cermep-bids-retro
lucie.chalet
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
cermep-bids-retro enables the formatting of retrospective PET and MRI data to BIDS standards from DICOM databases. When dealing with pre-clinical imaging modalities, numerous metadata are overlooked during acquisitions making it difficult to automatically format following BIDS guidelines. This python-based project is a flexible method for BIDS formatting with input csv files to compensate for lacking DICOM tags. And advanced data quality assessment.
Steps
cermep-bids-retro installation
Clone repository
In GitBash:
git clone https://gitlab.in2p3.fr/cermep/cermep-bids-retro.git
Create conda environment
cd cermep-bids-retro
conda env create -f environment.yml
conda activate bids-retro
Fill-in input files
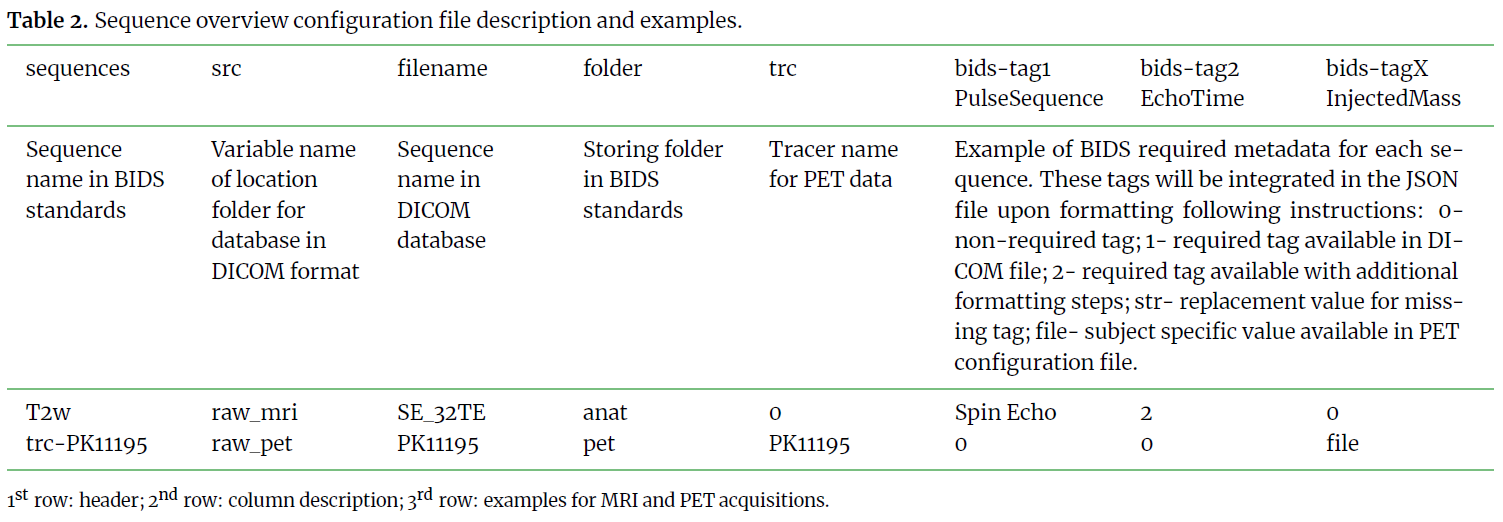
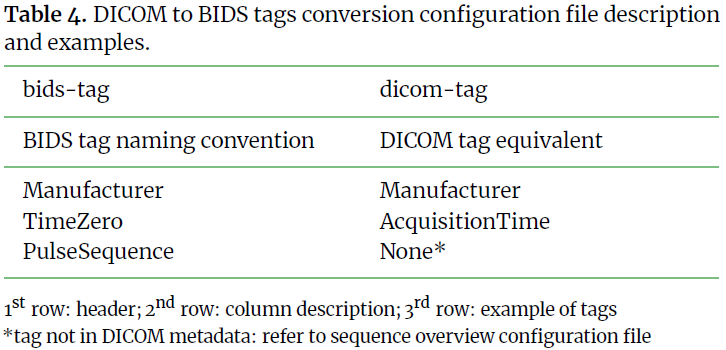
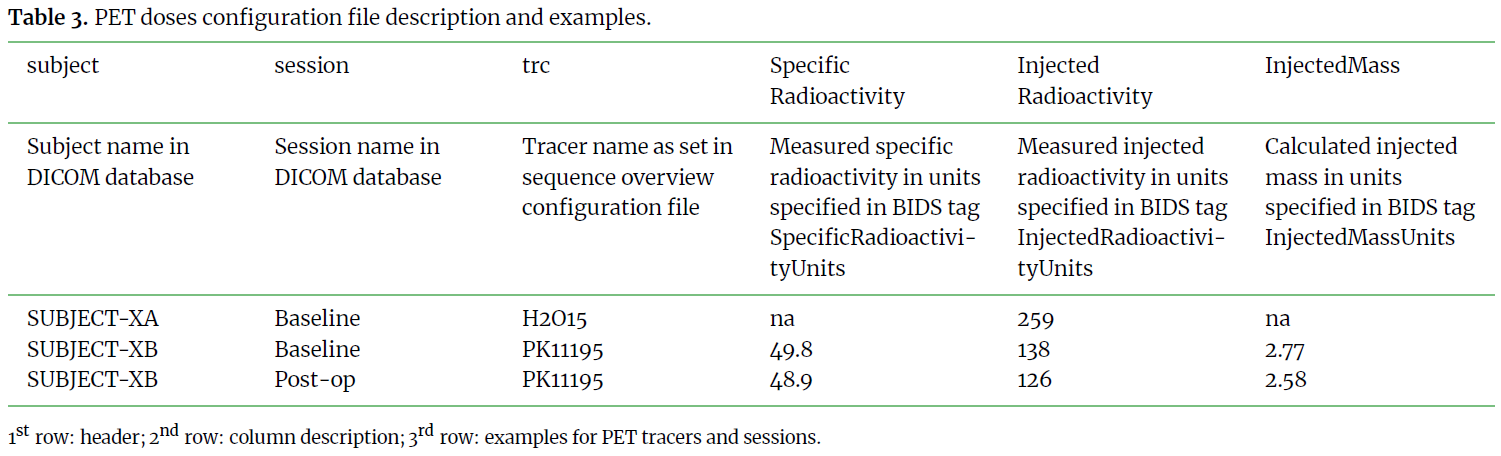
This step is key to a functional method. It requires a good knowledge
of the available tags and fields in the DICOM data and one can always refer
to the BIDS specifications to define all required tags for their modality.
Substeps below provide description and example for each column of the 3 input files.
Run formatting task
Replace argument values and run task from command line:
python bids_retro\\main.py \
--src_mri "path\to\modality" \
--src_pet "E:\PhD\temp\data_fabien\raw_dcm" \
--sequences_overview "inputs\seq_overview.csv" \
--pet_doses "inputs\pet_doses.csv" \
--dcm2bids_tags "inputs\dcm2bids_tags.csv" \
--species "name of species" \
--src_structure "A" \
--in_sub_dcm_list "subjectA,subjectB" \
--in_ses_dcm_list "session" \
--dest "path\to\bids\formatted\destination" \
--quality_testing 1 \
--results "path\to\save\quality_analysis\results" \
--include_dicom 0 --verbose 1