Using EMBL-EBI Services via Web Interface and Programmatically via Web Services
Fábio Madeira, Fábio Madeira, Nandana Madhusoodanan, Nandana Madhusoodanan, Joonheung Lee, Joonheung Lee, Alberto Eusebi, Alberto Eusebi, Ania Niewielska, Ania Niewielska, Adrian R. N. Tivey, Adrian R. N. Tivey, Stuart Meacham, Stuart Meacham, Rodrigo Lopez, Rodrigo Lopez, Sarah Butcher, Sarah Butcher
Abstract
The European Bioinformatics Institute (EMBL-EBI)’s Job Dispatcher framework provides access to a wide range of core databases and analysis tools that are of key importance in bioinformatics. As well as providing web interfaces to these resources, web services are available using REST and SOAP protocols that enable programmatic access and allow their integration into other applications and analytical workflows and pipelines. This article describes the various options available to researchers and bioinformaticians who would like to use our resources via the web interface employing RESTful web services clients provided in Perl, Python, and Java or who would like to use Docker containers to integrate the resources into analysis pipelines and workflows. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Retrieving data from EMBL-EBI using Dbfetch via the web interface
Alternate Protocol 1 : Retrieving data from EMBL-EBI using WSDbfetch via the REST interface
Alternate Protocol 2 : Retrieving data from EMBL-EBI using Dbfetch via RESTful web services with Python client
Support Protocol 1 : Installing Python REST web services clients
Basic Protocol 2 : Sequence similarity search using FASTA search via the web interface
Alternate Protocol 3 : Sequence similarity search using FASTA via RESTful web services with Perl client
Support Protocol 2 : Installing Perl REST web services clients
Basic Protocol 3 : Sequence similarity search using NCBI BLAST+ RESTful web services with Python client
Basic Protocol 4 : Sequence similarity search using HMMER3 phmmer REST web services with Perl client and Docker
Support Protocol 3 : Installing Docker and running the EMBL-EBI client container
Basic Protocol 5 : Protein functional analysis using InterProScan 5 RESTful web services with the Python client and Docker
Alternate Protocol 4 : Protein functional analysis using InterProScan 5 RESTful web services with the Java client
Support Protocol 4 : Installing Java web services clients
Basic Protocol 6 : Multiple sequence alignment using Clustal Omega via web interface
Alternate Protocol 5 : Multiple sequence alignment using Clustal Omega with Perl client and Docker
Support Protocol 5 : Exploring the RESTful API with OpenAPI User Inferface
INTRODUCTION
Since 1995, the European Bioinformatics Institute (EMBL-EBI) has provided access to a wide range of databases and analysis tools using web services technologies (Chojnacki et al., 2017; Li et al., 2015; Madeira et al., 2019; Madeira et al., 2022; Madeira et al., 2024; McWilliam et al., 2013). These comprise services to search, retrieve, and run analysis tools on the databases hosted at the institute and to explore the network of cross-references present in the data, e.g., EBI Search (Madeira et al., 2022; Park et al., 2017; Squizzato et al., 2015; Valentin et al., 2010). In this article, we introduce the reader to services used to retrieve entry data in various data formats and to access the data using specific fields [e.g., Dbfetch (Lopez et al., 2003)] and to analysis tool services, for example, sequence similarity search [SSS; e.g., FASTA (see Current Protocols article: Pearson, 2016; Pearson & Lipman, 1988) and NCBI BLAST+ (Altschul et al., 1997; Camacho et al., 2009; also see Current Protocols article: Ladunga, 2002)], multiple sequence alignment [MSA; e.g., Clustal Omega (see Current Protocols article: Sievers & Higgins, 2014; also see Sievers & Higgins, 2018; Sievers et al., 2011)], and pairwise sequence alignment and protein functional analysis [PFA; e.g., InterProScan (see Current Protocols article: Mulder & Apweiler, 2003; also see Jones et al., 2014)].
In addition to the web service technologies, Job Dispatcher now offers a new website (https://www.ebi.ac.uk/jdispatcher/) that serves as a central gateway for accessing several sequence analysis tool categories. This website simplifies the process of finding and selecting the appropriate tool for users. The homepage displays the status of submitted jobs and enables users to search for analysis results using the job identifier. Furthermore, the homepage provides the latest news, data-release updates, and the list of collaborators (Fig. 1).
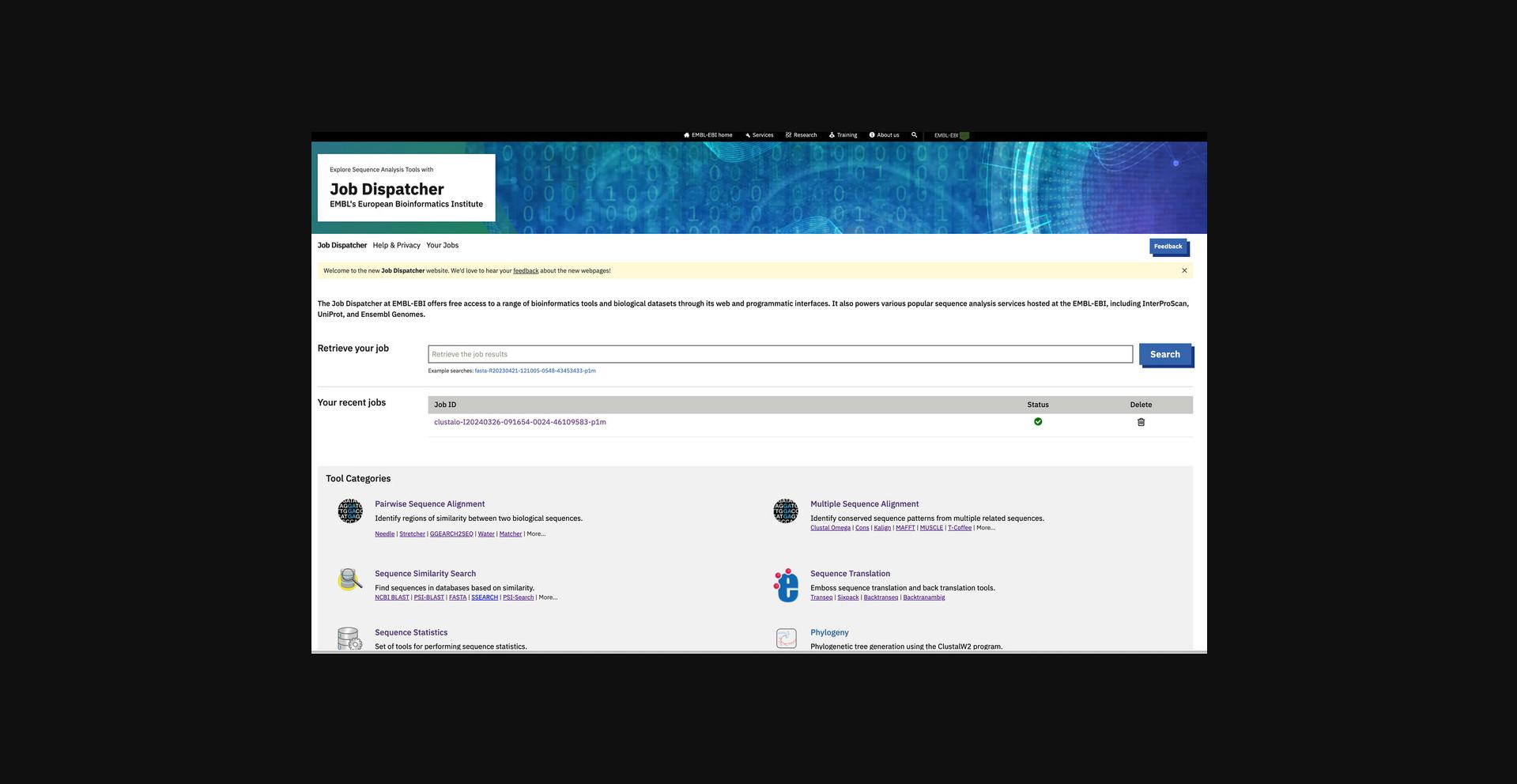
The “Help and Privacy” page offers comprehensive documentation, including links to webinars and other training materials. Additionally, it provides access to the Job Dispatcher's detailed documentation, available from https://www.ebi.ac.uk/jdispatcher/docs/.
To enhance user experience, all tool web forms have been redesigned. The new tool pages no longer offer email notifications from individual tool pages. Instead, users can find all submitted jobs under the “Your Jobs” page (https://www.ebi.ac.uk/jdispatcher/recentJobs).
The Representational State Transfer (REST) and Simple Object Access Protocol (SOAP) web services (https://www.ebi.ac.uk/jdispatcher/docs/webservices/) interfaces to these databases and tools allow their integration into other tools, applications, web portals, analysis pipelines, and workflows. Sample clients covering a range of popular bioinformatics programming languages (https://github.com/ebi-jdispatcher/webservice-clients/), a Docker container image with pre-installed sample clients (https://hub.docker.com/r/ebiwp/webservice-clients/), Common Workflow Language (CWL) descriptions for the sample clients (https://github.com/ebi-jdispatcher/webservice-cwl), and examples of usage are provided to help users get started using the EMBL-EBI's Job Dispatcher Sequence Analysis web services.
The following protocols describe how you can retrieve data from EMBL-EBI using Dbfetch (Basic Protocol 1, Alternate Protocols 1 and 2, and Support Protocol 1). You will also learn how to perform sequence similarity search using FASTA, NCBI BLAST+, and HMMER3 phmmer (Basic Protocols 2 to 4, Alternate Protocol 3, and Support Protocols 2 and 3), and how to perform protein functional analysis using InterProScan 5 (Basic Protocol 5, Alternate Protocol 4, and Support Protocol 4), as well as how to perform multiple sequence analysis using Clustal Omega (Basic Protocol 6 and Alternate Protocol 5). Finally, you will learn how to explore and navigate the Job Dispatcher RESTful API (Support Protocol 5).
STRATEGIC PLANNING
The most significant planning issues around the decision to use the SOAP and RESTful web services of EMBL-EBI services are detailed below.
Web services have several potential uses over and above normal web interface access to services: offering services behind or together with another service, systematic access to resources, and as a gateway to workflows. Although these needs can also be served by local installation of individual tools and databases, doing so comes with additional burdens on technical support and skills, for example, the requirement of keeping local software and databases up to date, as well as computational and storage burdens. Web services reduce these overheads by allowing a standardized interface to remotely managed servers (at EMBL-EBI in this instance) where the tools and database providers manage the software and database updating and also provide access to large compute resources and the management thereof.
Web services allow for programmatic access to services (for example, using scripts) and are thus suitable for mass/systematic analysis or for using the services as part of a wider workflow or as the backend to another service.
There are some situations where web services are not suitable:
- Where the analysis is time-critical: the nature of remote services necessarily adds some latency to the process.
- Where the data cannot leave the local computer/network for any reason: although web services use secure https protocols, license restrictions on datasets that you own may prevent their transmission in any form over the Internet.
Although using web services reduces the burden of maintaining software and data, it is important to note that the user still needs to be familiar with the tools used as well as programmatic concepts, though using a graphical workflow tool that interfaces with web services can alleviate some of the programming knowledge required.
Basic Protocol 1: RETRIEVING DATA FROM EMBL-EBI USING Dbfetch VIA THE WEB INTERFACE
In this protocol, we introduce the reader to commonly used biological sequence databases and how to retrieve data from them using services at EMBL-EBI.
A large number of databases exist that store biological data derived from experiments or computation. These aim to determine the order of nucleotides or amino acids (also known as the primary structure) and include methods such as Sanger sequencing (Sanger & Coulson, 1975), next-generation sequencing (NGS; Pettersson et al., 2009) for whole-genome and exome sequencing, peptide sequencing from C and N-terminal analysis (Edman et al., 1950), Edman degradation (Roberts & Murray, 1976), enzyme digestion (Hernandez et al., 2006), mass spectrometry, and X-ray crystallography of biomolecular structures (Franklin, 1956).
Nucleotide Sequences
The most commonly used nucleotide sequence database is the product of a trilateral agreement between EMBL-EBI, the National Center for Biological Information (NCBI), and the DNA Databank of Japan (DDBJ). These form the International Nucleotide Sequence Database Collaboration (INSDC). This collaborative database comprises the European Nucleotide Archive (Silvester et al., 2018; Yuan et al., 2024), GenBank (Benson et al., 2017; Sayers et al., 2024), and the DDBJ (Kodama et al., 2018; Tanizawa et al., 2023). These three centers collect and share data on a daily basis, forming perhaps the largest effort to exchange and share scientific data across the globe.
Genomes
NGS technology has evolved rapidly over recent years. The sequencing speeds afforded by traditional methods had been a limiting factor for obtaining whole genomes. Today, with NGS, it is possible to sequence a human genome in a single day and at a fraction of the cost of the older methods. This has led to an explosion in the number of genomes available for biomedical, agronomical, environmental, and computational research.
The largest collection of these genomes is spread across organism-specific databases, e.g., FlyBase (Gramates et al., 2017; Larkin et al., 2020), WormBase (Davis et al., 2022; Lee et al., 2018; also see Current Protocols article: Schwartz & Sternberg, 2004), SGD (Cherry et al., 2012; also see Current Protocols article: Skrzypek & Hirschman, 2011), Ensembl (Martin et al., 2023; Zerbino et al., 2018; also see Current Protocols article: Wolfsberg, 2007), and Ensembl Genomes (Kersey et al., 2018). Ensembl is a joint project between EMBL-EBI and the Wellcome Trust Sanger Institute and is primarily focused on genomes from vertebrate and other eukaryotic organisms. Ensembl Genomes is based on the Ensembl infrastructure and is divided across five websites that respectively focus on the genomes of bacteria, protists, fungi, plants, and invertebrate metazoa.
Protein Sequences
Amino acid sequences date back to the late 1940s, when Edman and Sanger developed methods for retrieving sequence from purified protein using a combination of biochemical methods. Just as with nucleotide sequences later, collecting and distributing these sequences became a task that would enable researchers to share and de-duplicate effort. The first such database was established in 1960s by the National Biochemical Research Foundation (NBRF) and was known as the Atlas of Protein Sequence and Structure, published by Margaret Dayhoff. Her group pioneered methods for the comparison of protein sequences using computational methods. The NBRF established the Protein Information Resource (PIR) in 1984 to produce and distribute the PIR–Protein Sequence Database (PIR-PSD; Wu & Nebert, 2004), the first international database that grew out from Dayhoff's Atlas of Protein Sequence and Structure. PIR, EMBL, and the Swiss Institute of Bioinformatics joined efforts to produce a single (and the largest) protein sequence database by unifying the PIR-PSD, TrEMBL, and Swiss-Prot (Bairoch et al., 2004) databases. This is known today as the UniProt Knowledgebase (UniProtKB; UniProt Consortium, 2019, 2023). This service provides access to sequences from multiple sources, including nucleotide translations and protein sequences derived from structures in the Protein Data Bank (PDB), as well as those from the Structural Genomics Consortium (SGC) initiative.
Retrieving Sequences from EMBL-EBI Using Dbfetch
Dbfetch (database fetch; Lopez et al., 2003) is a retrieval system specifically designed to provide a single point of access for biological data spread across multiple resources. Dbfetch has been in operation since 1995 and currently provides unified access to 58 databases (https://www.ebi.ac.uk/Tools/dbfetch/dbfetch/dbfetch.databases). Among the databases recently added to Dbfetch are the AlphaFold Protein Structure Database, COVID-19 Data Portal, European Nucleotide Archive (ENA) ribosomal RNA (ENA rRNA) browser, non-human Immunoglobulin-like Receptors (IPD-NHKIR) nucleotide coding sequence (CDS) database, IPD-NHKIR nucleotide genomic database, IPD-NHKIR protein database, PDB in Europe–Knowledge Base (PDBe-KB), and Electron Microscopy Data Bank (EMDB). Dbfetch uses multiple data sources to provide a range of data formats wider than that available from a single source and to mitigate the effect of a single data source being unavailable.
Alternate Protocol 1: RETRIEVING DATA FROM EMBL-EBI USING WSDbfetch VIA THE REST INTERFACE
Dbfetch provides three modes of access to the user. As described above in Basic Protocol 1, one is using a web browser and the CGI interface. The two others make use of data access standards called web services. Web services consist of two protocols, REST and SOAP, that complement each other and can be used to perform various data-retrieval tasks. Like Dbfetch, WSDbfetch (McWilliam et al., 2009) allows the user to retrieve entries. For the developer, the advantage of the REST interface is that it allows the functionality of Dbfetch to be integrated into an application, workflow, or process pipeline. Because the web services technologies are language agnostic, the developer can use the programming language of choice. EMBL-EBI provides fully working example clients written in a variety of common programming languages, including Perl, Python, and Java. These command-line interface (CLI) clients can be downloaded from https://github.com/ebi-jdispatcher/webservice-clients and give full access to the Dbfetch service from the command line (also known as the terminal, shell, or command prompt). The REST clients provide an easier-to-use interface that reports standard HTTP status codes (https://en.wikipedia.org/wiki/List_of_HTTP_status_codes). The REST interface can be operated using a web browser or common web retrieval utilities such as wget and curl. In the following examples, we will use RESTful URLs to demonstrate the WSDbfetch REST interface.
The fundamental syntax of the WSDbfetch REST interface is as follows:
where {db} is the database name (e.g., uniprotkb) and {id} is the identifier (e.g., WAP_MOUSE). The following shows how to fetch the mouse whey acidic protein (WAP) precursor from UniProtKB using the RESTful interface:
As described earlier, Dbfetch provides access to various formats and styles in which to download data. WSDbfetch provides the same functionality. To download WAP_MOUSE in the UniProtKB XML format (uniprotxml), the URL is as follows:
Likewise, to download WAP_MOUSE in UniProtKB flat-file format with HTML hyperlinks, the following URL would be used:
Dbfetch presently provides access to 58 databases. These are shown in Table 1 along with the acronyms used in Dbfetch and WSDbfetch as the database names.
| Database name | Database identifier |
|---|---|
|
AlphaFold Database COVID-19 Data Portal ChEMBL Targets |
afdb cdp chembl |
| EDAM | edam |
| ENA Coding | ena_coding |
| ENA Geospatial | ena_geospatial |
|
ENA Non-coding ENA rRNA |
ena_noncoding ena_rrna |
| ENA Sequence | ena_sequence |
| ENA Sequence Constructed | ena_sequence_con |
| ENA Sequence Constructed Expanded | ena_sequence_conexp |
| ENA/SVA | ena_sva |
| Ensembl Gene | ensemblgene |
| Ensembl Genomes Gene | ensemblgenomesgene |
| Ensembl Genomes Transcript | ensemblgenomestranscript |
| Ensembl Transcript | ensembltranscript |
| EPO Proteins | epo_prt |
| HGNC | hgnc |
| IMGT/HLA nucleotide cds | imgthlacds |
| IMGT/HLA nucleotide genomic | imgthlagen |
| IMGT/HLA protein | imgthlapro |
| IMGT/LIGM-DB | imgtligm |
| InterPro | interpro |
| IPD-KIR nucleotide cds | ipdkircds |
| IPD-KIR nucleotide genomic | ipdkirgen |
| IPD-KIR protein | ipdkirpro |
| IPD-MHC nucleotide cds | ipdmhccds |
| IPD-MHC nucleotide genomic | ipdmhcgen |
|
IPD-MHC protein IPD-NHKIR nucleotide cds IPD-NHKIR nucleotide genomic IPD-NHKIR protein |
Ipdmhcpro ipdnhkircds ipdnhkirgen ipdnhkirpro |
| IPRMC | iprmc |
| IPRMC UniParc | iprmcuniparc |
| JPO Proteins | jpo_prt |
| KIPO Proteins | kipo_prt |
| MEDLINE | medline |
| MEROPS-MP | mp |
| MEROPS-MPEP | mpep |
| MEROPS-MPRO | mpro |
| Patent DNA Non Redundant L1 | nrnl1 |
| Patent DNA Non Redundant L2 | nrnl2 |
| Patent Protein Non Redundant L1 | nrpl1 |
| Patent Protein Non Redundant L2 | nrpl2 |
| Patent Equivalents | patent_equivalents |
|
PDB PDBe Knowledge Base |
pdb pdbekb |
| Electron Microscopy Data Bank | emdb |
| RefSeq nucleotide | refseqn |
| RefSeq protein | refseqp |
| Taxonomy | taxonomy |
| UniParc | uniparc |
| UniProtKB | uniprotkb |
| UniRef100 | uniref100 |
| UniRef50 | uniref50 |
| UniRef90 | uniref90 |
| UniSave | unisave |
| USPTO Proteins | uspto_prt |
A listing of the available databases with a description of each database, details of the various available data formats, and result styles and example entry identifiers can be found at https://www.ebi.ac.uk/Tools/dbfetch/dbfetch/dbfetch.databases.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- The wget utility. For OS X, Linux, and UNIX systems, wget is commonly installed by default. If wget is not installed, it can be installed from the systems package manager or downloaded and installed from https://www.gnu.org/software/wget/. For MS Windows (versions 7SP1 and above, including 10), the iwr command is built-in inside powershell. The syntax is slightly different. One example is provided below. Alternatively, wget can be obtained in Cygwin (https://cygwin.com/).
Input
- Database entry identifiers in the format database name:database identifier supported by Dbfetch
1.Retrieve entry into a file.
2.Retrieve entry into a console or terminal.
3.Retrieve entry annotation.
4.Retrieve entry FASTA-format sequence.
5.Retrieve entry with cross-references and features.
Alternate Protocol 2: RETRIEVING DATA FROM EMBL-EBI USING Dbfetch VIA RESTful WEB SERVICES WITH PYTHON CLIENT
Dbfetch provides fully working RESTful web services clients (i.e., command-line applications) written in Perl and Python programming languages. For a full description of the Dbfetch RESTful web services, see https://www.ebi.ac.uk/Tools/dbfetch/syntax.jsp.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- Python (https://www.python.org/) with the xmltramp2 module installed
- EMBL-EBI Python RESTful web services clients (see Support Protocol 1 for how to download and install)
Input
- Database entry identifiers in the format database name:database identifier supported by Dbfetch
1.Display client usage. To do so, switch to the directory containing the downloaded Python client dbfetch.py. Run the script without specifying any parameters (or adding --help to the command shown below) to print a brief help message (Fig. 6).
- python dbfetch.py

2.Display a list of the databases supported by the service:
- python dbfetch.py getSupportedDBs
3.Display a list of the available formats associated with a particular database (e.g., uniprotkb):
- python dbfetch.py getDbFormats uniprotkb
4.Retrieve an entry.
5.Get the sequences of all the chains in the structure.
6.To get the sequence of a specific chain, instead of all the chains, use the chain identifier as suffix for the entry identifier.
7.Retrieve a set of entries from a database.
Support Protocol 1: INSTALLING PYTHON REST WEB SERVICES CLIENTS
Python is commonly used in bioinformatics and typically installed by default on UNIX and UNIX-like systems. Because many existing analytical pipelines are implemented in Python, the Python clients provide an option for integration of web services into existing pipelines.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- Python 3.5 or above (https://www.python.org/), pip (https://pypi.org/project/pip/), and xmltramp2 (https://pypi.org/project/xmltramp2/)
- A web browser, e.g., Google Chrome, Mozilla Firefox, Microsoft Edge, Safari, or Opera
1.Check that Python is installed on the system. In the Command Prompt or terminal, enter the following:
- python --version
If Python is not installed or the current version is not 3.5 or later, then download Python 3 and follow the instructions provided at https://www.python.org/downloads/.
2.Check that the xmltramp2 Python module has been installed:
-
python -c “import xmltramp2”
-
a.If an error message is returned, install the xmltramp2 module using pip:
-
pip install xmltramp2
-
b.
If pip is not installed, follow the instructions provided at https://pip.pypa.io/en/stable/installation/ on how to install it.
If Python version 2 is installed in your operating system, if available, pip will be linked to Python 2.After installing Python 3 and pip (for Python 3), replace all of the python and pip commands provided in this article by python3 and pip3.This ensures that you are running Python 3, which has the xmltramp2 dependency installed.
3.Open a web browser and go to the GitHub page for the EMBL-EBI web services clients at https://github.com/ebi-jdispatcher/webservice-clients.
4.Download the Python clients.
5.Test and run the clients. Within the Command Prompt or terminal, change to the directory that contains the client program downloaded in step 4.To test the program (e.g., dbfetch.py, retrieving sequences from UniProtKB in FASTA format), enter the following:
- python dbfetch.py fetchBatch uniprotkb P01174,P01173 fasta
Basic Protocol 2: SEQUENCE SIMILARITY SEARCH USING FASTA SEARCH VIA THE WEB INTERFACE
EMBL-EBI provides and maintains a comprehensive range of freely available analysis tools through web interfaces and web services (Chojnacki et al., 2017; Li et al., 2015; Madeira et al., 2019, Madeira et al., 2022, Madeira et al., 2024; McWilliam et al., 2013). The analysis services include 53 tools, divided into 10 categories. In this protocol, we aim to demonstrate how to run analysis tools and interpret results through the web interface.
Table 2 shows the analysis tools along with the categories and the URLs of their web interfaces. The popular categories include SSS (e.g., NCBI BLAST+ and FASTA), MSA (e.g., Clustal Omega), and PFA (e.g., InterProScan, Phobius).
| Tool category | Tools included | Main URL |
|---|---|---|
| Sequence similarity search | NCBI BLAST+, FASTA, FASTM/S/F, PSI-BLAST, PSI-Search, SSEARCH, GGSEARCH, GLSEARCH, PSI-Search2 | https://www.ebi.ac.uk/jdispatcher/sss/ |
| Multiple sequence alignment | Clustal Omega, Kalign, MAFFT, MUSCLE, T-Coffee, WebPRANK, MView | https://www.ebi.ac.uk/jdispatcher/psa/ |
| Protein function analysis | InterProScan 5, Phobius, Pratt, RADAR, HMMER 3 hmmscan, HMMER3 phmmer, PfamScan | https://www.ebi.ac.uk/jdispatcher/pfa/ |
| Sequence format conversion | Seqret, MView | https://www.ebi.ac.uk/jdispatcher/sfc/ |
| Phylogeny analysis | Simple Phylogeny | https://www.ebi.ac.uk/jdispatcher/phylogeny/ |
| Pairwise sequence alignment | Needle, Stretcher, Water, Matcher, LALIGN, GeneWise, GGSEARCH2SEQ, SSEARCH2SEQ | https://www.ebi.ac.uk/jdispatcher/psa/ |
| RNA analysis | Infernal cmscan, MapMi, R2DT | https://www.ebi.ac.uk/jdispatcher/rna/ |
| Sequence operation | SeqCksum | https://www.ebi.ac.uk/jdispatcher/so/ |
| Sequence translation | Transeq, Sixpack, Backtranseq, Backtransmbig | https://www.ebi.ac.uk/jdispatcher/st/ |
| Sequence Statistics | Pepinfo, Pepstats, Pepwindow, SAPS, Cpgplot, Newcpgreport, Isochore, Dotmatcher, Dotpath, Dottup, Polydot | https://www.ebi.ac.uk/jdispatcher/seqstats/ |
| EMBOSS tools | Needle, Stretcher, Water, Matcher, Transeq, Sixpack, Backtranseq, Backtransmbig, Pepinfo, Pepstats, Pepwindow, Cpgplot, Newcpgreport, Isochore, Seqret, Dotmatcher, Dotpath, Dottup, Polydot | https://www.ebi.ac.uk/jdispatcher/emboss/ |
In the following protocols, we will introduce the most commonly used sequence analysis tools using the web interface and REST web services client programs. EMBL-EBI provides freely available web services for analysis tools (https://www.ebi.ac.uk/jdispatcher/docs/webservices/), which mainly include SSS, MSA, PFA, Phylogeny Analysis, Pairwise Sequence Alignment (PSA), RNA Analysis, Sequence Format Convert (SFC), Sequence Statistics, Sequence Translation, and Sequence Operations (SO). Basic Protocol 2 demonstrates examples using web services for SSS, PFA, and MSA.
SSS is a method of searching sequence databases by using alignment to a query sequence. By statistically assessing how well database and query sequences match, one can infer homology and transfer information to the query sequence. The EMBL-EBI SSS web services contain the analysis tools of NCBI BLAST+, FASTA, FASTM, PSI-BLAST, and PSI-Search.
We use the FASTA service web interface to run and interpret a FASTA search job. The FASTA package provides a comprehensive set of similarity/homology searching programs, similar to those provided by NCBI BLAST+, and some additional programs for searching with short peptides and oligonucleotides.
Necessary Resources
Hardware
- Any Internet-connected computer
Software
- A web browser, e.g., Google Chrome, Mozilla Firefox, Microsoft Edge, Safari, or Opera
Input
- A plain text file containing a sequence in FASTA, EMBL, GenBank, GCG, PIR, NBRF, PHYLIP, or UniProtKB/Swiss-Prot format. If the file is not available, the entry identifier in the format database name:database identifier, e.g., UniProtKB:GSTM1_MOUSE, can be used as input, or a sequence in one of the formats mentioned above can be pasted into the form.
- This example uses the mouse protein “Glutathione S-transferase Mu 1” from the UniProtKB database as the input sequence. The entry details can be found at https://www.uniprot.org/uniprotkb/P10649/entry, and the FASTA-format sequence can be downloaded at https://rest.uniprot.org/uniprotkb/P10649.fasta.
1.Go to the SSS web page https://www.ebi.ac.uk/jdispatcher/sss/ using a web browser (Fig. 7).
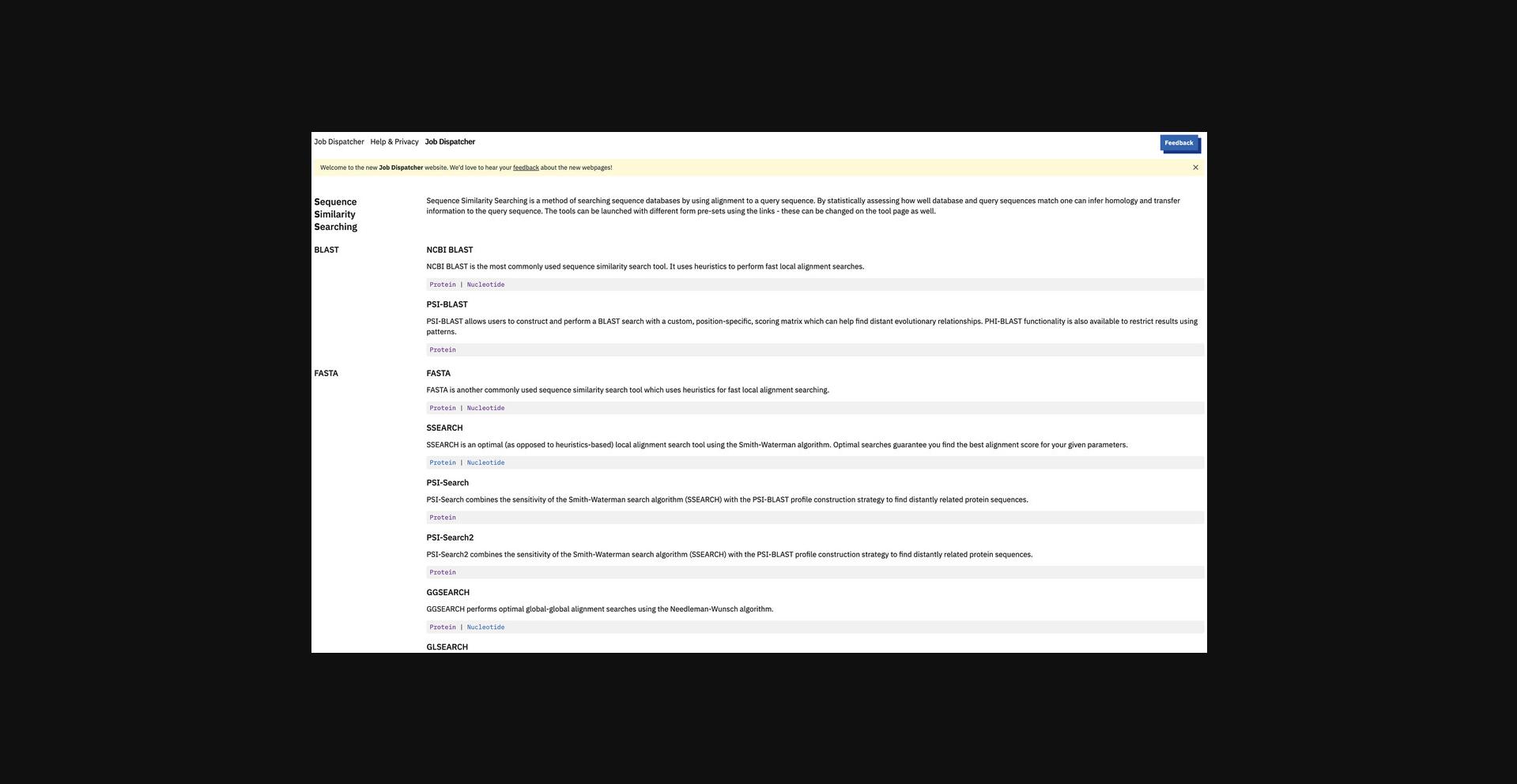
2.Click “Protein” search under the FASTA section or go directly to https://www.ebi.ac.uk/jdispatcher/sss/fasta/ (Fig. 8).
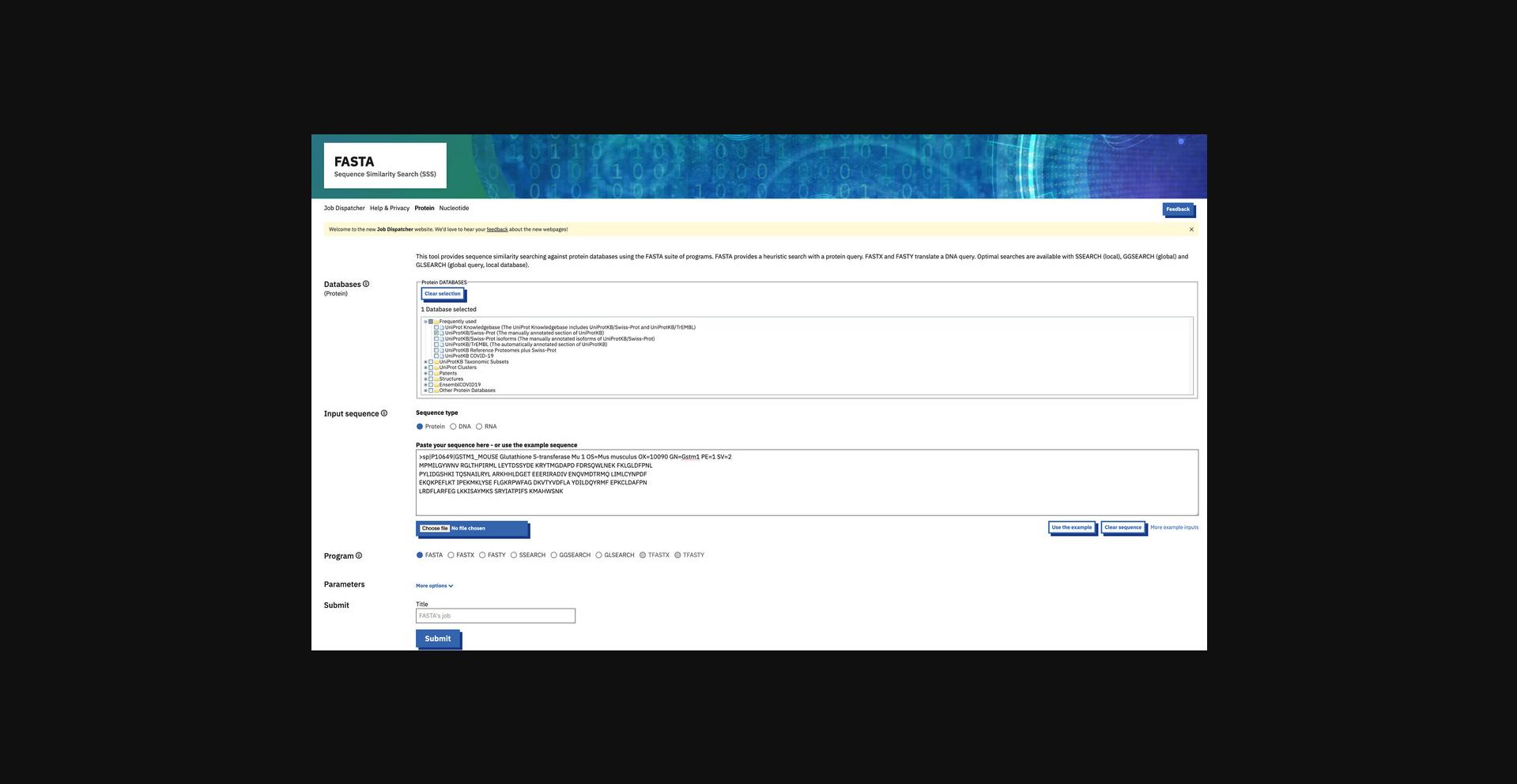
3.Select the databases to search. From the “Databases” section, click + or - to expand or collapse the available databases under the main database categories. Check or uncheck the boxes of the databases to select the appropriate databases.
4.Enter the input sequence by browsing and selecting the input sequence file. Alternatively, copy the sequence and paste it into the sequence box.
5.Set the parameters (Fig. 9). To do so, first, select the program to run (FASTA, FASTX, FASTY, SSEARCH, GLSEARCH, or GGSEARCH). Then, click on the “More options” button to expand the section for the advanced parameters (e.g., matrix, gap penalties, ktup, e-values, and output formats). Change the settings of the parameters according to need.
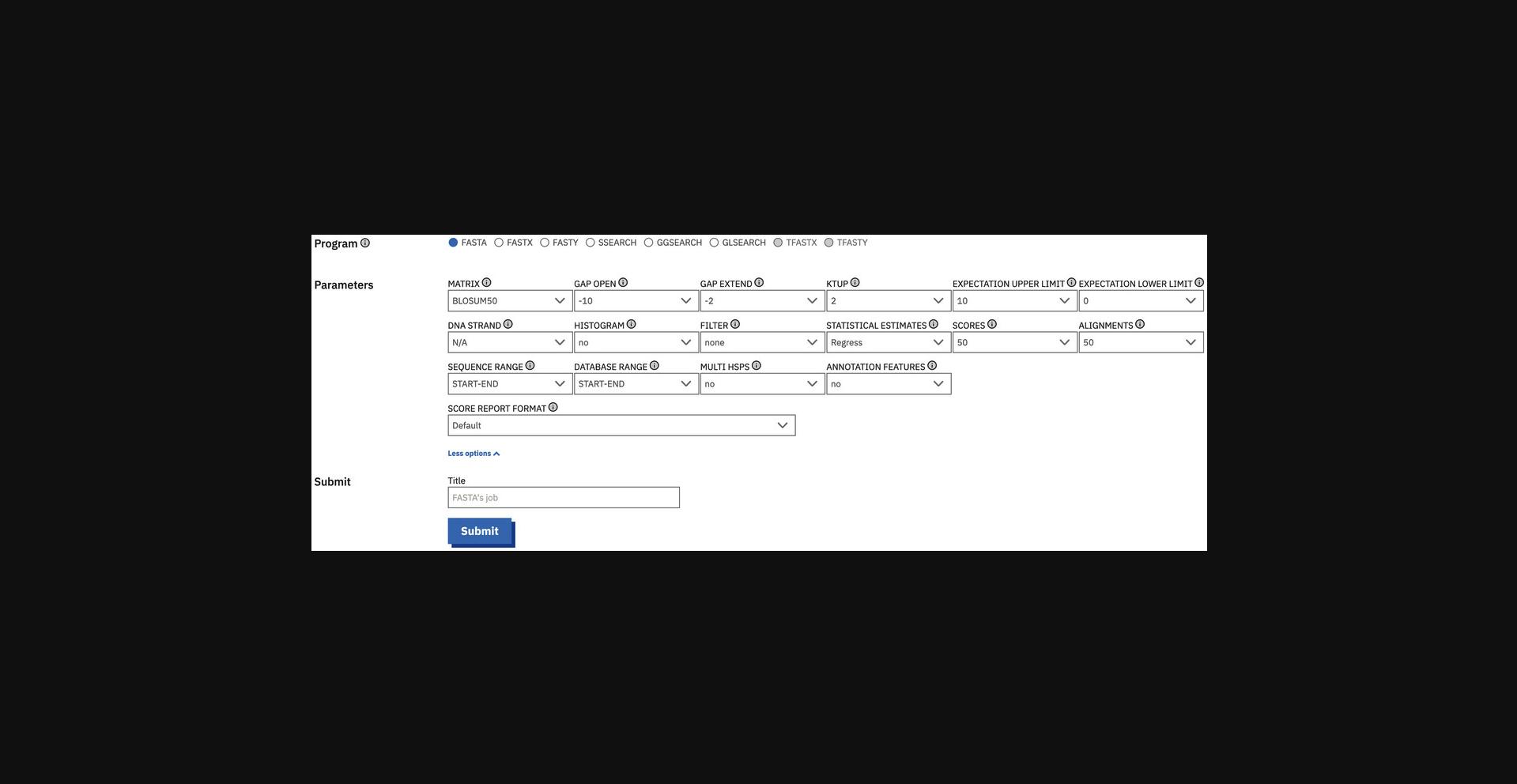
6.Submit the job. To do so, provide a job “Title” (optional) to briefly describe the job and click the “Submit” button.
7.View job result summary (Fig. 10).
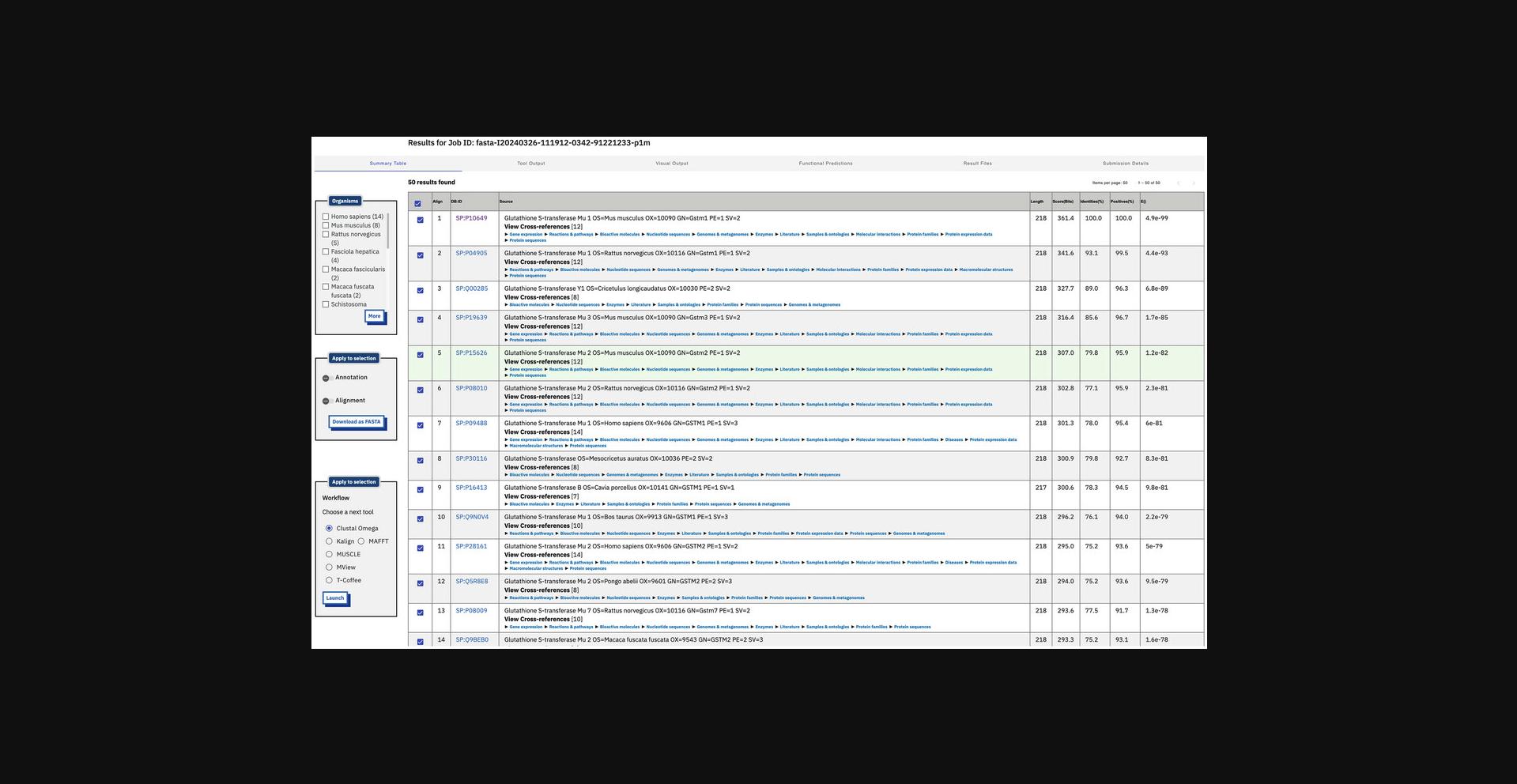
8.Display the tool raw output (Fig. 11) by clicking the “Tool Output” tab.
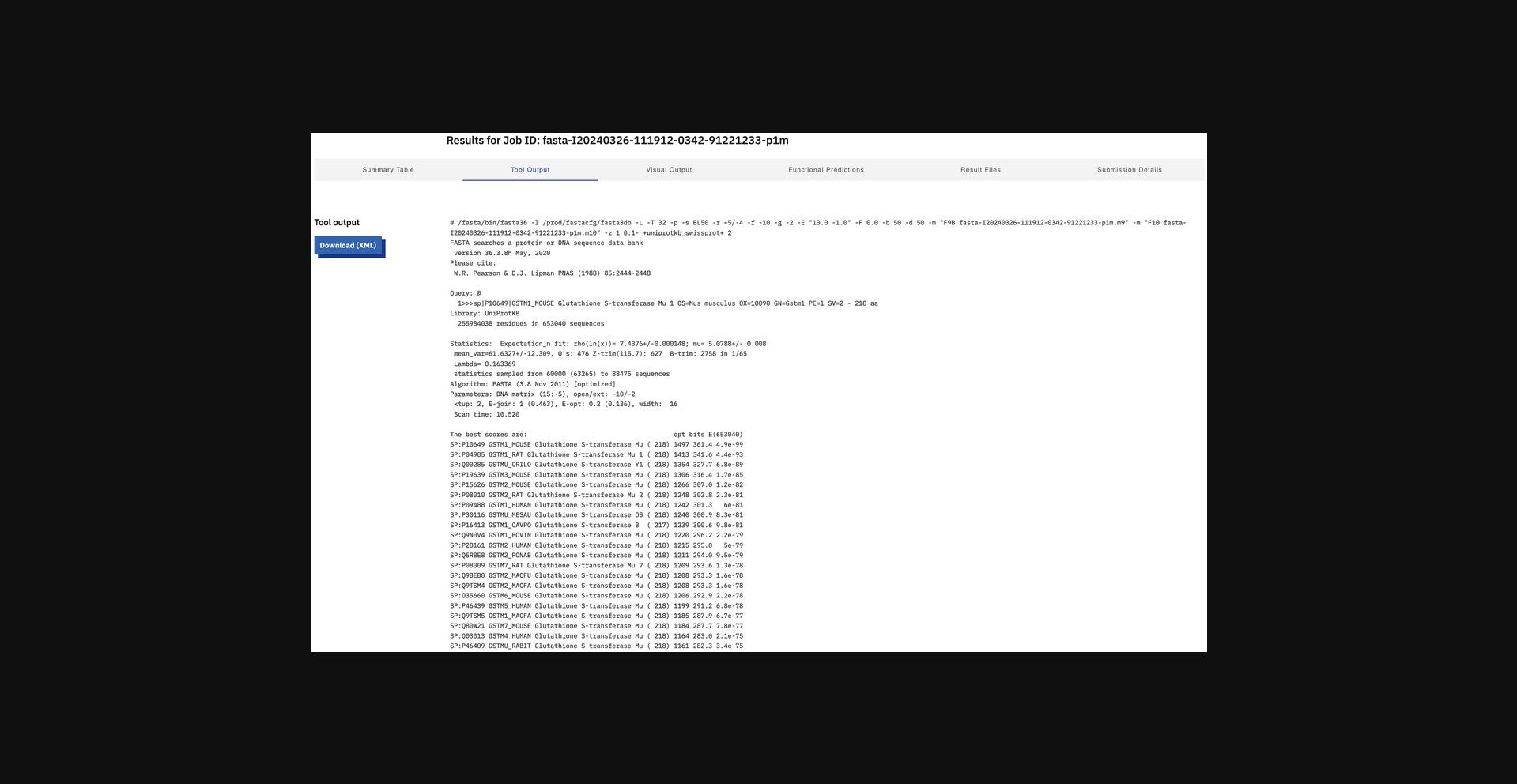
9.Visualize the result (Fig. 12) by switching to the “Visual Output” view.
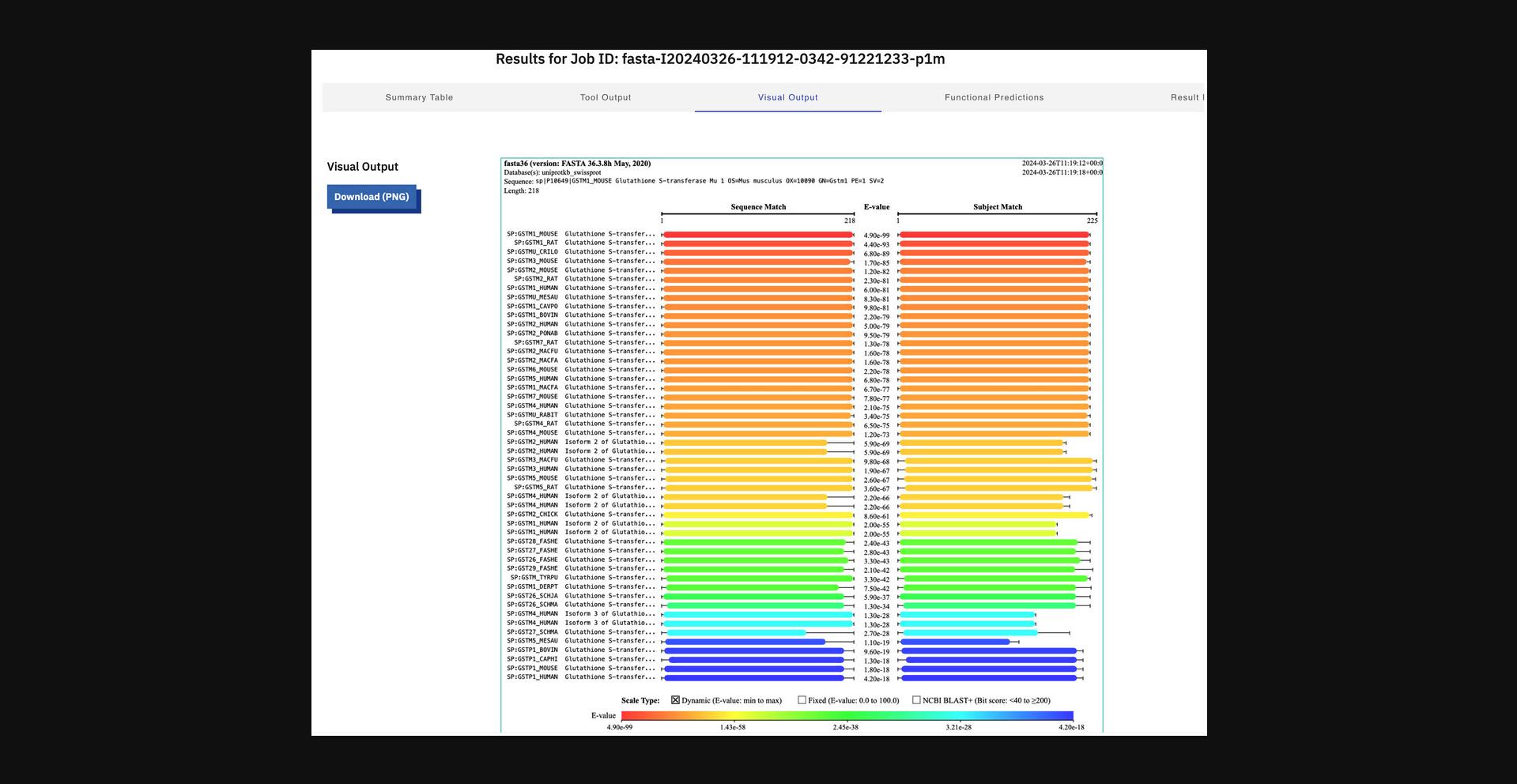
10.Display functional predictions (Fig. 13).
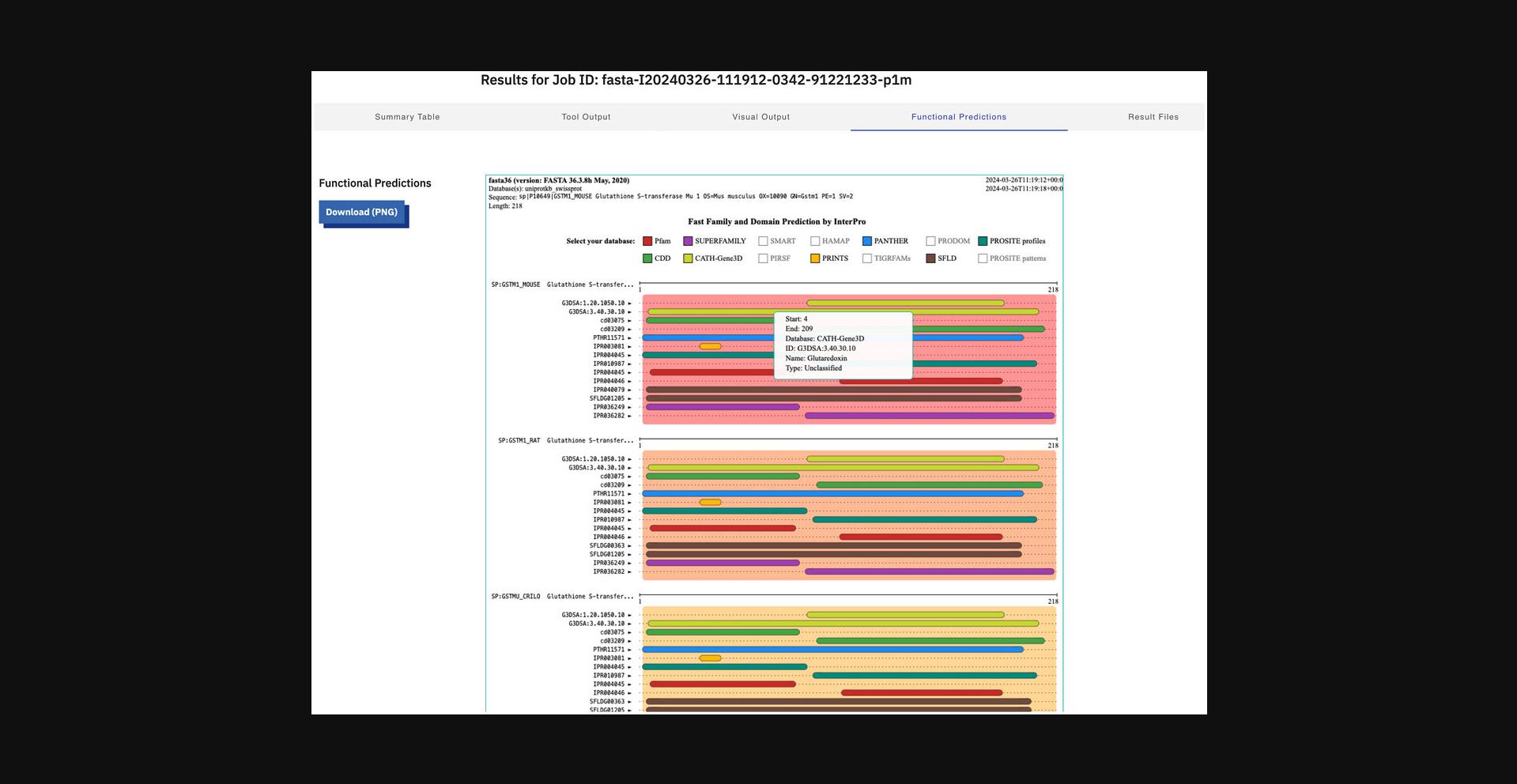
11.Click the “Result Files” tab to display all the result files the tool produces.
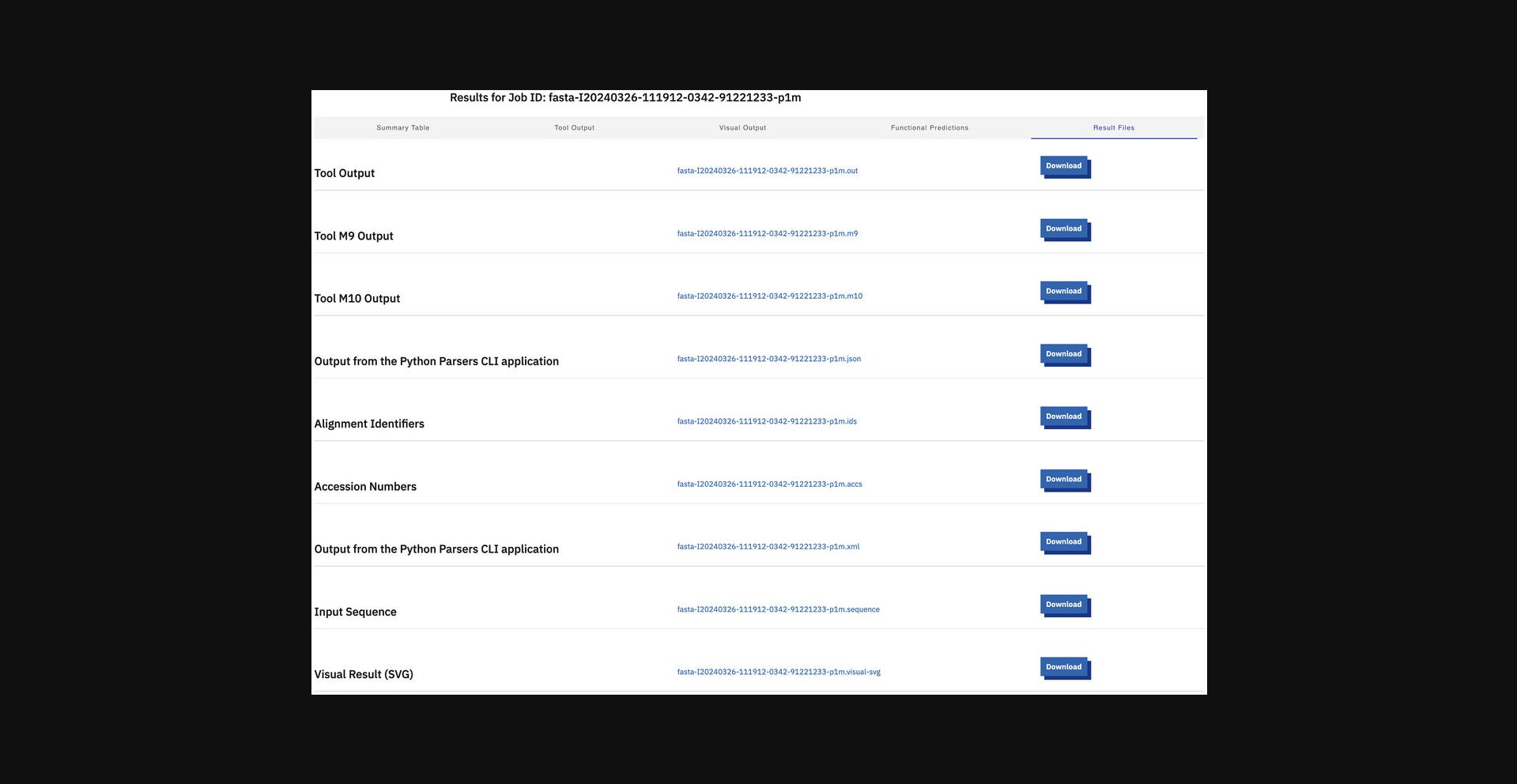
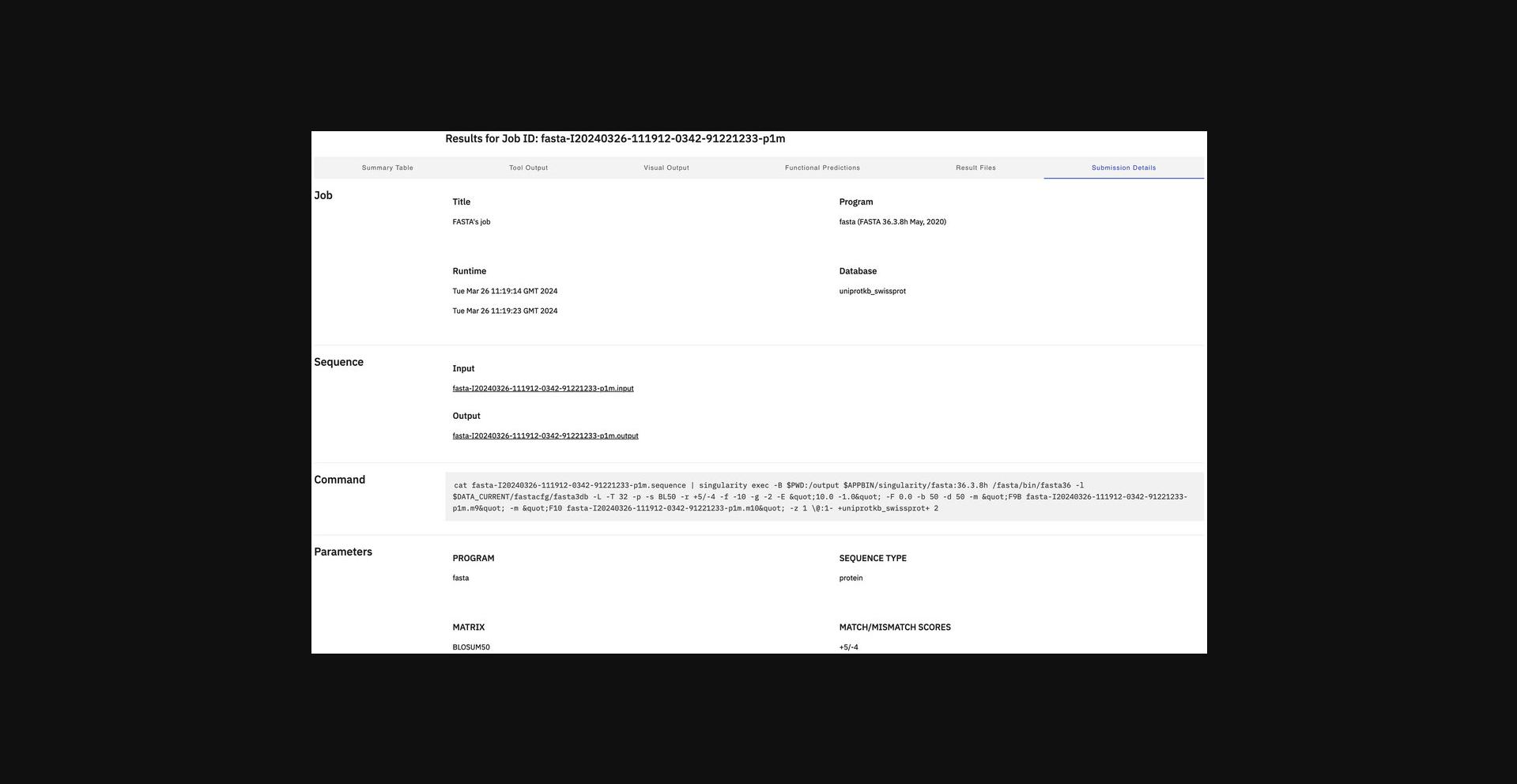
12.Display the submission details (Fig. 15) using the Submission Details view.
Alternate Protocol 3: SEQUENCE SIMILARITY SEARCH USING FASTA VIA RESTful WEB SERVICES WITH PERL CLIENT
Fully working RESTful web services clients written in Perl, Python, and Java programming languages are provided for FASTA SSS.
For a full description of the SSS RESTful web services, see https://github.com/ebi-jdispatcher/webservice-clients.
This protocol uses the Perl CLI client to run a FASTA search via the RESTful web services client.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- Perl (https://www.perl.org/) with the LWP and XML::Simple modules installed (see Support Protocol 2 for how to download and install the EMBL-EBI Perl RESTful web services clients)
Input
- A plain text file containing a sequence in FASTA, EMBL, or GenBank format
- GCG, PIR, NBRF, PHYLIP, and UniProtKB/Swiss-Prot or a database entry supported by EMBL-EBI in the format database name:database identifier (e.g., UniProtKB:GSTM1_MOUSE)
1.Display client usage. To do so, switch to the directory containing the downloaded client program fasta.pl. Run the script without specifying any parameters (or adding --help to the command shown below) to print a brief help message:
- perl fasta.pl --help
| Option | Type | Description |
|---|---|---|
| -h, --help | Show this help message and exit | |
| --asyncjob | Forces an asynchronous query | |
| --title | Str | Title for job |
| --status | Get job status | |
| --resultTypes | Get available result types for job | |
| --polljob | Poll for the status of a job | |
| --pollFreq | Int | Poll frequency in seconds (default 3 s) |
| --jobid | Str | JobId that was returned when an asynchronous job was submitted |
| --outfile | Str | File name for results (default is jobid; “-” for STDOUT) |
| --outformat | Str | Result format(s) to retrieve. It accepts comma-separated values. |
| --params | List input parameters | |
| --paramDetail | Str | Display details for input parameter |
| --quiet | Decrease output | |
| --verbose | Increase output | |
| --baseUrl | Str | Base URL. Defaults to https://www.ebi.ac.uk/Tools/services/rest/<tool_name>. |
| Option | Type | Description |
|---|---|---|
| Required (for job submission) | ||
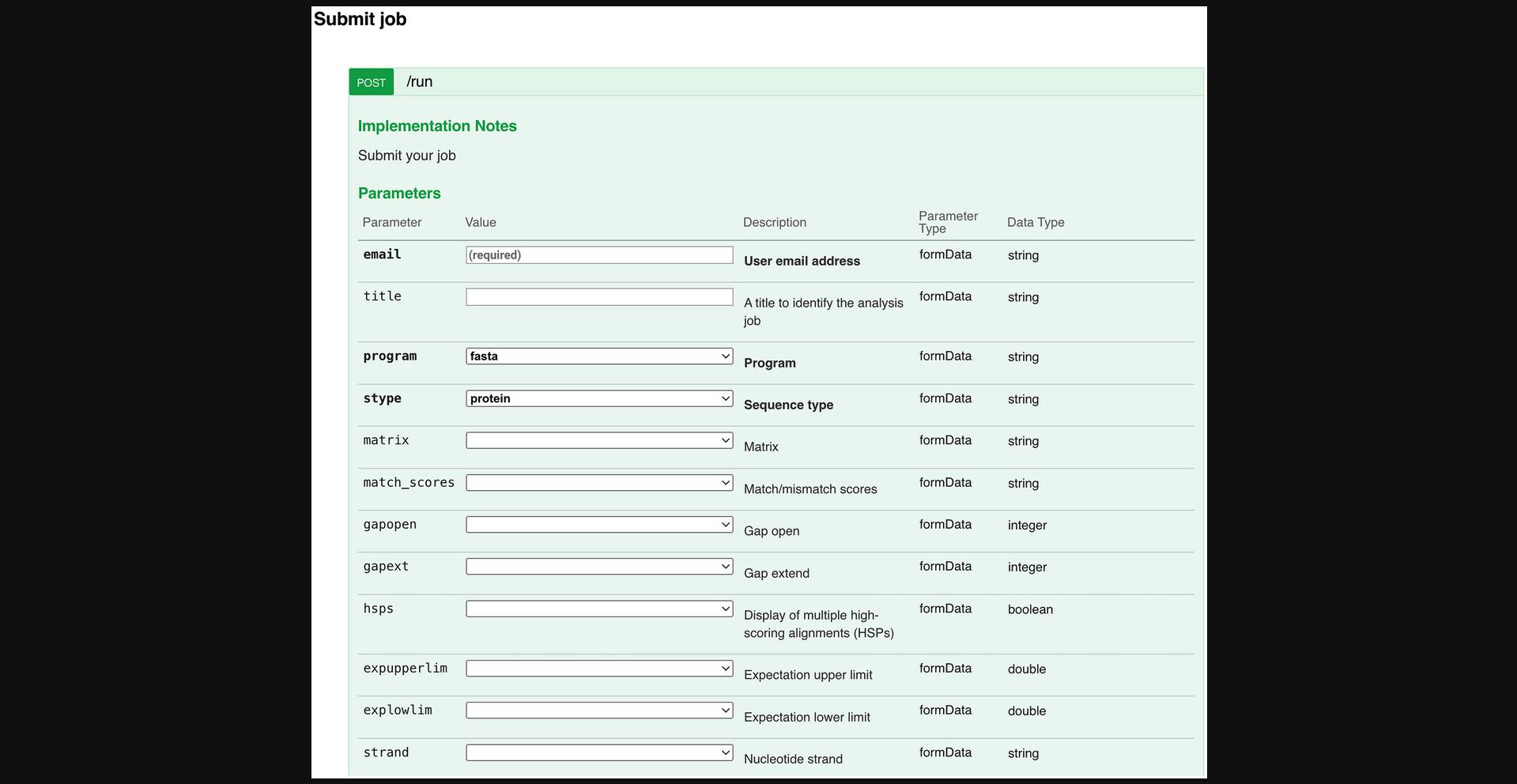
| str | E-mail address | |
| --program | str | The FASTA program to be used for the sequence similarity search |
| --stype | str | Indicates if the query sequence is protein, DNA, or RNA. Used to force FASTA to interpret the input sequence as specified type of sequence (via the -p, -n, or -U options), this prevents issues when using nucleotide sequences that contain many ambiguous residues. |
| --sequence | str | The query sequence can be entered directly into this form. The sequence can be in GCG, FASTA, EMBL (Nucleotide only), GenBank, PIR, NBRF, PHYLIP, or UniProtKB/Swiss-Prot (Protein only) format. A partially formatted sequence is not accepted. Adding a return to the end of the sequence may help certain applications understand the input. Note that directly using data from word processors may yield unpredictable results, as hidden/control characters may be present. |
| --database | str | The databases to run the sequence similarity search against multiple databases can be used at the same time. |
| Optional | ||
| --matrix | str | (Protein searches) The substitution matrix used for scoring alignments when searching the database. Target identity is the average alignment identity the matrix would produce in the absence of homology and can be used to compare different matrix types. Alignment boundaries are more accurate when the alignment identity matches the target identity percentage. |
| --match_scores | str | (Nucleotide searches) The match score is the bonus to the alignment score when matching the same base. The mismatch is the penalty when failing to match. |
| --gapopen | int | Score for the first residue in a gap |
| --gapext | int | Score for each additional residue in a gap |
| --hsps | bool | Turn on/off the display of all significant alignments between the query and library sequence |
| --expupperlim | float | Limits the number of scores and alignments reported based on the expectation value. This is the maximum number of times the match is expected to occur by chance. |
| --explowlim | float | Limit the number of scores and alignments reported based on the expectation value. This is the minimum number of times the match is expected to occur by chance. This allows closely related matches to be excluded from the results in favor of more distant relationships. |
| --strand | str | For nucleotide sequences, specify the sequence strand to be used for the search. By default, both upper (provided) and lower (reverse complement of provided) strands are used. For single-stranded sequences, searching with only the upper or lower strand may provide better results. |
| --hist | bool | Turn on/off the histogram in the FASTA result. The histogram gives a qualitative view of how well the statistical theory fits the similarity scores calculated by the program. |
| --scores | int | Maximum number of match score summaries reported in the result output |
| --alignments | int | Maximum number of match alignments reported in the result output |
| --scoreformat | str | Different score report formats |
| --stats | str | The statistical routines assume that the library contains a large sample of unrelated sequences. Options to select what method to use include regression, maximum likelihood estimates, shuffles, or combinations of these. |
| --annotfeats | bool | Turn on/off annotation features. Annotation features shows features from UniProtKB, such as variants, active sites, phospho-sites, and binding sites, that have been found in the aligned region of the database hit. To see the annotation features in the results after this has been enabled, select sequences of interest and click to “Show” alignments. This option also enables a new result tab (Domain Diagrams) that highlights domain regions. |
| --annotsym | str | Specify the annotation symbols |
| --dbrange | str | Specify the sizes of the sequences in a database to search against. For example, “100-250” will search all sequences in a database with length between 100 and 250 residues, inclusive. |
| --seqrange | str | Specify a range or section of the input sequence to use in the search. For example, specifying “34-89” in an input sequence of total length of 100 will tell FASTA to only use residues 34-89, inclusive. |
| --filter | str | Filter regions of low sequence complexity. This can avoid issues with low-complexity sequences where matches are found due to composition rather than meaningful sequence similarity. However, in some cases, filtering also masks regions of interest and so should be used with caution. |
| --transltable | int | Query genetic code to use in translation |
| --ktup | int | FASTA uses a rapid word-based lookup strategy to speed the initial phase of the similarity search. The KTUP is used to control the sensitivity of the search. Lower values lead to more sensitive but slower searches. |
2.Display parameter details. To display all parameters of the tool, run
- perl fasta.pl --params
- To see further details of the parameter, run with the argument --paramDetail
. - To see which FASTA programs are available, run
- perl fasta.pl --paramDetail program
- To see which FASTA databases are available, run
- perl fasta.pl --paramDetail database
3a. Run jobs in synchronous mode.
3b. Run jobs in asynchronous mode.
Support Protocol 2: INSTALLING PERL REST WEB SERVICES CLIENTS
Perl is commonly used in bioinformatics and typically installed by default on UNIX and UNIX-like systems. Because many existing analytical pipelines are implemented in Perl, the Perl clients provide an option for integration of EMBL-EBI's web services into existing pipelines.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- Perl 5 or above (https://www.perl.org/), LWP (https://metacpan.org/pod/LWP), and XML::Simple (https://metacpan.org/pod/XML::Simple)
- A web browser, e.g., Google Chrome, Mozilla Firefox, Microsoft Edge, Safari, or Opera
1.Check that Perl is installed on the system. To do so, in the Command Prompt or terminal, enter the following:
- perl --version
If Perl is not installed, download and follow the instructions provided at https://www.perl.org/get.html.
2.Check that the required LWP and XML::Simple Perl modules have been installed:
- perl --MLWP -e “print $LWP::VERSION;”
- If a “Can't locate LWP.pm” error message is returned, install the LWP Perl module.
The LWP Perl module can be installed via the operating system package manager on many Linux/UNIX systems. For example, on Debian-based Linux distributions (e.g., Bio-Linux, Linux Mint, and Ubuntu), the “lib-perl” package should be installed. The LWP Perl module can also be installed from the Comprehensive Perl Archive Network (CPAN); seehttps://www.cpan.org/for details:
perl -MXML::Simple -e “print$XML::Simple::VERSION;”
- If a “Can't locate XML/Simple.pm” error message is returned, install the XML::Simple Perl module.
The XML::Simple Perl module can be installed via the operating system package manager on many Linux/UNIX systems. For example, on Debian-based Linux distributions (e.g., Bio-Linux, Linux Mint, and Ubuntu), the “libxml-simpleperl” package should be installed. The XML::Simple Perl module can be installed from the CPAN; seehttps://www.cpan.org/for details.
3.Download the Perl clients (e.g., ncbiblast.pl) from https://github.com/ebi-jdispatcher/webservice-clients. Alternatively, download the NCBI BLAST+ client directly from GitHub with wget :
4.Test and run the client. Within the Command Prompt or terminal, change to the directory that contains the client program downloaded earlier. To test the program (e.g., ncbiblast.pl) , enter the following:
- perl ncbiblast.pl --help
Basic Protocol 3: SEQUENCE SIMILARITY SEARCH USING NCBI BLAST+ RESTful WEB SERVICES WITH PYTHON CLIENT
NCBI BLAST+ (Altschul et al., 1997; Camacho et al., 2009; also see Current Protocols article: Ladunga, 2002) emphasizes finding regions of sequence similarity, which will yield functional and evolutionary clues about the structure and function of your novel sequence.
EMBL-EBI provides web services clients written in Perl, Python, and Java programming languages for the NCBI BLAST+ SSS.
For a full description of the RESTful web services, see https://www.ebi.ac.uk/jdispatcher/docs/webservices/.
This protocol uses a Python client program to run NCBI BLAST+ via the RESTful web service CLI client.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- Python (https://www.python.org/) with the xmltramp2 module installed (see Support Protocol 1 for how to download and install the EMBL-EBI Python RESTful web services clients)
Input
- A plain text file containing a sequence in one of the formats of GCG, FASTA, EMBL, GenBank, PIR, NBRF, PHYLIP, or UniProtKB/Swiss-Prot or a database entry supported by EMBL-EBI in the format database name:database identifier (e.g., embl:x56957)
1.Display client usage by switching to the directory containing the downloaded Python client ncbiblast.py. For details of how to use the client, run the script with --help:
- python ncbiblast.py --help
2.Display parameter details. To display all parameters of the tool, run
- python ncbiblast.py --params
- To see further details of the parameter, run with the argument --paramDetail
. - To see the available BLAST programs, run
- python ncbiblast.py --paramDetail program
- To see the available BLAST databases, run
- python ncbiblast.py --paramDetail database
3a. Run jobs in synchronous mode.
3b. Run jobs in asynchronous mode.
Basic Protocol 4: SEQUENCE SIMILARITY SEARCH USING HMMER3 phmmer REST WEB SERVICES WITH PERL CLIENT AND DOCKER
SSS using profile hidden Markov models (HMMs) has become a common practice in biological sequence analysis. Profile HMMs are constructed from a set of related sequences, which can then be used to search large sequence databases. In addition to residue conservation, HMMs also incorporate rates of insertions and deletions. The sensitivity of profile HMMs is achieved by the position-specific probabilistic modeling of the MSA, which allows detection of even distantly related sequences (Eddy, 1998).
HMMER3 (Potter et al., 2018) is a popular software package for detecting sequence homology, comparing a profile HMM to either a single sequence or a database of sequences. HMMER3 phmmer is used to search a database of protein sequences with a protein sequence of interest.
For the full description of the REST web services, see https://www.ebi.ac.uk/jdispatcher/docs/webservices/.
This protocol uses Docker to run a pre-configured container that provides Perl, Python, and Java CLI clients. In this example, we use the Perl client to run HMMER3 phmmer via the REST web service interface.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation, running Docker
Software
- See Support Protocol 3 for instructions on downloading and installing Docker as well as the ebiwp/webservice-clients image that provides access to pre-configured Perl, Python, and Java REST Web Services CLI Clients
Input
- A plain text file containing a sequence in GCG, FASTA, EMBL, GenBank, PIR, NBRF, PHYLIP, or UniProtKB/Swiss-Prot format or a database entry supported by EMBL-EBI in the format database name:database identifier
1.Display client usage. To do so, call Perl, Python, or Java clients in the ebiwp/webservice-clients Docker image with docker run --rm ebiwp/webservice-clients. To see details of how to use the client and a detailed list of major command-line options, call the client with --help as follows:
- docker run --rm ebiwp/webservice-clients hmmer3_phmmer.pl --help
2.Display parameter details. To display all parameters of the tool, run
- docker run --rm ebiwp/webservice-clients hmmer3_phmmer.pl --params
- To see further details of the parameter, run with argument --paramDetail
. - To see which databases are available, run
- docker run --rm ebiwp/webservice-clients hmmer3_phmmer.pl --paramDetail database
3a. Run jobs in synchronous mode.
pwd:/results as command-line options to the Docker command. This will define /results as the path that result files will be written by the client (in the container). A -v, --volume mapping is used to provide the container access to the current working directory pwd to the container. See Support Protocol 3 for additional information about running EMBL-EBI clients with Docker.pwd:/results ebiwp/webservice-clients hmmer3_phmmer. pl --email your@email.com --database uniprotkb <SequenceFile.fasta>pwd:/results ebiwp/webservice-clients hmmer3_phmmer.pl --email your@email.com --database uniprotkb DB:Identifier3b. Run jobs in asynchronous mode.
pwd:/results ebiwp/webservice-clients hmmer3_phmmer.pl --asyncjob --email your@email.com --database uniprotkb <SequenceFile.fasta>pwd:/results ebiwp/webservice-clients hmmer3_phmmer.pl --polljob --outformat out --jobid pwd:/results ebiwp/webservice-clients hmmer3_phmmer.pl --polljob --jobid Support Protocol 3: INSTALLING DOCKER AND RUNNING THE EMBL-EBI CLIENT CONTAINER
Docker is based on an operating system–level virtualization technology known as “containerization” and a software platform that allows users to run pre-configure Docker containers. A Docker container contains software components along with all their dependencies, binaries, libraries, configuration files, scripts, and so forth. Containers are pre-configured and deployed in such a way that the contained programs are run in isolation. This greatly helps with reproducibility by leveraging the fact that the user does not need to worry about installation and configuration of specific versions of software and their dependencies.
A Docker image (ebiwp/webservice-clients) has been developed and is freely available to users. This provides users with pre-installed Perl, Python, and Java EMBL-EBI Web Service CLI Clients. The ebiwp/webservice-clients image can be pulled from the Docker Hub at https://hub.docker.com/r/ebiwp/webservice-clients/.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- Docker can be downloaded from https://www.docker.com/products/docker-desktop/
- A web browser, e.g., Google Chrome, Mozilla Firefox, Microsoft Edge, Safari, or Opera
1.Install Docker.
2.List available Docker images:
- docker image ls
3.Download and install the ebiwp/webservice-clients container by getting the latest tag of the required ebiwp/webservice-clients image by running the following:
- docker pull ebiwp/webservice-clients:latest
4.Run clients with the ebiwp/webservice-clients container.
5.Mount local directories to be accessible to the container.
pwd:/results as a command-line option to the Docker command. This will define /results as the path where result files will be written by the client (in the container). A --volume (-v) mapping is used to provide access to the current working directory pwd to the container:pwd:/results ebiwp/webservice-clients:latest <client.py|pl|jar> <options … >Basic Protocol 5: PROTEIN FUNCTIONAL ANALYSIS USING InterProScan 5 RESTful WEB SERVICES WITH THE PYTHON CLIENT AND DOCKER
InterProScan 5 (Jones et al., 2014) combines different protein signature recognition methods into one resource and allows the user to scan sequences for matches against the InterPro collection of protein signature databases. This example uses Docker and the Python client program to run an InterProScan 5 search via the REST web service interface.
More information about the InterProScan service is available at https://www.ebi.ac.uk/interpro/search/sequence/.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation, running Docker
Software
- See Support Protocol 3 for instructions on downloading and installing Docker as well as the ebiwp/webservice-clients image that provides access to pre-configured Perl, Python, and Java REST Web Services CLI Clients
Input
- A plain text file containing a sequence in GCG, FASTA, EMBL, GenBank, PIR, NBRF, PHYLIP, or UniProtKB/Swiss-Prot format or a database entry supported by EMBL-EBI in the format database name:database identifier
1.Display client usage. To do so, call Python clients in the ebiwp/webservice-clients Docker image with docker run --rm ebiwp/webservice-clients. To see details of how to use the client, a detailed list of major command-line options, how to run the client without any argument, or alternatively how to run the client with the argument --help, call the client as follows:
- docker run --rm ebiwp/webservice-clients iprscan5.py
2.Display parameter details. To display all parameters of the tool, run
- docker run --rm ebiwp/webservice-clients iprscan5.py --params
- To see the details of a parameter, use with the argument --paramDetail
. - To see which applications are available, run
- docker run --rm -w /results -v
pwd:/results ebiwp/webservice-clients iprscan5.py --paramDetail appl
3a. Run jobs in synchronous mode.
pwd:/results as command-line options to the Docker command. This will define /results as the path where result files will be written by the client (in the container). A -v, --volume mapping is used to provide the container access to the current working directory pwd. See Support Protocol 3 for additional information about running EMBL-EBI clients with Docker.pwd:/results ebiwp/webservice-clients iprscan5.py --email your@email.com<SequenceFile.fasta>pwd:/results ebiwp/webservice-clients iprscan5.py --email your@email.comDB:Identifierpwd:/results ebiwp/webservice-clients iprscan5.py --email your@email.com --goterms false --pathways false --sequence uniprot:gstm1_mouse3b. Run jobs in asynchronous mode.
pwd:/results ebiwp/webservice-clients iprscan5.py --asyncjob --email your@email.com uniprot:gstm1_mousepwd:/results ebiwp/webservice-clients iprscan5.py --polljob --outformat out --jobid pwd:/results ebiwp/webservice-clients iprscan5.py --polljob --outformat out,xml --jobid pwd:/results ebiwp/webservice-clients iprscan5.py --polljob --jobid Alternate Protocol 4: PROTEIN FUNCTIONAL ANALYSIS USING InterProScan 5 RESTful WEB SERVICES WITH THE JAVA CLIENT
This example uses a Java client program to run InterProScan 5 search via the REST web service interface.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- Java 8 or later runtime environment (https://www.java.com/)
- See Support Protocol 4 for instructions on downloading and installing the EMBL-EBI Java RESTful web services clients
Input
- A plain text file containing a sequence in one of the formats of FASTA, EMBL, GenBank, GCG, PIR, NBRF, PHYLIP, and UniProtKB/Swiss-Prot or a database entry supported by EMBL-EBI in the format database name:database identifier (e.g: UniProtKB:GSTM1_MOUSE)
1.Display client usage. To do so, switch to the directory containing the downloaded client program iprscan5.jar. For details of how to use the client, run it without any arguments:
- java -jar iprscan5.jar
Usage help will be shown on the screen. Alternatively, run it with the argument --help:
- java -jar iprscan5.jar --help
2.Display parameter details. To display all parameters of the tool, run
- java -jar iprscan5.jar --params
- To see the details of a parameter, use with the argument --paramDetail
. - To see which applications are available, run
- java -jar iprscan5.jar --paramDetail appl
3a. Run jobs in synchronous mode.
3b. Run jobs in asynchronous mode.
Support Protocol 4: INSTALLING JAVA WEB SERVICES CLIENTS
Commonly installed Java provides a platform-independent option for developing and deploying software.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- A Java 1.8 or above runtime environment, see https://www.java.com/
- A web browser, e.g., Google Chrome, Mozilla Firefox, Microsoft Edge, Safari, or Opera
1.Check that Java is installed in the system. In the Command Prompt or terminal, enter the following:
- java -version
2.If Java is not installed, download and follow the instructions provided at https://www.java.com/en/download/. If Java is installed in the system but the “java” command is not found, add Java to the PATH to try to solve the issue:
- a.
For MS Windows, check the location used to install Java using Explorer.
This will usually be something like C:\Program Files (x86)\Java\jre8. In the Command Prompt, add the location of the Java bin directory to the PATH by entering
set PATH=%PATH%;C:\Program Files (x86)\Java\jre8\bin
The “java” command should now be found.
- b.
On Linux, OS X, and UNIX systems, where the method to add a directory to the PATH depends on the shell being used, first locate the Java installation and then add the Java bin directory to the PATH.
For example, for a Java installation in /usr/lib/jvm/java-8-openjdk-amd64/, use the following commands:
1.For sh or bash shells :
export PATH=${PATH}:/usr/lib/jvm/java-8-openjdk-amd64/bin
2.For csh or tcsh shells :
setenv PATH ${PATH}:/usr/lib/jvm/java-8-openjdk-amd64/bin
3.Download the Java clients (e.g., clustalo.jar) from https://github.com/ebi-jdispatcher/webservice-clients. Alternatively, download the Clustal Omega client directly from GitHub with wget :
4.Test and run the clients. To do so, within the Command Prompt or terminal, change to the directory which contains the client program downloaded earlier. To test the program (e.g., clustalo.jar), enter
- java -jar clustalo.jar --help
Basic Protocol 6: MULTIPLE SEQUENCE ALIGNMENT USING CLUSTAL OMEGA VIA WEB INTERFACE
MSA is generally the alignment of three or more biological sequences. From the output, homology can be inferred and the evolutionary relationships between the sequences studied.
Clustal Omega (Sievers & Higgins, 2018; Sievers et al., 2011; also see Current Protocols article: Sievers & Higgins, 2014) is a fast, large-scale MSA program that uses seeded guide trees and HMM profile-profile techniques to generate alignments.
Necessary Resources
Hardware
- Any Internet-connected computer
Software
- A web browser, e.g., Google Chrome, Mozilla Firefox, Microsoft Edge, Safari, or Opera
Input
- A plain text file containing three or more sequences in FASTA, EMBL, GCG, PIR, NBRF, PHYLIP, GenBank, or UniProtKB/Swiss-Prot format or three or more database entries supported by EMBL-EBI in the format database name:database identifier
- This example uses a FASTA-format multiple sequence file containing a collection of WAP sequences. The example file can be downloaded from https://www.ebi.ac.uk/Tools/dbfetch/dbfetch?db=uniprotkb&id=wap_rat%2Cwap_mouse%2Cwap_pig&format=fasta&style=raw&Retrieve=Retrieve.
1.Optional : To view the range of MSA tools available at EMBL-EBI, point the browser to the MSA web page https://www.ebi.ac.uk/jdispatcher/msa.
2.Click “Launch Clustal Omega” under the Clustal Omega section or directly go to https://www.ebi.ac.uk/jdispatcher/msa/clustalo.
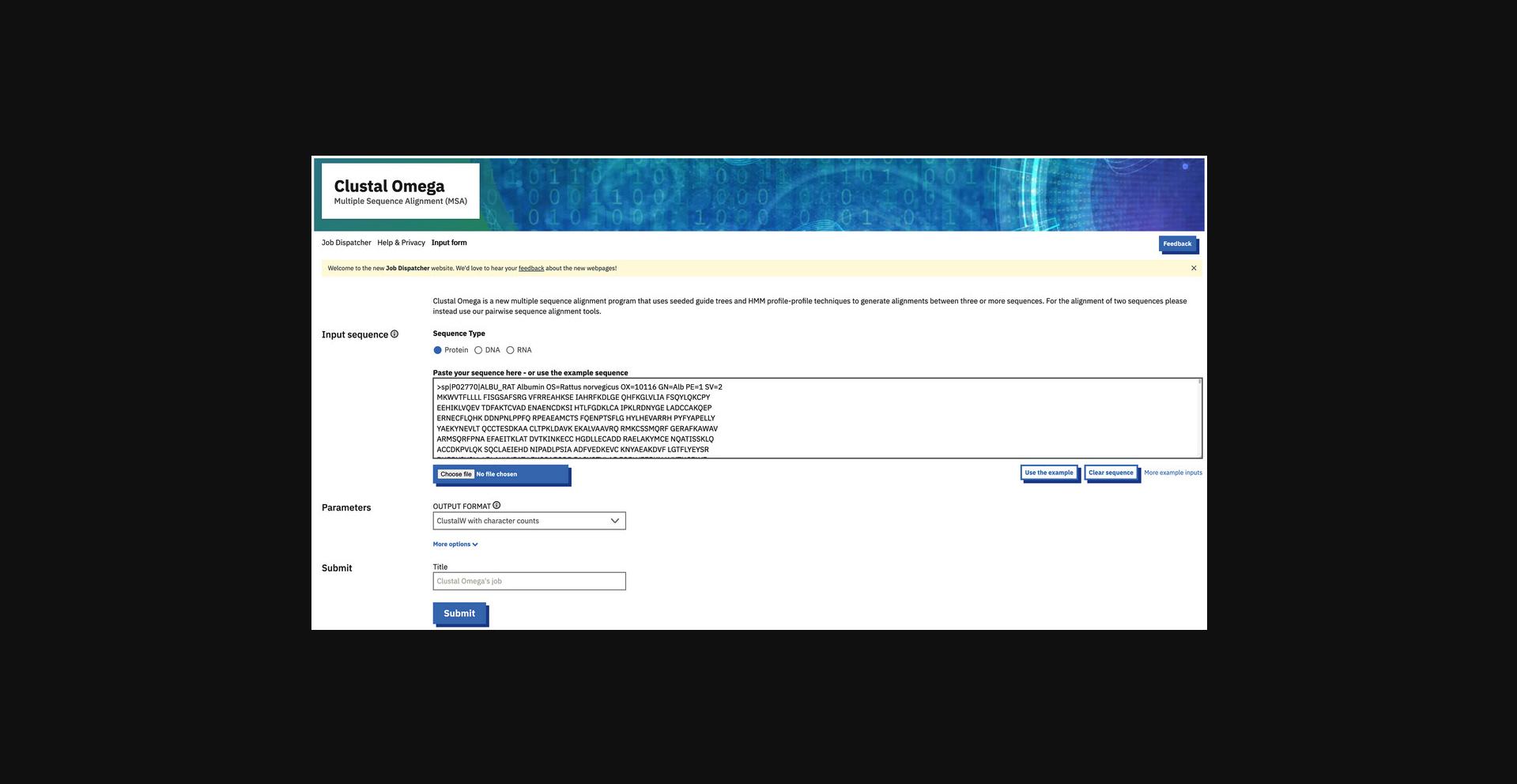
3.Enter the input sequences by browsing and selecting the input sequences file. Alternatively, copy the sequences and paste them into the sequence box. Select the correct input sequence type just above the input sequence box.
4.Set the parameters. To do so, first select the output format. To examine further options, click on the “More options” button to expand the section for the advanced parameters, which for Clustal Omega includes options to de-align input sequences, the number of iterations for the guide tree, and HMM stages, among others. Change the settings of the parameters according to needs.
5.Provide a job “Title” (optional) to briefly describe the job and click the “Submit” button.
6.View the results.
7.View the actual tool output.
8.View the guide and phylogenetic tree.
9.Display results viewers.
10.Display result files.
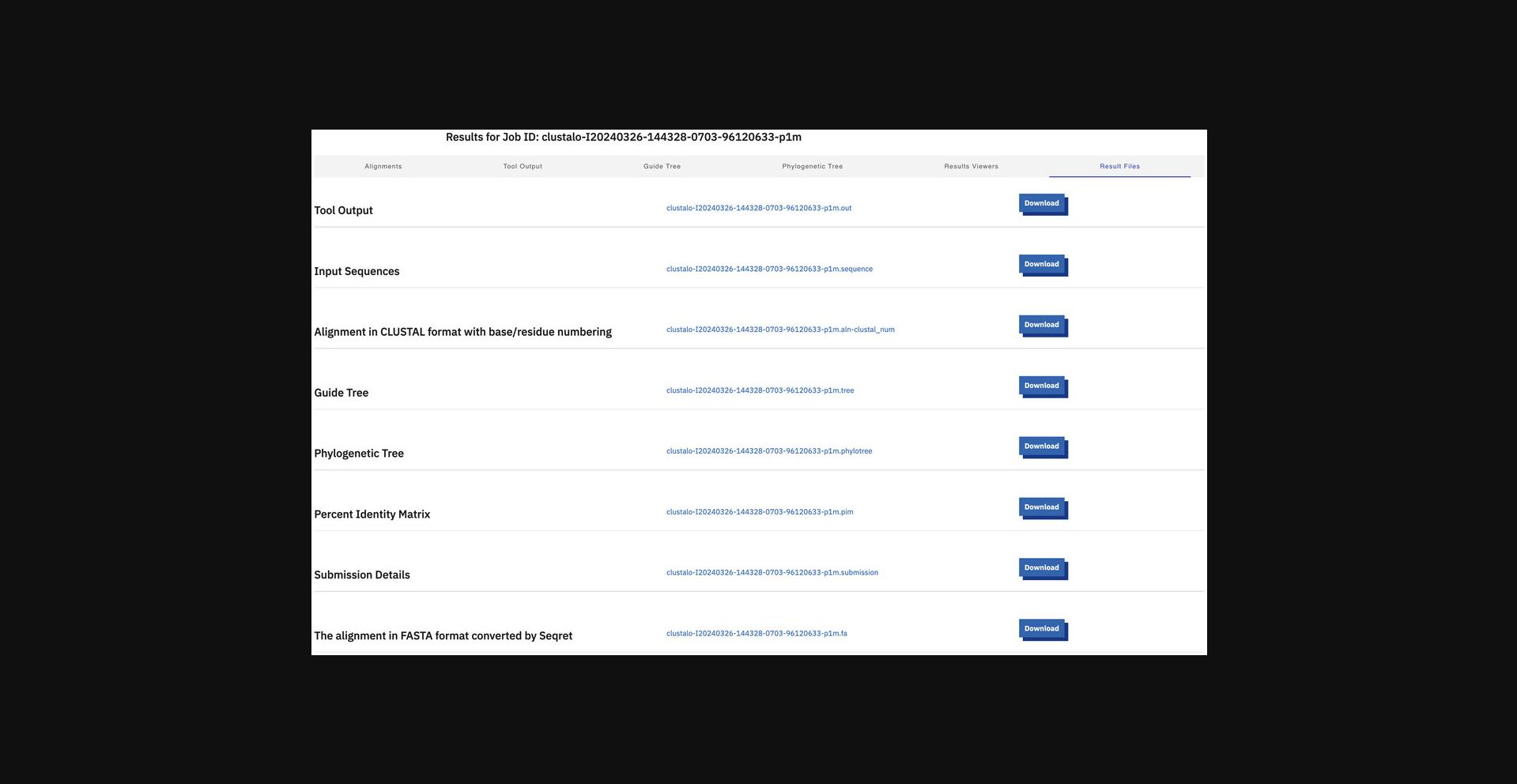
11.Display submission details.
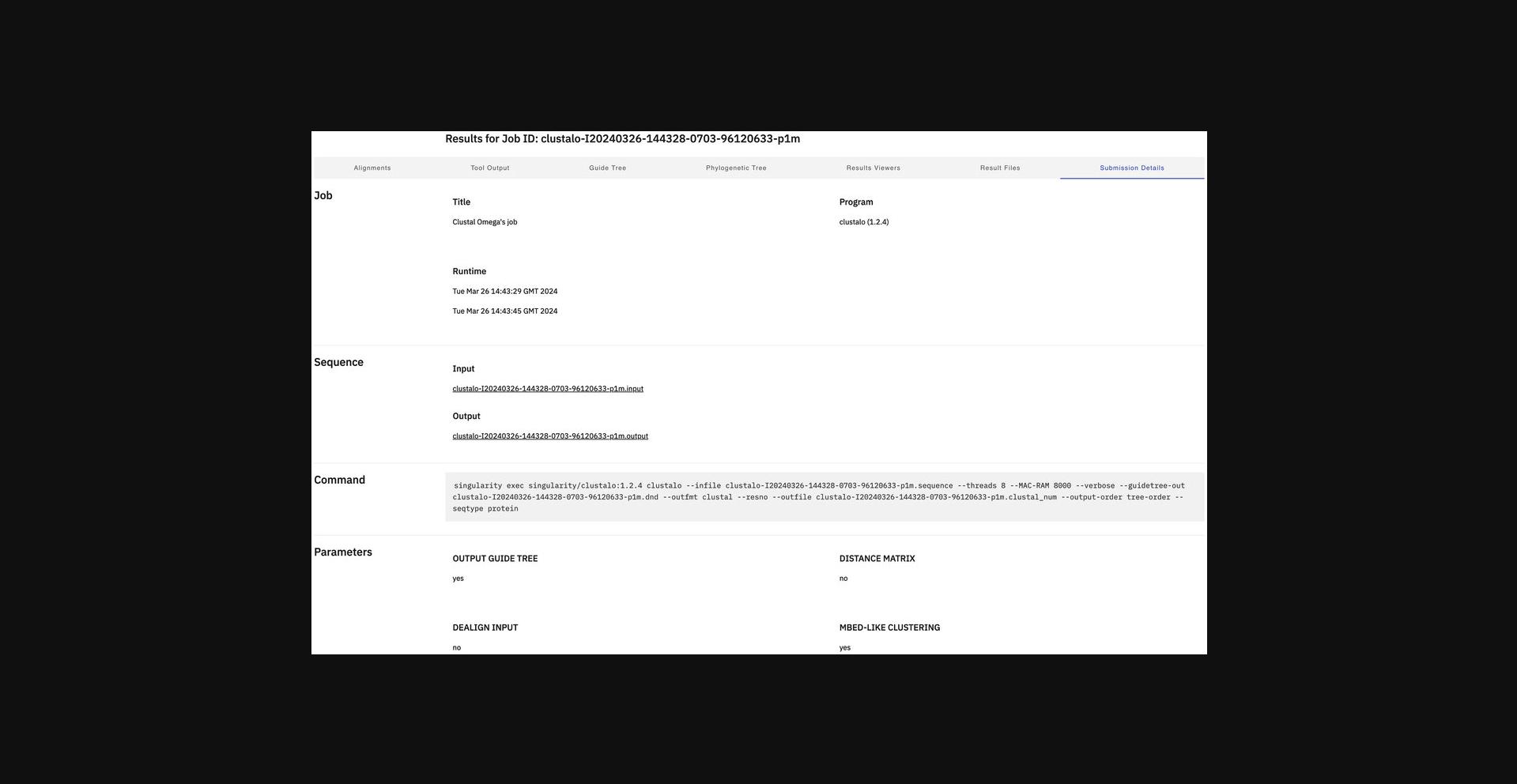
Alternate Protocol 5: MULTIPLE SEQUENCE ALIGNMENT USING CLUSTAL OMEGA WITH PERL CLIENT AND DOCKER
This protocol demonstrates a Clustal Omega MSA via web services using a Perl Client with Docker.
For the full description of the Clustal Omega REST web services, see https://www.ebi.ac.uk/jdispatcher/docs/webservices/.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation, running Docker
Software
- See Support Protocol 3 for instructions on downloading and installing Docker as well as the ebiwp/webservice-clients image that provides access to pre-configured Perl, Python, and Java REST Web Services CLI Clients
Input
- A plain text file containing three or more sequences in FASTA, EMBL, GCG, PIR, NBRF, PHYLIP, GenBank, or UniProtKB/Swiss-Prot format or three or more database entries supported by EMBL-EBI in the format database name:database identifier
- This example uses a FASTA-format multiple sequence file containing a collection of myosin sequences. The example file can be downloaded from https://www.ebi.ac.uk/Tools/examples/protein/sequence12.txt.
1.Display client usage by running Perl clients in the ebiwp/webservice-clients Docker image with docker run --rm ebiwp/webservice-clients.
2.Display parameter details. To display all parameters of the tool, run
- docker run --rm ebiwp/webservice-clients clustalo.pl --params
- To see further details of the parameter, run with the argument --paramDetail
. For example, to see what input types are available, run - docker run --rm ebiwp/webservice-clients clustalo.pl --paramDetail stype
3a. Run jobs in synchronous mode.
pwd:/results as command-line options to the Docker command. This will define /results as the path where result files will be written by the client (in the container), and a -v, --volume mapping is used to provide the container access to the current working directory pwd. See Support Protocol 3 for additional information about running EMBL-EBI clients with Docker.pwd:/results ebiwp/webservice-clients clustalo.pl --email your@email.com sequence12.txt3b. Run jobs in asynchronous mode.
pwd:/results ebiwp/webservice-clients clustalo.pl --asyncjob --email your@email.com sequence12.txtpwd:/results ebiwp/webservice-clients clustalo.pl --polljob --outformat out --jobid pwd:/results ebiwp/webservice-clients clustalo.pl --polljob --outformat out,xml --jobid pwd:/results ebiwp/webservice-clients clustalo.pl --polljob --jobid Support Protocol 5: EXPLORING THE RESTful API WITH OpenAPI USER INTERFACE
The EMBL-EBI RESTful web services can be explored with the aid of Swagger OpenAPI User Interface (UI). Swagger UI allows anyone to visualize, interact with, and explore the API's resources and endpoints. Documentation pages for all available bioinformatics web services are provided at https://www.ebi.ac.uk/jdispatcher/docs/webservices/. An accompanying Swagger UI is available for each tool at https://www.ebi.ac.uk/jdispatcher/docs/webservices/#openapi.
Necessary Resources
Hardware
- An Internet-connected UNIX, Linux, Mac, or Windows workstation
Software
- A web browser, e.g., Google Chrome, Mozilla Firefox, Microsoft Edge, Safari, or Opera
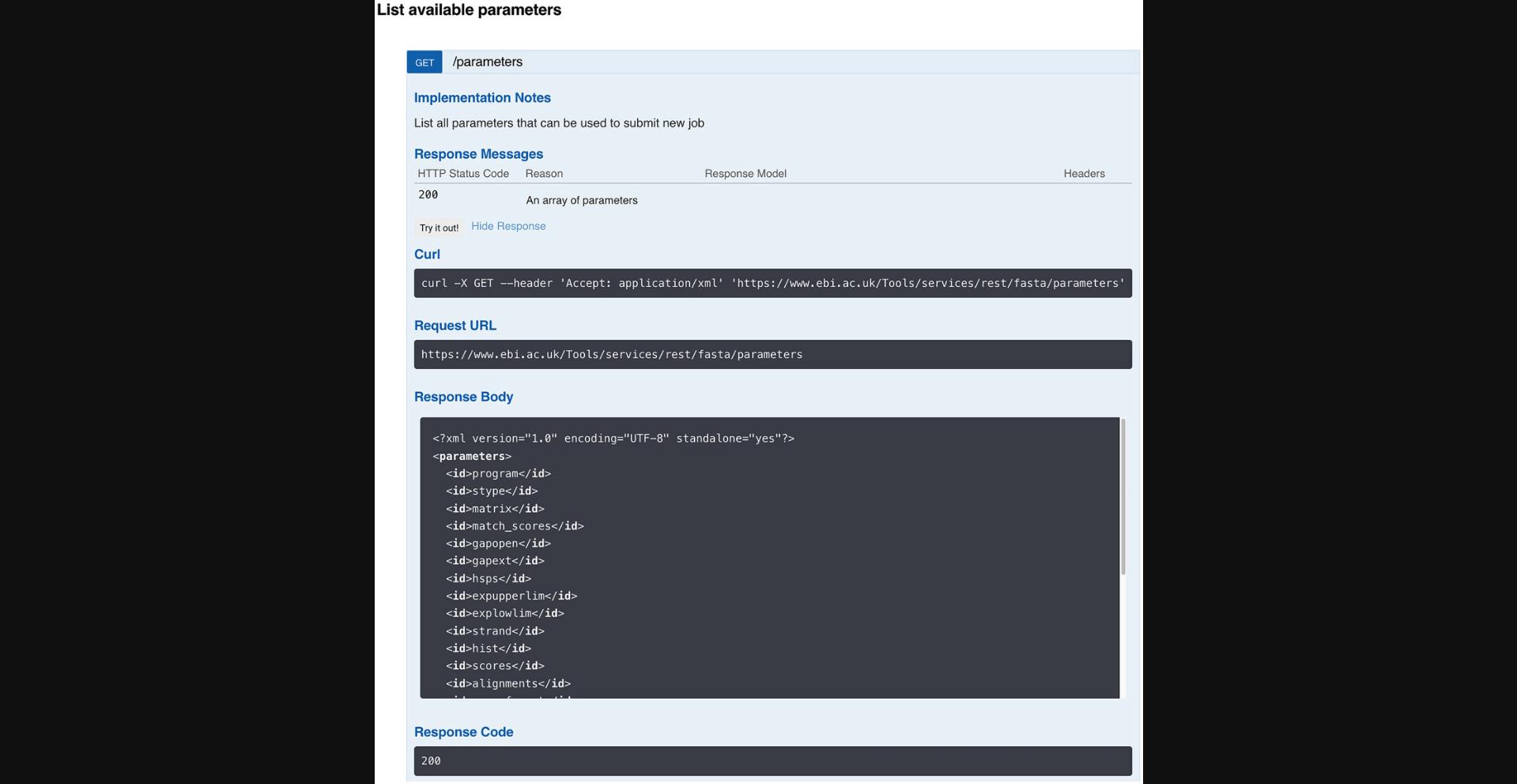
1.Choose a tool (e.g., FASTA). To do so, head to https://www.ebi.ac.uk/jdispatcher/docs/webservices/#openapi in a web browser. Then, in “STEP 1 - Choose a Tool,” click in the drop-down menu and select FASTA.
COMMENTARY
Understanding Results
The interpretation of the scientific results from the wide variety of tools that are available through the EMBL-EBI web interface and web services is beyond the scope of this article; however, in this section, we present some of the common outcomes from successful or unsuccessful uses of the services.
When a job is submitted through the web interface (Basic Protocol 2), a quick check on the input is carried out, and only after the data pass this validation check are the data submitted to the compute clusters where the actual request/analysis is executed. This check allows us to reduce the number of invalid submissions to the clusters and allows the user to quickly correct simple errors. If the input check is not passed, an error box appears on the web page with some detail about the error and what action the user can take to correct it (Fig. 27). If the check is passed, a temporary running page will be displayed with the JobId until the results are ready to be viewed (Fig. 28). The unique JobId currently consists of the name of the tool, the method of submission (I, E, R, or S, representing Interactive, Email, REST, and SOAP), the date and time of submission, and, finally, an identifier that is helpful to the administrators, internally relating to the running of jobs on our compute clusters.
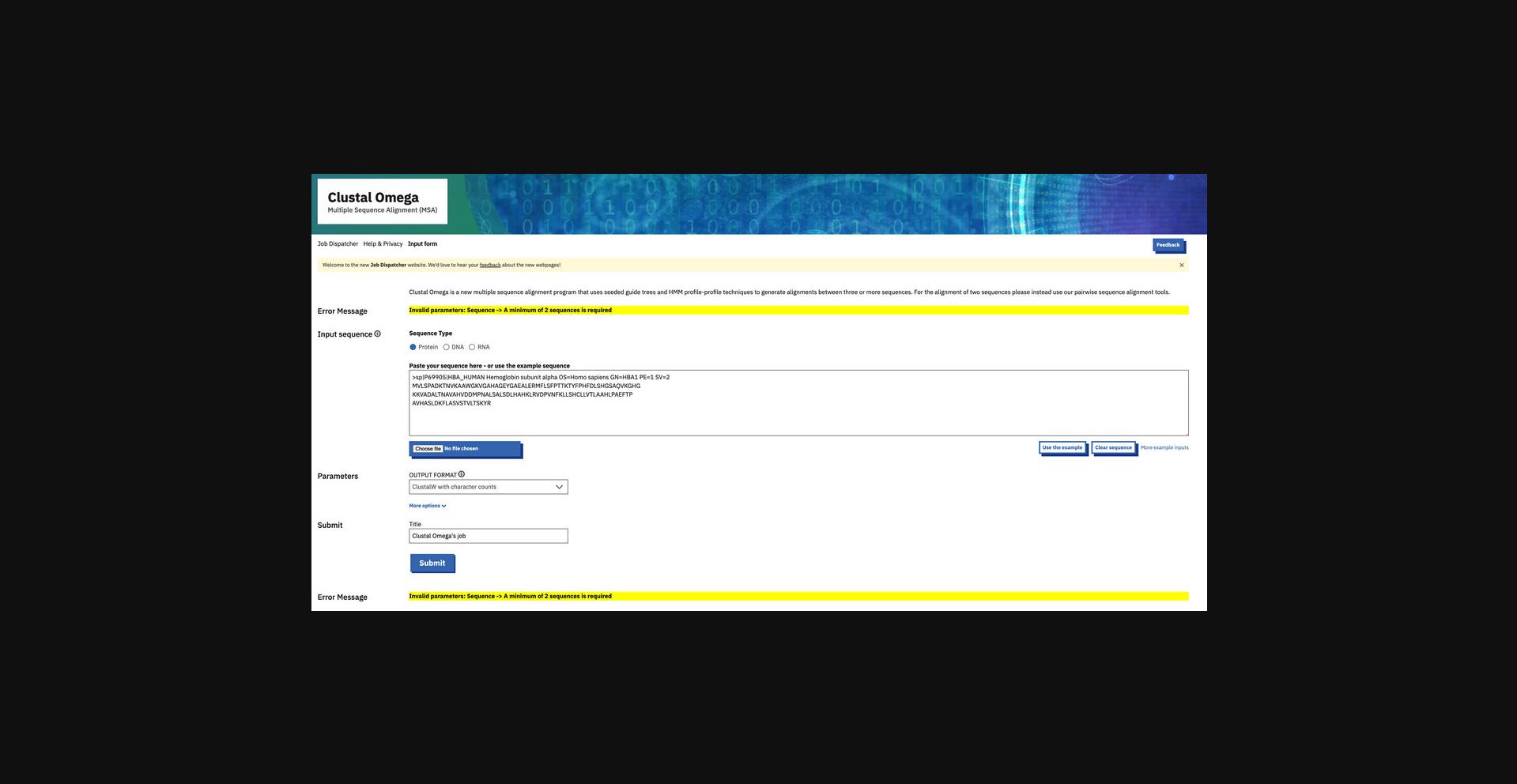
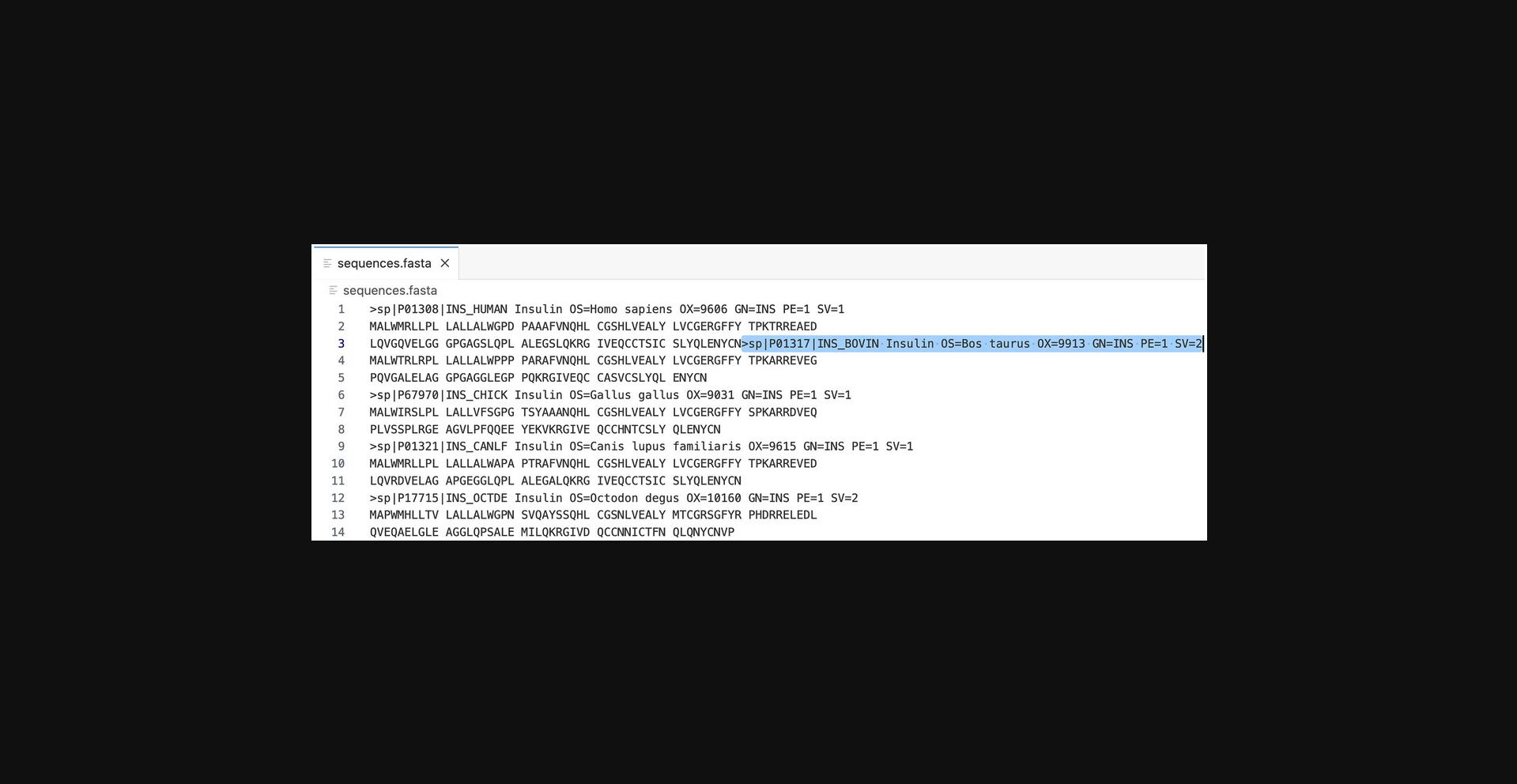
Causes for failing the validation check are usually simple user mistakes, such as failing to select a database to search against in the case of FASTA or accidently hitting the “Submit” button before a set of sequences has been uploaded or entered into the input box for Clustal Omega. Errors are also returned when the data input is too large. For popular tools, there are FAQs in the Documentation pages that address common causes of validation check failure.
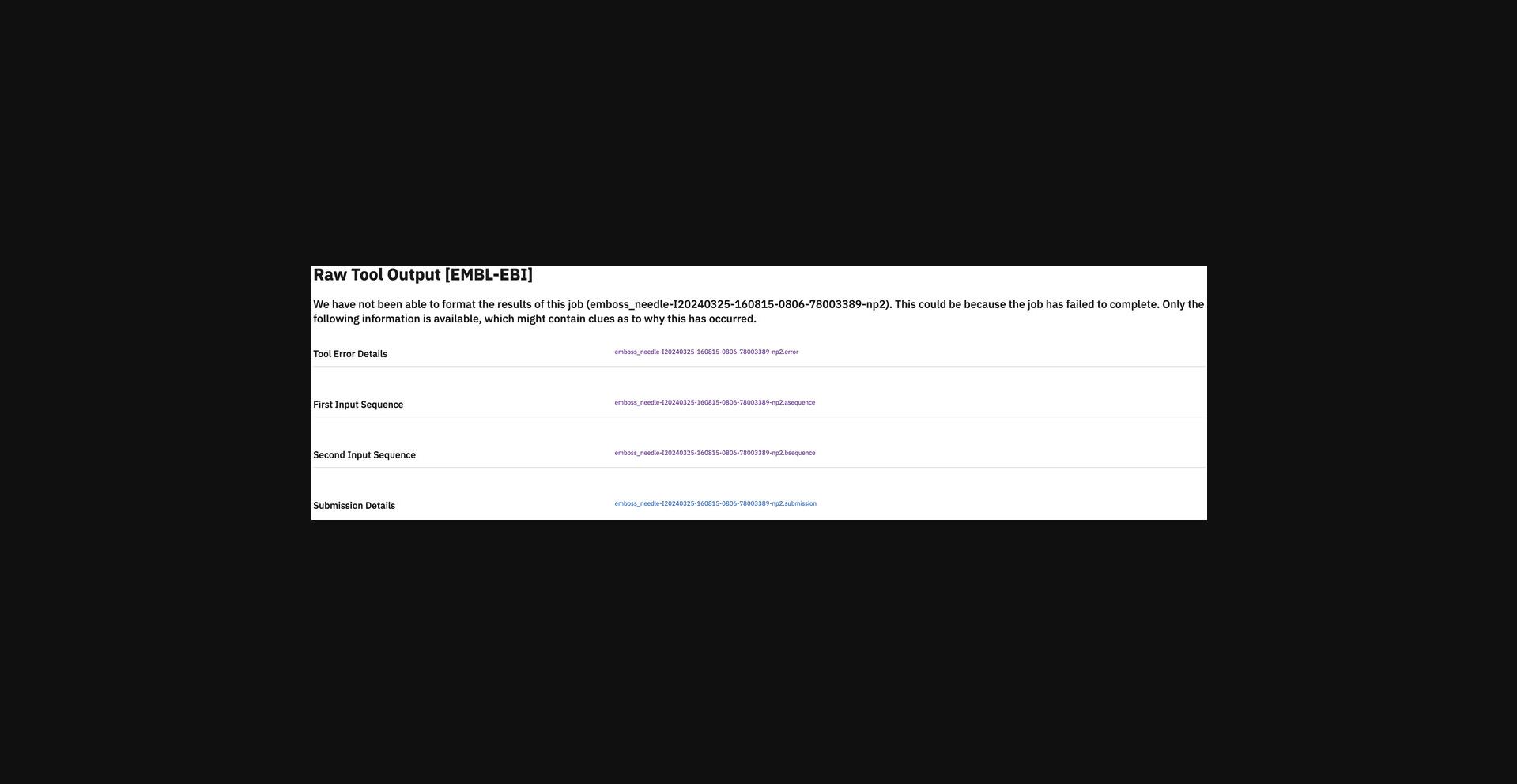
Unfortunately, passing the quick input validation check does not guarantee that the job will complete successfully, as there can be situations in which the underlying tool produces an error once it is run. An example is where a user has accidentally truncated the input for an MSA such that sequence file header text now appears in the middle of the sequence data for a different entry (Fig. 29). When we detect that a tool has failed to provide the expected results (or has produced an error), we highlight this to the user in place of the normal results pages and present links to the user that contain as much information as possible to help determine the cause of the error. In this case, the error file of EMBOSS Needle gives a message indicating the input sequence is too large and suggests another tool, EMBOSS Stretcher (Fig. 30). When encountering the error page, users should read any error messages from the tool and check their input carefully for errors. If help is still needed, the JobId should be sent to our help desk using the Feedback link at the top of the page or via https://www.ebi.ac.uk/about/contact/support/job-dispatcher-services.
Attempting to view the results of a job a long time after it was submitted may not succeed, as results are not kept indefinitely; currently, they are deleted after 7 days. Doing so generates a “job not available” page, as seen in Figure 31. To generate the results again, the user will need to carry out a new job submission.

The situation when using web services is similar. Incorrect usage of a command-line client, for example, supplying an incorrect parameter, returns an error such as “Unknown option:”. The user should run the client without any parameters to display correct usage and available parameters. Omission of data required for a job (for example, failing to select a database or supplying an input file for MSA that only contains one sequence) results an error being passed to the user in exactly the same terms as when the validation check fails on the website; behind the scenes, it is in fact the same check as for the web interface.
Successful web service requests result in a job status of “FINISHED;” this is analogous to the results page being displayed for web interface submissions. Problems with the running of the job (for example, due to server failure) result in a status of “ERROR” or “FAILURE.” Requests for an invalid JobId, either because the ID is incorrect or because the result has expired, return a status of “NOT FOUND” (Fig. 32).

If there is a problem and the tool generates an error, then error files are produced, together with your input and any standard output from the tool (Fig. 33). Error files can be identified by their suffix of .error and contain information about the error. These error files are of particular value when requesting assistance from our help desk. Common causes of errors include incorrect or missing parameters, using input that is incorrectly formatted or unsuitable for the tool, and attempted retrieval of results beyond the period when they are available.

Note that there are situations when an incorrect analysis has been requested yet the tool appears to run fine, for example, when a search is carried out against a protein database using DNA input. Correct usage would be to employ a tool such as FASTX to translate the DNA input; however, if the user incorrectly uses FASTA, the tool will still run and produce a result of sorts. This is because there are amino acids corresponding to the same single-letter characters used for DNA bases, so the program does not prevent the search. Another example might be the use of an MSA tool, such as Clustal Omega, for situations that it is not designed for, e.g., for pairwise alignment or to align short primers to a longer sequence. In general, if the standalone tool allows an analysis to be carried out, then we attempt to allow it at EMBL-EBI as well; it is up to the user to decide for what purposes they use the tools, and they should examine the results for the unexpected.
Please note that the results are stored for only 7 days, and it is recommended to download the results before the job expires.
We offer documentation and training courses (https://www.ebi.ac.uk/training/) to educate users on correct usage of the tools, and our help desk is available for further assistance at https://www.ebi.ac.uk/about/contact/support/job-dispatcher-services.
Acknowledgments
The EMBL-EBI services mentioned in this article are supported by core EMBL funding. EMBL-EBI is indebted to its funders, including the EMBL member states.
Author Contributions
Fábio Madeira: Writing—original draft; writing—review and editing. Nandana Madhusoodanan: Writing—original draft; writing—review and editing. Joonheung Lee: Writing—original draft; writing—review and editing. Alberto Eusebi: Writing—review and editing. Ania Niewielska: Writing—review and editing. Adrian R. N. Tivey: Writing—review and editing. Stuart Meacham: Writing—review and editing. Rodrigo Lopez: Writing—review and editing. Sarah Butcher: Writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data are openly available in a public repository that does not issue DOIs.
Literature Cited
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research , 25(17), 3389–3402. https://doi.org/10.1093/nar/25.17.3389
- Bairoch, A., Boeckmann, B., Ferro, S., & Gasteiger, E. (2004). Swiss-Prot: Juggling between evolution and stability. Briefings in Bioinformatics , 5, 39–55. https://doi.org/10.1093/bib/5.1.39
- Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., & Sayers, E. W. (2017). GenBank. Nucleic Acids Research , 45, D37–D42. https://doi.org/10.1093/nar/gkw1070
- Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., & Madden, T. L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics , 10, 421. https://doi.org/10.1186/1471-2105-10-421
- Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., Christie, K. R., Costanzo, M. C., Dwight, S. S., Engel, S. R., Fisk, D. G., Hirschman, J. E., Hitz, B. C., Karra, K., Krieger, C. J., Miyasato, S. R., Nash, R. S., Park, J., Skrzypek, M. S., … Wong, E. D. (2012). Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Research , 40, D&00–D705. https://doi.org/10.1093/nar/gkr1029
- Chojnacki, S., Cowley, A., Lee, J., Foix, A., & Lopez, R. (2017). Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Research , 45, W550–W553. https://doi.org/10.1093/nar/gkx273
- Davis, P., Zarowiecki, M., Arnaboldi, V., Becerra, A., Cain, S., Chan, J., Chen, W. J., Cho, J., da Veiga Beltrame, E., Diamantakis, S., Gao, S., Grigoriadis, D., Grove, C. A., Harris, T. W., Kishore, R., Le, T., Lee, R. Y. N., Luypaert, M., Müller, H. M., … Sternberg, P. W. (2022). WormBase in 2022-data, processes, and tools for analyzing Caenorhabditis elegans. Genetics , 220(4), iyac003. https://doi.org/10.1093/genetics/iyac003
- Eddy, S. R. (1998). Profile hidden Markov models. Bioinformatics , 14, 755–763. https://doi.org/10.1093/bioinformatics/14.9.755
- Edman, P., Högfeldt, E., Sillén, L. G., & Kinell, P.-O. (1950). Method for determination of the amino acid sequence in peptides. Acta Chemica Scandinavica , 4, 283–293. https://doi.org/10.3891/acta.chem.scand.04-0283
- Franklin, R. E. (1956). Structure of tobacco mosaic virus: Location of the ribonucleic acid in the tobacco mosaic virus particle. Nature , 177, 928–930. https://doi.org/10.1038/177928b0
- Gramates, L. S., Marygold, S. J., Santos, G. D., Urbano, J. M., Antonazzo, G., Matthews, B. B., Rey, A. J., Tabone, C. J., Crosby, M. A., Emmert, D. B., Falls, K., Goodman, J. L., Hu, Y., Ponting, L., Schroeder, A. J., Strelets, V. B., Thurmond, J., Zhou, P., & the FlyBase Consortium. (2017). FlyBase at 25: Looking to the future. Nucleic Acids Research , 45, D663–D671. https://doi.org/10.1093/nar/gkw1016
- Hernandez, P., Müller, M., & Appel, R. D. (2006). Automated protein identification by tandem mass spectrometry: Issues and strategies. Mass Spectrometry Reviews , 25, 235–254. https://doi.org/10.1002/mas.20068
- Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., McWilliam, H., Maslen, J., Mitchell, A., Nuka, G., Pesseat, S., Quinn, A. F., Sangrador-Vegas, A., Scheremetjew, M., Yong, S. Y., Lopez, R., & Hunter, S. (2014). InterProScan 5: Genome-scale protein function classification. Bioinformatics , 30, 1236–1240. https://doi.org/10.1093/bioinformatics/btu031
- Kersey, P. J., Allen, J. E., Allot, A., Barba, M., Boddu, S., Bolt, B. J., Carvalho-Silva, D., Christensen, M., Davis, P., Grabmueller, C., Kumar, N., Liu, Z., Maurel, T., Moore, B., McDowall, M. D., Maheswari, U., Naamati, G., Newman, V., Ong, C. K., … Yates, A. (2018). Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Research , 46, D802–D808. https://doi.org/10.1093/nar/gkx1011
- Kodama, Y., Mashima, J., Kosuge, T., Kaminuma, E., Ogasawara, O., Okubo, K., Nakamura, Y., & Takagi, T. (2018). DNA data bank of Japan: 30th anniversary. Nucleic Acids Research , 46, D30–D35. https://doi.org/10.1093/nar/gkx926
- Ladunga, I. (2002). Finding homologs to nucleotide sequences using network BLAST searches. Current Protocols in Bioinformatics , 00, 3.3.1–3.3.25. https://doi.org/10.1002/0471250953.bi0303s00
- Larkin, A., Marygold, S. J., Antonazzo, G., Attrill, H., Dos Santos, G., Garapati, P. V., Goodman, J. L., Gramates, L. S., Millburn, G., Strelets, V. B., Tabone, C. J., Thurmond, J., & FlyBase Consortium (2020). FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Research , 49, D899–D907. https://doi.org/10.1093/nar/gkaa1026
- Lee, R. Y. N., Howe, K. L., Harris, T. W., Arnaboldi, V., Cain, S., Chan, J., Chen, W. J., Davis, P., Gao, S., Grove, C., Kishore, R., Muller, H. M., Nakamura, C., Nuin, P., Paulini, M., Raciti, D., Rodgers, F., Russell, M., Schindelman, G., … Sternberg, P. W. (2018). WormBase 2017: Molting into a new stage. Nucleic Acids Research , 46, D869–D874. https://doi.org/10.1093/nar/gkx998
- Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., Park, Y. M., Buso, N., & Lopez, R. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Research , 43, W580–W584. https://doi.org/10.1093/nar/gkv279
- Lopez, R., Duggan, K., Harte, N., & Kibria, A. (2003). Public services from the European Bioinformatics Institute. Briefings in Bioinformatics , 4, 332–340. https://doi.org/10.1093/bib/4.4.332
- Madeira, F., Madhusoodanan, N., Lee, J., Eusebi, A., Niewielska, A., Tivey, A. R. N., Lopez, R., & Butcher, S. (2024). The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Research , gkae241. https://doi.org/10.1093/nar/gkac241
- Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., Basutkar, P., Tivey, A. R. N., Potter, S. C., Finn, R. D., & Lopez, R. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research , 47, W597–600. https://doi.org/10.1093/nar/gkz268
- Madeira, F., Pearce, M., Tivey, A. R. N., Basutkar, P., Lee, J., Edbali, O., Madhusoodanan, N., Kolesnikov, A., & Lopez, R. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Research , 50, W276–W279. https://doi.org/10.1093/nar/gkac240
- Martin, F. J., Amode, M. R., Aneja, A., Austine-Orimoloye, O., Azov, A. G., Barnes, I., Becker, A., Bennett, R., Berry, A., Bhai, J., Bhurji, S. K., Bignell, A., Boddu, S., Branco Lins, P. R., Brooks, L., Ramaraju, S. B., Charkhchi, M., Cockburn, A., Da Rin Fiorretto, L., … Flicek, P. (2023). Ensembl 2023. Nucleic Acids Research , 51, D933–D941. https://doi.org/10.1093/nar/gkac958
- McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., Cowley, A. P., & Lopez, R. (2013). Analysis tool web services from the EMBL-EBI. Nucleic Acids Research , 41, W597–600. https://doi.org/10.1093/nar/gkt376
- McWilliam, H., Valentin, F., Goujon, M., Li, W., Narayanasamy, M., Martin, J., Miyar, T., & Lopez, R. (2009). Web services at the European Bioinformatics Institute-2009. Nucleic Acids Research , 37, W6–W10. https://doi.org/10.1093/nar/gkp302
- Mulder, N. J., & Apweiler, R. (2003). The InterPro database and tools for protein domain analysis. Current Protocols in Bioinformatics , 2, 2.7.1–2.7.19. https://doi.org/10.1002/0471250953.bi0207s02
- Park, Y. M., Squizzato, S., Buso, N., Gur, T., & Lopez, R. (2017). The EBI search engine: EBI search as a service—Making biological data accessible for all. Nucleic Acids Research , 45, W545–W549. https://doi.org/10.1093/nar/gkx359
- Pearson, W. R. (2016). Finding protein and nucleotide similarities with FASTA. Current Protocols in Bioinformatics , 53, 3.9.1–3.9.25. https://doi.org/10.1002/0471250953.bi0309s53
- Pearson, W. R., & Lipman, D. J. (1988). Improved tools for biological sequence comparison. Proceedings of the National Academy of Sciences , 85, 2444–2448. https://doi.org/10.1073/pnas.85.8.2444
- Pettersson, E., Lundeberg, J., & Ahmadian, A. (2009). Generations of sequencing technologies. Genomics , 93, 105–111. https://doi.org/10.1016/j.ygeno.2008.10.003
- Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., & Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Research , 46, W200–W204. https://doi.org/10.1093/nar/gky448
- Roberts, R. J., & Murray, K. (1976). Restriction endonuclease. Critical Reviews in Biochemistry and Molecular Biology , 4, 123–164. https://doi.org/10.3109/10409237609105456
- Sanger, F., & Coulson, A. R. (1975). A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. Journal of Molecular Biology , 94, 441k–448. https://doi.org/10.1016/0022-2836(75)90213-2
- Sayers, E. W., Cavanaugh, M., Clark, K., Pruitt, K. D., Sherry, S. T., Yankie, L., & Karsch-Mizrachi, I. (2024). GenBank 2024 update. Nucleic Acids Research , 52(D1), D134–D137. https://doi.org/10.1093/nar/gkad903
- Schwartz, E. M., & Sternberg, P. W. (2004). Searching WormBase for information about Caenorhabditis elegans. Current Protocols in Bioinformatics , 6, 1.8.1–1.8.44. https://doi.org/10.1002/0471250953.bi0108s6
- Shank, S. D., Weaver, S., & Pond, S. L. K. (2018). phylotree.js—A JavaScript library for application development and interactive data visualization in phylogenetics. BMC Bioinformatics , 19, 276. https://doi.org/10.1186/s12859-018-2283-2
- Sievers, F., & Higgins, D. G. (2014). Clustal Omega. Current Protocols in Bioinformatics , 48, 3.13.1–3.13.16. https://doi.org/10.1002/0471250953.bi0313s48
- Sievers, F., & Higgins, D. G. (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Science , 27, 135–145. https://doi.org/10.1002/pro.3290
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D., & Higgins, D. G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology , 7, 539. https://doi.org/10.1038/msb.2011.75
- Silvester, N., Alako, B., Amid, C., Cerdeño-Tarrága, A., Clarke, L., Cleland, I., Harrison, P. W., Jayathilaka, S., Kay, S., Keane, T., Leinonen, R., Liu, X., Martínez-Villacorta, J., Menchi, M., Reddy, K., Pakseresht, N., Rajan, J., Rossello, M., Smirnov, D., … Cochrane, G. (2018). The European nucleotide archive in 2017. Nucleic Acids Research , 46, D36–D40. https://doi.org/10.1093/nar/gkx1125
- Skrzypek, M. S., & Hirschman, J. (2011). Using the Saccharomyces Genome Database (SGD) for analysis of genomic information. Current Protocols in Bioinformatics , 35, 1.20.1–1.20.23. https://doi.org/10.1002/0471250953.bi0120s35
- Squizzato, S., Park, Y. M., Buso, N., Gur, T., Cowley, A., Li, W., Uludag, M., Pundir, S., Cham, J. A., McWilliam, H., & Lopez, R. (2015). The EBI Search engine: Providing search and retrieval functionality for biological data from EMBL-EBI. Nucleic Acids Research , 43, W585–W588. https://doi.org/10.1093/nar/gkv316
- Tanizawa, Y., Fujisawa, T., Kodama, Y., Kosuge, T., Mashima, J., Tanjo, T., & Nakamura, Y. (2023). DNA Data Bank of Japan (DDBJ) update report 2022. Nucleic Acids Research , 51, D101–D105. https://doi.org/10.1093/nar/gkac1083
- UniProt Consortium. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research , 47(D1), D506–D515. https://doi.org/10.1093/nar/gky1049
- UniProt Consortium. (2023). UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Research , 51, D523–D531. https://doi.org/10.1093/nar/gkac1052
- Valentin, F., Squizzato, S., Goujon, M., McWilliam, H., Paern, J., & Lopez, R. (2010). Fast and efficient searching of biological data resources-using EB-eye. Briefings in Bioinformatics , 11, 375–384. https://doi.org/10.1093/bib/bbp065
- Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., & Barton, G. J. (2009). Jalview version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics , 25, 1189–1191. https://doi.org/10.1093/bioinformatics/btp033
- Wolfsberg, T. G. (2007). Using the NCBI map viewer to browse genomic sequence data. Current Protocols in Bioinformatics , 16, 1.5.1–1.5.22. https://doi.org/10.1002/0471250953.bi0105s16
- Wu, C., & Nebert, D. W. (2004). Update on genome completion and annotations: Protein information resource. Human Genomics , 1, 229–233. https://doi.org/10.1186/1479-7364-1-3-229
- Yuan, D., Ahamed, A., Burgin, J., Cummins, C., Devraj, R., Gueye, K., Gupta, D., Gupta, V., Haseeb, M., Ihsan, M., Ivanov, E., Jayathilaka, S., Kadhirvelu, V. B., Kumar, M., Lathi, A., Leinonen, R., McKinnon, J., Meszaros, L., O'Cathail, C., … Cochrane, G. (2024). The European nucleotide archive in 2023. Nucleic Acids Research , 52(D1), D92–D97. https://doi.org/10.1093/nar/gkad1067
- Zerbino, D. R., Achuthan, P., Akanni, W., Amode, M. R., Barrell, D., Bhai, J., Billis, K., Cummins, C., Gall, A., Girón, C. G., Gil, L., Gordon, L., Haggerty, L., Haskell, E., Hourlier, T., Izuogu, O. G., Janacek, S. H., Juettemann, T., To, J. K., … Flicek, P. (2018). Ensembl 2018. Nucleic Acids Research , 46, D754–D761. https://doi.org/10.1093/nar/gkx1098