Working with Worms: Caenorhabditis elegans as a Model Organism
Philip M. Meneely, Philip M. Meneely, Caroline L. Dahlberg, Caroline L. Dahlberg, Jacqueline K. Rose, Jacqueline K. Rose
Abstract
Since its introduction as a laboratory organism 50 years ago, the nematode worm Caenorhabditis elegans has become one of the most widely used and versatile models for nearly all aspects of biological and genomic research. Many experiments in C. elegans begin with the generation and analysis of mutants that affect a specific biological process, so genetic techniques are the foundation of worm research. Many different aspects of biology are being studied in C. elegans , and three different recent Nobel Prizes have recognized six researchers working with worms. In addition, C. elegans was the first multicellular organism to have its genome sequenced, so many of the standard genomic methods have also been pioneered in C. elegans. In fact, many novel techniques and ideas are initially tested in C. elegans because of its versatility as a research organism. It is also appropriate for introducing undergraduate students to research, and some of its strengths and challenges for this purpose are discussed. © 2019 The Authors.
INTRODUCTION
The nematode worm Caenorhabditis elegans has become one of the most widely used model organisms for nearly every aspect of biology. Among the features that make C. elegans an appealing and effective model organism are that it is easy to work with in the lab, with minimal nutritional and growth requirements, that it produces a large number of offspring by self-fertilization within a few days, and that it has been thoroughly studied. It was also the first genome from a multicellular organism to be sequenced. Just as significantly, the community of researchers working with worms is highly interactive and collaborative, which makes it attractive to newcomers to the field, including undergraduate researchers. We provide a general background to working with worms, including a selective list of some of the more significant research results so far and of the many current research topics being studied in worms; we also include some general laboratory recipes and handling tips from our own experiences with beginning research students. Most importantly, links and literature that will allow a researcher to learn about different topics in C. elegans research are introduced and discussed.
OVERVIEW, HISTORY, AND PRINCIPLES
The nematode worm Caenorhabditis elegans was intentionally chosen for research based on its properties as a laboratory organism. The choice can be precisely dated, and the early years of C. elegans research have been very well documented (Brenner, 2009; Corsi, Wightman, & Chalfie, 2015, 2018; White, 2018). In 1963, the eminent molecular biologist Sydney Brenner, working at the Medical Research Council in Cambridge, England, decided to turn his research attention from bacteria and phage to an animal. Brenner believed in the power of genetic analysis, that is, in the use of mutations that disrupt a biological process as a tool to understand the normal process. Brenner, along with others, had used genetic analysis in demonstrating that the genetic code consists of non-overlapping triplets of nucleotides with stop codons, and in investigating the role of mRNA and the circuitry of the lac operon. Thus, when looking for an animal to serve as a model organism for eukaryotic processes, particularly for behavior and neurobiology, he carefully considered and tested a number of different organisms in his laboratory before deciding to work with C. elegans , familiarly referred to as “the worm.”
C. elegans is a free-living (that is, non-parasitic) nematode found around the world (Brusca, 2016). Some relevant information about worms is summarized as “fast facts” in Table 1. Adult animals are approximately 1 mm long, so all genetic experiments can be done by examining the worms with a dissecting microscope using sub-stage illumination, as shown in Figure 1. As we will discuss below, the natural history of C. elegans is much less studied than is the case for other model organisms, and its ecological significance is largely unknown, though nematodes are found in terrestrial and aquatic ecosystems (Brusca, 2016; Kiontke et al., 2011). Its primary importance has been as a domesticated, laboratory species, and laboratory strains differ from wild strains in a variety of ways, just as laboratory mice differ from mice in the wild. The virtues of C. elegans for genetic analysis include that it is easily grown in the laboratory on agar plates, that it eats a standard strain of non-pathogenic E. coli , that it reproduces rapidly to produce approximately 300 offspring within a reproductive cycle of only 3.5 days at room temperature, and that it is capable of both internal self-fertilization and cross-fertilization.
| Length (adult hermaphrodite) | ∼1 mm |
| Sexes | Male (XO), hermaphrodite (XX) |
| Chromosomes | Five autosomes (I, II, III, IV, V), one sex chromosome (X) |
| Life cycle (fertilized egg to adult) | ∼3 days, at 20°C |
| Life span | ∼2.5 weeks |
| Laboratory food source | E. coli (OP-50, OP-51, HB 101) |
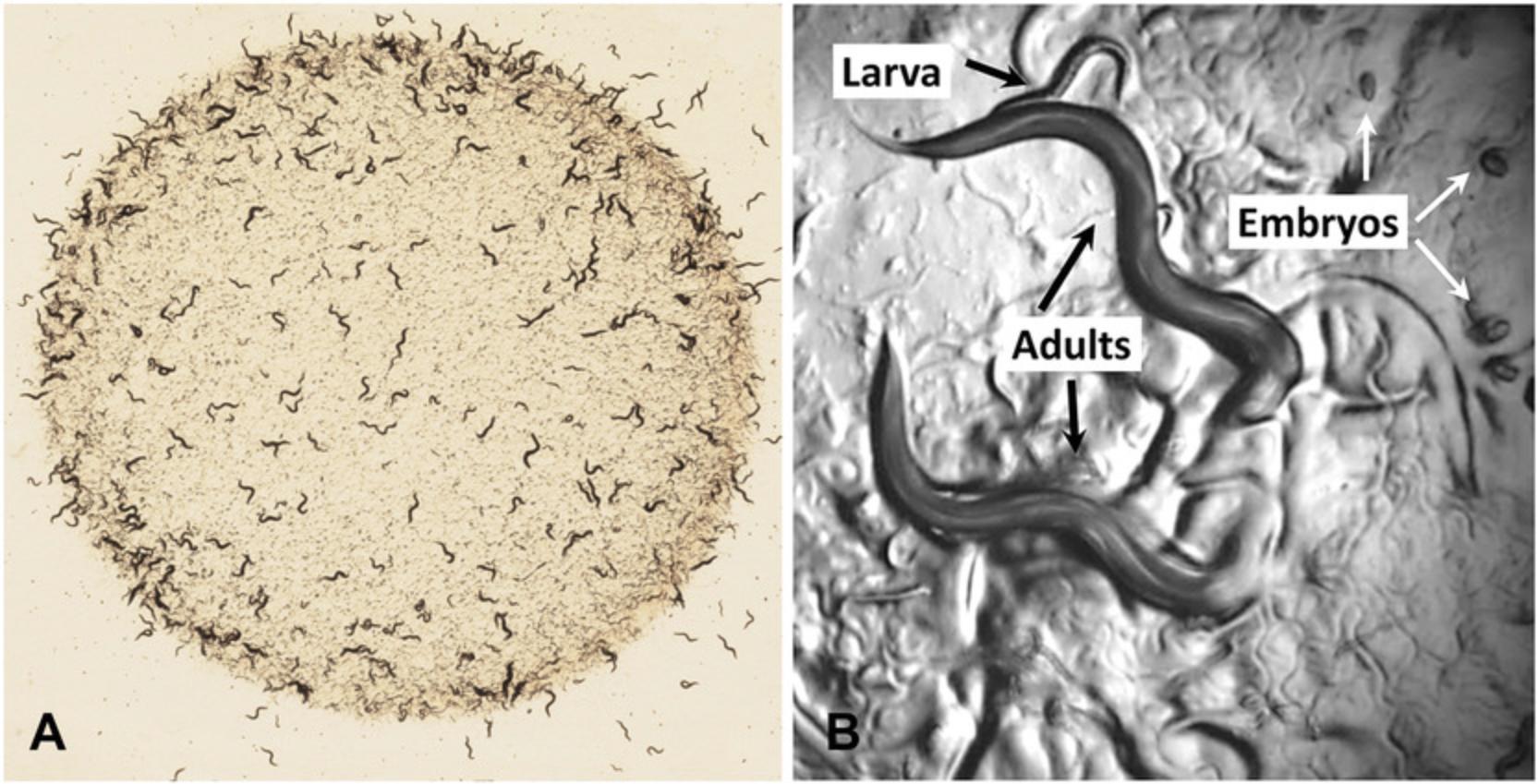
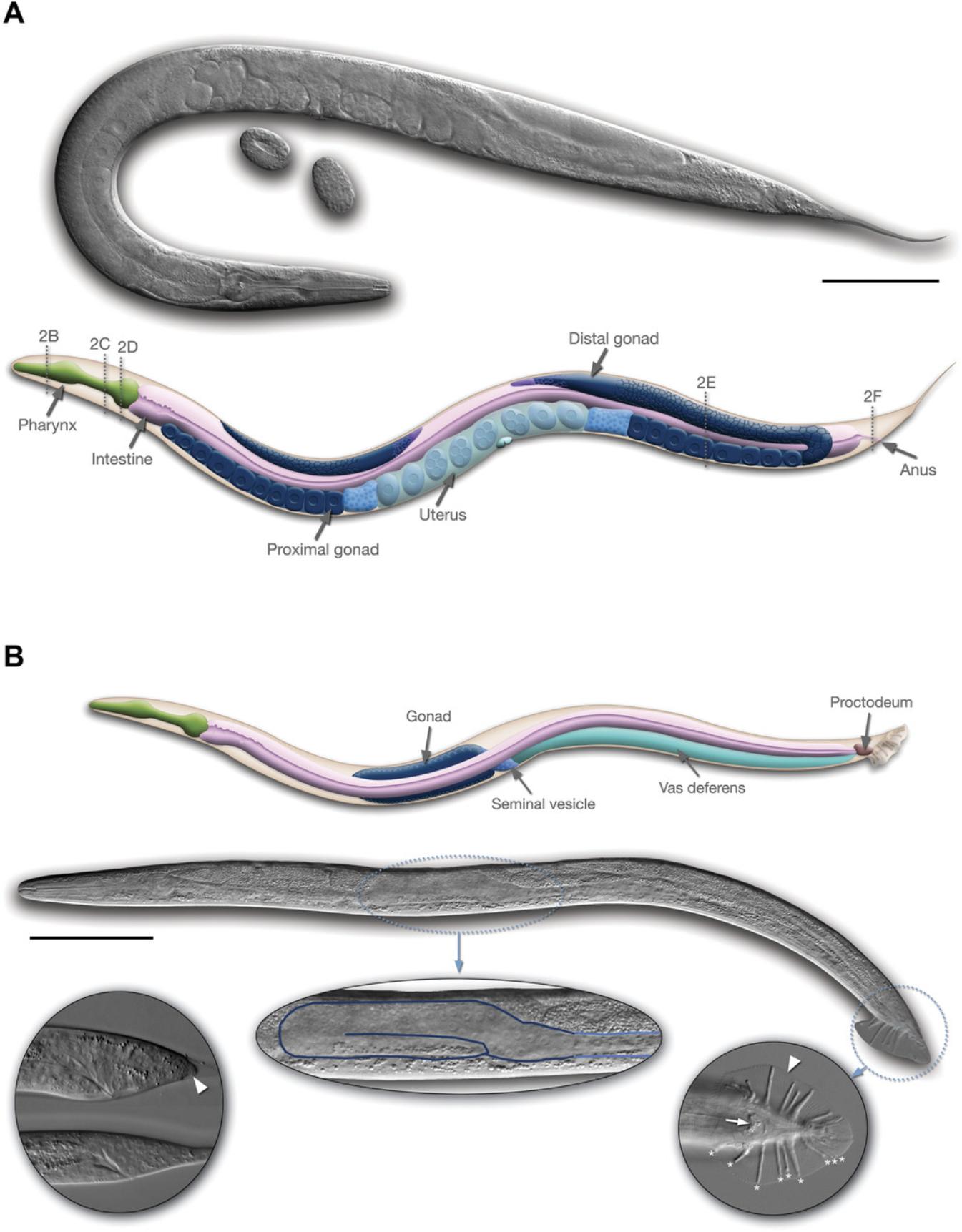
This last characteristic is particularly important for genetic research with worms. C. elegans has two sexes, males with a single X chromosome and hermaphrodites with two X chromosomes (Fig. 2). Related species of nematode such as C. briggsae have males and non-hermaphroditic females; the females of these species closely resemble the hermaphrodites of C. elegans in overall morphology, so C. elegans hermaphrodites can be thought of as females that make some sperm. Indeed, when meiosis begins in the germline, the first 300 or so gametes produced are tail-less sperm that use multi-directional crawling to move around the gonad; after sperm are produced for about an hour, spermatogenesis is shut off and oogenesis begins and continues for as long as sperm are present. In hermaphrodites, sperm are stored in an organ known as the spermatheca, through which mature ova pass and are fertilized to become a zygote and an embryo before being laid as an egg by the hermaphrodite.
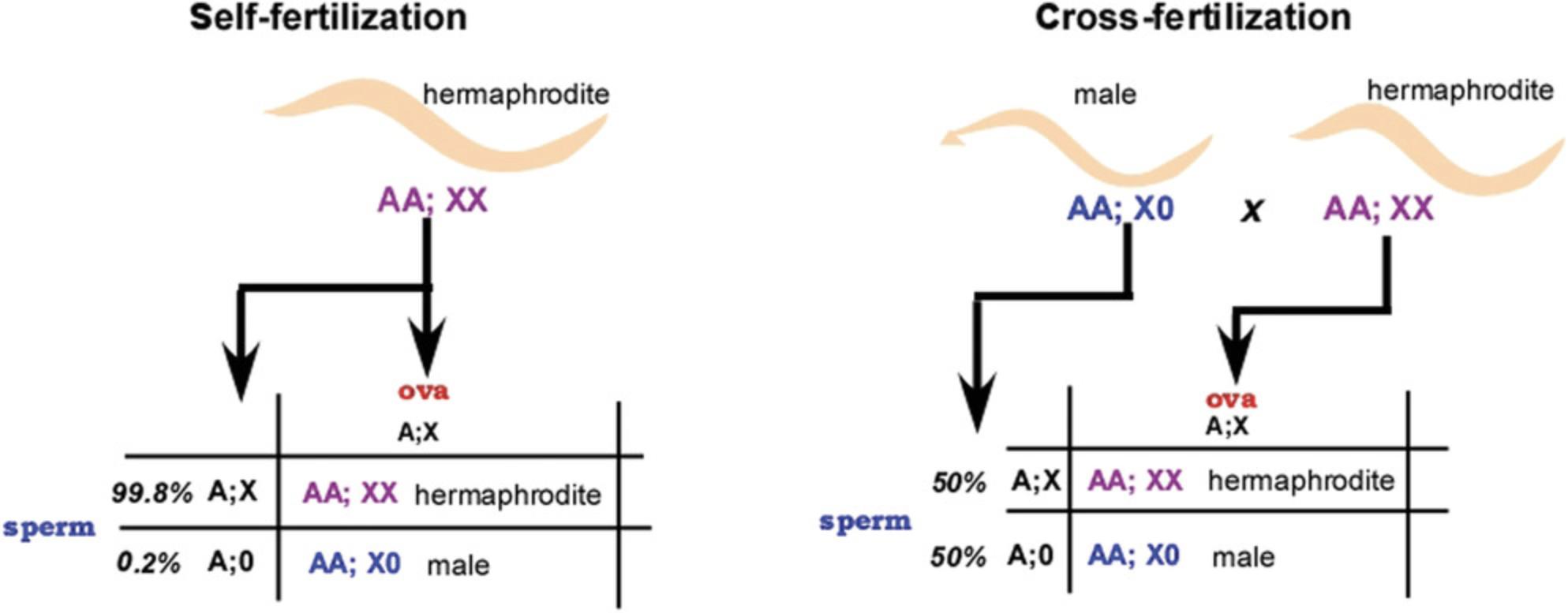
Self-fertilization by a hermaphrodite results in highly inbred strains and makes maintenance of homozygous strains in the laboratory very easy, even for mutants that cannot move or mate; the investigator simply places a young hermaphrodite on an agar plate to produce the next generation. Males arise spontaneously by the loss of one X chromosome (there is no Y chromosome in nematodes) at a rate of about one worm in 600. Males are easily distinguished from hermaphrodites by the shape of their tails and their behavior (Fig. 2), and a male strain can be maintained by mating a male to a hermaphrodite, where half of the cross progeny will be males (Fig. 3). The brood of a mated hermaphrodite will always include both cross-progeny and self-progeny. Thus, males can introduce new alleles into hermaphrodite strains, but the F1 and subsequent generations can be propagated by self-fertilization of the appropriate hermaphrodites. The frequency of spontaneous males (that is, of X chromosome meiotic loss) increases when a hermaphrodite at the beginning of gametogenesis is grown at a slightly higher than normal temperature for several hours (see Resources, Recipes, and Protocols, below). Resulting males are crossed to hermaphrodites to produce more males or a sustainable male-producing strain.
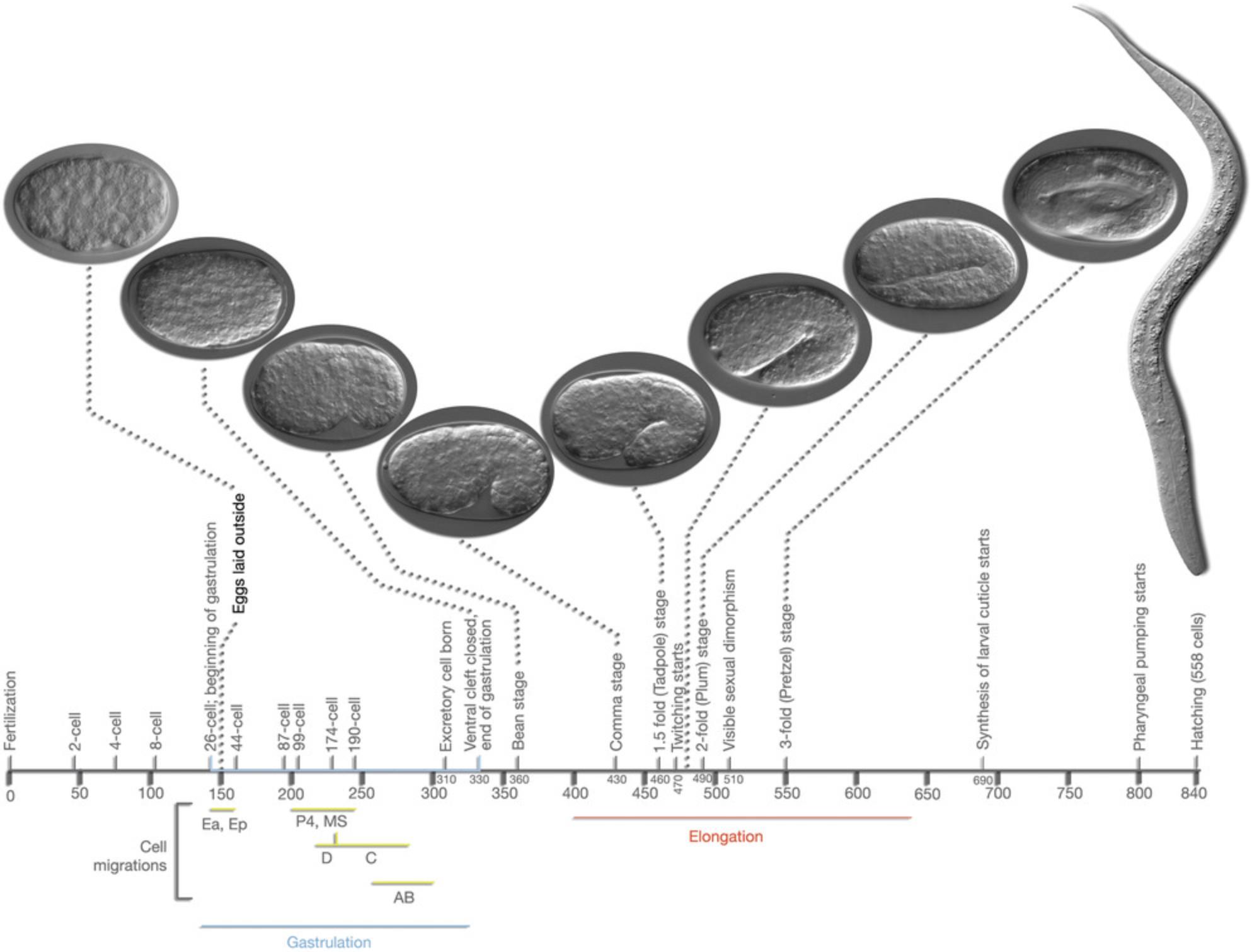
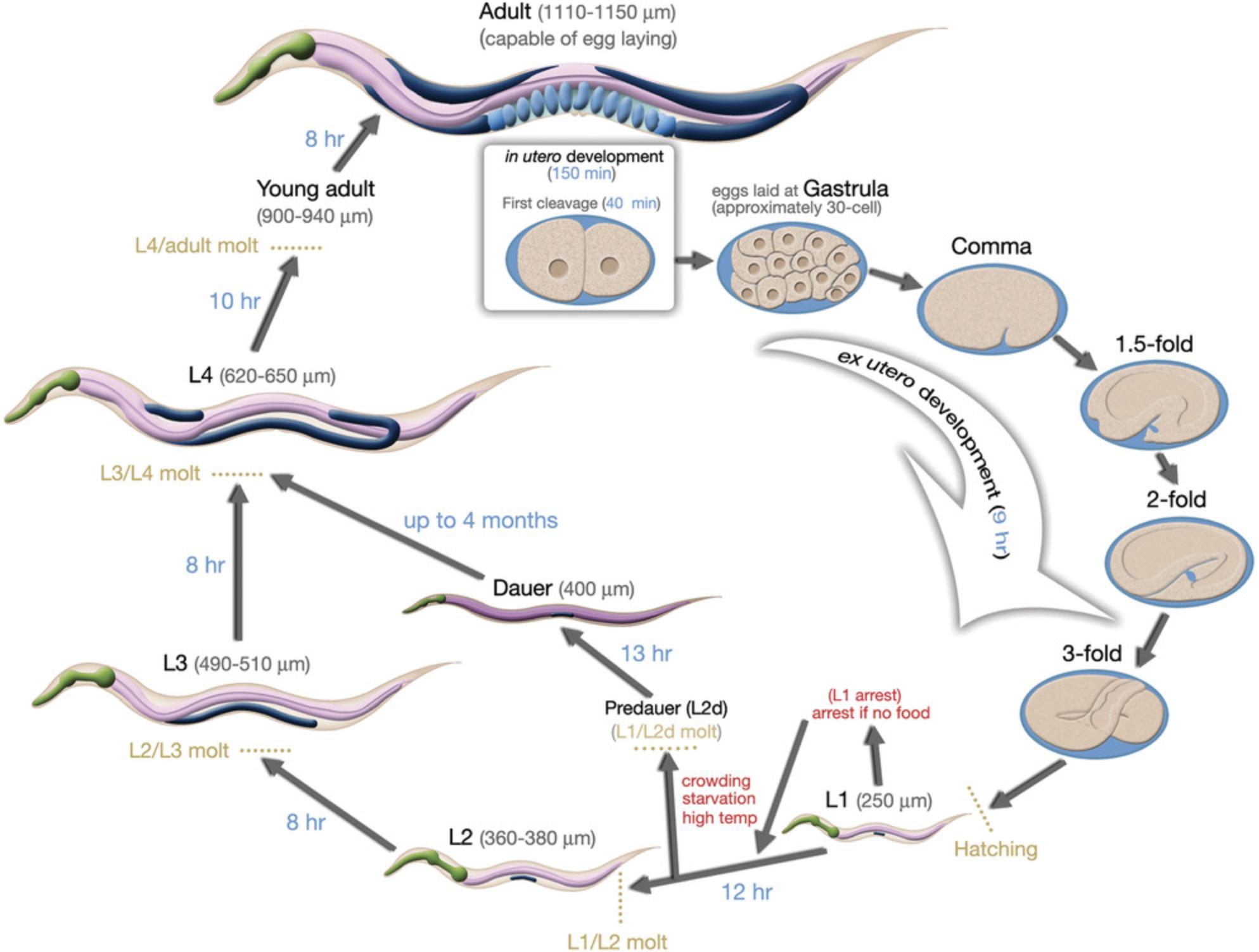
After fertilization, an eggshell forms around the egg, and mitotic divisions (called cleavages) begin (Fig. 4). Depending on conditions, an egg will be laid about 2 hr after fertilization. The worm hatches from the egg at about 12 hr after being laid, having completed embryogenesis (Fig. 5). The nearly hatched L1 larva is about 1/6 the length and width of a young adult, and is barely visible with a standard dissecting microscope, in part because it is nearly transparent. It will grow and molt (replacing its cuticle) to become an L2 larva, and then an L3 and L4 larva. The final molt is from an L4 larva to an adult hermaphrodite, which then begins laying its own eggs 12 hr later. The life cycle from egg to egg takes about 3.5 days at room temperature in the lab. Under stress conditions such as crowding, the L2 worms will molt into a distinct stage called a dauer larva, which is thinner and more darkly colored (since it has a resistant cuticle) than the alternative L3 stage. When conditions improve, the dauer larvae molt to produce a L4 stage, so the L3 stage is skipped. Dauer larvae can persist for months, and most wild isolates are likely to be dauer larvae, but this stage might not be observed with well-fed worms in the lab.
The “Worm Community”
While C. elegans itself has many virtues as a model organism for genetic research, one of the other significant advantages for working with worms is more sociological. The worm research community is well-known as an exceptionally interactive and collaborative group. This community spirit likely arose because nearly all of the early researchers who adopted C. elegans worked at the MRC Cambridge, often with each other, and conveyed this attitude to their research students. In the early days, the spirit was to understand every aspect of how a worm “works” using mutant analysis, a task that was obviously much larger than any individual could imagine undertaking. Information, genetic strains, tips on techniques, and so on were freely and informally shared, often well before the papers themselves were published. The Worm Breeder's Gazette, an informal newsletter published two or three times a year, encouraged sharing of ideas or unusual findings, as well as work before publication; archived editions of the Gazette can be found at WormBook.org (described below).
While the worm community has grown significantly in numbers since those early days, much of that collaborative attitude persists. As a result, there are extensive community resources that any worm worker consults regularly. We list some of these in the Resources section, but particular attention should be drawn to WormBase (https://wormbase.org), the genetic and genomic database for essentially all information about C. elegans and some related nematodes. WormBase includes links to the WormBook (https://wormbook.org), a regularly updated series of definitive review articles about many different research topics in C. elegans ; to the WormAtlas (https://wormatlas.org), which includes images and illustrations of tissues, organs, cells, and life cycle stages; and to the Caenorhabditis Genetics Center (http://cgc.umn.edu), which distributes strains and monitors information on gene names, laboratory contacts, and so on; as well as additional links. All of these sites have links to many other resources, which get updated regularly. Each of these websites also includes extensive tutorials and introductions to working with worms, which can also be consulted for more information.
Basic Biology of C. elegans
This section represents a very brief and necessarily incomplete overview of information; many more details can be found in WormBook and WormAtlas.
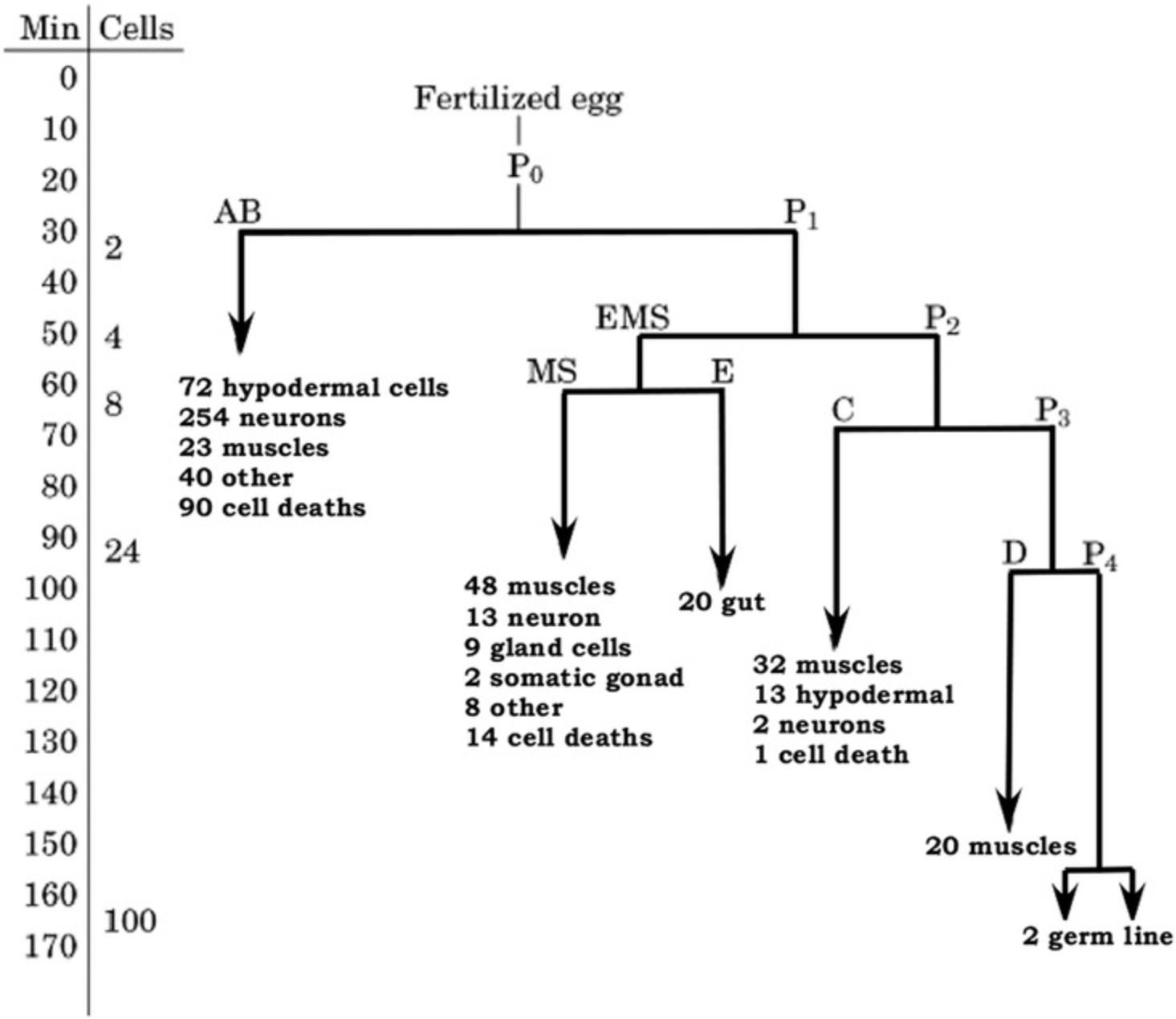
C. elegans offers relatively simple examples of animal biology. Therefore, nearly all aspects of basic animal biology can, and are, studied using the worm (Corsi et al., 2015; Shen, Yue, Zheng, & Park, 2018). Detailed reviews can be found as entries on WormBook, which were written by leaders in their fields. The 959 somatic cells in a hermaphrodite represent a wide variety of tissues from all three germ layers, and the developmental biology of C. elegans has been traced from zygote to adult. Among the tools available for these studies are nuclear (histone) markers, fluorescent cell lineage reporters, and transcriptome sequencing.
A strong, partially permeable cuticle surrounds the pseudocoelemic fluid. Six coelemocytes act as primitive innate immune cells, scavenging waste products from the pseudoceolem. Because of their requirements for engulfing and degrading foreign materials, they have been widely studied by researchers interested in secretion, endocytosis, and related biochemical and physiological pathways.
Roughly one-third to one-half of the body volume of the hermaphrodite is taken up by the bi-lobed gonad that produces oocytes, which mature and move through the spermatheca where amoeboid sperm fertilize the eggs prior to being laid through the vulva on the ventral side of the animal. Sex-specific gene expression and cell fates are long-standing areas of interest to C. elegans researchers. Recent research takes advantage of the large body of sex-determination data to investigate chromatin patterns and remodeling and to directly research sex-specific behaviors and patterns of gene expression.
Getting Started
While worms are very easy to work with, a novice researcher who wants to begin using C. elegans is generally well-advised to spend a few weeks working in an established worm lab to become familiar with the animal, the methods, the nomenclature, the equipment, the life cycle (and identifying life stages of individuals), and so on. A Transparent Window into Biology: A Primer on Caenorhabditis elegans (Corsi et al., 2015) also provides helpful discussions on gene names, anatomy, and tools for those just beginning their work with worms. While the short life cycle is an advantage for those engaged in research full-time, it can be more challenging for those working with less flexibility in their laboratory schedules (for example, laboratories that depend on undergraduate researchers), since worms differing in age by even a few hours might be at different stages of their life cycle. A somewhat synchronized population of L1 worms, which will become adults in about 2 days, can be obtained by bleaching, as described in Resources, Recipes, and Protocols, below.
NOTABLE DISCOVERIES
To date, two different Nobel Prizes have been awarded to scientists working solely with C. elegans , and a third was shared among a C. elegans researcher and others. In addition, many notable discoveries in many different fields of biology have had significant contributions, even pioneering studies, from people working with worms. It seems likely that the combination of the virtues of C. elegans , charismatic leaders such as Sydney Brenner and others who attracted very talented younger scientists, and the collaborative spirit pervading the worm community has contributed to this highly successful endeavor. We cannot make a comprehensive list of all of the notable discoveries that can be tracked back to research in C. elegans , so some distinguished contributions cannot be included. The WormBook and other resources could certainly broaden this list.
Cell lineages and apoptosis
One of the key biological features of nematodes is that their development is highly determinant. That is, cells divide at predictable times and follow precise and nearly invariant lineages, both embryonically and post-embryonically. Because the egg shell and the worm itself are transparent, an investigator with sufficient patience can sit at a microscope equipped with differential interference phase-contrast microscopy and observe the entire development of a worm, from fertilization until its lays is first egg. In the hands of an experienced observer, every cell division occurs on schedule and every cell can be tracked. A 4-min video of embryogenesis is shown in the WormAtlas at https://www.wormatlas.org/embryo/introduction/EIntroframeset.html.
The cell numbers for nematodes are thus small and precise. An adult hermaphrodite has exactly 959 cells (a male has 1048), which are always found at the same location and carrying out the same role. The limited variation is also defined—i.e., if Cell A adopts role 1, then Cell B will adopt role 2; if on the other hand Cell A adopts role 2, then Cell B will adopt role 1. There is essentially no cell replacement for damaged cells. Thus, cells destroyed in a living worm (such as by laser ablation or cell-specific expression of lethal gene products) are missing thereafter, and the morphological and behavioral effects of the missing cell and its mitotic descendants can be observed; confocal microscopy arose in part from such laser ablation experiments in worms. Even for those investigators who have not themselves followed the cell lineages, this cell-by-cell knowledge of worm anatomy is an important research resource.
A typical cell division pattern showed the prevalence of stem cells especially among the embryonic cell lineages; stem cells were commonly referred to in classical developmental biology as blast cells. In embryogenesis, and in some post-embryonic cell lineages, an undifferentiated cell forms two cells via mitosis (Fig. 6). One of these daughter cells (in embryogenesis, it is often the anterior cell) then carries out a series of cell divisions, and all of its descendants become differentiated cells of a particular type or a few specific types, such as neurons or other ectodermal cells. The other daughter cell remains undifferentiated as a blast cell or stem cell; its subsequent mitosis repeats the pattern that one daughter cell becomes the progenitor of a number of differentiated cells while the other daughter cell remains an undifferentiated blast or stem cell. The presence of undifferentiated blast or stem cells in animals was known to classical embryologists (some of whom studied other nematodes) long before any work was done on the cell lineages in C. elegans. The work in C. elegans showed that blast cells were a specific developmental outcome rather than cells that simply had not differentiated yet; this work further identified genes and molecules necessary both for the differentiation to occur and for the blast or stem cell to be maintained as undifferentiated.
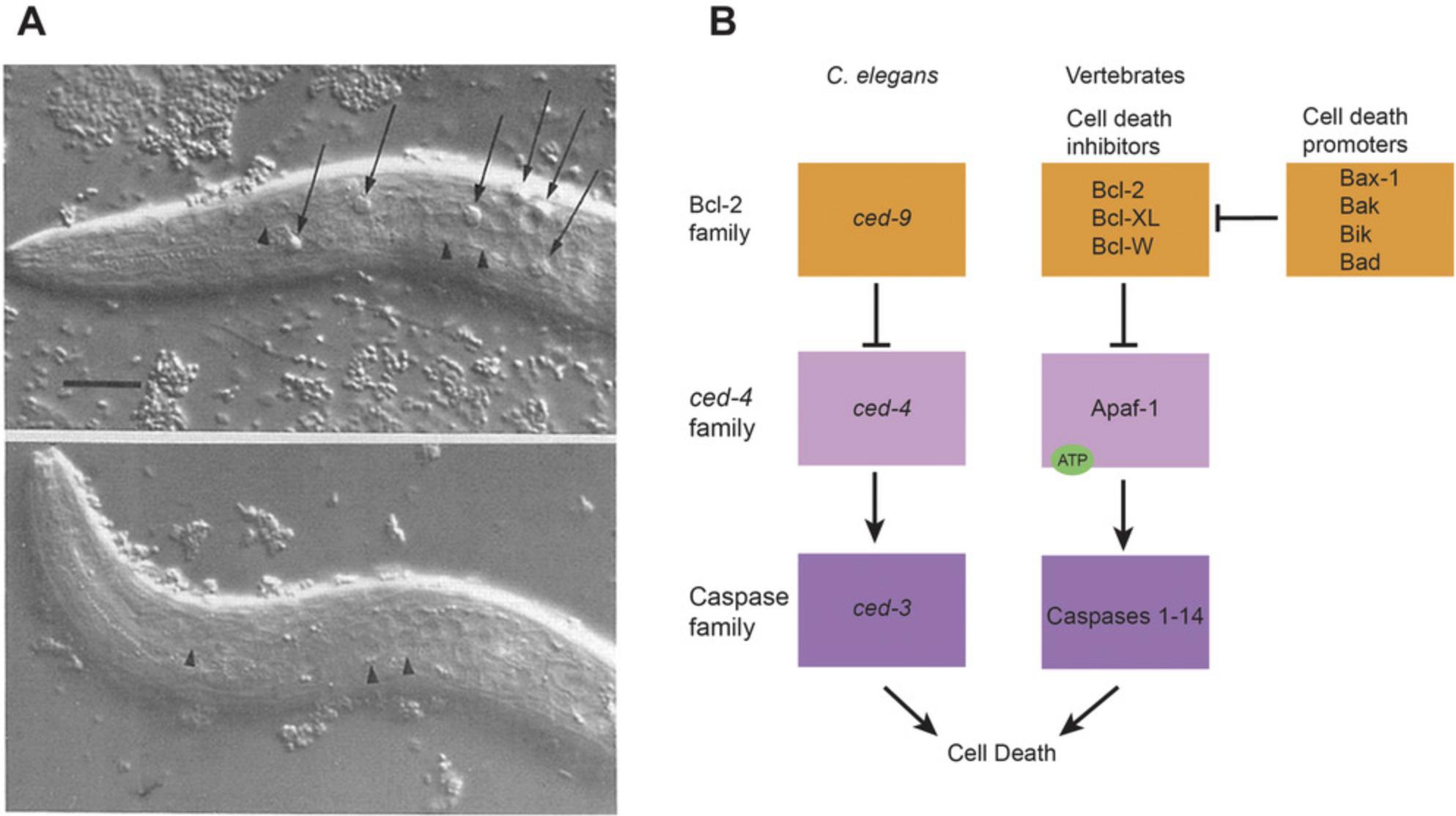
One of the most important insights arising from the precise cell lineages was the role of apoptosis or programmed cell death as part of normal biological processes (Fig. 7A). Cell death had certainly been seen in many organisms prior to the work with worm lineages, but the precise lineages showed that apoptosis occurs in specific cells, at specific times, by a specific biochemical process under very tight genetic control. The genes responsible for apoptosis are evolutionarily conserved among animals (Fig. 7B), and the apoptosis pathway (or its misregulation) has been implicated in many human diseases, including cancers, autoimmune diseases, and neurodegenerative diseases; the apoptosis pathway is frequently activated by chemotherapy and radiation therapy. This work on the cell lineages and apoptosis, and the development of C. elegans as a model organism for genetic analysis, resulted in the Nobel Prize for Physiology and Medicine being awarded in 2002 to Sydney Brenner, John Sulston, and H. Robert Horvitz.
Signaling pathways
Nearly every signal transduction pathway that occurs in animals was found using mutants that affected normal embryonic or post-embryonic processes in C. elegans , with the work in worms typically being among the earliest research in unraveling the genetic and molecular bases for these pathways. Among the signal transduction pathways whose early analysis included worm mutants are the RTK/Ras pathway, the Wnt pathway, the Smad pathway, the Notch pathway, and others. The introduction to signal transduction in the WormBook provides an overview of these pathways, and many of the individual pathways have their own chapter (Greenwald, 2005). Most cancers in humans have been found to have mutations in the corresponding pathways, so understanding their role in fundamental biological process in worms has been an important contribution to cancer biology as well. In addition, more recent studies into neural signaling (calcium, neuropeptides, neurotransmitters) and insulin signaling pathways have been uncovered in the worm, which is an attractively simple model for studying complex and evolutionarily conserved pathways.
Aging and longevity
The average lifespan for a worm grown under laboratory conditions is 14 to 21 days, or about 1 to 2 weeks after egg laying ceases. Mutants that extended the lifespan by as much as 1 week or even 2 (that is, 50% to 100% lifespan extension) have been found by a variety of means, and a number of genes responsible for longevity in worms have been identified (Denzel, Lapierre, & Mack, 2019; Kenyon, 2010). Nearly all of these genes are evolutionarily conserved among animals, and most if not all of them have also been implicated in extended life spans in other animals including mice. Aging in humans is a complicated process affecting nearly every organ system, so it was initially somewhat surprising to find that the underlying biological process is similar in diverse animals that have widely different average lifespans. Nonetheless, the studies in worms have shown that longevity and senescence have some central genetic controls, in addition to the stochastic events that occur during the life of every organism to affect its lifespan.
The mind of a worm
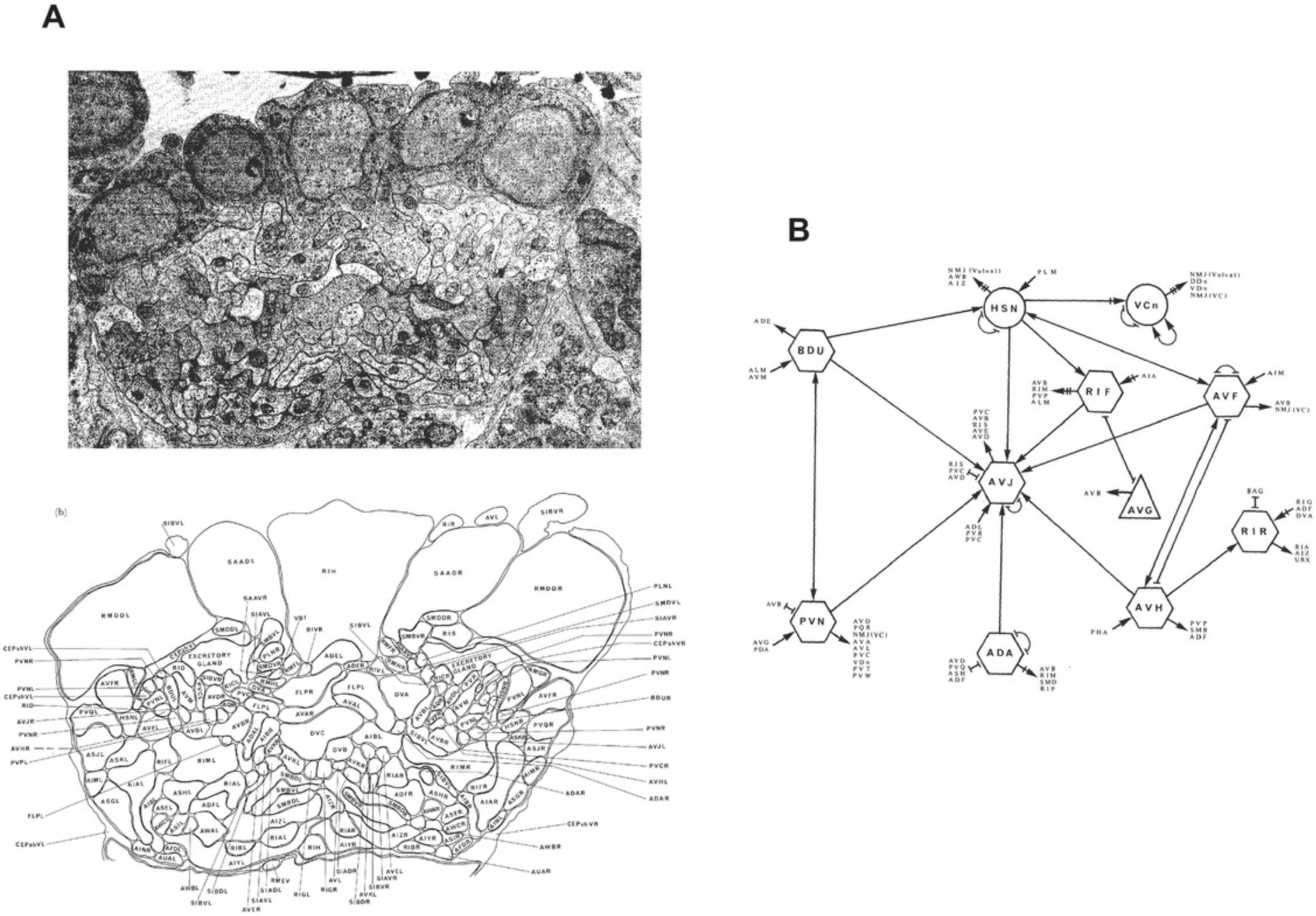
Because there is very little cell migration post-embryonically in worms, and the migrations that occur are predictable, a particular cell will occupy that same position in different individual worms, and will have cell-cell contacts with the same neighboring cells. This is an important feature of the nervous system—one that initially attracted Brenner's interest. An adult hermaphrodite worm has 302 neurons and 56 glia. By EM reconstructions, as well as other methods such as cell markers, the entire nervous system has been reconstructed (Cook et al., 2019; Hall & Russell, 1991; Ward, Thomson, White, & Brenner, 1975; White, Southgate, Thomson, & Brenner, 1986). Body wall muscle is innervated by motor neurons. The mouth of the animal is lined with chemosensory neurons, some ensheathed by glia, and some directly contacting the exterior environment. Other sensory neural subtypes detect and differentiate between changes in touch, light, temperature, salt, and other conditions. Sensory neurons initiate circuits that are modulated by interneurons which, in turn, directly synapse onto motor neurons to control muscle contraction.
As the final output of an organism's nervous system, C. elegans behavior was initially seen as being limited to very basic processes: feeding, egg-laying, and locomotion (Brenner, 1974). Essentially, the worm was seen as a moving tube, so basic phenotypes could be described as anything that visibly altered the appearance of the animal (e.g., dumpy or long) or impaired the locomotion of the worm (e.g., uncoordinated or roller; Brenner, 1974). This list quickly expanded into what we know now as an array of behaviors that can be measured and are often both finely tuned and plastic, some of which are described below. What followed was a series of studies identifying the individual neurons thought to play a role in each of these behaviors, essentially resulting in a functional connectome of the organism. This wiring diagram has been referred to The Mind of the Worm (Fig. 8), and has been a valuable resource in understanding behavioral circuits.
GFP as a cell marker
Martin Chalfie shared the Nobel Prize for Chemistry in 2008 for demonstrating that green fluorescent protein (GFP) could be used to monitor gene expression and protein localization in living organisms (Chalfie, Tu, Euskirchen, Ward, & Prasher, 1994). Chalfie did his work on touch sensitivity in worms, a behavioral response controlled by a relatively simple neuronal circuit involving only a few cells. The circuitry was identified using mutants that failed to respond to a gentle touch, as well as laser-ablation studies of candidate neurons. Chalfie identified a large number of touch-insensitive mutants, characterized the structures of their touch neurons, and then used GFP in reporter gene constructs to examine their expression. The invariant circuitry, as well as the transparency of the worm and the collection of mutants, were important aspects of this research. Current research across cell types uses the entire range of other fluorescent proteins, as well as pH-sensitive gene-encoded fluorophores as reporters and fusions, to monitor protein localization and abundance, cell fate, and gene expression, among other biological phenomena. Many of these research applications have their origins in mutant worms that failed to respond when stroked with an eyelash (Chalfie et al., 1985; Hart, Sims, & Kaplan, 1995; Maricq, Peckol, Driscoll, & Bargmann, 1995).
Heterochronic mutants, developmental timing, microRNAs, and RNAi
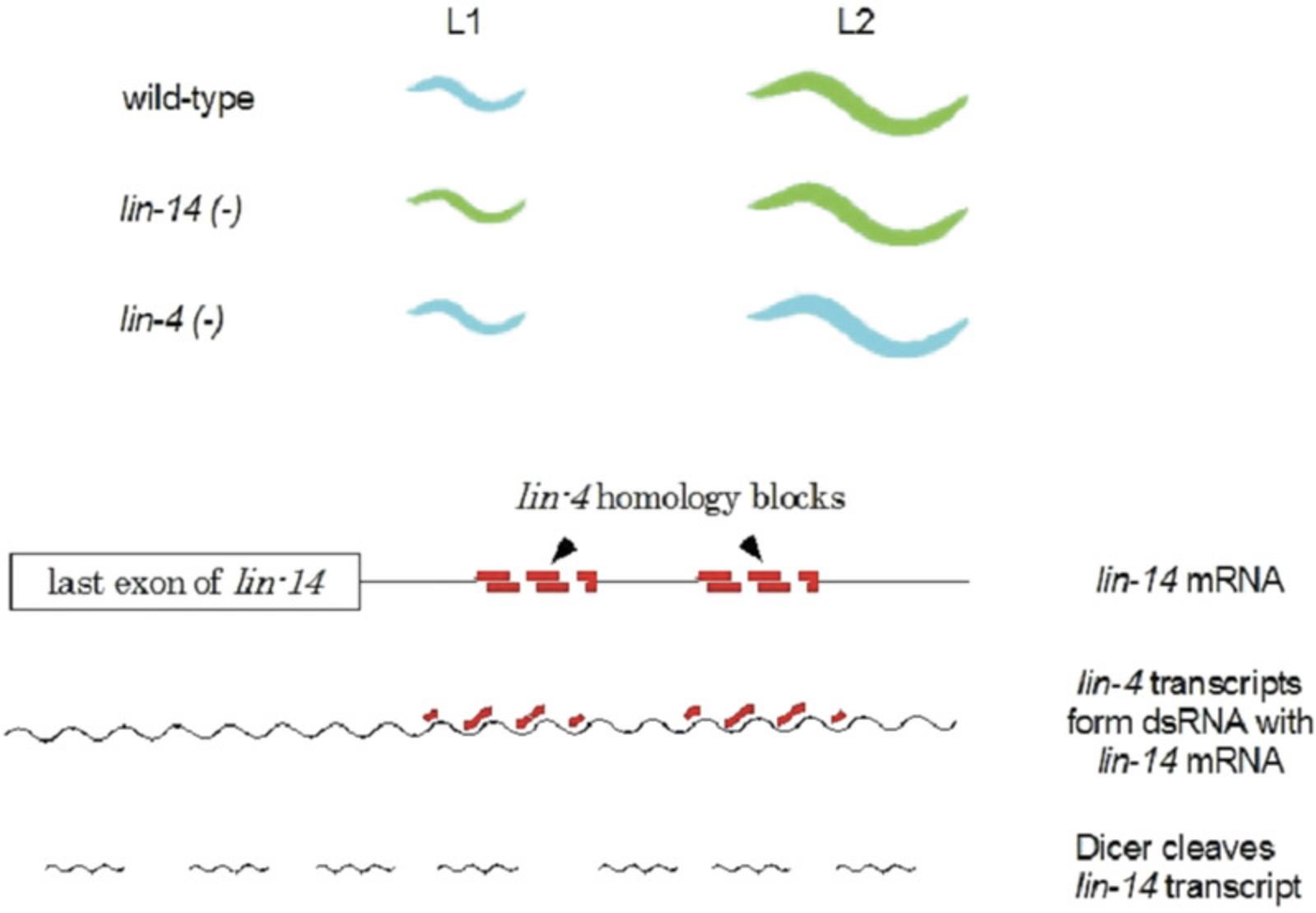
The life cycle of C. elegans has four distinct larval stages leading to the adult; each larval stage, as well as the dauer larva and the adult, makes a cuticle with a specific structure composed of some stage-specific collagens. Mutations that altered the timing of the developmental stages (heterochronic) were identified by several different genetic screens; curiously, none of the original screens was focused on looking for mutants affecting the developmental stages or the cuticle pattern. The mutant strains fall into two categories, those that skipped over a particular developmental stage (as determined in part by the analysis of the cuticle or the cell division patterns) and those that continued to re-iterate an earlier developmental stage. For example, as summarized in Figure 9, animals harboring mutations in the lin-4 gene reiterate the L1 stage, suggesting that the wild-type lin-4 + gene product is needed to exit the L1 stage and enter the L2 stage. Mutants in the lin-14 gene had the opposite effect, skipping the L1 stage and initiating the other stages precociously, suggesting that wild-type lin-14+ product must be necessary for the L1 stage to occur. When the two genes were cloned and sequenced, the lin-4 gene was found to lack any sustained open reading frame that would allow it to encode a polypeptide (the lin-14 gene is a conventional protein-coding gene). Instead, lin-4 encodes a small RNA transcript, initially about 70 nt in length, and processed to a final length of 22 nt (Lee, Feinbaum, & Ambros, 1993). The sequence of the mature lin-4 transcript is the complement of 22 base sequences in the 3′untranslated region of the lin-14 mRNA, as summarized in Figure 9. These findings led to the understanding that lin-4 encodes a microRNA that shuts off lin-14 expression by blocking translation combined with mRNA degradation; lin-4 was the first gene demonstrated to encode a microRNA. A second gene, let-7 , was identified about the same time by mutant analysis and was shown later to also encode a distinct microRNA, with many target genes, including some that regulate genes involved in the later larval stages. These two worm genes set the foundation for recognizing that genes encoding microRNAs are found in the genomes of every multicellular organism; the worm genome has about 260 such microRNA genes, while the human genome has approximately 1800 recognized microRNA genes, including orthologs of let-7. Thus, these microRNAs revealed a broadly important mechanism for regulating gene expression in all multicellular organisms that had been previously unsuspected.
At about the same time as the microRNA genes were found, and often involving researchers in the same labs, the use of RNA inference (RNAi) to inhibit gene expression was developed in C. elegans. RNAi had been encountered in petunias, where it was named post-transcriptional gene silencing, but the amenability of C. elegans has been critical in understanding the mechanism of RNAi and its experimental parameters, as well as its uses and limitations (Ahringer, 2006). RNAi has proven to be broadly useful in many organisms, in large part because its effect is exerted by the same molecular mechanism by which microRNAs regulate gene expression, so all multicellular organisms have the machinery for RNAi in their genomes. Andrew Fire and Craig Mello shared the 2006 Nobel Prize in Physiology and Medicine for their work unraveling the mechanism of RNAi and showing its broad applicability as a research tool.
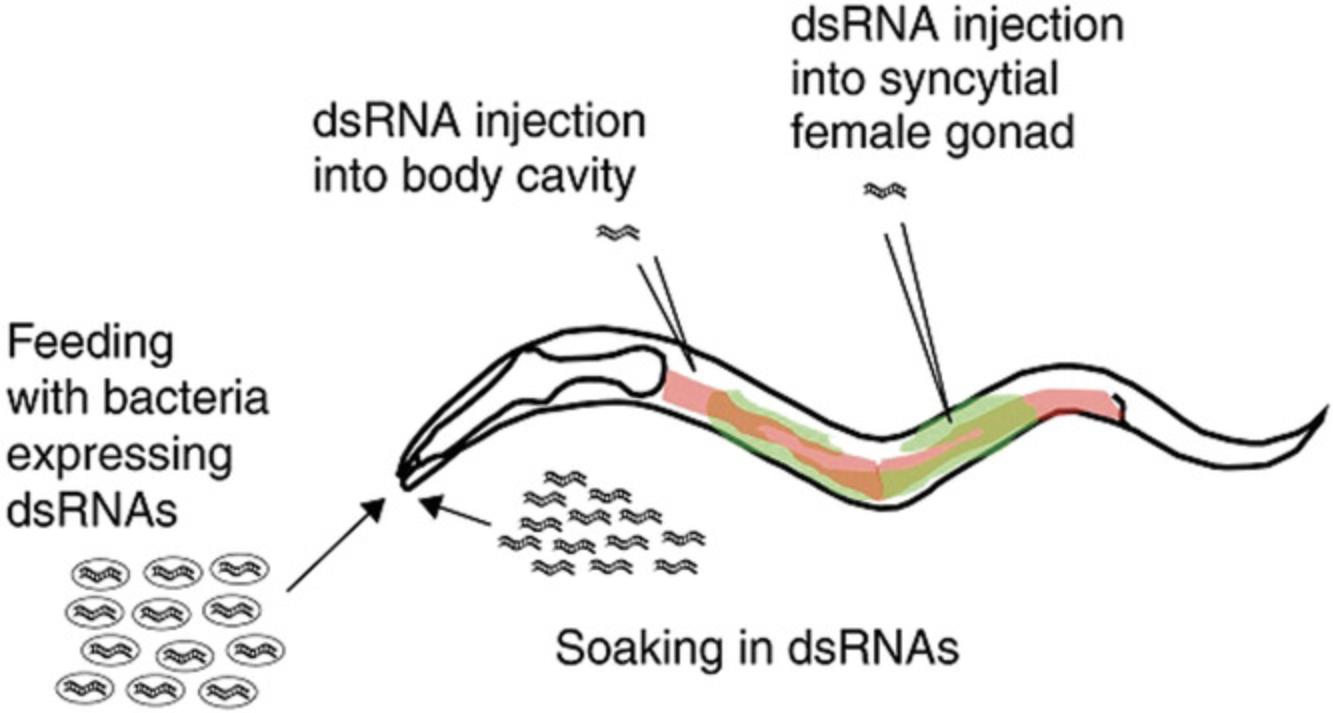
RNAi experiments can be performed in worms using several different protocols (Fig. 10). In the first experiments, a double-stranded RNA corresponding to any part of the coding region of a worm gene was produced by in vitro transcription from each strand of the gene, annealed in vitro, and microinjected into the hermaphrodite gonad. The dsRNA is taken up by ova and persists in the embryos, blocking the expression of the corresponding gene via the same mechanism as microRNAs. Worms could also be soaked in a solution that contained the dsRNA at high concentration, with the offspring showing the effect, demonstrating that ingested dsRNA was somehow moved into the germ line in C. elegans. The ingested effect is a fortuitous peculiarity of C. elegans and depends on two (or possibly a few more) genes in the C. elegans genome; these genes are absent from the genomes of nearly all other organisms, including the closely related nematode C. briggsae. RNAi is therefore done by microinjection or by transfection into cell lines in organisms other than C. elegans.
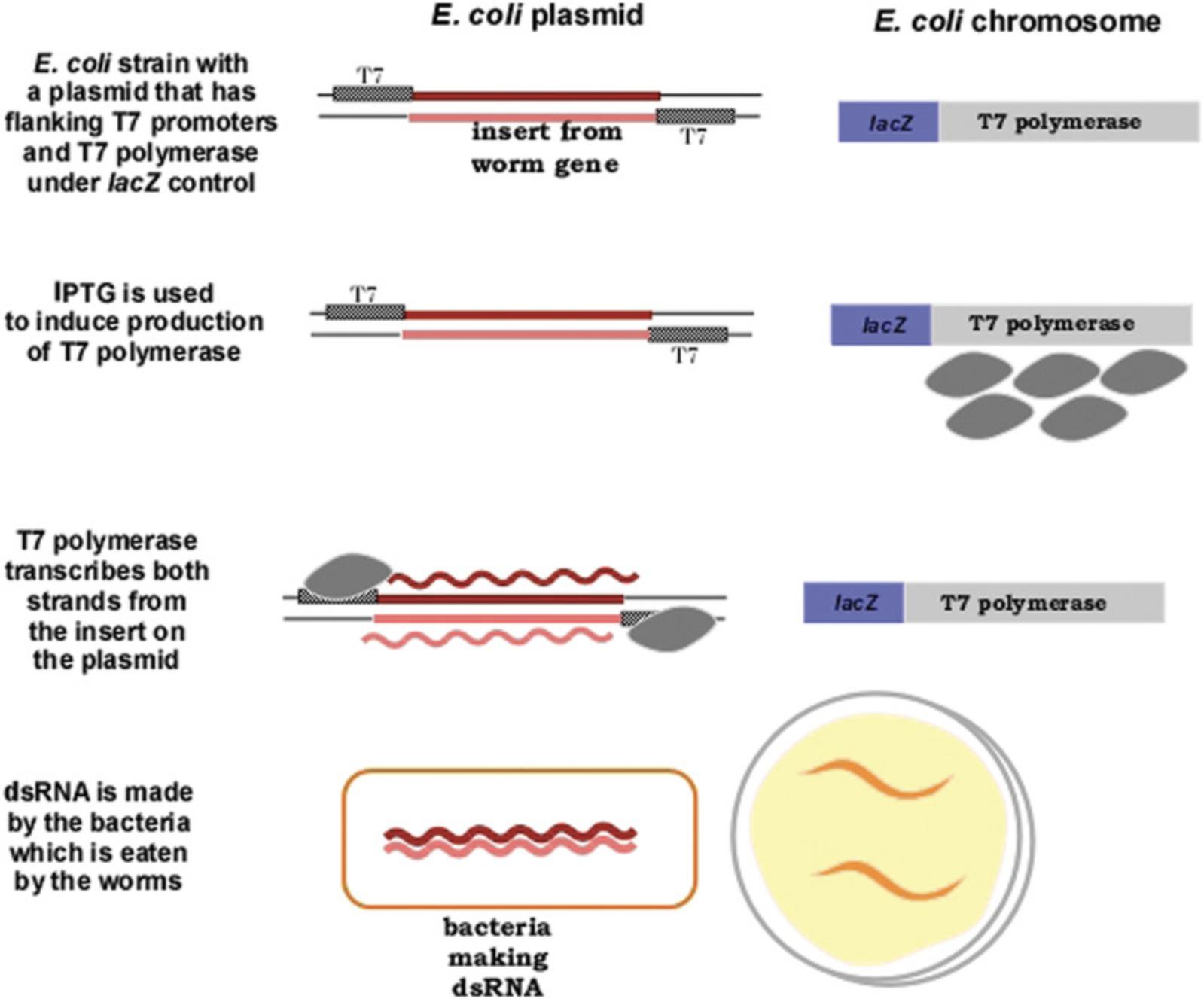
The finding that ingested dsRNA could induce RNAi led to the most widely used technique for carrying out RNAi in C. elegans. A plasmid is made in vitro with any part of the coding region a worm gene; flanking the plasmid insert are (commonly) T7 promoters (Fig. 11). The plasmid is transformed into an E. coli strain that has T7 RNA polymerase under inducible control, commonly induced by IPTG which induces transcription of the lac promoter in E. coli. In this way, the E. coli strain will make the dsRNA corresponding to the insert from the worm gene. When C. elegans eats this E. coli strain, the dsRNA is ingested and RNAi is induced in the offspring of the feeding worms (Fig. 11). Libraries of E. coli strains that have plasmid inserts from every known worm gene have been constructed and are commercially available (http://www.sourcebioscience.com). Thus, any lab equipped for culturing E. coli and worms can readily carry out RNAi experiments for any gene, and potentially use RNAi in a screen of every gene in the genome. Many RNAi “mutant” screens have been done using this method. In most organisms, the effects of RNAi are transient and seen only in the first generation; C. elegans is unusual in that the RNAi effects can persist for several generations. RNAi screens are easily done in undergraduate research and teaching labs, so we provide detailed protocols for preparing the media, culturing the bacteria, and so on. One caveat is that neurons are less susceptible to RNAi (especially via feeding), so screens using neurons may require rearing animals for two generations on RNAi, and may still yield high numbers of false-negative results.
Genome sequencing
No technology has changed genetics more than the ability to obtain, annotate, and apply the DNA sequence of an organism's genome. C. elegans was the first multicellular organism to have its genome sequenced, with the sequence completed in 1998 (C. elegans Sequencing Consortium, 1998), only 2 years after the genome of the yeast Saccharomyces cerevisiae was sequenced. The worm genome then provided the template for other genome sequencing projects, including the Human Genome Project, completed in 2002 (and involving many of the same researchers who had worked on the worm genome). In addition to extensive insight about the structure of eukaryotic genes and genomes, the C. elegans genome project yielded information about sequence procedures, cloning vectors, assembly procedures, data sharing and storage, community interactions, and much more. Although the manual methods used to sequence the worm genome (and the human genome) have been supplanted by automated methods, the overall strategy and philosophy used for the worm genome has been widely adopted by nearly all other genome sequencing projects. The annotation of the worm genome is ongoing, and current information is available on WormBase.org. In addition, since researchers using C. elegans have had access to complete genome sequence for more than 20 years, many novel experimental strategies enabled by knowing the DNA sequence beforehand have been developed in worms. A revolution in biology has come from having the gene and genome sequence as a research reagent rather than a result to be obtained experimentally—research in C. elegans has been in the front lines of this revolution for the past two decades. In fact, the title of the paper reporting the genome sequence (see Literature Cited) reveals this perspective; the genome sequence has become a platform for investigating many aspects of biology.
NEW WAYS WORMS ARE BEING USED
C. elegans has been at the forefront of technological innovations since the beginning of research in worms; this has continued, and is likely to continue, for the foreseeable future. C. elegans provides an easily manipulated experimental system, with thousands of worms being grown in a small space in a short time to provide results quickly. As a result, nearly every new experimental method, tool, or strategy, successful or not, has been tried out in C. elegans. Our examples here are a very brief summary of some of the vibrant and novel research being done in worms.
Genetic mutants, transgenics, and gene editing
Because of its tractability as a genetic system, forward genetic screens have been used almost from the beginning to help define and refine genetic pathways in C. elegans. Male animals can be mutagenized using EMS, ENU, or UV irradiation and mated to wild-type hermaphrodites, and the resulting offspring can be isolated and screened for phenotypes of interest. The MMP (Million Mutation Project) augments the mutational screens held by individual laboratories, and complements the deletion strains engineered and maintained by other consortium labs. These result can be found at https://shigen.nig.ac.jp/c.elegans/mutants/index.xhtml and https://cgc.umn.edu/ (C. elegans Deletion Mutant Consortium, 2012; Thompson et al., 2013). The combined power of whole-genome sequencing (see above) and forward genetics resulted in a 2007 strain “library” of genetic mutant animals with an average of over 400 homozygous mutations per strain. This Million Mutation Project represents the allelic series for most protein-coding genes, based mostly on single-nucleotide changes.
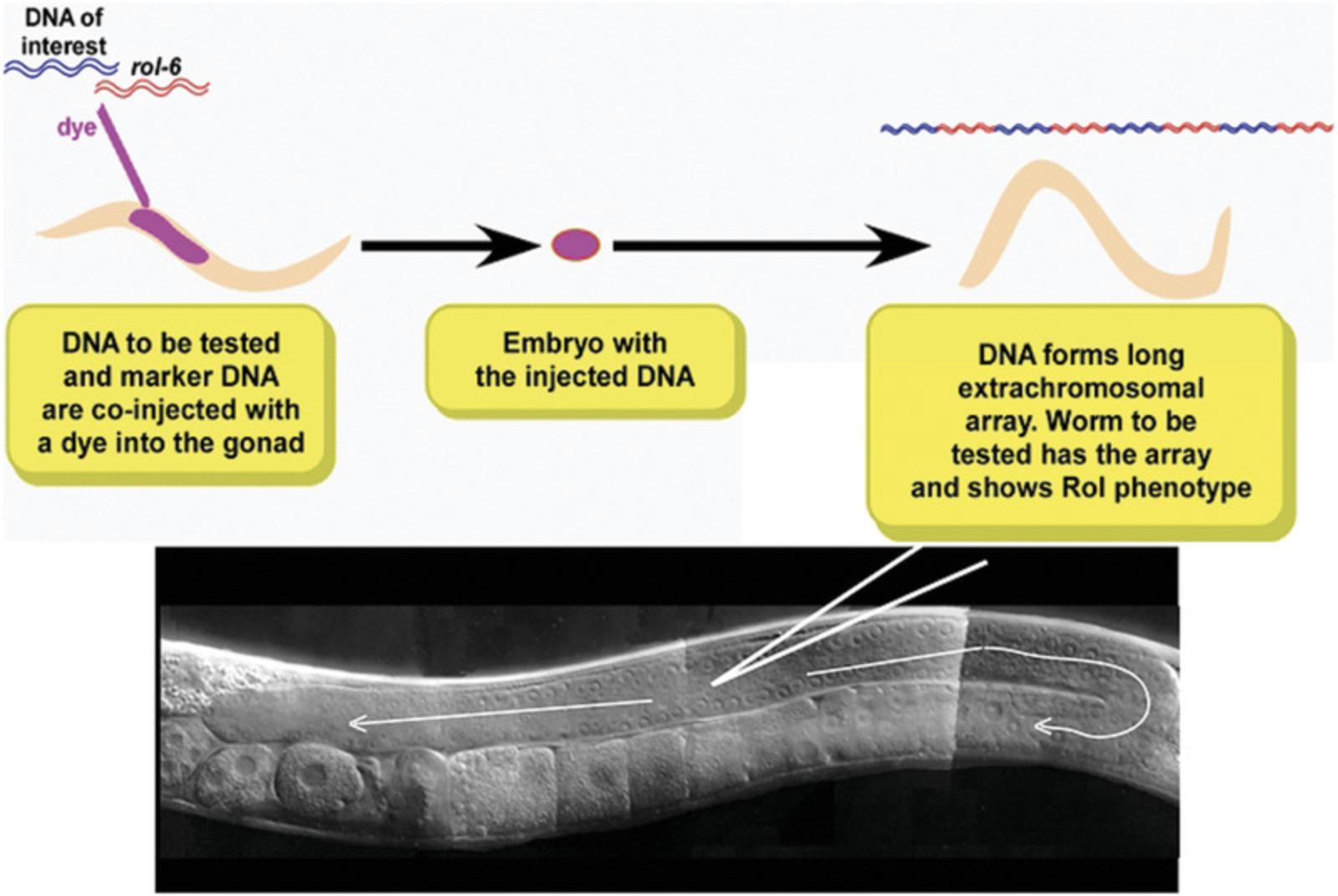
Developing oocytes in the hermaphrodite gonad exist in a syncytium prior to cellularization and fertilization. Thus, direct injection of plasmid DNA into the gonad can result in the uptake of DNA into germ cells prior to fertilization and egg laying (Fig. 12). Plasmid DNA concatenates within the germ cell and can be passed on (with variable penetrance) to future generations as extrachromosomal arrays. For many experiments, maintaining the gene as part of an extrachromosomal array is adequate. For others, particularly ones in which gene dosage or stable inheritance is important, the arrays can be integrated into the chromosome. Extrachromosomal arrays can be integrated into the C. elegans genome by inducing double strand-breaks in both the array and the chromosome. At low frequency, repair of the chromosomal break results in the integration of the plasmid DNA as transgene (Corsi et al., 2015).
Traditional transgenesis in worms results in the presence of multiple copies of the transgene in the form of extrachromosomal arrays or integrated concatemers. In order to reduce overexpression or transcription factor depletion phenotypes, researchers can take advantage of the mos-1 transposon, whose insertion site has been engineered into inbred strains of C. elegans. Injection of plasmid DNA that encodes genes of interest flanked by the mos-1 transposable element can be induced to “jump” into the genome at defined sites, resulting in single-copy transgenes at known locations in the genome (Frøkjær-Jensen et al., 2014). Libraries of plasmids and strains for this technique are publicly available on Addgene and through the CGC (http://www.wormbuilder.org/).
Because of the relative ease of engineering transgenic animals through injection, and because molecular machinery to repair double-strand breaks are encoded in the worm genome, the C. elegans community was an early user of CRISPR technology, and improvements in CRISPR techniques are routinely being reported. Initially, laboratories injected plasmids encoding the Cas9 nuclease, sgRNA, and repair templates (Dickinson, Pani, Heppert, Higgins, & Goldstein, 2015; Norris, Kim, Colaiácovo, & Calarco, 2015). Various protocols now incorporate variations, including injection of the Cas9 protein and/or purified sgRNA (Kim & Colaiácovo, 2016). Innovations and adaptations of CRISPR/Cas9 technologies comprise the inclusion of LoxP/Cre sites on recombination templates (to allow removal of selection markers from established knock-in/out lines) and the SapTrap system derived from GoldenGate cloning, which provides a simple way to make a variety of knock-in/out repair constructs (Au et al., 2019; Schwartz & Jorgensen, 2016). CRISPR interference and CRISPR activation (CRISPRi and CRISPRa, respectively) have also been successful in C. elegans (Long et al., 2015; Savell et al., 2019).
Evolutionary biology and natural variation
Despite being known as “soil-dwelling” nematodes, C. elegans are most often found foraging on rotting fruit or vegetables (Kiontke et al., 2011). Apple orchards and other fruit farms are rich sources of wild nematodes, where the animals thrive on the bacteria that grow on decaying fruit. In the absence of a rich food source, wild worms survive as dauers. A growing body of research has focused on the evolution, development, and biology of wild isolates, including how the microbiomes of the wild animals differ from the inbred strains used in laboratories. EM and light microscopy show that C. elegans can maintain fungal and some bacterial cells in its intestine (Frézal & Félix, 2015; Jiang & Wang, 2018; Schulenburg & Félix, 2017), despite the double pharyngeal system that precedes it. Wild isolates are also the focus of “citizen science” initiatives (for an example, see https://www.wpi.edu/people/faculty/jsrinivasan), which have expanded the breadth of characterized C. elegans strains, as well as serving to identify new isolates and strains of other nematode species.
Wild C. elegans have been collected from all over the world, and a strain isolated from Hawaii has been an especially important tool for mapping genes using SNP mapping. Generally, C.elegans have fairly low genetic variability, approximately 10-8/basepair/generation (Barriere & Felix, 2005). Thus, there is fairly low genetic diversity across wild strains—despite their being globally widespread—and evidence of low rates of crossing over suggest that most animals are the offspring of hermaphrodite self-fertilization (Barriere, 2005; Brusca, 2016; Kiontke, 2006). Nonetheless, wild isolates of C. elegans do have measurable phenotypic differences that include longevity, frequency of males, age of egg-laying, olfactory and other chemosensation or chemoactivity, body size, and sensitivity to toxins/pheromones; these have been reviewed in Barriere (2005). How these relate to developmental and evolutionary patterns are areas of current research that are aided by the ever-easier accessibility to video/imaging technology and whole-genome sequencing.
Historically, among other nematodes of the Rhabditids clade, C. elegans was considered most closely related to C. briggsae , C. ramenei , and C. elegans n. sp. (represented by different strains, including CB5161). These four make up the Elegans group, whose closest phylogenetic relative (outside of the group) is C. japonica. Only C. elegans and C. briggsae exist as hermaphrodites, with other rhabitids being gonochoristic (require mating of male and female for reproduction). For comparison, the genetic distance between C. briggsae and C. elegans is approximately the same as that between human and mouse. More recent collections of C. elegans and other species of closely related nematodes have given a fuller picture of these relationships, as well as providing a more detailed description of the global versus isolated population genetics (Félix et al., 2013; Kiontke et al., 2011; Schulenburg & Félix, 2017).
Neural networks and behavior
The nervous system of C. elegans is well mapped (see The Mind of the Worm, above), making it a strong model for current studies of neural specification, development, function, and connectome analysis; both the WormAtlas and the WormBook have sections devoted to these subjects. Because of the relatively small number of neurons, and because the connections between cells are known, computer modeling of the worm connectome is an appealing model for defining minimal circuits and modeling complex behaviors in silico (see, for example, http://openworm.org/). Increasingly, the C. elegans nervous system is used as a model for the cellular basis of human neural diseases and disorders, including an increased focus on the physiology and function of glia in C. elegans (Singhvi & Shaham, 2019) and its possible connection to human diseases. Neuron-specific promoters and reporters, optogenetics, and electrophysiology are all regularly used to dissect and trace the physiology and connections between neurons and other cells; neural cultures can also be isolated and matured in vitro.
Neural circuits and behavior
When White et al. (1986) reported a comprehensive description of the anatomy of a single worm captured by serial dissection and electron microscopy, this provided a complete wiring diagram for the entire nervous system. Knowledge of the worm connectome has allowed researchers to identify individual neurons involved in detecting environmental stimuli including mechanosensation, chemosensation, and thermosensation. Describing these specific circuits at the single-neuron level was initially accomplished using laser-ablation techniques (Chalfie et al., 1985; Fang-Yen, Gabel, Samuel, Bargmann, & Avery, 2012; Gray, Hill, & Bargmann, 2005; see Fang-Yen et al., 2012, for methods and extensive review). Genetically targeted cell-death transgenes have also been utilized for “genetic ablation” of target neurons (Chelur & Chalfie, 2007; Harbinder et al., 1997), while targeting of phototoxic proteins (e.g., Killer Red, which produces reactive oxygen species upon green-light illumination) allows for a type of “photoablation” (Kobayashi et al., 2013). Finally, optogenetic rhodopsin channels can also be genetically targeted to some individual neurons, and activation/inhibition driven by these channels can temporarily render a neuron “offline” (Bergs et al., 2018). Thus, there is a wealth of tools available to dissect neuron function within an intact C. elegans neural circuit.
Neural circuits have also been identified that mediate detection and response to additional sensory stimuli including UV light, O2 and CO2 concentrations, electric fields, and more (Bretscher et al., 2011; Chang, Chronis, Karow, Marletta, & Bargmann, 2006; Gabel et al., 2007; Ward, Liu, Feng, & Xu, 2008). To supplement the connectome to add greater functionality, C. elegans researchers have gradually uncovered the identity of neurons that release specific neurotransmitters or express particular receptors; for example, neurons throughout the C. elegans nervous system have been identified for acetylcholine and GABA transmitter signaling (Gendrel, Atlas, & Hobert, 2016; Pereira et al., 2015; see http://www.openworm.org resource to visualize). Clear definitions of behavioral output, combined with anatomical wiring and synaptic communication, has allowed for proposed models of overall nervous system organization; for instance, coincident detection and/or a hub-and-spoke model (Ghosh, Nitabach, Zhang, & Harris, 2017). These well-described behaviors, coupled with detailed neuroanatomy, allow for validation of experimental manipulations as behavioral measures and can be used to read the final output of the nervous system at the organismal level.
In addition to a well-described nervous system, there are additional advantages to using C. elegans behavior as a measure of neuron plasticity. Over the last 20 years, it has been well established that worm neurons function in many of the same ways as neurons in other animals to associate environmental stimuli to certain internal conditions that then determine future responsiveness to the same environmental stimuli. For instance, if a chemical or odorant is consistently present when the E. coli food is absent, worms will actively avoid that chemical or odorant (Hart, 2006; Saeki, Yamamoto, & Iino, 2001). Originally, the well-described nervous system of the worm was seen as being largely determined and invariant between individual worms. However, if features of a worm's environment can modulate their responsiveness based on experience, this is seen as evidence that C. elegans synapses are modifiable, not just in the case of neuron gene mutation (Ardiel et al., 2016; Cherra & Jin, 2015). Together, this work highlights both the plasticity and functionality within the C. elegans system.
Stress in worms
Worms are also a well-established model for examining the effects of environmental stress, and by extension, for examining the measures affected by different forms of stress including longevity, development, etc. (see review in Rodriguez, Snoek, De Bono, & Kammenga, 2013). Several different forms of stress have been reported in C. elegans including heat shock, hypoxia, oxidative stress, osmotic stress, ER stress, and others. A myriad of “stress” genes has been identified as well as a means to measure the outcome of different stress responses such as fertility, brood size, lifespan, body size, etc. Also, with available stress markers that include fluorophore reporter constructs fused to stress-induced protein expression and biochemical assays (ROS, hsp-2 ::GFP, etc.), researchers are equipped to identify environmental conditions and experimental manipulations that trigger stress responses in the whole animal.
C. elegans and disease
Although the extent to which C. elegans can be used as a direct indicator of human disease processes was once considered limited, one strength that lends the worm to this line of research is that human genes, and by extension human gene variants, can be expressed in worms. This has allowed for investigation into the roles of these genes in basic cellular processes, providing insight into the possible function of these genes in human cells. One example is the kindlin-1 gene; in C. elegans the protein product of the C. elegans homolog of that gene interacts with integrins, which led to the discovery of a similar disease process in humans (Siegel et al., 2003). When the human gene presenilin-1 , implicated in early-onset Alzheimer's disease, was expressed in worms, researchers reported both a behavioral phenotype (thermotaxis defective) and a putative cellular mechanism involving Notch signaling (Levitan et al., 1996; Wittenburg et al., 2000). The flexibility of transgenesis in C. elegans allows for the discovery of cellular mechanisms through the expression of disease-related genes. Researchers have recently taken advantage of the genetic tools available with C. elegans to begin to identify orthologs of genes involved in human autism-spectrum disorder (McDiarmid et al., 2018; Wong et al., 2019), further expanding the possibilities for investigation of human disease gene functions.
Newer Technologies for Studying Worm Behavior
Bulk/population behaviors (Worm Trackers) and an organismal readout
One challenge to reliably measuring and analyzing worm behavior is that, as a microscopic organism, very small perturbations in the environment can influence the behavioral output (Hart, 2006). Also, small differences between experimenter raters can result in large differences in measured outcomes. As a result, behavioral assays typically require that a large number of animals be observed and measured to account for this potential for data variability. Collecting behavioral data from a large number of worms can be time-consuming, and is often viewed as a rate-limiting step to an experimental behavior procedure. To increase both the rate and reliability of behavioral measures, researchers sought to automate the process of behavior capture and analysis. As a result, many individual labs took it upon themselves to design software, Worm Trackers, programmed to measure behaviors specific to that lab's research interests. Most have made their Worm Tracker freely available online. Some Trackers require hardware (e.g., automated microscope stage) and typically run off another software platform (e.g., MATLAB). The outcome of this endeavor is that there are several worm behavior trackers available for research; these are summarized by Husson, Costa, Schmitt, & Gottschalk (2012) in WormMethods at https://wormbook.org. The table in Husson et al. (2012) is by no means an exhaustive list, as many other labs have devised smaller versions of analysis software for worm behavior analysis. Many of these trackers are freely available online at sites like GitHub (http://github.com). However, most trackers require some knowledge of coding in order to tailor the tracker to the needs of the lab. If coding is not within a researcher's skill set, there is at least one commercially available worm tracker (MicroBrightField; https://www.mbfbioscience.com/wormlab).
Microfluidics
When measuring the behavioral output of a worm, a significant source of variability comes from measuring behavior from a moving animal. However, the use of microfluidics allows for measurements to be taken from a relatively static sample. For instance, certain neuron activation reporters or genetically encoded calcium indicators (see below) require that a stimulus be presented to a static animal so that neuron activation can be measured as a change in fluorescence of the fluorophore indicator. This is difficult to do in a behaving animal, and so researchers have devised means whereby stimulus presentation can be conducted in a uniform fashion while recording activity of neurons using calcium indicators to identify neurons activated during stimulus events. For instance, microfluidics chambers with micro-tunnels large enough for a worm to pass are used to capture neuron activation data during locomotion, while other types of microfluidic chamber devices hold the worm in place in order to perform olfactory or chemical stimulus presentation while simultaneously imaging neuron activation (Chronis, Zimmer, & Bargmann, 2007). Finally, other microfluidic devices allow for high-throughput single-worm behavior protocols (Fig. 13).
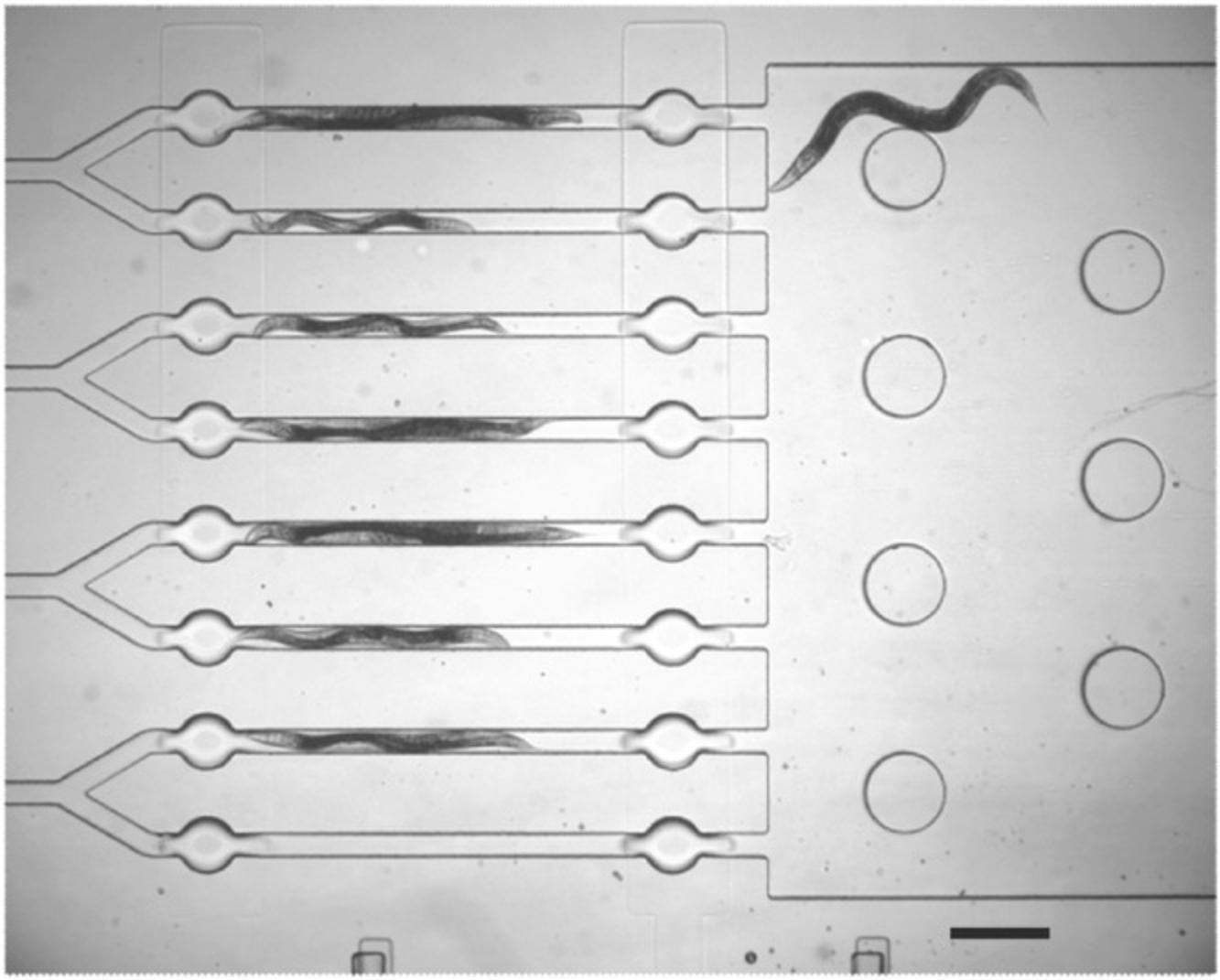
To build a microfluidic device, liquid polydimethylsiloxane (PDMS) can be molded into micro chambers allowing for the passage of single worms into confined yet visible spaces. Valves can be included in the chamber design to allow the passage of air as well as chemicals. Some researchers use these microfluidic devices in order to increase throughput of single-worm behavioral assays, as movement of worms in and out of the experiment arena can be accomplished by changing the air or volume pressure within the chamber at the flip of a switch, instead of the time-consuming manual transfer of worms from plate to plate. Instructions for designing and building your own microfluidic chamber can found in WormBook (San-Miguel & Lu, 2013).
Optogenetics and chemical imaging
Due to the transparent outer cuticle, the internal structures of the animal are visible using a basic light microscope. This transparency confers a special advantage upon the worm as a model organism in that proteins fused to fluorescence reporter proteins can be visualized within the intact organism, as indicated earlier. In fact, almost any genetically encoded reporter can be visualized through the cuticle of the worm. Beyond GFP, other genetically encoded reporters include calcium indicators (GECI) that express a calcium-sensitive protein that changes conformation in the presence of calcium, bringing together components that will fluoresce when in proximity to one another. Some GECIs emit one color wavelength under low calcium conditions that changes to another color wavelength in the presence of calcium via fluorescence resonance emission transfer (FRET; Kerr et al., 2000), while other GECI's are non-fluorescing under low calcium conditions, with the appearance of fluorescence indicating that calcium levels have increased (e.g., GCaMP). As a result, researchers can monitor in vivo calcium levels to identify which neurons become active, and also monitor the dynamic activation of identified neurons during an event (Akerboom et al., 2012).
In addition to detecting neuron activation, researchers can also take advantage of expressing genetically encoded light-sensitive channels in the worm: for example, channelrhodopsin (ChR2) or halorhodopsin (NpHR). These channels were originally isolated from green algae and archaea, respectively. When expressed in the neurons of a worm, exposure to particular wavelengths of light will open the ChR2 cation channel and depolarize the neuron or open the NpHR chloride channel and hyperpolarize the neuron. As such, genetic targeting of these light-sensitive channels allows for controlled depolarization or hyperpolarization of a neuron subset, or even a single neuron (Husson, Gottschalk, & Leifer, 2013). Interestingly, activating optogenetic channels in C. elegans does not necessarily require expensive hardware (Yu, McDiarmid, Ardiel, & Rankin, 2019). Initially, researchers noted that the light power from an arc lamp on a fluorescent microscope was sufficient to activate optogenetic channels. Since then, other researchers have reported success in generating activation of these light-sensitive channels using relatively inexpensive wavelength-specific LEDs (Ardiel et al., 2016; Ezcurra, Tanizawa, Swoboda, & Schafer, 2011; Singaram et al., 2011). There are multiple optogenetic strains available from the CGC. With these strains, researchers have been able to investigate the physiology of neuron pathways in the intact animal.
A few tips for using optogenetic C. elegans strains: when using channelrhodopsin strains, cover microscope white-light sources with orange transparent lighting filters (e.g., R23 Orange Cinegel Supergel, Roscolux) to prevent additional blue-wavelength light exposure of ChR2-expressing worms. Additionally, like other systems, channelrhodopsin requires all-trans retinal (ATR; Nagel et al., 2005) to form functional channels, so incorporate ATR into E. coli immediately prior to seeding colony plates (add 1 µl of 100 mM ATR dissolved in ethanol to 250 µl of E. coli OP50 suspension immediately prior to seeding). Limit light exposure to ChR2 worm plates; this can be accomplished by wrapping colony plates in aluminum foil, for instance. Finally, C. elegans express an endogenous blue light-sensitive receptor (LITE-1), so to ensure that effects of a manipulation are due to optogenetics, be sure the opto-transgene is expressed in a lite-1 mutant background.
RESOURCES
The C. elegans community is extraordinarily collaborative, and online databases and repositories exemplify the cooperation of researchers interested in different elements of worm biology. WormBase, WormBook, and WormAtlas represent core information hubs for researchers looking for information about genes, phenotypes, protocols, history, and anatomy. Below, we describe each of these, and list several other helpful sites set up and maintained by the worm community; this list is by no means exhaustive.
WormBase (https://wormbase.org) is the primary repository for all information about C. elegans and some related nematodes. There are many strategies to access this information, but one of the most common is to begin with the gene name in the search box in the upper right corner. WormBase is detailed and extensive, but an FAQ link and a User's Guide accessible from the Support section at the bottom of the main page provide some guidance. The curators at WormBase have a reputation for being exceptionally responsive and helpful. Links to other resources can be found under Community–Resources from the WormBase main page. A brief student introduction to WormBase, which we have used with undergraduate students, is given below.
Review articles that cover many different aspects of worm biology can be found in the WormBook (https://wormbook.org). The Introduction by Corsi, Wightman, and Chalfie (2018) is written by authors who work with undergraduates routinely and provides an update-to-date and thorough background. The WormBook is divided into sections by general biological topic (e.g., Developmental Control, Neurobiology and Behavior, Evolution, and Ecology), each of which contain subheadings that include literature reviews and some compendia of relevant genes and gene functions. New chapters are added regularly and are highlighted at the bottom of the WormBook homepage.
The WormBook also includes WormMethods and WormHistory. WormMethods includes chapters on maintenance, genetics, transgenics, gene expression constructs, microscopy, and more techniques and methods; several of these provide links out to videos showing the method. Worm strains can be frozen in liquid nitrogen and recovered years later; the procedure for this important but occasionally finicky technique is described in the Maintenance chapter of WormMethods. We also offer a protocol below. WormHistory includes all of the Nobel Lectures given by those working with C. elegans including videos, and articles offering an historical perspective on many aspects of worms. The WormBook, WormMethods, and WormHistory are open-access and provide the single best source of information for anyone who is new to worms altogether or wants to learn more about any topic regarding worms.
An extensive collection of images, photographs, drawings, videos, and much other useful information about worm morphology are found at the WormAtlas (https://wormatlas.org), a compendium of anatomical data and illustrations about C. elegans throughout development. The WormAtlas features several handbooks of anatomy based on imaging data, each one with an introduction and one or more clickable “chapters” of visual information. Other features of WormAtlas include interactive collections of EM images in the “slidable worm” and wiring diagrams based on the Mind of the Worm (discussed elsewhere). There are also in-depth Resources for investigating cell lineages, viewing digital movies of development, or describing individual neurons, among others.
Strains can be obtained, for a small fee, from the Caenorhabditis Genetics Center (CGC) at the University of Minnesota (https://cgc.umn.edu/). The CGC also has a very brief introduction to C. elegans , as well as information on gene names and laboratories.
Some Other Online Resources
WormWeb (http://www.wormweb.org) : This website is not actively maintained currently, but includes a neural connectome viewer and an intron-exon mapping tool that is useful for making images of gene structure based on sequences downloaded from WormBase (see above).
WormBuilder (http://www.wormbuilder.org/) : This website includes information about MosSCI insertions for single-copy transgenesis (see description of transgenics and gene modification). It also includes useful information about plasmids, selection markers, optimized fluorophores, and existing strains, many with direct links to suppliers.
MMP (http://genome.sfu.ca/mmp/): The Million Mutation project produced over 2000 homozygous mutagenized strains that result in an average of nine novel mutations per protein-coding gene in wild-type (N2) background. Thus, allelic series are available for most non-lethal genes. There are also wild isolates represented.
GExplore (http://genome.sfu.ca/gexplore/gexplore_search_mutations.html): This website features a search tool to aid in looking for specific (or all) mutations in a gene of interest. Alleles generated through the Million Mutation Project as well as other consortia are searchable.
Worms in Teaching Laboratories
We have often used C. elegans in teaching and research labs with undergraduate students. The sections below are taken and adapted from our lab protocol manuals and include the recipes, methods, and tips that we include for working with worms in undergraduate labs.
The recipes for nematode growth medium below for routine cultures of worms are based on using the E. coli strain OP50 (a uracil auxotroph) as the food source, which we prefer. Some labs include streptomycin in the growth media and use the strep-resistant E. coli strain OP51 to reduce bacterial contamination. Both OP50 and OP51 are available as stocks from the CGC.
STRATEGIC PLANNING: HANDLING AND CULTURING WORMS
Bacterial strain for feeding worms
Streak E. coli strain OP50 on an LB plate (Riley, Woodman, Holt, & Stevenson, 2017) and grow overnight at 37°C. To prepare bacteria for seeding plates, pick a single OP50 colony into a 30-ml conical tube of LB broth (Riley et al., 2017) and set the culture shaking at 37°C overnight. Alternatively, OP51 (OP50-1) can be streaked on LB+strep plates (see Riley et al., 2017, for preparation of antibiotic plates), and plates with streptomycin can be used in subsequent culture steps. Bacterial cultures in broth can be used for up to 1 week after growth, but fresh is better.
RNAi feeding bacterial clones
Bacterial strains for RNAi are available commercially, as described. Streak bacteria with RNAi clone on Carb/Tet LB plates as prepared above (see Table 2) and grow overnight at 37°C. To make bacteria for seeding plates, pick a single OP50 colony into a 30-ml conical tube of LB broth plus Carb (see Table 2) and set the culture shaking at 37°C overnight.
| Solution | Concentration | Weight | Volume | Sterilized? | Aliquot |
|---|---|---|---|---|---|
| Cholesterol | 5 mg/ml | 250 mg | 50 ml 95% ethanol | No | No |
| Carbenicillin | 25 mg/ml | 1250 mg | 50 ml diH2O | Yes, by filtering | 1.5-ml tubes, 1010 µl/tube |
| IPTG | 1 M | 5.97 g | 25 ml diH2O | Yes, by filtering | 1.5-ml tubes, 1010 µl/tube |
| Tetracycline | 12.5 mg/ml | 625 mg | 50 ml diH2O | Yes, by filtering | 1.5-ml tubes, 1010 µl/tube |
| Streptomycin | 100 mg/ml | 5 g | 50 ml diH2O | Yes, by filtering | 1.5-ml tubes, 1010 µl/tube |
| CaCl2 | 1 M | 73.505 g | 500 ml diH2O | Yes, both filtered and autoclaved | 50-ml tubes |
| MgSO4 | 1 M | 123.23 g | 500 ml diH2O | Yes, both filtered and autoclaved | 50-ml tubes |
| Levamisole | 100 mg/ml | 10 g | 100 ml diH2O | Yes, by filtering | 1.5-ml tubes, 1010 µl/tube |
Seeding plates
Using a sterile pipette, drop an appropriate volume (2 to 5 drops depending on the size of the plate) onto the center of an agar plate. Tip the plate gently to spread the bacteria around, but try to avoid having bacteria at the edges of the plate, directing the worms toward the center of the plate. Do not damage the surface of the agar with the pipette tip; worms will then burrow under the surface. Let culture grow overnight at 37°C or at room temperature for 2 or 3 days after seeding to form a bacterial lawn before using. Seeded plates can be saved at 4°C in a sealed container for ∼1 week if not to be used immediately.
Contamination
Plates of animals can be contaminated by non-E. coli bacteria, as well as fungi. Contaminating microorganisms can be removed from a population by bleaching gravid hermaphrodites as follows. Prepare 400 μl of 5 N NaOH, 500 μl of bleach, and 100 μl of water. Pick as many gravid animals as possible onto a single pick, and release them into a 10 to 20 μl pool of bleaching solution on a clean, seeded plate; the bleaching solution should be deposited away from the E. coli lawn. The maternal animal and the contaminating microbes will die, but the eggshell will protect developing larvae, which will go on to hatch.
Mites can also colonize plates occasionally, and will prey on animals. There are commercial treatments available (anti-mite papers and powders), but some of these can also affect worm growth and fertility. Mites can be hard to remove once an incubator is infested. Regular use of soap and bleach water to wipe down the interior of incubators will reduce the number of mites.
Making a platinum wire pick for transferring individual worms
Cut a 2- to 3-in. long piece of platinum wire. Use a hammer or pliers to flatten out the very end of the ∼32-G platinum wire. If desired, bend the flat section so that it is at an angle to the rest of the wire. Finally, place the non-flattened end inside a short Pasteur pipette and seal into the glass over a Bunsen burner. About 2 cm of wire should protrude from the glass, but personal preferences vary. Eventually, the end of the pick wears out, and the end can be cut off and a new end flattened and bent.
Transferring worms
Worms live and reproduce between 15° and 25°C, and are often grown at room temperature. Bacteria are spread as a lawn on the surface of an agar plate, and the worms crawl on the surface grazing on the bacteria. Bacteria are ingested and pumped back to a muscular pharynx. The pharynx consists of two lobes connected by an isthmus; the second lobe is known as the grinder.
An adult worm is approximately a millimeter long, barely visible with the naked eye. It is easiest to observe them using a dissecting microscope with substage illumination. The worms crawl and eat continuously, so until you get used to their movement, it seems like they are really scooting. Once you get used to it, the nice smooth waves have a certain calming effect on the worm researcher. Hermaphrodites typically move forward, and only back up if they bump into something. Males are more active than hermaphrodites, and back up frequently. The worm is transparent, so you may be able to make out the fertilized eggs lined up inside the gonad of an adult hermaphrodite.
Two methods for picking worms
Transferring individual worms takes practice and a bit of patience. As with many protocols, there are numerous online videos describing different ways to pick worms (see YouTube—keyword search: “picking worms; elegans”). Every person develops an individual method and has an individual type of wire pick that they like best. Here are two suggested methods.
Method 1 : To move a worm to another plate, use a platinum-wire worm pick. Some people prefer a slightly flattened pick, formed like a scoop, while others prefer a pointed tip. Tap the worm on the snout and watch it back up. When you are ready to pick one up, tuck the pick under its middle and quickly lift it up. As quickly as possible, remove the lid from the other petri dish and allow the worm to crawl off your pick. This takes practice, but you must be able to do this without poking a hole in the surface of the agar plate. Worms will crawl down any hole in the surface and will be lost to your experiment. This will seem difficult or impossible at first, but keep at it.
Some tips are:
- • move as quickly as possible so that the worm stays near the tip
- • keep your wrist as flat as possible so that you are picking at a flat trajectory
- • practice
Method 2 : To pick up worms, flame the wire to sterilize it, and drag the pick along a bacterial lawn to coat the underside of the pick with bacteria. Goopy, several day old bacterial lawns are the best for this; some people keep old but non-contaminated plates next to the microscope just for coating their picks. Gently dab the worm to be picked up so that it sticks to the bacteria on the pick. To set the worm down, gently brush against the new plate, and allow the worm to crawl off. The worm should be immediately active on the new plate if it is undamaged. It is possible to pick up many worms onto your pick at a time; however, you don't want the worms to spend too much time on the pick, as they will dry out.
PROTOCOLS
Basic Protocol 1: FREEZING AND THAWING WORMS
Sydney Brenner wrote: “ everything survives freezing—it's the thawing that typically does the damage” (Brenner, 2009). C. elegans are remarkably robust when it comes to thawing, which means that freezing animals is a straightforward way to maintain strains for the long term.
Materials
-
Worms to be frozen
-
11 plates, seeded for worm growth as described in Strategic Planning (see recipe for NGM medium in Reagents and Solutions)
-
Freezing solution (see recipe in Reagents and Solutions)
-
10× M9 buffer (see recipe)
-
Cryovials
-
Small metal spatula, such as for weighing chemicals, sterilized
Cryopreservation
1.Set up cultures of about five worms each on 10 seeded plates. The 11th plate is needed for recovery.
2.Allow worms to grow to high density on the seeded plates; do not transfer them to new plates. When worm cultures are at high density but not starved, continue with the next steps. Starved worms will stop laying eggs.
3.Melt freezing solution and keep 2 ml melted in a heat block at 37°C.
4.Wash the worms off of their plates into a 15-ml conical tube using 4 to 5 ml of 1× M9 buffer. The buffer will be absorbed into the plates, so the final volume will only be about 2 ml. Wash the plates serially, so that the worms stay concentrated.
5.Put the conical tube with worms (in M9) on ice for 15 min.
6.Mix the 2 ml of freezing solution into the worm/M9 mixture and divide the mixture into two labeled cryovials.
7.Put the cryovials at −80°C, and allow the worms to freeze down slowly.
Recovery
8.Use a metal spatula to scrape about 250 μl of frozen worm slurry from a cryotube.
9.Deposit the frozen material on a seeded plate by scraping the spatula against the side of the plate. The frozen worms and freezing solution should drop onto the NGM agar.
10.Allow the mass to thaw with the lid slightly ajar.
11.The plate can be inverted and stored as usual after about 2 hr at room temperature.
12.Check for emergent L1 animals as soon as 4 hr post-thaw, though you may need to wait as long as 4 days if there are few of them and/or they are difficult to find before adulthood.
Basic Protocol 2: BLEACHING C. elegans TO OBTAIN EGGS AND L1s
Bleach solution will kill worms, but will not destroy eggs because they contain a protective eggshell. The eggs retained in a worm are within ∼2 hr of fertilization, so bleaching can be used to synchronize populations of animals.
CAUTION : While handling bleach, wear gloves, goggles, and a lab coat.
Materials
-
Bleach
-
Sodium hydroxide (NaOH)
-
1× M9 buffer That is, dilute 10× M9 1 to 10 before using (see recipe for 10×), sterile (2 to 3 L may be needed for washes)
-
Adult gravid hermaphrodites (several plates are recommended)
-
100-mm seeded plates for recovery (see Strategic Planning; see recipe for NGM medium in Reagents and Solutions)
-
15-ml conical tubes
-
Standard benchtop centrifuge
-
Pasteur pipettes to remove excess liquids
-
Rotator or shaker
-
Compound microscope and dissecting microscope
-
Roller platform
-
Coverslips
1.Make a fresh solution of bleach and NaOH, as below. Do not use if >1 week old.
| 5 ml | 10 ml | 50 ml | |
| Sterile H2O | 3.35 ml | 6.7 ml | 33.5 ml |
| 8.25% bleach | 1.45 ml | 2.9 ml | 14.5 ml |
| 50% (w/w) NaOH | 200 μl | 400 μl | 2.0 ml |
Commercial bleach (from the grocery store) is usually about 8.25%, but different brands vary in their concentration and effectiveness. In general, the cheaper brands without scent-masking additives work a little better.
2.Wash gravid worms from the plates with 1× M9 buffer into the 15-ml conical tubes.
3.Spin in the benchtop centrifuge for 1 min 3000 rpm (set brakes on centrifuge to HIGH).
4.Remove excess liquid using Pasteur pipettes, leaving about 1 ml in tube
5.Wash one or two additional times with 1× M9 until the solution is no longer cloudy with food.
6.Aspirate last wash down to 3 ml worms in M9.
7.Add 3 ml of bleach/NaOH mix from step 1.Shake vigorously by hand and monitor adult lysis under the microscope. Adults should break open and dissolve at ∼7 min. Be careful about the time, since over-bleaching will damage the eggs, while under-bleaching will cause bacterial contamination of the egg prep.
8.As soon as adult bodies are all broken open and starting to dissolve, add 6 to 8 ml of 1× M9.
9.Immediately spin the worms in bleach/M9 mixture (step 8) for no more than 1 min at 3000 rpm, then brake rapidly. Aspirate as much liquid as possible without disturbing egg pellet at bottom of 15-ml tube.
10.Resuspend pellet in 1× M9, then shake until pellet is entirely resuspended.
11.Spin 1 min at 3000 rpm.
12.Repeat wash and spin twice.
13.Suspend embryos in 1× M9 to 1 to 3 ml, depending on number of animals. Shake hard to break apart embryos sticking together.
14.Examine eggs under microscope. In a good preparation, the egg shell will be shiny and smooth.
15.Put 15-ml tube on a roller platform overnight.
16.The next day, dispense 5 to 10 µl of the preparation onto a coverslip and count the number of L1 larvae under a dissecting microscope to estimate survival, if desired
17.Dispense animals on to larger growth plates, or into liquid culture for growth.
Basic Protocol 3: GENERATING MALES BY HEAT SHOCK
Males arise spontaneously from hermaphrodite self-fertilization at about 1 in 500 eggs laid, which is often too infrequently to be useful for performing genetic crosses. The frequency of self-progeny males can be increased by employing heat shock.
Materials
- Six or more plates with at least five L4 hermaphrodites of the desired genotype on each
- Incubator at 30°C
1.Incubate the plates with the L4 hermaphrodites for 4 to 6 hr at 30°C. Longer times can be used but result in fewer progeny.
2.Return the plates to 20°C.
3.After 3 days, screen the plates for males. These can be recognized by the distinctive morphology of the tail and by their behavior, as shown in Figure 2 and described above. There will probably be many eggs that do not hatch, but there should be a few males per plate in the F1.
4.To establish a culture with many males that can be used for genetic crosses, the newly arisen males are crossed to hermaphrodites of the desired genotype. Since the newly arisen males from the heat shock often have low fertility, it is helpful to put an excess of these males (as many as 8 to 10) with a few L4 hermaphrodites of the same genotype. The next generation should have many males that arise from cross-fertilization. These males can then be used for subsequent genetic crosses, or for establishing a strain that routinely produces a high number of males.
Basic Protocol 4: LYSIS OF SINGLE WORMS FOR PCR ANALYSIS
Materials
-
PCR lysis buffer (see recipe; at least 1 μl for each worm to be analyzed)
-
Proteinase K
-
Cultures of worms to be analyzed
-
PCR tubes
-
Worm pick for transfer
-
Microcentrifuge for PCR tubes
-
−80°C freezer
-
Thermal cycler
1.Add proteinase K to the lysis buffer for a final concentration of 200 μg/ml. Keep mixture on ice.
2.Put 1 μl (or more if desired) of the lysis buffer/proteinase K mixture into the cap of a PCR tube.
3.Transfer a worm into the drop in the cap of the tube and quickly cap the tube.
4.Spin the lysis solution with the worm down to the bottom of the PCR tube using a microcentrifuge.
5.Freeze PCR tubes at −80°C for 10 to 15 min.
6.Put the PCR tube in a thermocycler: 60°C for 60 min, then 95°C for 15 min. This allows the proteinase K to digest the worm proteins, and then inactivates the enzyme.
7.This lysate may be stored at −20°C until needed. When ready to begin PCR, add the PCR reagents to the tube with the worm lysate.
Basic Protocol 5: IMMOBILIZING C. elegans FOR CONFOCAL MICROSCOPY
The cellular anatomy of worms is best analyzed by confocal microscopy.
Materials
-
Melted 3% agarose in H2O
-
10 mM levamisole (Table 2; dilute 100 mM levamisole stock in H2O)
-
Worm culture
-
Pasteur pipet and bulb
-
Worm pick
-
Microscope slides and coverslips
1.Using a Pasteur pipet, place a drop of melted agarose on a microscope slide.
2.Immediately place another slide on top of the first at a 90° angle. After about a minute, slide the slides apart. This will leave a small pad of agarose on one of the slides.
3.Pipet a 3 µl drop of 10 mM levamisole onto the agarose pad.
4.Pick animals into the levamisole as quickly as possible (in <3 min). Do not jab the agarose pad, and transfer as few bacteria as possible.
5.Once the worms appear immobilized, gently place a coverslip over the agarose pad and observe the worms using confocal microscopy. It is generally difficult to recover worms from the agarose pad, so these are typically discarded after use.
Basic Protocol 6: WormBase TUTORIAL
This tutorial was designed for intermediate-level biology students with no C. elegans experience and very little experience with bioinformatics. The questions can guide a user through WormBase as a way to help familiarize them with some of the tools of the site. As WormBase is updated and changed, this tutorial may become out of date, so an instructor will want to try it out before assigning it.
1.Choose one of the following gene IDs for your subsequent analysis. Use the entire DNA sequence, including both the uppercase and lowercase letters.
- C25E10.12
- D1022.1
- F55A11.3
2.To determine what information is already available on these genes, go to the BLAST website (http://blast.ncbi.nlm.nih.gov/Blast.cgi), enter your sequence and determine the results.
3.The sequences above are the entire unspliced transcripts. Look for the best mRNA/cDNA match for the following questions:
-
How far down the list of matches do you need to go in order to find this match?
-
C. elegansgenes are named with three to four lowercase letters, followed by a dash and one to three numerals. Based on the gene results, what are the names of these genes (Unnamed genes are designated by a longer string of letters and numbers that describe a genes position relative to a short portion of the genome.)
-
What is the e-value of the top BLAST hit that you obtained with the gene you chose?
4.From this point forward, choose ONE gene to follow up on.
5.Copy the corresponding Alignment information, accession number, and the description of your BLAST hit below. Locate the information for the following questions.
-
If you click on the Sequence ID of your BLAST hit, you will be redirected to the NCBI resource page for that gene. Are there any publications listed for this protein (i.e., has it been studied before)? How could that information be useful for your future studies of this gene?
-
What is the predicted protein sequence for your gene (include it below)? Include the number of amino acids in your protein.
-
Click on the GeneID link (located under FEATURES within the “gene” description section). What is the gene immediately upstream of your gene?
-
Click on “Protein” (right hand link under “Related Information”). You can then use the “Related information” menu to find Conserved Domains. Are there any conserved domains present in your protein? What function does/do it/they serve? This may require a bit of searching (including looking at publications listed on the NCBI resource page).
There are two ways to find out more information about your gene (Method 1: Genomic sequence search, and Method 2: Protein sequence search)
6.Method 1 : Genomic sequence search.
- Click on the WormBase ID following the text/db_xref=Wormbase:in the NCBI resources text under gene or CDS.
This will redirect you to a gene-information page at WormBase, which is a C. elegans sequence database. WormBase has large variety of collapsible/expandable sections under the Page Content heading on the left-hand side of the page (i.e., Location, Reagents, Phenotypes, Sequences, etc.). Take a few minutes to navigate through the page and get comfortable manipulating the show/hide options for each collapsible section.
What is the WormBase ID for your gene?Cut and paste the informational blurb about your gene here.In the box to the right of the general information, there are some important notes.What species is your gene from (full name)?Does your gene have other names?
-
Click on “Genetics.” What kinds of alleles are currently available for your gene?
-
Click on “Reagents.” What kinds of reagents are available for studying this gene (i.e., transgenes, primers, antibodies, etc.)?If you click on a transgene, what kind of information can you find?If you click on a mutant allele, what kind of information can you find?
-
Click on “Expression.” Paste an image that described the expression of your gene. What do you think this pattern means?On which chromosome is the gene located?Based on the gene's placement in the Genome Browser (“Location”, is it in the FWD or REV direction on the chromosome (i.e., check the direction of the ORF within the chromosome, which is displayed in the 5′→3′ direction)?Under Gene Model and Products, the Genomic Browser will show intron sequences as linear regions between exons (which are shown as rectangular boxes) under “Transcript.” Based on this initial schematic of your gene and the surrounding sequences, does your gene have any introns? If so, how many? How many exons does it have?
-
Click on the “Sequences” menu.How many mRNA transcripts does this gene encode?Click on one transcript and display the “spliced transcript + UTR” by clicking on the small arrow. Select the sequence of the spliced transcript not including the 3′ and 5′ UTRs and paste it here.What do the colored texts represent?Which of the sequences shown will be most useful for predicting amplicon sizes for a genomic PCR genotyping experiment?
-
Click on “Homology.” What sorts of organisms have homologous proteins encoded in their genomes? Does your gene have any paralogs?
-
Under Sequences, click on Protein.What does WormBase suggest is the best human ortholog to your gene? Where do you think that this information came from?How big is your protein, in kDa?
7.Method 2: Protein sequence search.
- Enter yourproteinsequence into BLAST and do a non-redundant protein sequences (nr) search. Whatnon-Caenorhabditisproteins come up? List the top three non-Caenorhabditishits (include host species, protein name and their accession number but try to pick three hits from three different organisms).
Do not lose track of these pages.
Does your protein seem to be conserved? How do you know?Go to CLUSTAL OMEGA (http://ebi.ac.uk/Tools/msa/clustal/) and perform an alignment on yourC. elegansprotein sequence along with your sequences from other organisms. Paste the CLUSTAL alignment (with numbers) below.How highly conserved do these proteins look? Where do you think the highest level of conservation lies? How does that correlate with specific protein domains?How likely is it that your three non-Caenorhabditishits have the same biochemical function (i.e., same ligand binding and/or enzymatic activities) or same cellular function (i.e., involved in the same cellular process—such as transcriptional control) as yourCaenorhabditisprotein?
REAGENTS AND SOLUTIONS
Laboratory stock solutions to keep on hand are listed in Table 2.
Freezing solution
Mix the following:
| 200 ml | 1 M NaCl | (20 ml) |
| 100 ml | 1 M potassium phosphate, pH 6 | (10 ml) |
| 600 ml | Glycerol | (60 ml) |
| 2 L | Deionized water | 200 ml (110 ml H2O) |
Aliquot into 200-ml bottles and autoclave. This can be stored at 4°C indefinitely.
Before use:
- Add 60 μl 1 M MgSO4 to each 200-ml aliquot.
- Add 0.4 g agar to 100-ml aliquots of the freezing solution, and autoclave.
M9 buffer, 10×
- 59.6 g Na2HPO4
- 29.9 g KH2PO4
- 12.8 g NaCl
- 2.5 MgSO4
- 1 L diH2O
Add the above ingredients one at a time while stirring 750 ml diH2O. When dissolved, bring the volume to 1 L. Filter the M9 and store at room temperature. Dilute 1:10 and autoclave. M9 buffer is also commercially available at various concentrations.
NGM (nematode growth medium) for pouring plates
| NaCl | Peptone | Agar | H2O | |
| NGM | 3 g | 2.5 g | 17 g | 1 L |
After autoclaving, put the medium on a stir plate at 70°C and a stir at a moderate speed.
Let medium cool until you can keep a hand on the side of the flask for >5 s. This should take about an hour, and will obviously take longer for increased volumes.
In a laminar flow hood or under aseptic conditions add the following to each liter of medium.
| Stock solution (concentration) | Volume to add to 1 L medium |
| Cholesterol (5 mg/ml in ethanol; see Table 2) | 1 ml |
| CaCl2 (1 M) (sterile) | 1 ml |
| MgSO4 (1 M) (sterile; see Table 2) | 1 ml |
| KH2PO4 (1 M) (pH = 6.0) (sterile) | 25 ml |
Add the cholesterol slowly so that it will not precipitate in the medium. Once all the cholesterol is added, let it mix into solution. Slowing down the stir bar helps the cholesterol dissipate, but return the bar to normal speed when you add the remaining chemicals. CaCl2, MgSO4, and KH2PO4 can be added at a normal rate, since they do not precipitate.
If desired, add 1 ml of streptomycin (100 mg/ml; see Table 2) per liter.
Many C. elegans labs use 60-mm petri dishes for routine worm transfers and experiments, and larger (100-mm) petri dishes for long-term maintenance and preparing worm cultures for freezing. The 60-mm plates are best poured with a pump that delivers a uniform volume. The plate should be roughly two-thirds full of agar. The 100-mm plates can be poured by hand or with a pump. Plates that will not be used within 3 days can be stored in a sealed container at 4°C for at least 1 month.
For making RNAi plates, add 1 ml of 1 M IPTG (see Table 2) and 1 ml of carbenicillin (see Table 2; 25 mg/ml) per liter.
PCR lysis buffer
| Stock | Volume | Final concentration |
| 1 M KCl | 2.5 ml | 50 mM KCl |
| 1 M Tris, pH 8.8 | 500 μl | 10 mM Tris |
| 1 M MgCl2 | 125 μl | 2.5 mM MgCl2 |
| 10% Tween 20 | 2.25 ml | 0.45% Tween 20 |
| 10% NP-40 | 2.25 ml | 0.45% NP-40 |
| H2O | 42.4 ml | |
| 50 ml |
The KCl, Tris, MgCl2, and H2O should be sterilized by autoclaving. Volumes > 50 ml can be prepared and stored at 4°C indefinitely.
Potassium phosphate buffer (KH2PO4)
Make the following two solutions and mix them to obtain the correct pH 6.0:
1 M KH2PO4 (monobasic): Add 136 g to 750 ml water. After it has gone into solution, bring the final volume to 1 L.
1 M KH2PO4 (dibasic): Add 174 g to 750 ml water. After it has gone into solution, bring the final volume to 1 L.
Add the 1 M dibasic solution to the 1 M monobasic solution to bring the pH up from ∼4 to 6.0. This should require ∼150 ml of dibasic solution for 500 ml of monobasic.
Vacuum filter the solution and store it at room temperature.
Literature Cited
- Ahringer, J. (2006). Reverse genetics. WormBook. Available at https://static.yanyin.tech/literature/current_protocol/10.1002/cpet.35/attachments/introreversegenetics.pdf.
- Akerboom, J., Chen, T.-W., Wardill, T. J., Tian, L., Marvin, J. S., Mutlu, S., … Looger, L. L. (2012). Optimization of a GCaMP calcium indicator for neural activity imaging. Journal of Neuroscience , 32, 13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012.
- Ardiel, E. L., Giles, A. C., Yu, A. J., Lindsay, T. H., Lockery, S. R., & Rankin, C. H. (2016). Dopamine receptor DOP-4 modulates habituation to repetitive photoactivation of a C. elegans polymodal nociceptor. Learning Memory , 23, 495–503. doi: 10.1101/lm.041830.116.
- Au, V., Li-Leger, E., Raymant, G., Flibotte, S., Chen, G., Martin, K., … Moerman, D. G. (2019). CRISPR/Cas9 methodology for the generation of knockout deletions in Caenorhabditis elegans. G3 (Bethesda, MD) , 9, 135–144.
- Barriere, A., & Felix, M. A. (2005). Natural variation and population genetics of Caenorhabditis elegans. WormBook. Available at http://www.wormbook.org/chapters/www_naturalvariationgenetics/naturalvariationgenetics.html.
- Bergs, A., Schultheis, C., Fischer, E., Tsunoda, S. P., Erbguth, K., Husson, S. J., … Liewald, J. F. (2018). Rhodopsin optogenetic toolbox v2.0 for light-sensitive excitation and inhibition in Caenorhabditis elegans. PloS One , 13, e0191802. doi: 10.1371/journal.pone.0191802.
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics , 77, 71–94.
- Brenner, S. (2009). In the beginning was the worm. Genetics , 182, 413–415. doi: 10.1534/genetics.109.104976.
- Bretscher, A. J., Kodama-Namba, E., Busch, K. E., Murphy, R. J., Soltesz, Z., Laurent, P., & de Bono, M. (2011). Temperature, oxygen, and salt-sensing neurons in C. elegans are carbon dioxide sensors that control avoidance behavior. Neuron , 69, 1099–1113. doi: 10.1016/j.neuron.2011.02.023.
- Brusca, R. (2016). Invertebrates. Oxford, New York: Oxford University Press.
- C. elegans Deletion Mutant Consortium. (2012). Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda, MD) , 2, 1415–1425. doi: 10.1534/g3.112.003830.
- C. elegans Sequencing Consortium. (1998). Genome sequence of the nematode C. elegans : A platform for investigating biology. Science , 282(5396), 2012–2018. doi: 10.1126/science.282.5396.2012.
- Chalfie, M., Sulston, J. E., White, J. G., Southgate, E., Thomson, J. N., & Brenner, S. (1985). The neural circuit for touch sensitivity in Caenorhabditis elegans. Journal of Neuroscience , 5, 956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985.
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., & Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science , 263, 802–805. doi: 10.1126/science.8303295.
- Chang, A. J., Chronis, N., Karow, D. S., Marletta, M. A., & Bargmann, C. I. (2006). A distributed chemosensory circuit for oxygen preference in C. elegans. PloS Biology , 4, e274. doi: 10.1371/journal.pbio.0040274.
- Chelur, D. S., & Chalfie, M. (2007). Targeted cell killing by reconstituted caspases. Proceedings of the National Academy of Sciences , 104, 2283–2288. doi: 10.1073/pnas.0610877104.
- Cherra, S. J., & Jin, Y. (2015). Advances in synapse formation: Forging connections in the worm. Wiley Interdisciplinary Reviews: Developmental Biology , 4, 85–97. doi: 10.1002/wdev.165.
- Chronis, N., Zimmer, M., & Bargmann, C. I. (2007). Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nature Methods , 4, 727–731. doi: 10.1038/nmeth1075.
- Cook, S. J., Jarrell, T. A., Brittin, C. A., Wang, Y., Bloniarz, A. E., Yakovlev, M. A., … Emmons, S. W. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature , 571, 63–71. doi: 10.1038/s41586-019-1352-7.
- Corsi, A. K., Wightman, B., & Chalfie, M. (2015). A transparent window into biology: A primer on Caenorhabditis elegans. Genetics , 200, 387–407. doi: 10.1534/genetics.115.176099.
- Corsi, A. K., Wightman, B., & Chalfie, M. (2018). A Transparent window into biology: A primer on Caenorhabditis elegans (WormBook). Available at https://static.yanyin.tech/literature/current_protocol/10.1002/cpet.35/attachments/celegansintro.pdf.
- Denzel, M. S., Lapierre, L. R., & Mack, H. I. D. (2019). Emerging topics in C. elegans aging research: Transcriptional regulation, stress response and epigenetics. Mechanisms of Ageing and Development , 177, 4–21. doi: 10.1016/j.mad.2018.08.001.
- Dickinson, D. J., Pani, A. M., Heppert, J. K., Higgins, C. D., & Goldstein, B. (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics , 200, 1035–1049. doi: 10.1534/genetics.115.178335.
- Ellis, H. M., & Horvitz, H. R. (1986). Genetic control of programmed cell death in the nematode. C. elegans. Cell , 44(6), 817–829.
- Ezcurra, M., Tanizawa, Y., Swoboda, P., & Schafer, W. R. (2011). Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO Journal , 30, 1110–1122. doi: 10.1038/emboj.2011.22.
- Fang-Yen, C., Gabel, C. V., Samuel, A. D. T., Bargmann, C. I., & Avery, L. (2012). Laser microsurgery in Caenorhabditis elegans. Methods in Cell Biology , 107, 177–206. doi: 10.1016/B978-0-12-394620-1.00006-0.
- Félix, M. A. (2008). RNA interference in nematodes and the chance that favored Sydney Brenner. Journal of Biology , 7(9), 34. doi: 10.1186/jbiol97.
- Félix, M.-A., Jovelin, R., Ferrari, C., Han, S., Cho, Y. R., Andersen, E. C., … Braendle, C. (2013). Species richness, distribution and genetic diversity of Caenorhabditis nematodes in a remote tropical rainforest. BMC Evolutionary Biology , 13, 10. doi: 10.1186/1471-2148-13-10.
- Frézal, L., & Félix, M.-A. (2015). C. elegans outside the Petri dish. ELife , 4, e05849. doi: 10.7554/eLife.05849.
- Frøkjær-Jensen, C., Davis, M. W., Sarov, M., Taylor, J., Flibotte, S., LaBella, M., … Jorgensen, E. M. (2014). Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nature Methods , 11, 529–534. doi: 10.1038/nmeth.2889.
- Gabel, C. V., Gabel, H., Pavlichin, D., Kao, A., Clark, D. A., & Samuel, A. D. T. (2007). Neural circuits mediate electrosensory behavior in Caenorhabditis elegans. Journal of Neuroscience , 27, 7586–7596. doi: 10.1523/JNEUROSCI.0775-07.2007.
- Gendrel, M., Atlas, E. G., & Hobert, O. (2016). A cellular and regulatory map of the GABAergic nervous system of C. elegans. ELife , 5, e17686. doi: 10.7554/eLife.17686.
- Ghosh, D. D., Nitabach, M. N., Zhang, Y., & Harris, G. (2017). Multisensory integration in C. elegans. Current Opinion in Neurobiology , 43, 110–118. doi: 10.1016/j.conb.2017.01.005.
- Gray, J. M., Hill, J. J., & Bargmann, C. I. (2005). A circuit for navigation in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America , 102, 3184–3191. doi: 10.1073/pnas.0409009101.
- Greenwald, I. (2005). Introduction to signal transduction (WormBook). Available at http://wormbook.org/chapters/www_introsigtrans/introsignaltransd.html.
- Hall, D. H., & Russell, R. L. (1991). The posterior nervous system of the nematode Caenorhabditis elegans : Serial reconstruction of identified neurons and complete pattern of synaptic interactions. Journal of Neuroscience , 11, 1–22. doi: 10.1523/JNEUROSCI.11-01-00001.1991.
- Harbinder, S., Tavernarakis, N., Herndon, L. A., Kinnell, M., Xu, S. Q., Fire, A., & Driscoll, M. (1997). Genetically targeted cell disruption in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America , 94, 13128–13133. doi: 10.1073/pnas.94.24.13128.
- Hart, A. C. (2006). Behavior (WormBook). http://wormbook.org/chapters/www_behavior/behavior.html.
- Hart, A. C., Sims, S., & Kaplan, J. M. (1995). Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature , 378, 82–85. doi: 10.1038/378082a0.
- Husson, S. J., Costa, S. W., Schmitt, C., & Gottschalk, S. J. (2012). Keeping track of worm trackers. Available at: http://www.wormbook.org/chapters/www_tracking/tracking.html.
- Husson, S. J., Gottschalk, A., & Leifer, A. M. (2013). Optogenetic manipulation of neural activity in C. elegans : From synapse to circuits and behaviour. Biologie Cellulaire , 105, 235–250. doi: 10.1111/boc.201200069.
- Jiang, H., & Wang, D. (2018). The microbial zoo in the C. elegans intestine: Bacteria, fungi and viruses. Viruses , 10(2), 85.
- Kenyon, C. J. (2010). The genetics of ageing. Nature , 464, 504–512. doi: 10.1038/nature08980.
- Kerr, R., Lev-Ram, V., Baird, G., Vincent, P., Tsien, R. Y., & Schafer, W. R. (2000). Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron , 26, 583–594. doi: 10.1016/S0896-6273(00)81196-4.
- Kim, H.-M., & Colaiácovo, M. P. (2016). CRISPR-Cas9-guided genome engineering in C. elegans. Current Protocols in Molecular Biology , 115, 31.7.1–31.7.18. doi: 10.1002/cpmb.7.
- Kiontke, K. (2006). Ecology of Caenorhabditis species (WormBook). Available at http://wormbook.org/chapters/www_ecolCaenorhabditis/ecolCaenorhabditis.html.
- Kiontke, K. C., Félix, M.-A., Ailion, M., Rockman, M. V., Braendle, C., Pénigault, J.-B., & Fitch, D. H. A. (2011). A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evolutionary Biology , 11, 339. doi: 10.1186/1471-2148-11-339.
- Kobayashi, J., Shidara, H., Morisawa, Y., Kawakami, M., Tanahashi, Y., Hotta, K., & Oka, K. (2013). A method for selective ablation of neurons in C. elegans using the phototoxic fluorescent protein, KillerRed. Neuroscience Letters , 548, 261–264. doi: 10.1016/j.neulet.2013.05.053.
- Lee, R. C., Feinbaum, R. L., & Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell , 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y.
- Levitan, D., Doyle, T. G., Brousseau, D., Lee, M. K., Thinakaran, G., Slunt, H. H., … Greenwald, I. (1996). Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proceedings of the National Academy of Sciences , 93, 14940–14944. doi: 10.1073/pnas.93.25.14940.
- Long, L., Guo, H., Yao, D., Xiong, K., Li, Y., Liu, P., … Liu, D. (2015). Regulation of transcriptionally active genes via the catalytically inactive Cas9 in C. elegans and D. rerio. Cell Research , 25, 638–641. doi: 10.1038/cr.2015.35.
- Maricq, A. V., Peckol, E., Driscoll, M., & Bargmann, C. I. (1995). Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature , 378, 78–81. doi: 10.1038/378078a0.
- McDiarmid, T. A., Au, V., Loewen, A. D., Liang, J., Mizumoto, K., Moerman, D. G., & Rankin, C. H. (2018). CRISPR-Cas9 human gene replacement and phenomic characterization in Caenorhabditis elegans to understand the functional conservation of human genes and decipher variants of uncertain significance. Disease Models & Mechanisms, 11(2), pii: dmm036517.
- Nagel, G., Brauner, M., Liewald, J. F., Adeishvili, N., Bamberg, E., & Gottschalk, A. (2005). Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Current Biology , 15, 2279–2284. doi: 10.1016/j.cub.2005.11.032.
- Norris, A. D., Kim, H.-M., Colaiácovo, M. P., & Calarco, J. A. (2015). Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics , 201, 449–458. doi: 10.1534/genetics.115.180679.
- Pereira, L., Kratsios, P., Serrano-Saiz, E., Sheftel, H., Mayo, A. E., Hall, D. H., … Hobert, O. (2015). A cellular and regulatory map of the cholinergic nervous system of C. elegans. ELife , 4, e12432. doi: 10.7554/eLife.12432.
- Riley, S. P., Woodman, M. E., Holt, J., & Stevenson, B. (2017). Culture of E. coli and related bacteria. Current Protocols in Microbiology , 15, 4.2.1–4.2.30. doi: 10.1002/cpet.17.
- Rodriguez, M., Snoek, L. B., De Bono, M., & Kammenga, J. E. (2013). Worms under stress: C. elegans stress response and its relevance to complex human disease and aging. Trends in Genetics , 29, 367–374. doi: 10.1016/j.tig.2013.01.010.
- Saeki, S., Yamamoto, M., & Iino, Y. (2001). Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. Journal of Experimental Biology , 204, 1757–1764.
- San-Miguel, A., & Lu, H. (2013). Microfluidics as a tool for C. elegans research (WormBook). Available at http://www.wormbook.org/chapters/www_microfluidics/microfluidics.html.
- Savell, K. E., Bach, S. V., Zipperly, M. E., Revanna, J. S., Goska, N. A., Tuscher, J. J., … Day, J. J. (2019). A neuron-optimized CRISPR/dCas9 activation system for robust and specific gene regulation. eNeuro , 6(1), pii: ENEURO.0495-18.2019. doi: 10.1523/ENEURO.0495-18.2019.
- Schulenburg, H., & Félix, M.-A. (2017). The natural biotic environment of Caenorhabditis elegans. Genetics , 206, 55–86. doi: 10.1534/genetics.116.195511.
- Schwartz, M. L., & Jorgensen, E. M. (2016). SapTrap, a toolkit for high-throughput CRISPR/Cas9 gene modification in Caenorhabditis elegans. Genetics , 202, 1277–1288. doi: 10.1534/genetics.115.184275.
- Shen, P., Yue, Y., Zheng, J., & Park, Y. (2018). Caenorhabditis elegans : A convenient in vivo model for assessing the impact of food bioactive compounds on obesity, aging, and Alzheimer's disease. Annual Review of Food Science and Technology , 9, 1–22. doi: 10.1146/annurev-food-030117-012709.
- Siegel, D. H., Ashton, G. H. S., Penagos, H. G., Lee, J. V., Feiler, H. S., Wilhelmsen, K. C., … Epstein, E. H. (2003). Loss of kindlin-1, a human homolog of the Caenorhabditis elegans actin-extracellular-matrix linker protein UNC-112, causes Kindler syndrome. American Journal of Human Genetics , 73, 174–187. doi: 10.1086/376609.
- Singaram, V. K., Somerlot, B. H., Falk, S. A., Falk, M. J., Sedensky, M. M., & Morgan, P. G. (2011). Optical reversal of halothane-induced immobility in C. elegans. Current Biology , 21, 2070–2076. doi: 10.1016/j.cub.2011.10.042.
- Singhvi, A., & Shaham, S. (2019). Glia-neuron interactions in Caenorhabditis elegans. Annual Review of Neuroscience , 42, 149–168. doi: 10.1146/annurev-neuro-070918-050314.
- Stirman, J. N., Brauner, M., Gottschalk, A., & Lu, H. (2010). High-throughput study of synaptic transmission at the neuromuscular junction enabled by optogenetics and microfluidics. Journal of Neuroscience Methods , 191(1), 90–93. doi: 10.1016/j.jneumeth.2010.05.019 .
- Thompson, O., Edgley, M., Strasbourger, P., Flibotte, S., Ewing, B., Adair, R., … Waterston, R. H. (2013). The million mutation project: A new approach to genetics in Caenorhabditis elegans. Genome Research , 23, 1749–1762. doi: 10.1101/gr.157651.113.
- Ward, A., Liu, J., Feng, Z., & Xu, X. Z. S. (2008). Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nature Neuroscience , 11, 916–922. doi: 10.1038/nn.2155.
- Ward, S., Thomson, N., White, J. G., & Brenner, S. (1975). Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. Journal of Comparative Neurology , 160, 313–337. doi: 10.1002/cne.901600305.
- White, J. (2018). Obituary: John Sulston (1942-2018). Development , 145, dev166538. doi: 10.1242/dev.166538.
- White, J. G., Southgate, E., Thomson, J. N., & Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences , 314, 1–340. doi: 10.1098/rstb.1986.0056.
- Wittenburg, N., Eimer, S., Lakowski, B., Röhrig, S., Rudolph, C., & Baumeister, R. (2000). Presenilin is required for proper morphology and function of neurons in C. elegans. Nature , 406, 306–309. doi: 10.1038/35018575.
- Wong, W.-R., Brugman, K. I., Maher, S., Oh, J. Y., Howe, K., Kato, M., & Sternberg, P. W. (2019). Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Human Molecular Genetics , 28(13), 2271–2281.
- Yu, A. J., McDiarmid, T. A., Ardiel, E. L., & Rankin, C. H. (2019). High-throughput analysis of behavior under the control of optogenetics in caenorhabditis elegans. Current Protocols in Neuroscience , 86, e57. doi: 10.1002/cpns.57.
Citing Literature
Number of times cited according to CrossRef: 56
- Lara Areal-Hermida, Pedro Coelho, Ángeles Pichardo-Gallardo, Cristina Prudêncio, Carmen Sieiro, Potential probiotic and functional properties of Brettanomyces strains isolated from kombucha tea, Frontiers in Microbiology, 10.3389/fmicb.2024.1415616, 15 , (2024).
- Anna Frolova, Irina Milentyeva, Anastasiya Fedorova, Ekaterina Miller, Sergey Luzyanin, Effect of Biologically Active Substances on Thermal and Oxidative Stress in Caenorhabditis elegans Models, Food Processing: Techniques and Technology, 10.21603/2074-9414-2024-3-2530, 54 , 3, (571-584), (2024).
- S. K. Abilev, E. M. Machigov, S. V. Smirnova, M. V. Marsova, Nematode Caenorhabditis elegans as an Object for Testing the Genotoxicity of Chemical Compounds, Russian Journal of Genetics, 10.1134/S1022795424700674, 60 , 9, (1148-1152), (2024).
- Marissa Duckett, Megan N Taylor, Claire Bowman, Nic M Vega, Parallel evolution of alternate morphotypes of Chryseobacterium gleum during experimental evolution with Caenorhabditis elegans , FEMS Microbiology Ecology, 10.1093/femsec/fiae039, 100 , 5, (2024).
- Tanoy Dutta, Barsha Chakraborty, Aditya Nigam, Shilpi Minocha, Apurba Lal Koner, A small-molecule probe to decipher stress-induced ER microenvironments and ER-Golgi communication, Journal of Materials Chemistry B, 10.1039/D4TB00572D, 12 , 32, (7848-7857), (2024).
- Ahmed Al Harraq, Min Feng, Hashir M. Gauri, Ram Devireddy, Ankur Gupta, Qing Sun, Bhuvnesh Bharti, Magnetic Control of Nonmagnetic Living Organisms, ACS Applied Materials & Interfaces, 10.1021/acsami.4c02325, 16 , 14, (17339-17346), (2024).
- Eva-Maria S. Collins, Ellen V.S. Hessel, Samantha Hughes, How neurobehavior and brain development in alternative whole-organism models can contribute to prediction of developmental neurotoxicity, NeuroToxicology, 10.1016/j.neuro.2024.03.005, 102 , (48-57), (2024).
- Fivos Borbolis, Konstantinos Palikaras, Identifying therapeutic compounds for autosomal dominant optic atrophy (ADOA) through screening in the nematode C. elegans, Animal Models of Disease - Part B, 10.1016/bs.mcb.2024.04.004, (89-108), (2024).
- Joykishan Sharma Hanjabam, Oinam Sangita Devi, Awaiga Collins, Lutrika Moirangthem, Nidhi Brahmacharimayum, Maharabam Anandi Devi, Khuraijam Mrinalini Devi, Analysis of Molecular Circuitry Integrated to Lethargus State of Caenorhabditis elegans: A Review, Proceedings of the Zoological Society, 10.1007/s12595-024-00524-6, 77 , 2, (155-163), (2024).
- Lucía E. Fernandez-Hubeid, Paula A. Albrecht, Michael Aschner, Miriam B. Virgolini, Enduring Ethanol-Induced Behavioral Alterations in Caenorhabditis elegans After Developmental Lead Exposure, Teratogenicity Testing, 10.1007/978-1-0716-3625-1_15, (307-316), (2024).
- Jia‐Jun Tu, Zhen‐Zhen Yu, Mei‐Ling Ou, Jin‐Xiong Cen, Kun Xue, Jing Zhou, Shao‐Jun Li, Guo‐Dong Lu, Differential impacts of nonsteroidal anti‐inflammatory drugs on lifespan and healthspan in aged , Journal of Applied Toxicology, 10.1002/jat.4655, 44 , 10, (1528-1539), (2024).
- Fariba Heydari, David Rodriguez-Crespo, Chantal Wicky, The New Nematicide Cyclobutrifluram Targets the Mitochondrial Succinate Dehydrogenase Complex in Caenorhabditis elegans, Journal of Developmental Biology, 10.3390/jdb11040039, 11 , 4, (39), (2023).
- Hai-Jun Fu, Xing-Yue Zhou, Ya-Ping Li, Xue Chen, Yan-Ni He, Da-Lian Qin, Lu Yu, Chong-Lin Yu, Jian-Ming Wu, An-Guo Wu, Xiao-Gang Zhou, The Protective Effects of Reineckia carnea Ether Fraction against Alzheimer’s Disease Pathology: An Exploration in Caenorhabditis elegans Models, International Journal of Molecular Sciences, 10.3390/ijms242216536, 24 , 22, (16536), (2023).
- Wiebke Burkhardt, Carina Salzinger, Jennie Fischer, Burkhard Malorny, Matthias Fischer, Istvan Szabo, The nematode worm Caenorhabditis elegans as an animal experiment replacement for assessing the virulence of different Salmonella enterica strains, Frontiers in Microbiology, 10.3389/fmicb.2023.1188679, 14 , (2023).
- E. A. Machigov, S. K. Abilev, E. V. Igonina, M. V. Marsova, Study of the Genotoxicity of Beta-Propiolactone Using E. coli lux Biosensors and Nematode Caenorhabditis elegans , Генетика, 10.31857/S0016675823040070, 59 , 5, (507-516), (2023).
- Arne Skorping, Rundormer (Nematoda) – sannsynligvis den mest artsrike dyregruppen på kloden, Naturen, 10.18261/naturen.147.1.5, 147 , 1, (37-42), (2023).
- Ali Tarihi, Mojtaba Tarihi, Taki Tiraihi, Cell‐based allometry: an approach for evaluation of complexity in morphogenesis, Quantitative Biology, 10.15302/J-QB-022-0319, 11 , 2, (183-203), (2023).
- Khameeka N. Kitt, Using RNAi to Examine the Connection between Phenotype and Genotype in Caenorhabditis elegans to Enhance the Undergraduate Research Experience , The American Biology Teacher, 10.1525/abt.2023.85.2.111, 85 , 2, (111-116), (2023).
- Mathieu Legras, Giulia Ghisleni, Léna Regnard, Manon Dias, Rabouant Soilihi, Enzo Celmar, Guillaume Balavoine, Fast cycling culture of the annelid model Platynereis dumerilii, PLOS ONE, 10.1371/journal.pone.0295290, 18 , 12, (e0295290), (2023).
- Anastasia A. Teterina, John H. Willis, Matt Lukac, Richard Jovelin, Asher D. Cutter, Patrick C. Phillips, Genomic diversity landscapes in outcrossing and selfing Caenorhabditis nematodes, PLOS Genetics, 10.1371/journal.pgen.1010879, 19 , 8, (e1010879), (2023).
- E. A. Machigov, S. K. Abilev, E. V. Igonina, M. V. Marsova, Study of the Genotoxicity of Beta-Propiolactone Using Lux Biosensors E. coli and the Nematode Caenorhabditis elegans, Russian Journal of Genetics, 10.1134/S1022795423040075, 59 , 5, (432-440), (2023).
- Xiang Wang, Zhihao Li, Shuxu Wang, Koki Sano, Zhifang Sun, Zhenhua Shao, Asuka Takeishi, Seishiro Matsubara, Dai Okumura, Nobuyuki Sakai, Takayoshi Sasaki, Takuzo Aida, Yasuhiro Ishida, Mechanical nonreciprocity in a uniform composite material, Science, 10.1126/science.adf1206, 380 , 6641, (192-198), (2023).
- Hillel T Schwartz, Chieh-Hsiang Tan, Jackeline Peraza, Krystal Louise T Raymundo, Paul W Sternberg, Molecular identification of a peroxidase gene controlling body size in the entomopathogenic nematode Steinernema hermaphroditum , GENETICS, 10.1093/genetics/iyad209, 226 , 2, (2023).
- Benjamin Kirchweger, Julia Zwirchmayr, Ulrike Grienke, Judith M. Rollinger, The role of Caenorhabditis elegans in the discovery of natural products for healthy aging , Natural Product Reports, 10.1039/D3NP00021D, 40 , 12, (1849-1873), (2023).
- Douglas S. Portman, Carlos A. Díaz-Balzac, Developmental biology: A hole in the matrix, Current Biology, 10.1016/j.cub.2023.08.082, 33 , 19, (R1016-R1018), (2023).
- Anjaneyulu Jalagam, Ashwini Godbole, Parkinson's Disease models of Caenorhabditis elegans to study mechanism of action of Ayurvedic nootropics, Ayurvedic Herbal Preparations in Neurological Disorders, 10.1016/B978-0-443-19084-1.00010-7, (487-520), (2023).
- Chengming Hu, Wenlong Yang, Alternatives to animal models to study bacterial infections, Folia Microbiologica, 10.1007/s12223-023-01084-6, 68 , 5, (703-739), (2023).
- Mitchell B. Lee, Benjamin Blue, Michael Muir, Matt Kaeberlein, The million-molecule challenge: a moonshot project to rapidly advance longevity intervention discovery, GeroScience, 10.1007/s11357-023-00867-6, 45 , 6, (3103-3113), (2023).
- Samantha Hughes, Nathaniel Szewczyk, Using the Model Organism Caenorhabditis elegans to Explore Neuromuscular Function, Neuromuscular Assessments of Form and Function, 10.1007/978-1-0716-3315-1_14, (275-297), (2023).
- Hyun-Min Kim, Yebin Hong, Jiani Chen, A Decade of CRISPR-Cas Gnome Editing in C. elegans, International Journal of Molecular Sciences, 10.3390/ijms232415863, 23 , 24, (15863), (2022).
- Kaisar Ahmad Bhat, Rakeeb Ahmad Mir, Asmat Farooq, Madhiya Manzoor, Ammarah Hami, Kaisar Ahmad Allie, Shaheen Majeed Wani, M. N. Khan, R. Z. Sayyed, Peter Poczai, Waleed Hassan Almalki, Sajad Majeed Zargar, Ali Asghar Shah, Advances in Nematode Identification: A Journey from Fundamentals to Evolutionary Aspects, Diversity, 10.3390/d14070536, 14 , 7, (536), (2022).
- Nicole M. E. Valle, Mariana P. Nucci, Arielly H. Alves, Luiz D. Rodrigues, Javier B. Mamani, Fernando A. Oliveira, Caique S. Lopes, Alexandre T. Lopes, Marcelo N. P. Carreño, Lionel F. Gamarra, Advances in Concentration Gradient Generation Approaches in a Microfluidic Device for Toxicity Analysis, Cells, 10.3390/cells11193101, 11 , 19, (3101), (2022).
- Lucie Larigot, Daniel Mansuy, Ilona Borowski, Xavier Coumoul, Julien Dairou, Cytochromes P450 of Caenorhabditis elegans: Implication in Biological Functions and Metabolism of Xenobiotics, Biomolecules, 10.3390/biom12030342, 12 , 3, (342), (2022).
- P. A. Albrecht, L .E. Fernandez-Hubeid, R. Deza-Ponzio, M. B. Virgolini, The intertwining between lead and ethanol in the model organism Caenorhabditis elegans, Frontiers in Toxicology, 10.3389/ftox.2022.991787, 4 , (2022).
- Micaela R. Pribic, Aristide H. Black, Asia D. Beale, Jessica A. Gauvin, Lisa N. Chiang, Jacqueline K. Rose, Association of Two Opposing Responses Results in the Emergence of a Novel Conditioned Response, Frontiers in Behavioral Neuroscience, 10.3389/fnbeh.2022.852266, 16 , (2022).
- Cyril Poupet, Étienne Rifa, Sébastien Theil, Muriel Bonnet, Philippe Veisseire, Guillaume Cardin, Élise Guéret, Stéphanie Rialle, Christophe Chassard, Adrien Nivoliez, Stéphanie Bornes, In vivo investigation of Lcr35® anti-candidiasis properties in Caenorhabditis elegans reveals the involvement of highly conserved immune pathways, Frontiers in Microbiology, 10.3389/fmicb.2022.1062113, 13 , (2022).
- Mark Louie D. Lopez, Ya‐ying Lin, Stephan Q. Schneider, Chih‐hao Hsieh, Fuh‐Kwo Shiah, Ryuji J. Machida, Allometric scaling of interspecific RNA transcript abundance to extend the use of metatranscriptomics in characterizing complex communities, Molecular Ecology Resources, 10.1111/1755-0998.13711, 23 , 1, (52-63), (2022).
- Nolan Frey, Utku M. Sönmez, Jonathan Minden, Philip LeDuc, Microfluidics for understanding model organisms, Nature Communications, 10.1038/s41467-022-30814-6, 13 , 1, (2022).
- Max K. Steinbach, Jan Leipert, Christine Blurton, Matthias Leippe, Andreas Tholey, Digital Microfluidics Supported Microproteomics for Quantitative Proteome Analysis of Single Caenorhabditis elegans Nematodes , Journal of Proteome Research, 10.1021/acs.jproteome.2c00274, 21 , 8, (1986-1996), (2022).
- Xingli Yu, Javier A. Ceja-Navarro, Xiaoqin Wu, Sublethal Toxicity of Fluorine-Free Firefighting Foams in Soil Invertebrate Caenorhabditis elegans , Environmental Science & Technology Letters, 10.1021/acs.estlett.2c00219, 9 , 6, (561-566), (2022).
- Yali Wang, Yidan Sun, Xingguo Wang, Yue Wang, Langxing Liao, Yonghui Zhang, Baishan Fang, Yousi Fu, Novel antioxidant peptides from Yak bones collagen enhanced the capacities of antiaging and antioxidant in Caenorhabditis elegans, Journal of Functional Foods, 10.1016/j.jff.2022.104933, 89 , (104933), (2022).
- Guoye Guan, Zhongying Zhao, Chao Tang, Delineating the mechanisms and design principles of Caenorhabditis elegans embryogenesis using in toto high-resolution imaging data and computational modeling, Computational and Structural Biotechnology Journal, 10.1016/j.csbj.2022.08.024, 20 , (5500-5515), (2022).
- Jacob Smoot, Stephanie Padilla, Aimen K. Farraj, The utility of alternative models in particulate matter air pollution toxicology, Current Research in Toxicology, 10.1016/j.crtox.2022.100077, 3 , (100077), (2022).
- Ayush Ranawade, Erel Levine, Primer on Mathematical Modeling in C. elegans, C. elegans, 10.1007/978-1-0716-2181-3_21, (375-386), (2022).
- Qing-Hua Liu, Jia-Wei Tang, Peng-Bo Wen, Meng-Meng Wang, Xiao Zhang, Liang Wang, From Prokaryotes to Eukaryotes: Insights Into the Molecular Structure of Glycogen Particles, Frontiers in Molecular Biosciences, 10.3389/fmolb.2021.673315, 8 , (2021).
- Diego Francisco Benítez-Chao, Angel León-Buitimea, Jordy Alexis Lerma-Escalera, José Rubén Morones-Ramírez, Bacteriocins: An Overview of Antimicrobial, Toxicity, and Biosafety Assessment by in vivo Models, Frontiers in Microbiology, 10.3389/fmicb.2021.630695, 12 , (2021).
- Serpen Durnaoglu, Sun-Kyung Lee, Joohong Ahnn, Human Endogenous Retroviruses as Gene Expression Regulators: Insights from Animal Models into Human Diseases, Molecules and Cells, 10.14348/molcells.2021.5016, 44 , 12, (861-878), (2021).
- Mihiri Munasinghe, Abdullah Almotayri, Jency Thomas, Deniz Heydarian, Markandeya Jois, Early Exposure is Necessary for the Lifespan Extension Effects of Cocoa in C. elegans , Nutrition and Metabolic Insights, 10.1177/11786388211029443, 14 , (2021).
- Caroline L. Dahlberg, Christian A. Grove, Heino Hulsey-Vincent, Samiya Ismail, Student Annotations of Published Data as a Collaboration between an Online Laboratory Course and the C. elegans Database, WormBase, Journal of Microbiology & Biology Education, 10.1128/jmbe.v22i1.2331, 22 , 1, (2021).
- Anastasia S Khodakova, Daniela Vidal Vilchis, Dana Blackburn, Ferdinand Amanor, Buck S Samuel, Population scale nucleic acid delivery to Caenorhabditis elegans via electroporation , G3 Genes|Genomes|Genetics, 10.1093/g3journal/jkab123, 11 , 7, (2021).
- Marlon R. Schneider, Michael Oelgeschlaeger, Tanja Burgdorf, Peter van Meer, Peter Theunissen, Anne S. Kienhuis, Aldert H. Piersma, Rob J. Vandebriel, Applicability of organ-on-chip systems in toxicology and pharmacology, Critical Reviews in Toxicology, 10.1080/10408444.2021.1953439, 51 , 6, (540-554), (2021).
- Balasubramanian Chellammal Muthubharathi, Boopathi Balasubramaniam, Dilawar Ahmad Mir, Velayutham Ravichandiran, Krishnaswamy Balamurugan, Physiological and Metabolite Alterations Associated with Neuronal Signals of Caenorhabditis elegans during Cronobacter sakazakii Infections , ACS Chemical Neuroscience, 10.1021/acschemneuro.1c00559, 12 , 22, (4336-4349), (2021).
- Janneke Wit, Clayton M. Dilks, Erik C. Andersen, Complementary Approaches with Free-living and Parasitic Nematodes to Understanding Anthelmintic Resistance, Trends in Parasitology, 10.1016/j.pt.2020.11.008, 37 , 3, (240-250), (2021).
- Mark Greener, What has Caenorhabditis elegans ever done for us?, Prescriber, 10.1002/psb.1955, 32 , 11-12, (29-32), (2021).
- Ilke Sen, Alan Kavšek, Christian G. Riedel, Chromatin Immunoprecipitation and Sequencing (ChIP‐seq) Optimized for Application in Caenorhabditis elegans, Current Protocols, 10.1002/cpz1.187, 1 , 7, (2021).
- Jan M. Gebauer, Alexandra Naba, The Matrisome of Model Organisms: From In-Silico Prediction to Big-Data Annotation, Extracellular Matrix Omics, 10.1007/978-3-030-58330-9_2, (17-42), (2020).
