Unveiling Prolonged COVID Variants: A Protocol for Clustering and Phylogenetic Analysis
Vidya Niranjan, Lavanya C, Spoorthi R Kulkarni, Baishali Garai
Abstract
Prolonged COVID-19 has emerged as a significant concern globally, with a subset of individuals experiencing persistent symptoms long after the acute phase of the infection. Understanding the genomic basis of prolonged COVID-19 can provide crucial insights into its pathophysiology and aid in the development of targeted therapeutic strategies. In this study, we retrieved genome sequences of COVID-19 from the National Center for Biotechnology Information (NCBI) database and employed a comprehensive pipeline to identify single nucleotide polymorphisms (SNPs). The primary objective was to investigate the clustering similarity of different variants of COVID-19 prevalent in the Indian population. By focusing solely on Indian population genome sequences, we aimed to capture the unique genetic landscape of COVID-19 variants circulating in this demographic. Our analysis revealed distinct SNP patterns across the Indian population, indicative of genetic diversity within the viral strains. Furthermore, we plan to map these identified SNPs to relevant pathways to elucidate their potential functional significance in the context of prolonged COVID-19.
Steps
SAMPLE COLLECTION AND REFERENCE GENOME
The genomic sequences utilized in this protocol were sourced from the NCBI Virus Database, which encompasses region-specific genome sequences. Focused on the Indian population, the protocol primarily analyzes sample sequences provided in FASTA format, alongside reference genomes obtained from the NCBI Virus Database.
REFERENCE GENOME INDEXING
Index the reference genome of COVID-19 by executing the 'bwa index' command followed by the name of the FASTA file containing the reference genome. This step facilitates efficient alignment of sequencing reads to the reference genome during subsequent analyses
#Indexing the reference genome
bwa index Reference_genome.fasta
ALIGNMENT AND MAPPING
The bwa command is used to align the sequencing reads to the indexed reference genome. This command efficiently aligns the reads, taking into account possible mismatches, insertions, deletions, and sequencing errors. SAMtools is a suite of programs for interacting with high-throughput sequencing data in SAM/BAM format.
#Aligning the reference genome with sample genome
bwa mem Reference_genome.fasta sample_ID.fasta > sample_ID.sam
#SAM to BAM conversion using SAM tools
samtools view -@ 4 -Sb -o sample_ID.bam sample_ID.sam
VARIANT CALLING AND SNP GENERATION USING SAMtools MPILEUP AND BCFTOOLS CALL
This protocol step involves variant calling and single nucleotide polymorphism (SNP) generation using SAMtools mpileup and BCFtools call. The SAMtools mpileup command is used to generate a pileup format file from multiple BAM files aligned to a reference genome. This file contains information about the alignment of sequencing reads to the reference genome at each genomic position. The BCFtools call command then analyzes the pileup data to identify variants, including SNPs, insertions, deletions, and complex variants. The output is generated in variant call format (VCF), providing detailed information about the detected variants, including their genomic coordinates, allele frequencies, and quality scores. This protocol step is crucial for identifying genetic variations and understanding the genomic landscape of the samples under investigation.
#VCF generation
samtools mpileup -uf Reference_genome.fasta sample_ID.bam | bcftools call -O v -mv -o 1_output.vcf
VARIANT GENERATION USING MULTIPLE SEQUENCE ALIGNMENT
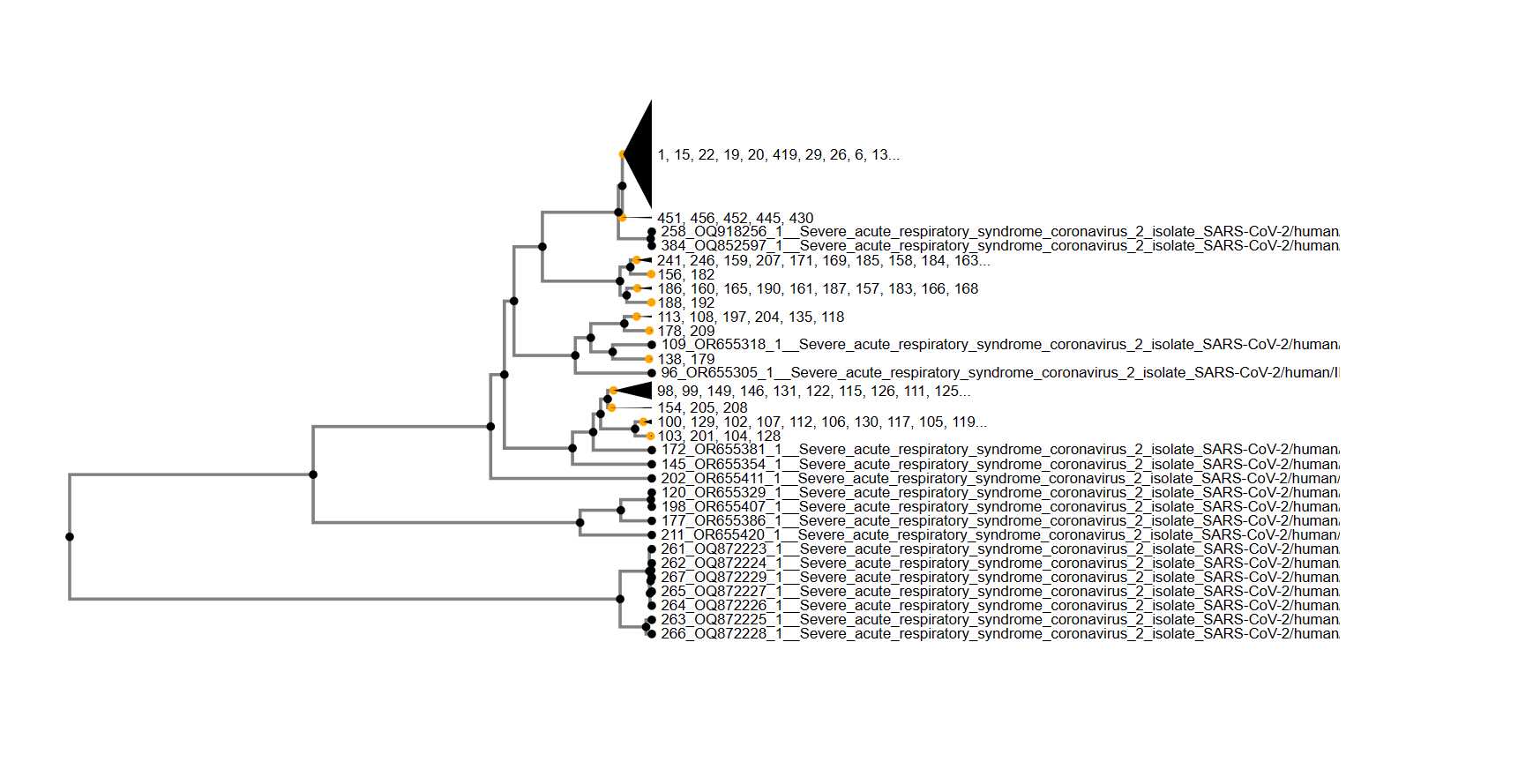
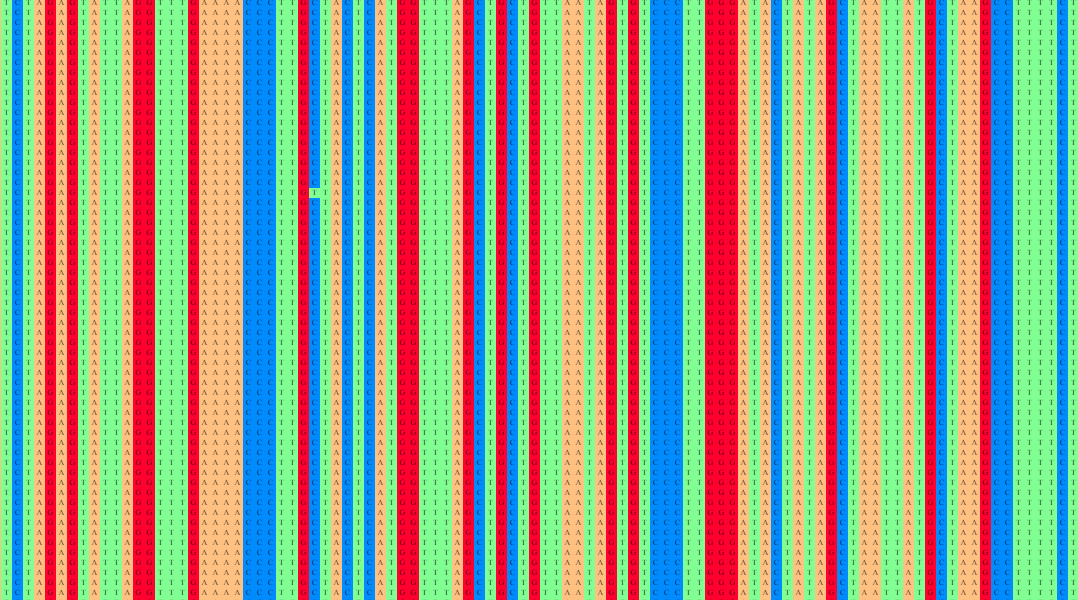
The Multiple Sequence Alignment (MSA) was conducted using the MAFFT version 7 tool, leveraging the alignment parameters specifically chosen for the analysis. The FASTA files, obtained from earlier downloads, served as input for the MSA procedure. The primary objective of this step was to align the genomic sequences to visualize shared single nucleotide polymorphisms (SNPs) and subsequently generate a phylogenetic tree and similarity score clusters.
#MAFFT version 7 web server
https://mafft.cbrc.jp/alignment/server/
Clustering and Phylogenetic Tree Analysis