTACTCTG | 2905 bp | eDA226* |
| LHRE (IN) (BsaI) | F336-CGGTCTCCtcagGACTTACCTCGTTTTAACTTAGTCGG | R350-GGGTCTCCctacACAAGTTTAACTAAGCGTCAGCC | 892 bp | eDA9 |
| HygR-LHRZ (BsaI) | F306-CCTGTCGACGGTCTCAgtagGACATGGAGGCCCAGAATACCC | R317-GGGGTACCGGTCTCAcgcgGCGATCGCCCTGCAGGTTGAGTTGGCGAAGGTGCG | 2583 bp | eDA115** |
- see 1.1 for prepartion (below)
**link https://www.protocols.io/view/preparation-of-parts-hygr-lhrz-and-zeor-lhrz-cqujvwun
Prestep - Generation of pPICZA with POAX1PDIhis - (eDA226)
Plasmid pPICZalphaPDI eDA143 (reference citation)
volumes in microliter
| A | B |
|---|---|
| water | up to 50 |
| Q5 HF enzyme | 0.7 |
| eDA143 100pg/uL | 1 |
| betaine 5M | 10 |
| oligo447/ oligo448 | 2.5/2.5 |
| 10mM dNTP (TAKARA) | 4 |
| 5xQ5 buffer | 10 |
| A | B | C | D |
|---|---|---|---|
| Td-initial | 98oC | 30sec | |
| Td | 98oC | 10sec | 33 cycles |
| Ta | 55oC | 20sec | |
| Te | 72oC | 1 min 40sec | |
| Final extension | 72oC | 2 min | |
| hold | 4oC | hold |
| A | B | C |
|---|---|---|
| F447 | PDI part (MfeI) | CCCCAATTGACAAGCTTTTGATTTTAACG |
| R448 | PDI part (NotI) introducing HIS-tag-TAA stop codon (italics) | TTTTTGCGGCCGCTTAATGATGATGGTGGTGATGCAACTCATCGTGAGCATCAGCTTC |
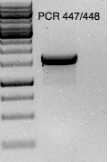
PCR clean-up
RE digested with MfeI/NotI
pPICZA Thermo Fisher vectors
| A | B | C |
|---|---|---|
| 1 | 2 | |
| pPICZA 100ng/uL | 13 | |
| PCR product of 447/448 | 48 | |
| MfeI-Hf (20u/ul) | 1.5 | 1 |
| NotI-Hf (20u/ul) | 1.5 | 1 |
| rcutsmart | 8 | 6 |
| water | up to 80 | up to 60 |
| after digestion additional dephosorylation reation | ||
| + 7ul AP buffer + 2uL Antarctic Phosphatase |
60 uL of pPICZA MfeI/NotI digested mixed with 7 ul AP buffer and 2 uL 0h 30m 0s
37°C
PCR product and pPICZA (MfeI/NotI) cleanup
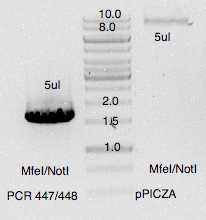
Ligation reactions T4 DNA ligase (NEB)
| A | B |
|---|---|
| pUC B/K BamHI/KpnI | 1 |
| T4 DNA ligase | 1 |
| Paox1PDI Mfe/NotI | 1 |
| 10 X T4 buffer | 6 |
| water | up to 10 -> |
| pPICZA MfeI/NotI | 1 |
E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB +100ug/mL 37°C
Several colonies obtained on both agar plates. Single colonies transferred on new LB +100ug/mL
see protocol colony PCR
Material loaded on agarose gel for verification. 5 colonies verified positively using oligo 24/241.
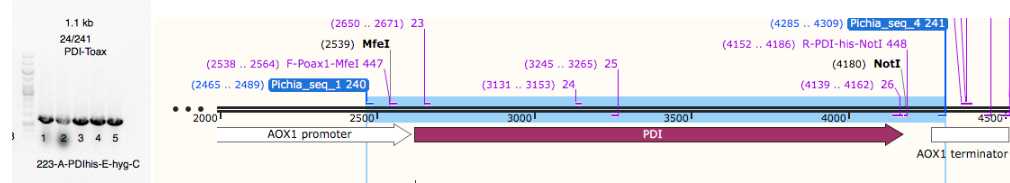
Positively verified clones re-cultured in LB + 100 ug/ml carbenicillin, plasmid eDA226 was extracted by miniprep (plasmid preparation)
STEP 1 - PCR amplification of parts and digestion with suitable IIS RE
Parts amplified by PCR (using ProFlex PCR system, Applied Biosystems). Thermal Cycler
volumes in microliter
see oligo tables step 1 (above)
| A | B | C | D | E |
|---|---|---|---|---|
| 5xQ5 buffer | 10 | 10 | 10 | 10 |
| betaine 5M | 10 | 10 | 10 | 10 |
| 10mM dNTP (TAKARA) | 4 | 4 | 4 | 4 |
| oligo 301/ oligo 315 | 2.5/2.5 | |||
| oligo 302/ oligo 303 | 2.5/2.5 | |||
| oligo 336/ oligo 350 | 2.5/2.5 | |||
| oligo 306/ oligo 317 | 2.5/2.5 | |||
| eDA8 100ng/ul | 1 | |||
| eDA226 (see 1.1) 100ng/ul | 1 | |||
| eDA9 100ng/ul | 1 | |||
| eDa115 100ng/ul | 1 | |||
| Q5 HF enzyme | 0.7 | 0.7 | 0.7 | 0.7 |
| water | up to 50 uL | up to 50 | up to 50 | up to 50 |
| A | B | C | D |
|---|---|---|---|
| Td-initial | 98oC | 30sec | |
| Td | 98oC | 10sec | 33 cycles |
| Ta | 58oC | 20sec | |
| Te | 72oC | 2min 30sec | |
| Final extension | 72oC | 1 min | |
| hold | 4oC | hold |
All PCR products purified by
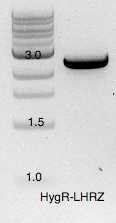
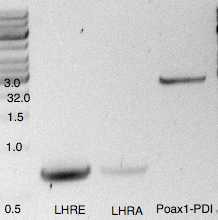
| A | B | C | D |
|---|---|---|---|
| size (bp) | concentration (ng/ul) | Molar concentration (nM) | |
| LHRA | 883 | 28 | 48 |
| Poax1PDI | 2905 | 64 | 34 |
| LHRE | 892 | 82 | 142 |
| HygR-LHRZ | 2583 | 101 | 60 |
Digestion with
| A | B | C | D | E |
|---|---|---|---|---|
| LHRA 315/301 | 46 | |||
| Paox1PDI 302/303 | 46 | |||
| LHRE 336/335 | 46 | |||
| HygR-LHRZ 306/317 | 46 | |||
| BsaI HF (20U/ul) | 2 | 2 | 2 | |
| Esp3I (10U/ul)) | 3 | |||
| buf10x cutsmart | 6 | 6 | 6 | 6 |
| water | up to 60uL | up to 60uL | up to 60uL | up to 60uL |
All PCR products purified by
DNA molar concentration calculation before ligation
| A | B | C | D |
|---|---|---|---|
| size (bp) | concentration (ng/ul) | Molar concentration (nM) | |
| LHRA | 883 | 28 | 48 |
| Poax1PDI | 2905 | 64 | 34 |
| LHRE | 892 | 82 | 142 |
| HygR-LHRZ | 2583 | 101 | 60 |
STEP 2 - preparation of semi-products, a partial ligated arrays
| A | B | C |
|---|---|---|
| expected size ~3.7kb | expected size ~3.47kb | |
| 2x T7 ligase buffer | 15 | 15 |
| HygR-LHRZ | - | 10 |
| LHRa | 5.7 | - |
| LHRE | - | 4 |
| Poax1PDI | 8.3 | - |
| T7 DNA ligase | 1 | 1 |
| water | - | - |
| ul | ul |
Ligation reaction
1h 0m 0s 25°C
All ligation mixtures loaded on 1% agarose gel and separated to excise a corresponding size band
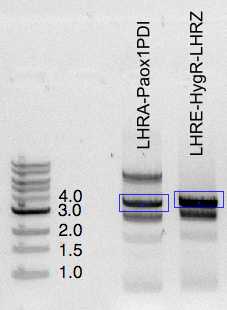
DNA bands representing expected size were excised and purified using
DNA concentration calculated by DeNovix DS-11
| A | B | C |
|---|---|---|
| ng/uL | Molar concentration (nM) | |
| LHRA-Paox1PDI | 10 | 4 |
| LHRE-HygR-LHRZ | 16 | 7 |
pUC19 digestion with Esp3I
Digestion with
| A | B |
|---|---|
| pUC19 500ng/uL | 5 uL |
| Esp3I | 2 ul |
| rCutsmart 10X | 4 ul |
| water | up to 40 uL |
Digestion mixture cleanup
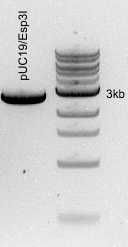
| A | B | C |
|---|---|---|
| ng/uL | Molar concentration (nM) | |
| pUC19/Esp3I | 35 | 19 |
STEP 3 - the final ligation of pre-assembled parts with Esp3I-linearized pUC19, E.coli transformation and colony PCR verification
| A | B | C |
|---|---|---|
| nM | ml | |
| T4 DNA ligase | 1 | |
| 10x T4 ligase buffer | 1.5 | |
| LHRA-Paox1PDI (4nM) | 4 | 7 |
| LHRE-HygR-LHRZ (7nM) | 7 | 4 |
| pUC19/Esp3I (19nM) | 19 | 1.5 |
| water | - |
16h 0m 0s 16°C
E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB +100ug/mL + 37°C
Several colonies obtained on both agar plates. Single colonies transferred on new LB agar plates +100 ug/mL carbanicillin + 150 ug/mL hygromycin B
Growth overnight and a single colonies (8 colonies) submitted for bacterial colony PCR.
see protocol colony PCR
Material loaded on agarose gel for verification. 2 of 8 colonies verified positively using oligo 24/241. (expected 1.1kb amplicon) and 42/394 (0.91kb amplicon).
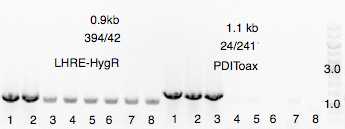
Colony 1 and 2 inoculated into LB + carbanicillin + hygromycin and growth
Plasmids isolated by plasmid isolation (miniprep) Qiagen miniprep 1h 0m 0s
Sanger sequencing confirmed the sequence of eDA227
Preparation of double-LHR integration array LHRE-PtefGFP-ZeoR-LHRZ
DNA parts preparation by PCR reactions LHRE+PtefGFP (1st pre-assembly ligation mixture)
List of DNA parts
| A | B | C | D | E |
|---|---|---|---|---|
| LHRE (OUT) (BsaI) | F335-CGGTCTCCgggaGCGATCGCGACTTACCTCGTTTTAACTTAGTCGG | R338-GGGTCTCCtgtcACAAGTTTAACTAAGCGTCAGCC | 900 bp | eDA9 |
| PtefGFP (BsmBI) | F302a-CCGTCTCCgacaAACATCCAAAGACGAAAGGTTG | R304a-CCGTCTCCctacGCCTTCGAGCGTCCC | 1442 bp | eDA189* |
| ZeoR-LHRZ (BsmBI) | F308-CCTGTCGACGGTCTCAgtagGACATGGAGGCCCAGAATACCC | R318-GGGGTACCGGTCTCAcgcgGCGATCGCCCTGCAGGTTGAGTTGGCGAAGGTGCG | 2171 bp | eDA105** |
- eDA189 map .dna file eda189.dna
**link https://www.protocols.io/view/preparation-of-parts-hygr-lhrz-and-zeor-lhrz-cqujvwun
STEP 1 - PCR amplification of parts LHRE(out)+PtefGFP and digestion with suitable IIS RE
Parts amplified by PCR (using ProFlex PCR system, Applied Biosystems). Thermal Cycler
volumes in microliter
| A | B | C | D |
|---|---|---|---|
| Q5 HF enzyme | 0.7 | 0.7 | 0.7 |
| 10mM dNTP (TAKARA) | 4 | 4 | 4 |
| water | up to 50 uL | up to 50 | up to 50 |
| oligo 302a/ oligo 304a | 2.5/2.5 | ||
| oligo 335/ oligo 338 | 2.5/2.5 | ||
| oligo 308/ oligo 318 | 2.5/2.5 | ||
| eDA189 100ng/ul | 1 | ||
| eDA9 100ng/ul | 1 | ||
| eDa105 100ng/ul | 1 |
| A | B | C | D |
|---|---|---|---|
| Td-initial | 98oC | 30sec | |
| Td | 98oC | 10sec | 33 cycles |
| Ta | 58oC | 20sec | |
| Te | 72oC | 2min 10sec | |
| Final extension | 72oC | 1 min | |
| hold | 4oC | hold |
All PCR products purified by

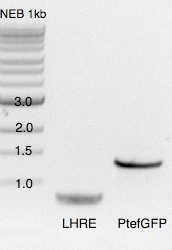
Digestion with
| A | B | C | D |
|---|---|---|---|
| LHRE 335/338 | 46 | ||
| PtefGFP 302a/304a | 46 | ||
| ZeoR-LHRZ 308/318 | 46 | ||
| BsaI HF (20U/ul) | 2 | ||
| Esp3I (10U/ul)) | 3 | 3 | |
| buf10x cutsmart | 6 | 6 | 6 |
| water | up to 60uL | up to 60uL | up to 60uL |
Reaction 37°C 6h 0m 0s
All PCR products purified by
DNA molar concentration calculation before ligation
| A | B | C | D |
|---|---|---|---|
| size (bp) | concentration (ng/ul) | Molar concentration (nM) | |
| LHRE out | 900 | 66 | 113 |
| PtefGFP | 1442 | 92 | 98 |
| ZeoR-LHRZ | 2171 | 111 | 78 |
STEP 2 - preparation of semi-product, a partial ligated array LHRE-PtefGFP
| A | B |
|---|---|
| expected size ~2.3kb | |
| 2x T7 ligase buffer | 15 |
| LHRE (out) | 6.5 |
| PtefGFP | 7.5 |
| T7 DNA ligase | 1 |
| water | - |
| ul |
Ligation reaction
1h 0m 0s 25°C
Ligation mixture loaded on 1% agarose gel and separated to excise a corresponding size band

DNA band representing expected size was excised and purified using
DNA concentration calculated by DeNovix DS-11
| A | B | C | D | E |
|---|---|---|---|---|
| size kbp | ng/uL | Molar concentration (nM) | ||
| LHRA-PtefGDP | ~2.3 | 55 | 37 | |
| ZeoR-LHRZ* | 2.17 | 101 | 72 |
STEP 3 - the final ligation of pre-assembled parts with Esp3I-linearized pUC19, E.coli transformation and colony PCR verification
| A | B | C |
|---|---|---|
| uL | nM | |
| T4 DNA ligase | 1 | |
| 10x T4 ligase buffer | 1 | |
| LHRE-PtefGFP | 2.3 | 37 |
| ZeoR-LHRZ | 1.2 | 78 |
| pUC19/Esp3I | 4.5 | 19 |
16h 0m 0s 16°C
E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB +100ug/mL carbanicillin + 50ug/mL zeocin
37°C
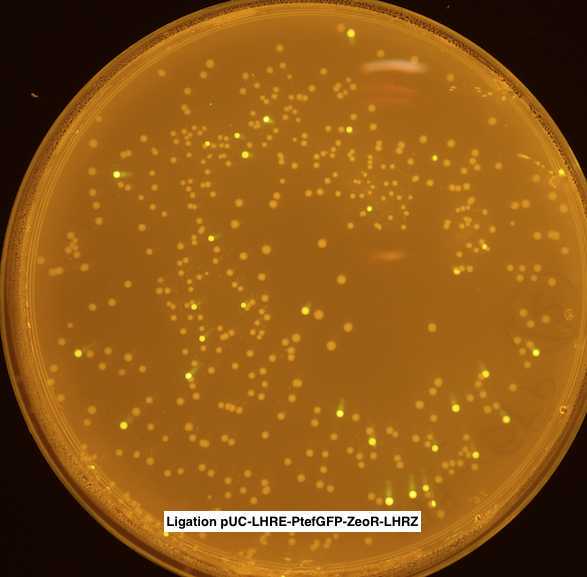
Several fluorescent colonies obtained on both agar plates. Single colonies transferred on new LB agar plates +100 ug/mL carbanicillin + 50 ug/mL zeocin
Single fluorescent colonies inoculated on fresh LB + carbanicillin/zeocin and growth
Next day a pasmid isolation (miniprep) Qiagen miniprep 1h 0m 0s
Sanger sequencing confirmed the sequence of eDA197
Preparation of double-LHR insertion array LHRE-PtefMCH-HygR-LHRZ
DNA parts preparation by PCR reactions
List of DNA parts
| A | B | C | D | E |
|---|---|---|---|---|
| DNA part | Forward oligo | Revers oligo | product | template |
| LHRE (OUT) (BsaI) | F335-CGGTCTCCgggaGCGATCGCGACTTACCTCGTTTTAACTTAGTCGG | R338-GGGTCTCCtgtcACAAGTTTAACTAAGCGTCAGCC | 900 bp | eDA9 |
| PtefMCh (BsmBI) | F302a-CCGTCTCCgacaAACATCCAAAGACGAAAGGTTG | R304a-CCGTCTCCctacGCCTTCGAGCGTCCC | 1442 bp | eDA191* |
| HygR-LHRZ (BsaI) | F306-CCTGTCGACGGTCTCAgtagGACATGGAGGCCCAGAATACCC | R317-GGGGTACCGGTCTCAcgcgGCGATCGCCCTGCAGGTTGAGTTGGCGAAGGTGCG | 2583 bp | eDA115** |
- eDA191 map .dna file eda191.dna
**link https://www.protocols.io/view/preparation-of-parts-hygr-lhrz-and-zeor-lhrz-cqujvwun
STEP 1 - PCR amplification of parts
LHRE(out) (done in section 2, step 2.1) with oligos 335/338
HygR-LHRZ (done in section 1, step 1.2) with oligos 306/317
PtefMCh amplification and digestion with suitable IIS RE
Parts amplified by PCR (using ProFlex PCR system, Applied Biosystems). Thermal Cycler
volumes in microliter
| A | B |
|---|---|
| Q5 HF enzyme | 0.7 |
| 10mM dNTP (TAKARA) | 4 |
| water | up to 50 |
| oligo 302a/ oligo 304a | 2.5/2.5 |
| eDA191 100ng/ul | 1 |
volume in microliters
| A | B | C | D |
|---|---|---|---|
| Td-initial | 98oC | 30sec | |
| Td | 98oC | 10sec | 33 cycles |
| Ta | 58oC | 20sec | |
| Te | 72oC | 2min 10sec | |
| Final extension | 72oC | 1 min | |
| hold | 4oC | hold |
PCR product purified by
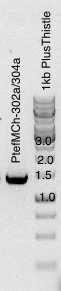
Digestion with
| A | B |
|---|---|
| Esp3I (10U/ul)) | 3 |
| PtefMCh 302a/304a | 46 |
| buf10x cutsmart | 6 |
| water | up to 60uL |
Reaction 37°C 6h 0m 0s
All PCR products purified by
DNA molar concentration calculation before ligation
| A | B | C | D |
|---|---|---|---|
| size (bp) | concentration (ng/ul) | Molar concentration (nM) | |
| LHRE out | 900 | 66 | 113 |
| PtefMCh | 1442 | 92 | 98 |
| HygR-LHRZ | 2583 | 101 | 60 |
STEP 2 - preparation of semi-product, a partial ligated array LHRE-PtefMCh
| A | B |
|---|---|
| ul | |
| expected size ~2.3kb | |
| PtefGFP | 7.5 |
| LHRE (out) | 6.5 |
| 2x T7 ligase buffer | 15 |
| T7 DNA ligase | 1 |
| water | - |
Ligation reaction (in duplicate)
1h 0m 0s 25°C
Ligation mixture loaded on 1% agarose gel and separated to excise a corresponding size band
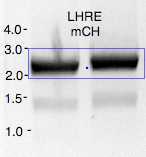
DNA band representing expected size was excised and purified using
DNA concentration calculated by DeNovix DS-11
| A | B | C | D |
|---|---|---|---|
| size kbp | ng/uL | Molar concentration (nM) | |
| LHRE-PtefMCH | ~2.3 | 96 | 64 |
| HygR-LHRZ* | 2.5 | 101 | 72 |
- HygR-LHRZ from step 1.2
STEP 3 - the final ligation of pre-assembled parts with Esp3I-linearized pUC19, E.coli transformation and colony PCR verification
| A | B | C |
|---|---|---|
| uL | nM | |
| T4 DNA ligase | 1 | |
| 10x T4 ligase buffer | 1 | |
| LHRE-PtefMCH | 1.6 | 64 |
| ZeoR-LHRZ | 1.4 | 72 |
| pUC19/Esp3I | 5 | 19 |
16h 0m 0s 16°C
E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB +100ug/mL carbanicillin + 150ug/mL hygromycin B
growth at 37°C
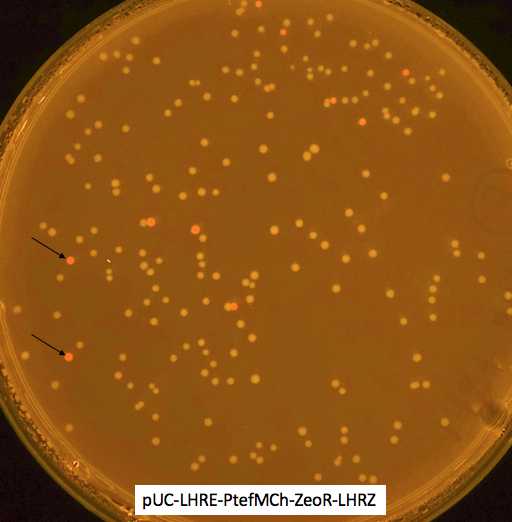
Several fluorescent colonies obtained on both LB agar plates. Single colonies transferred on new LB agar plates +100 ug/mL carbanicillin + 50 ug/mL zeocin
Single fluorescent colonies inoculated on fresh LB + carbanicillin/hygromycin and growth
Next day a pasmid isolation (miniprep) Qiagen miniprep 1h 0m 0s
Sanger sequencing confirmed the sequence of eDA199
Preparation of triple-LHR insertion array LHRA-GFP:CFH-LHRE-ZeoR-LHRZ
DNA parts preparation by PCR reactions
List of DNA parts
| A | B | C | D | E |
|---|---|---|---|---|
| LHRE (OUT) (BsaI) | F335-CGGTCTCCgggaGCGATCGCGACTTACCTCGTTTTAACTTAGTCGG | R338-GGGTCTCCtgtcACAAGTTTAACTAAGCGTCAGCC | 900 bp | eDA9 |
| PAOX1GFP:CFH (BsmBI) | F302-CCGTCTCCgacaAACATCCAAAGACGAAAGGTTG | R303-GCGTCTCCctgaTCTCACTTAATCTTCTGTACTCTG | 5991bp | eDA89* |
| LHRD (IN) (BsmBI) | F304-CCGTCTCCtcagGACAGCAACCTAACCGAC | R305-GCGTCTCCctacCAGTCCTCGTGAAAGACGAG | 1105 bp | eDA9 |
| ZeoR-LHRZ (BsmBI) | F308-CCTGTCGACGGTCTCAgtagGACATGGAGGCCCAGAATACCC | R318-GGGGTACCGGTCTCAcgcgGCGATCGCCCTGCAGGTTGAGTTGGCGAAGGTGCG | 2171 bp | eDA105** |
- map eDA89.dna plasmid prepared as described in the paper (DOI.....PBarlow,DAbramczyk)
**link https://www.protocols.io/view/preparation-of-parts-hygr-lhrz-and-zeor-lhrz-cqujvwun
STEP 1 - PCR amplification of parts and digestion with suitable IIS RE
Parts amplified by PCR (using ProFlex PCR system, Applied Biosystems). Thermal Cycler
volumes in microliters
| A | B | C | D | E |
|---|---|---|---|---|
| 5xQ5 buffer | 10 | 10 | 10 | 10 |
| betaine 5M | 10 | 10 | 10 | 10 |
| 10mM dNTP (TAKARA) | 4 | 4 | 4 | 4 |
| oligo 335/ oligo 338 | 2.5/2.5 | |||
| oligo 302/ oligo 303 | 2.5/2.5 | |||
| oligo 304/ oligo 305 | 2.5/2.5 | |||
| oligo 308/ oligo 318 | 2.5/2.5 | |||
| eDA89 100ng/uL | 1 | |||
| eDA9 100ng/ul | 1 | 1 | ||
| eDa105 100ng/ul | 1 | |||
| Q5 HF enzyme | 0.7 | 0.7 | 0.7 | 0.7 |
| water | up to 50 uL | up to 50 | up to 50 | up to 50 |
Part LHRE (out) generated as described above in protocol 2.1
Part ZeoR-LHRZ generated as described above in protocol 2.1
| A | B | C | D |
|---|---|---|---|
| Td-initial | 98oC | 30sec | |
| Td | 98oC | 10sec | 33 cycles |
| Ta | 58oC | 20sec | |
| Te | 72oC | *1min 10 sec (for LHRD) ** 5min 30 sec for Poax1GFP:CFH | |
| Final extension | 72oC | 1 min | |
| hold | 4oC | hold |
All PCR products purified by
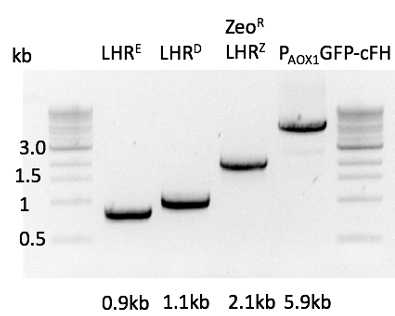
Digestion with
| A | B | C | D | E |
|---|---|---|---|---|
| LHRE 335/338 | 46 | |||
| PaoxGFP:CFH 302/303 | 46 | |||
| LHRD 304/305 | 46 | |||
| ZeoR-LHRZ 308/318 | 46 | |||
| BsaI HF (20U/ul) | 2 | |||
| Esp3I (10U/ul)) | 3 | 3 | 3 | |
| buf10x cutsmart | 6 | 6 | 6 | 6 |
| water | up to 60uL | up to 60uL | up to 60uL | up to 60uL |
All PCR products purified by
DNA molar concentration calculation before ligati
| A | B | C | D |
|---|---|---|---|
| size (bp) | concentration (ng/ul) | Molar concentration (nM) | |
| LHRE out | 900 | 66 | 48 |
| PaoxGFPCFH | 5991 | 123 | 34 |
| LHRD | 1105 | 120 | 142 |
| ZeoR-LHRZ | 2171 | 111 | 78 |
STEP 2 - preparation of semi-products, a partial ligated arrays
| A | B | C |
|---|---|---|
| ZeoR-LHRZ | - | 9 |
| LHRD | - | 5 |
| water | - | - |
| T7 DNA ligase | 1 | 1 |
| 2x T7 ligase buffer | 15 | 15 |
| LHRE out | 5.8 | - |
| PoaxGFP:CFH | 8.2 | - |
| expected size ~6.9kb | expected size ~3.2kb | |
| ul | ul |
Ligation reaction
1h 0m 0s 25°C
All ligation mixtures loaded on 1% agarose gel and separated to excise a corresponding size band
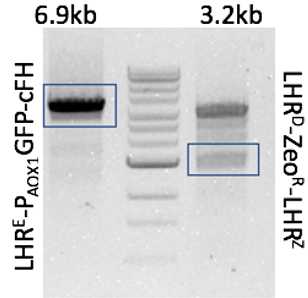
DNA band representing expected size was excised and purified using
DNA concentration calculated by DeNovix DS-11
| A | B | C |
|---|---|---|
| ng/uL | Molar concentration (nM) | |
| LHRE-PaoxGFPCFH | 22 | 5 |
| LHRD-ZeoR-LHRZ | 9 | 4 |
STEP 3 - the final ligation of pre-assembled parts with Esp3I-linearized pUC19, E.coli transformation and colony PCR verification
| A | B | C |
|---|---|---|
| nM | ml | |
| T4 DNA ligase | 1 | |
| 10x T4 ligase buffer | 1.5 | |
| LHRE-PaoxGFPCFH | 5 | 5.0 |
| LHRD-ZeoR-LHRZ | 4 | 6.2 |
| pUC19/Esp3I (19nM) | 19 | 1.3 |
| water |
16h 0m 0s 16°C
E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB +100ug/mL + 50ug/mL
and growth at 37°C
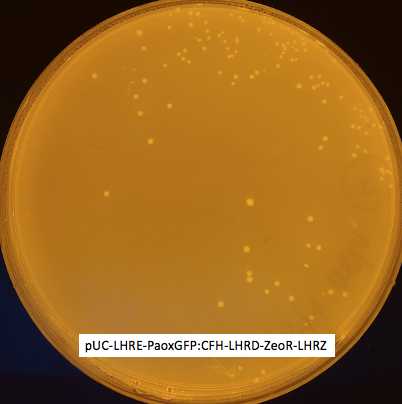
Several colonies obtained on both agar plates. Single colonies transferred on new LB agar plates +100 ug/mL carbanicillin + 50 ug/mL zeocin B.
Two of them submitted for bacterial colony PCR.
see protocol colony PCR
| A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|
| 352/353 | 274/278 | 355/354 | 345/40 | 351/133 | 374/370 | ||
| puc-E 0.45kb | GFP-FH 0.75kb | Aox-D 0.29kb | D-zeo 0.9kb | zeoc-C 0.6kb | 0.7kb contr+ | ||
| 1 | clone1 (eDA250) | OK | OK | OK | OK | OK | OK |
| 2 | clone2 | none | none | poor | none | none | none |
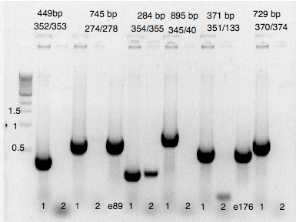
Material loaded on agarose gel for verification. 1 of 2 colonies verified positively verified
The colony inoculated on fresh LB + carbanicillin/zeocin and growth
Next day a pasmid isolation (miniprep) Qiagen miniprep 1h 0m 0s
Sanger sequencing confirmed the sequence of eDA250
Plasmidsaurus sequencing confirms the correct sequence (added to .dna file)

