Three-Dimensional Vessels-on-a-Chip Based on hiPSC-derived Vascular Endothelial and Smooth Muscle Cells
Valeria V. Orlova, Valeria V. Orlova, Christine L. Mummery, Christine L. Mummery, Merve Bulut, Merve Bulut, Marc Vila Cuenca, Marc Vila Cuenca, Mees de Graaf, Mees de Graaf, Francijna E. van den Hil, Francijna E. van den Hil
differentiation
endothelial cells
human induced pluripotent stem cells
neural crest
vascular smooth muscle cells
vessels-on-a-chip
Abstract
Blood vessels are composed of endothelial cells (ECs) that form the inner vessel wall and mural cells that cover the ECs to mediate their stabilization. Crosstalk between ECs and VSMCs while the ECs undergo microfluidic flow is vital for the function and integrity of blood vessels. Here, we describe a protocol to generate three-dimensional (3D) engineered vessels-on-chip (VoCs) composed of vascular cells derived from human induced pluripotent stem cells (hiPSCs). We first describe protocols for robust differentiation of vascular smooth muscle cells (hiPSC-VSMCs) from hiPSCs that are effective across multiple hiPSC lines. Second, we describe the fabrication of a simple microfluidic device consisting of a single collagen lumen that can act as a cell scaffold and support fluid flow using the viscous finger patterning (VFP) technique. After the channel is seeded sequentially with hiPSC-derived ECs (hiPSC-ECs) and hiPSC-VSMCs, a stable EC barrier covered by VSMCs lines the collagen lumen. We demonstrate that this 3D VoC model can recapitulate physiological cell-cell interaction and can be perfused under physiological shear stress using a microfluidic pump. The uniform geometry of the vessel lumens allows precise control of flow dynamics. We have thus developed a robust protocol to generate an entirely isogenic hiPSC-derived 3D VoC model, which could be valuable for studying vessel barrier function and physiology in healthy or disease states. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Differentiation of hiPSC-VSMCs
Support Protocol 1 : Characterization of hiPSC-NCCs and hiPSC-VSMCs
Support Protocol 2 : Preparation of cryopreserved hiPSC-VSMCs and hiPSC-ECs for VoC culture
Basic Protocol 2 : Generation of 3D VoC model composed of hiPSC-ECs and hiPSC-VSMCs
Support Protocol 3 : Structural characterization of 3D VoC model
INTRODUCTION
Blood vessels are lumenized tubes composed of endothelial cells (ECs), which form the inner vessel wall lining, and surrounding mural cells that cover the outer endothelium. Mural cells are subdivided into pericytes and vascular smooth muscle cells (VSMCs) depending on their location within the vascular bed in vivo. Crosstalk between ECs and mural cells is essential for proper function and integrity of blood vessels (Gaengel, Genové, Armulik, & Betsholtz, 2009) and ensures vessel stability. Aberrant interactions between ECs and mural cells have been implicated in many pathological states, including congenital vascular diseases, diabetic retinopathy, atherosclerosis, and coronary artery disease (Owens, Kumar, & Wamhoff, 2004; Sweeney & Foldes, 2018).
Microphysiological systems integrating human vascular cells, also referred as Vessel-on-Chip (VoC) devices, are considered good representations of human vasculature in vitro because microfluidic flow promotes the recapitulation of key physiological aspects of vascular biology. Although the majority of current VoC platforms incorporate primary human cells, because of the batch-to-batch and genetic variability of primary vascular cells, as well as their restricted availability, there is a growing interest in utilizing human induced pluripotent stem cell (hiPSC)-derived vascular cells in VoC platforms (Cochrane et al., 2019). With the emergence of reproducible methods for directed differentiation, hiPSCs generated from heathy individuals and patients with various conditions have already demonstrated value as a renewable source of vascular cells for disease modeling and drug discovery (Passier, Orlova, & Mummery, 2016; Samuel, Duda, Fukumura, & Jain, 2015; Shi, Inoue, Wu, & Yamanaka, 2016). In this regard, we and others have generated ECs from hiPSC lines and described robust protocols for the purification and cryopreservation of hiPSC-ECs, allowing the derivation and storage of large batches of cells from the same donor and with features similar to those of primary ECs (Orlova et al., 2014a, 2014b; Patsch et al., 2015; Rufaihah et al., 2013; Zhang et al., 2017). VSMCs are also phenotypically highly dependent on their cell or lineage of origin (Majesky, 2007). We and others have previously derived hiPSC-VSMCs from the neural crest (NC), the primary source of VSMCs in the cerebral vasculature (Cheung, Bernardo, Trotter, Pedersen, & Sinha, 2012; Dash et al., 2016; Granata et al., 2016; Halaidych, Cochrane, van den Hil, Mummery, & Orlova, 2019a; Wanjare, Kuo, & Gerecht, 2013, 2014).
Here, we describe two detailed protocols for (1) differentiating VSMCs from hiPSCs (Basic Protocol 1) and (2) incorporating hiPSC-derived ECs and VSMCs into 3D-engineered VoCs (Basic Protocol 2); the method described is a refinement of one we have used previously (de Graaf et al., 2019; Vila Cuenca et al., 2021). Our method is a robust and reproducible approach to generating 3D multi-cell-type VoC models entirely based on hiPSC that enable functional assays integrating flow dynamics for vascular disease modeling. Prior to incorporation in the 3D VoC model, hiPSC-derived neural crest cells (hiPSC-NCCs) and VSMCs are differentiated and cryopreserved in large batches following Basic Protocol 1. The differentiation efficiency of hiPSC-NCCs and VSMCs is assessed by following Support Protocol 1. hiPSC-ECs are derived and cryopreserved following a previously published protocol (Orlova et al., 2014b). Cryopreserved vials of hiPSC-ECs and VSMCs are thawed and cultured for 4 days (Support Protocol 2) before being integrated into the microfluidic devices. The devices are fabricated using the soft-lithography technique, and collagen type I lumenized scaffolds are created using an optimized viscous finger patterning (VFP) method for subsequent cell seeding (Basic Protocol 2). The structure of the 3D vessel barrier thus generated has a consistent geometry over its entire length, and its quality can be assessed following Support Protocol 3.
Basic Protocol 1: DIFFERENTIATION OF hiPSC-VSMCs
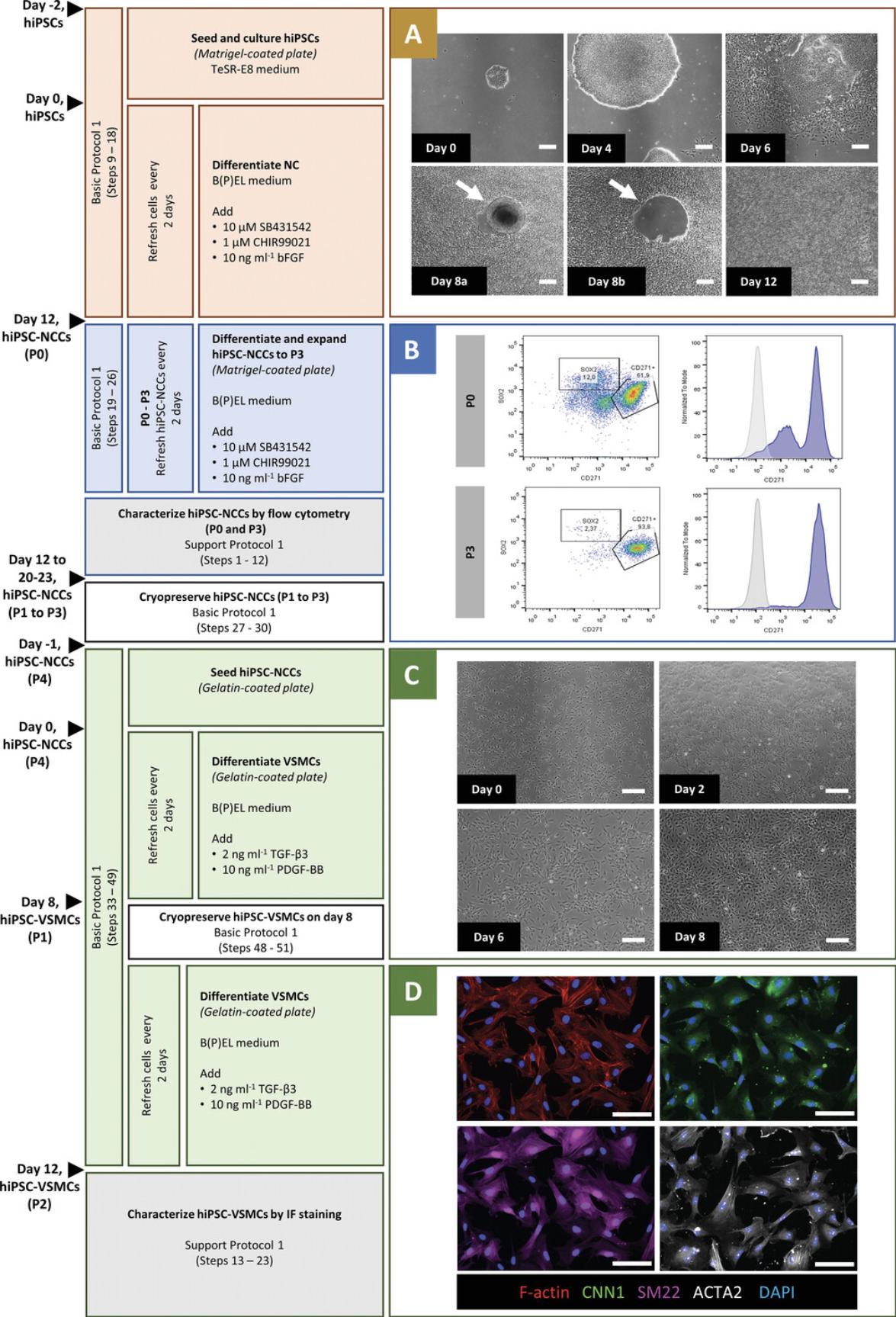
This protocol provides a method for efficient derivation of hiPSC-VSMCs through NC intermediates. It has previously proven robust and reproducible across multiple independent hiPSC lines (Halaidych et al., 2019a; Vila Cuenca et al., 2021). VSMC differentiation from hiPSCs is divided into three steps (Fig. 1). The first step is seeding and culture of hiPSCs followed by NC induction for 12 days (Fig. 1A). The initial seeding density of hiPSCs is critical for the differentiation efficiency. NC formation is induced by combining the inhibitor of glycogen synthase kinase 3, CHIR99021, to activate canonical WNT signaling with basic fibroblast growth factor and activin/nodal inhibitor SB431542.On day 12 (passage 0, P0), the differentiation efficiency can be assessed by flow cytometry analysis for reduced expression of the pluripotency marker SOX2 and increased expression of the NC marker CD271 (Support Protocol 1). The second step is the further differentiation and expansion of the hiPSC-NC cells (hiPSC-NCCs) from P0 to P3 for 8-11 days using the same (defined) medium used for differentiation. At P3, flow cytometry analysis should show a high total yield of CD271+ NC cells and high reproducibility across different lines and batches (Fig. 1B). At this stage, the cells can be cryopreserved and thawed for further differentiation to VSMCs. The third step is VSMC induction for 12 days (Fig. 1C). Both fresh and cryopreserved hiPSC-NCCs (P3) can be used to differentiate VSMCs. The initial seeding density of hiPSC-NCCs is important for a high differentiation yield. Differentiation to VSMCs is induced using PDGF-BB and TGF-β3 for 12 days. Ideally, hiPSC-VSMCs are cryopreserved on day 8 of VSMC induction to make a large batch of cells from the same differentiation batch that can be used over long periods in multiple experiments. Cryopreserved vials can later be thawed for further differentiation until day 12 (Support Protocol 2) for use in functional assays and characterization (Support Protocol 1).
Materials
-
Vitronectin-coated 6-well plate (see recipe)
-
TeSR™-E8™ Kit for hESC/hiPSC (Stemcell Technologies, cat. no. 05990)
-
Human induced pluripotent stem cells (hiPSCs):
- LUMC0054 (LU54, generated from kidney epithelial cells isolated from cells in urine, http://hpscreg.eu/cell-line/LUMCi001-A; Halaidych et al., 2018)
- Allen Cell Collection line AICS-0016 (ACTB-mEGFP, generated from skin fibroblasts, https://hpscreg.eu/cell-line/UCSFi001-A-3; Roberts et al., 2017)
-
Gentle Cell Dissociation Reagent (GCDR; Stemcell Technologies, cat. no. 07174)
-
Cell Adhere™ dilution buffer (Stemcell Technologies, cat. no. 07183)
-
Dulbecco's modified Eagle's medium/nutrient mixture F-12 (DMEM/F12) medium (Gibco™; Thermo Fisher Scientific, cat. no. 31331028)
-
Matrigel-coated 6-well plate (see recipe)
-
B(P)EL medium (see recipe)
-
NC differentiation medium (see recipe)
-
Dulbecco's phosphate-buffered saline (DPBS; Life Technologies, cat. no. 14190)
-
TrypLE™ Express Enzyme (1×), no phenol red; Gibco™, Thermo Fisher Scientific, cat. no. 12604021)
-
CryoStor® CS10 cryopreservation medium (Stemcell Technologies, cat. no. 07930)
-
Gelatin-coated 6-well plate (see recipe)
-
VSMC differentiation medium (see recipe)
-
6-well suspension plate (Greiner Bio-One, cat. no. 657185)
-
Tissue culture incubator, 37°C, 5% CO2
-
28-cm cell scraper, blue (Greiner Bio-One, cat. no. 541070)
-
15-ml tubes (Greiner Bio-One, cat. no. 188271)
-
Stereomicroscope (Leica MZ6 or equivalent)
-
6-well culture plate (Greiner Bio-One, cat. no. 657160)
-
Cryovials (Greiner Bio-One, cat. no. 123263)
-
Mr. Frosty™ Freezing Container
Passaging of hiPSCs for maintenance
Passage cells once a week when hiPSC colonies become relatively big and start to contact each other (∼90% confluency).
1.Prewarm a vitronectin-coated 6-well suspension plate, or freshly coat two wells of a 6-well suspension plate with vitronectin (see recipe). Prewarm TeSR-E8 to room temperature.
2.Before beginning cell dissociation, remove regions of spontaneous differentiation by scraping them off with a 200-μl pipet tip. Aspirate TeSR-E8 containing differentiated parts of the culture.
3.Add 1 ml GCDR to each well of a fresh 6-well plate and incubate 3-6 min at room temperature.
4.Aspirate the GCDR and add 1 ml TeSR-E8 to each plate well. Gently detach cell colonies by scraping them with a cell scraper.
5.Transfer cell aggregates into a 15-ml tube using a 1000-µl pipet tip. Pipet up and down gently once or twice with a 1000-µl pipet tip to break up large aggregates.
6.Wash the vitronectin-coated 6-well plate (from step 1) with 1 ml Cell Adhere dilution buffer per well and then add 2 ml TeSR-E8.
7.Add the small aggregates from step 5 to each well at a split ratio of 1:20-1:40.Move the plate in a cross-like pattern to distribute cells evenly, and place it the incubator at 37°C and 5% CO2.
8.Refresh cells with 2 ml TeSR-E8 once a day starting 24 hr after passaging.
Passaging of hiPSCs for NC differentiation (day –2)
Passage hiPSCs when they reach 90% confluency, as with the passaging for maintenance above.
9.Prewarm a Matrigel-coated 6-well plate, or freshly coat two wells of a 6-well plate (see recipe). Prewarm TeSR-E8 medium to room temperature.
10.Dissociate hiPSC colonies as described in steps 3-5.
11.Aspirate Matrigel and add 2 ml TeSR-E8 to each well of 6-well plate.
12.Add 20-25 μl of cell suspension (step 10) to each well at a split ratio of 1:30-1:40. Distribute cells evenly by moving the plate in a cross-like pattern, and place it in the incubator at 37°C and 5% CO2.
13.Refresh each well with 2 ml TeSR-E8 after 24 hr (day –1).
Differentiation of hiPSC-NCCs (from day 0 to 12)
IMPORTANT NOTE : Always prewarm medium to room temperature and add growth factors to the medium right before refreshing cells. Replace the cells in the 37°C, 5% CO2 incubator after refreshing.
14.NC induction: 48 hr after passaging (step 12), aspirate TeSR-E8 medium and add 2 ml NC differentiation medium in each well of the 6-well plate (day 0).
15.Refresh each well with 2 ml NC differentiation medium every 2 days until day 12.
16.Gently remove the cells in the centers of initial colonies by scraping them with a 200-μl pipet tip (day 8 or 9). This can be done under a stereomicroscope.
17.After removing the cells in the center of the colonies, refresh each well with 2 ml NC differentiation medium (day 8 or 9). Refresh each well again 2 days after mechanical elimination (day 10 or 11).
18.Passage the cells with a split ratio of 1:2 when they reach 80% confluency (day 12) for further differentiation and expansion following steps 19-25.
Differentiation and expansion of hiPSC-NCCs P0–P3 (from day 12 to day 20 or 23)
19.Prewarm a Matrigel-coated 6-well plate, or freshly coat four wells of a 6-well plate (see recipe). Prewarm B(P)EL medium to room temperature and freshly add the growth factors to prepare NC differentiation medium (see recipe).
20.Aspirate NC differentiation medium and add 2 ml DPBS to each well of a 6-well plate.
21.Aspirate DPBS and add 0.5 ml TrypLE™ to each well. Incubate for 3 min at room temperature.
22.Add 1.5 ml of B(P)EL to each well to dilute TrypLE. Transfer cell suspension to a 15-ml tube using a 1000-µl pipet tip. Pipet up and down gently once or twice with a 1000-µl pipet tip to break up the cell aggregates.
23.Centrifuge the tube 3 min at 300 × g , room temperature. Aspirate the supernatant.
24.Resuspend the cell pellet in freshly prepared 8 ml NC differentiation medium.
25.Aspirate Matrigel from four wells of 6-well plate and add 2 ml hiPSC-NCCs from step 24 to each well (for a split ratio of 1:2). Distribute the cells evenly by moving the plate in a cross-like pattern, and place it in the incubator at 37°C and 5% CO2.
26.Follow the same steps for passaging to P2 and to P3.
Cryopreservation of hiPSC-NCCs P1-P3
NOTE : It is best to cryopreserve hiPSC-NCCs at P1-P3 in order to create batches from the same differentiation.
27.Prechill a Mr. FrostyTM Freezing Container to 4°C. Label cryovials and place on ice.
28.Dissociate hiPSC-NCCs as described in steps 20-23.
29.Resuspend the cell pellet in cold CryoStor® CS10 cryopreservation medium (1 ml per 20 cm2 growth surface). Transfer 500 µl of cell suspension to each cryovial (10 cm2 per cryovial).
30.Place all cryovials into a Mr. Frosty™ Freezing Container and leave at −80°C for 24 hr. Transfer all cryovials into liquid nitrogen for prolonged storage.
Differentiation of VSMCs from hiPSC-NCCs in 12 days (days 0-12)
NOTE: VSMC differentiation steps can be followed by using either a cryopreserved vial or fresh hiPSC-NCCs at P3 from step 26.
31.On day –1, prewarm a 0.1% gelatin-coated 6-well plate, or freshly coat two wells of a 6-well plate. Prewarm B(P)EL medium to room temperature and add the growth factors to prepare NC or VSMC differentiation medium.
32.If using cryopreserved hiPSC-NCCs, remove cryovial (P3) from liquid nitrogen and thaw in a water bath at 37°C until only a small ice crystal remains. Transfer the cells into a 15-ml tube containing 5 ml B(P)EL. Wash cryovial once with 1 ml B(P)EL to collect remaining cells.
33.Gently homogenize the cell suspension with a 5-ml pipet and take a small aliquot to use in counting cells. Centrifuge the 15-ml tube 3 min at 300 × ɡ , room temperature. Aspirate supernatant.
34.Resuspend the cell pellet in NC differentiation medium to reach 1.25 × 105 cells/ml.
35.Aspirate gelatin from two wells of a 6-well plate and add 2 ml cell suspension to each well. Distribute the cells evenly by moving the plate in a cross-like pattern, and place it in the incubator at 37°C and 5% CO2 (day –1).
36.At 24 hr after seeding, start VSMC differentiation. Refresh each well with 2 ml VSMC differentiation medium (day 0).
37.On day 2, refresh each well with 2 ml VSMC differentiation medium.
38.Passage the cells at a split ratio of 1:4 (day 4). Prewarm 0.1% gelatin-coated 6-well suspension plates, or freshly coat eight wells.
39.Aspirate VSMC differentiation medium and add 2 ml DPBS to each well of a 6-well plate.
40.Aspirate DPBS and add 0.5 ml TrypLE™ to each well. Incubate 3 min at room temperature.
41.Add 1.5 ml B(P)EL to each well to dilute TrypLE™. Transfer cell suspension to a 15-ml tube using a 1000-µl pipet tip. Pipet up and down gently once or twice with a 1000-µl pipet tip to dissociate the large aggregates.
42.Centrifuge the tube 3 min at 300 × g , room temperature. Aspirate the supernatant.
43.Resuspend the cell pellet in 16 ml VSMC differentiation medium.
44.Aspirate gelatin from eight wells of 6-well suspension plates and add 2 ml cell suspension to each well (at a split ratio of 1:4). Distribute the cells evenly by moving the plates in a cross-like pattern, and place them in the incubator at 37°C and 5% CO2.
45.On day 6, refresh each well with 2 ml VSMC differentiation medium.
46.Alternatively, on day 8, passage VSMCs as described in steps 39-44.
47.On day 10, refresh each well with 2 ml VSMC differentiation medium.
Cryopreservation of VSMCs (day 8)
48.Prechill a Mr. FrostyTM Freezing Container to 4°C. Label cryovials from each differentiation and place on ice.
49.Dissociate hiPSC-VSMCs as described in steps 39-42.
50.Resuspend the cell pellet in cold CryoStor® CS10 cryopreservation medium to get a final concentration of 20 cm2/ml. Transfer 500 µl of cell suspension to each cryovial (10 cm2 per cryovial).
51.Place all cryovials in a Mr. Frosty™ Freezing Container and leave at −80°C for 24 hr. Then transfer all cryovials into liquid nitrogen for prolonged storage.
Support Protocol 1: CHARACTERIZATION OF hiPSC-NCCs and hiPSC-VSMCs
This protocol describes characterization of hiPSC-NCCs and hiPSC-VSMCs. On day 12 of NC induction (P0), the efficiency of differentiation can be determined through flow cytometry by assessing the expression of pluripotency markers (e.g., SOX2) and NC markers (e.g., CD271). More than 50% of the cells should be CD271+ NCCs. The analysis should be repeated at P3 (day 20 or 23), when the total CD271+ yield is expected to be higher (>90%) and the expression of SOX2 lower than at P0 (Fig. 1B). hiPSC-NCCs at P3 can be cryopreserved and thawed for VSMC induction. On day 12 of VSMC induction, hiPSC-VSMCs are fully differentiated and ready for functional characterization. At this stage, hiPSC-VSMCs exhibit increased expression of contractile proteins calponin 1 (CNN1), α-smooth muscle actin (αSMA; ACTA2), and SM22 (TAGLN), which can be assessed by immunofluorescence staining (Fig. 1D).
Materials
-
P3 hiPSC-NCCs (Basic Protocol 1, step 24)
-
Dulbecco's phosphate-buffered saline (DPBS; Life Technologies, cat. no. 14190)
-
TrypLE™ Express Enzyme (1×), no phenol red (Gibco™; Thermo Fisher Scientific, cat. no. 12604021)
-
Fluorescence-activated cell sorting buffer (FACSB; see recipe)
-
BD Cytofix/Cytoperm kit (BD Biosciences, cat. no. 554714)
-
Fluorescent-conjugated antibodies:
- Anti-human CD271, BV421 conjugated (used at 1:100 dilution; BD Biosciences, cat. no. 562562)
- Anti-human SOX2, A488 conjugated (used at 1:50 dilution; eBiosciences, cat. no. 53-9811-80)
-
Day 12 hiPSC-VSMCs (Basic Protocol 1, step 47, or Support Protocol 2, step 6)
-
B(P)EL (see recipe)
-
4% (w/v) PFA (see recipe)
-
0.1% (v/v) Triton X-100 (see recipe)
-
1% (w/v) BSA in DPBS (see recipe)
-
Primary antibodies:
- Anti-human Calponin, Mouse (used at 1:200 dilution; Sigma-Aldrich, cat. no. C2687)
- Anti-human Smooth Muscle Actin 1A4, Mouse (used at 1:200 dilution; Sigma-Aldrich, cat. no. A2547)
- Anti-human SM22 (TAGLIN/Transgelin), Rabbit (used at 1:200 dilution; Abcam, cat. no. AB14106)
-
100-μm sterile filters (CellTrics, cat. no. 04-004-2328)
-
5-ml round-bottom FACS tube (BD Biosciences, cat. no. 352058)
-
MACSQuant® VYB Flow Cytometer (Miltenyi Biotec, cat. no. 130-096-116)
-
Tissue culture incubator, 37°C, 5% CO2
-
96-well plate
-
Falcon® 96-well Black/Clear Imaging Microplate (Corning Life Sciences, cat. no. 353219)
-
Sealing film, 10 cm × 38 m (Parafilm M, cat. no. 291-1213)
-
EVOS M7000 imaging system (Thermo Fisher Scientific, cat. no. AMF7000)
Characterization of hiPSC-NCCs by flow cytometry (P0 and P3)
1.Aspirate NC differentiation medium from hiPSC-NCCs (P0 or P3) in 6-well plates and add 2 ml DPBS to each well.
2.Aspirate DPBS and add 0.5 ml TrypLE™ to each well. Incubate 3 min at room temperature.
3.Add 1 ml FACSB to each well to stop the dissociation, and wash the cells by pipetting up and down once or twice.
4.Place a 100-μm CellTrics sterile filter on top of a 5-ml FACS tube. Transfer an aliquot of hiPSC-NCC suspension into a 5-ml FACS tube for analysis. Centrifuge the tube 3 min at 300 × g , room temperature.
5.Wash the cell suspension with 1 ml FACSB and centrifuge the tube 3 min at 300 × g , room temperature. Aspirate the supernatant and leave around 50 µl FACSB with the cell pellet.
6.Add fluorescent-conjugated FACS antibodies to the cell suspension to the desired working concentration in FACSB. Resuspend cells by flicking the bottom of the tube. Incubate in the dark for 60 min at 4°C.
7.Turn off the light of the cell culture hood. Wash the cell suspension with 1 ml FACSB and centrifuge the tube 3 min at 300 × g , room temperature.
8.Aspirate the supernatant but leave ∼50 μl in the tube. Add 250 μl Fixation/Permeabilization solution (from BD Cytofix/Cytoperm kit). Resuspend the cells by flicking the bottom of the tube. Incubate in the dark for 20 min at 4°C.
9.Wash the cells once with 1 ml 1× BD Perm/Wash™ buffer (from kit) and centrifuge the tube 3 min at 300 × g , room temperature. Aspirate the supernatant but leave ∼50 μl in the tube.
10.Add fluorescent-conjugated FACS antibodies to the cell suspension to the desired working concentration in 1× BD Perm/Wash™ buffer. Resuspend the cells by flicking the bottom of the tube. Incubate in the dark for 20 min at 4°C.
11.Wash the cells once with 1 ml 1× BD Perm/Wash™ buffer and centrifuge the tube for 3 min at 300 × g , room temperature. Resuspend the cells in 1 ml 1× BD Perm/Wash™ buffer.
12.Analyze the samples by flow cytometry immediately or the next day.
Characterization of hiPSC-VSMCs using immunofluorescence staining
13.Freshly coat the required number of wells of a 96-well Black/Clear Imaging Microplate with fibronectin (see recipe). Prewarm B(P)EL medium to room temperature.
14.Dissociate hiPSC-VSMCs (day 12) as described in Basic Protocol 1, steps 39-42.Resuspend the cell pellet in B(P)EL to 7.5 × 104 cells per ml.
15.Aspirate fibronectin and add 200 µl cell suspension to each well. Distribute the cells evenly by moving the plate in a cross-like pattern and place in the incubator at 37°C and 5% CO2.
16.After 24 hr, aspirate B(P)EL and add 200 µl of 4% (w/v) PFA to each well to fix the cells. Incubate for 10 min at room temperature.
17.Wash the cells three times with DPBS, 5 min per wash, at room temperature.
18.Add 200 µl 0.1% (v/v) Triton-X 100 in DPBS to each well to permeabilize the cells. Incubate 5 min at room temperature.
19.Add 200 µl 1% (w/v) BSA in DPBS to each well to block nonspecific sites. Incubate 1 hr at room temperature.
20.Dilute the primary antibodies in 1% (w/v) BSA in DPBS and add it to the corresponding wells. Incubate overnight at 4°C.
21.Wash the cells three times with DPBS for 10 min per wash at room temperature.
22.Dilute the secondary antibodies in 1% (w/v) BSA in DPBS and add to the corresponding wells. Incubate 2 hr at room temperature.
23.Wash the cells three times with DPBS for 15 min per wash at room temperature. Seal the plates with Parafilm and store in the dark at 4°C until microscopic imaging.
Support Protocol 2: PREPARATION OF CRYOPRESERVED hiPSC-VSMCs AND hiPSC-ECs for VoC CULTURE
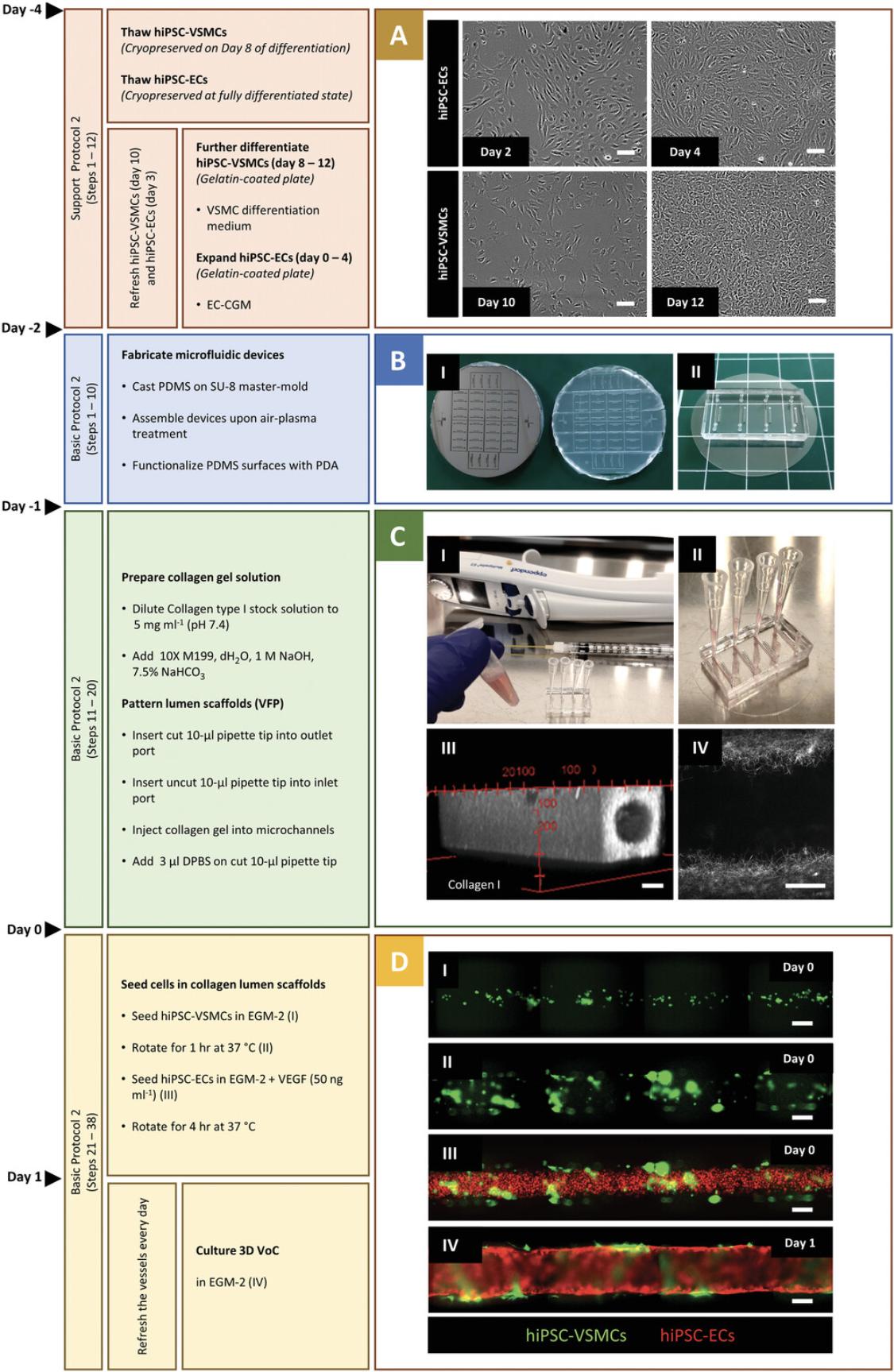
Vascular cells can be derived from hiPSCs using the protocols above and cryopreserved for further functional experiments. It is important to generate large batches of cells from the same differentiation for use in multiple VoC experiments. In this protocol, we describe how to prepare hiPSC vascular cells prior to VoC culture and functional assays. We previously described the cryopreservation of hiPSC-VSMCs on day 8 of differentiation. Cryopreserved hiPSC-VSMCs are thawed and cultured in VSMC differentiation medium for 4 days to reach the fully differentiated state (Fig. 2A). Additionally, hiPSC-ECs are produced efficiently and cryopreserved in their fully differentiated state in large batches using the robust protocol we reported previously (Orlova et al., 2014b). hiPSC-ECs are thawed and expanded in Endothelial Cell Complete Growth Medium (EC-CGM) for 4 days. Both cell types reach >90% confluency after 4 days and are then ready to be integrated in the collagen lumenized scaffolds created following Basic Protocol 2.
Materials
-
Gelatin-coated 6-well plate (see recipe)
-
Cryopreserved hiPSC-derived VSMCs on day 8 (Basic Protocol 1, step 51)
-
B(P)EL (see recipe)
-
VSMC differentiation medium (see recipe)
-
Cryopreserved hiPSC-derived ECs (Orlova et al., 2014b)
-
Endothelial Cell Complete Growth Medium (EC-CGM; see recipe)
-
6-well culture plate (Greiner Bio-One, cat. no. 657160)
-
15-ml tubes (Greiner Bio-One, cat. no. 188271)
-
Tissue culture incubator, 37°C, 5% CO2
Further differentiation of hiPSC-VSMCs in 4 days (from day 8 to 12)
1.Prewarm a 0.1% gelatin-coated 6-well plate, or freshly coat four wells of a 6-well plate. Prewarm B(P)EL to room temperature, and freshly add the growth factors to prepare VSMC differentiation medium.
2.Thaw the cryopreserved vial from day 8 of differentiation in a water bath at 37°C by swirling the cryovial until only a small ice crystal remains.
3.Transfer all cell suspension in the cryovial to a 15-ml tube containing 5 ml B(P)EL. Centrifuge 3 min at 300 × g , room temperature. Aspirate the supernatant.
4.Resuspend the cell pellet in 8 ml VSMC differentiation medium.
5.Aspirate 0.1% gelatin from four wells of the 6-well plate and add 2 ml cell suspension to each well (with a split ratio of 1:4). Distribute the cells evenly by moving the plate in a cross-like pattern and place the plates in the incubator at 37°C and 5% CO2 (day 8).
6.Refresh each well with 2 ml VSMC differentiation medium (day 10).
Expansion of hiPSC-ECs in 4 days (from day 0 to 4)
7.Prewarm a 0.1% gelatin-coated 6-well plate, or freshly coat six wells of a 6-well plate (see recipe). Prewarm EC-SFM and EC-CGM to room temperature.
8.Thaw the vial of cryopreserved cells in a water bath at 37°C by swirling the cryovial until only a small ice crystal remains.
9.Transfer all cell suspension in the cryovial into a 15-ml tube containing 8 ml EC-SFM. Centrifuge 3 min at 300 × g , room temperature. Aspirate supernatant.
10.Resuspend the cell pellet in 12 ml EC-CGM.
11.Aspirate 0.1% gelatin and add 2 ml cell suspension to each well (with a split ratio of 1:6). Distribute the cells evenly by moving the plate in a cross-like pattern and place the plates in the incubator at 37°C and 5% CO2 (day 0).
12.Refresh the cells with EC-CGM (day 3).
Basic Protocol 2: GENERATION OF 3D VoC MODEL COMPOSED OF hiPSC-ECs AND hiPSC-VSMCs
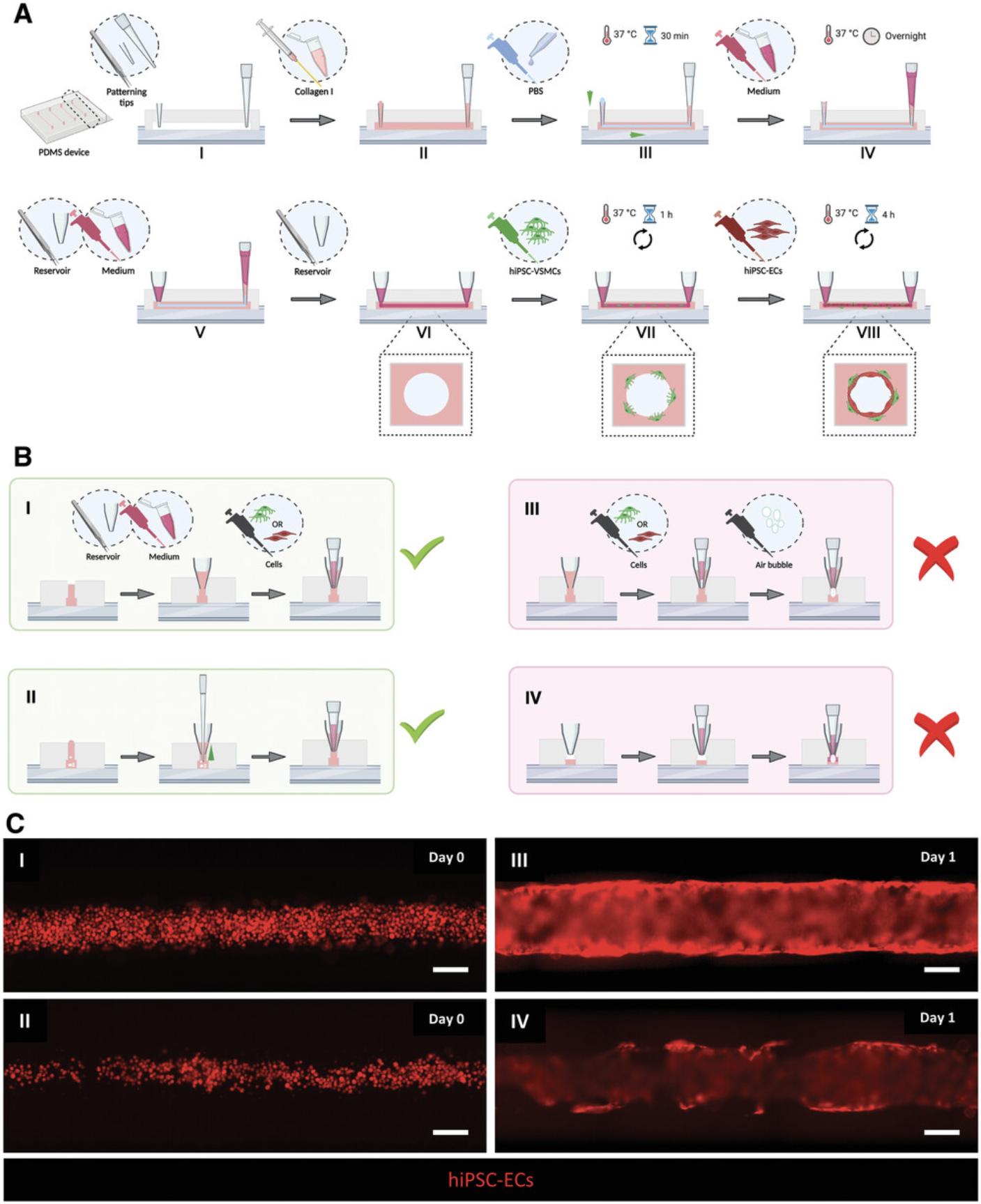
This protocol describes the generation of 3D-engineered vessels composed of hiPSC-ECs and hiPSC-VSMCs inside a PDMS microfluidic chip. The protocol is divided into three steps (Fig. 2). The first step is fabricating the devices by casting polydimethylsiloxane (PDMS) silicone elastomer on a customized SU-8 master mold using soft lithography. The master mold serves as a template to generate four straight microfluidic channels with fixed dimensions 0.5 mm (w) × 0.5 mm (h) × 10 mm (l) (Fig. 2B and 2I). After curing, PDMS is carefully peeled off, single devices are cut out, and the inlet/outlet ports of the microchannels are punched using biopsy puncher (1.2 mm) (Fig. 2B, II). To achieve watertight microfluidic channels, single devices are contact bonded with PDMS-coated coverslips and then treated with air plasma. Subsequently, the PDMS surface is functionalized by polydopamine (PDA) to promote the binding of collagen type I. The second step is patterning the lumens inside the collagen type I in the microfluidic channels using viscous finger patterning (VFP; de Graaf et al., 2019), which is illustrated in Figure 3A. The success rate of the second step is >95% if the details outlined in the Critical Parameters are applied exactly. If necessary, 3D collagen scaffolds can be imaged at this point and analyzed using two-photon second-harmonic generation (2P-SHG; Fig. 2C, III, and IV). The last step is the sequential seeding of hiPSC-derived vascular cells into the collagen I lumen scaffolds, illustrated in Figure 3A. hiPSC-VSMCs and hiPSC-ECs are prepared as described in Support Protocol 2 (Fig. 2A) and subsequently seeded by injecting the cells into the lumen via the inlet port (Fig. 2D, I-III). After each seeding step, the devices are slowly rotated to promote even coverage of the lumen scaffold. On day 1, a confluent hiPSC-EC monolayer is formed that is surrounded by abluminal hiPSC-VSMCs (Fig. 2D, IV). Functional assays can be performed on day 2 or 3 after cell seeding. Continuous and controlled perfusion can be done upon connection to the microfluidic pumps. Structural analysis in 3D is performed following Support Protocol 3, which reveals cell-cell interactions in 3D (Fig. 4). More details about expected results are described in the Understanding Results section.
Materials
-
Polydimethylsiloxane (PDMS) SYLGARD™ 184 Silicone Elastomer Kit (Dow Corning, cat. no. 101697)
-
2 mg/ml (w/v) poly(dopamine) (PDA; see recipe)
-
Distilled water (dH2O; Gibco™; Thermo Fisher Scientific, cat. no. 15230089)
-
5 mg/ml collagen type I, pH ∼7.4 (see recipe)
-
Dulbecco's phosphate-buffered saline (DPBS; Life Technologies, cat. no. 14190)
-
Endothelial Cell Growth Medium 2 Kit (EGM-2; Promocell, cat. no. C-22111; see recipe)
-
Day 12 hiPSC-VSMCs (Support Protocol 2, step 6), derived from Allen Cell Collection hiPSC line AICS-0016 (ACTB-mEGFP, generated from skin fibroblasts, https://hpscreg.eu/cell-line/UCSFi001-A-3)
-
Day 4 hiPSC-ECs (Support Protocol 2, step 12), derived from NCRM1 hiPSC line (NIH Center for Regenerative Medicine hiPSC line, https://hpscreg.eu/cell-line/CRMi003-A) engineered to express mCherry fluorescent protein under the control of the human cytomegalovirus (hCMV) early enhancer/chicken β-actin (CAG) promoter (de Graaf et al., 2019)
-
TrypLE Express Enzyme (1×), no phenol red (Gibco™; Thermo Fisher Scientific, cat. no. 12604021)
-
Human VEGF (165) IS, premium grade (Miltenyi Biotec, cat. no. 130-109-386)
-
Vacuum desiccator (Fisher Scientific, cat. no. 467-0090)
-
Custom-made SU-8 master-mold
-
Oven (80°C)
-
Round cover glass, ø 35 mm #1 (Thermo Fisher Scientific, 11887902)
-
Laurell WS-650HZ-23NPP/UD2 spin coater
-
Razor blades (GEM Scientific, cat. no. 233-1094)
-
Harris Uni-Core punch Ø: 1.2 mm plus cutting mat (Merck, cat. no. WHAWB100028)
-
3M Scotch Magic Tape 810 (Lyreco, 3339099)
-
CUTE Vacuum Plasma System 220-240V (Femto Science, serial no. FT20-045, cat. no. CUTE-1MPR)
-
Petri dish (100 mm; Greiner Bio-One, cat. no. 664160)
-
Pressured nitrogen-gas flow
-
CoolCLAVE Plus Ozone and UV Personal Sterilizer, 220V (Amsbio, E330220)
-
Laminar-flow hood
-
Tissue culture incubator, 37°C, 5% CO2
-
P10 pipet tips (Greiner Bio-One, cat. no. 741015)
-
Cut 10-μl pipet tips (7 mm; see recipe)
-
1.5-ml reaction tube (Greiner Bio-One, cat. no. 616201)
-
Luer-Lok™ 1-ml syringe (BD, cat. no. 309628), prechilled on ice
-
Plastic needle, 20 AWG × 1.5 in., yellow (OK International Ltd., cat. no. TS20P-1-1/2PK), prechilled on ice
-
Electronic multi-dispenser pipet (Multipette® E3; Eppendorf, cat. no. 4987000010)
-
Combitips® advanced, Biopur®, 0.2 ml (Eppendorf, cat. no. 0030089626)
-
Forceps, blunt tip
-
P1000 pipet tip (Greiner Bio-One, cat. no. 740295)
-
Medium reservoirs for microchannels (cut 1000-μl pipet tip; see recipe)
-
20-µl Gel-Load filter pipet tips (Greiner Bio-One, cat. no. 775288)
-
Grant Instruments™ PTR-35 rotator (Fisher Scientific, cat. no. 11481068)
Fabrication of PDMS devices (day –2 or earlier)
1.Mix PDMS elastomer base and curing agent thoroughly in a 10:1 ratio and degas in a vacuum desiccator at room temperature.
2.Pour uncured PDMS mixture onto the SU-8 master mold to a thickness of ∼5 mm.
3.Degas the PDMS until air bubbles are completely removed. Cure overnight in oven at 80°C.
4.Coat the cover glasses with a thin layer of PDMS using a spin-coater (5 s at 1000 rpm, 30 s at 4000 rpm). Cure for at least 3 hr at 80°C.
5.Cool the mold to room temperature. Remove PDMS carefully in a direction parallel to the microchannels.
6.Cut out the individual chips using razor blades. Punch inlet and outlet ports at the opposite ends of the microchannels using a 1.2-mm biopsy punch. Clean the PDMS surface with 3M Scotch® tape and store dust-free.
7.Place PDMS device (channel side up) and spin-coated cover glass (coated side up) in a vacuum plasma system. Treat the surfaces with air-plasma for 45 s at 50 W, 50 kHz.
8.Immediately bond the air-plasma-treated surfaces by pressing gently and evenly with fingers. Avoid breaking the cover glass.
Surface functionalization of the microfluidic channels to promote attachment of collagen I (day –2 or earlier)
9.Inject 10 µl PDA solution into the microchannels. Incubate 1 hr at room temperature. Wash the microchannels thoroughly with dH2O in a Petri dish. Dry completely using pressured nitrogen-gas flow.
10.Sterilize the devices in CoolClave Ozone+UV chamber using the “UV” setting (30 min). The devices can be stored dry and in the dark up to 1 week at room temperature.
Patterning of collagen I lumens using VFP technique (day –1)
NOTE : Follow aseptic techniques under sterile laminar hood. Use sterilized microfluidic devices and pipet tips.
11.Place 5 ml EGM-2 medium in a slightly opened 15-ml tube in the incubator at 37°C with 5% CO2 for at least 2 hr.
12.Insert a cut 10-μl pipet tip (7 mm) into the outlet port and an uncut 10-μl pipet tip into the inlet port of the microchannel. Press the tips lightly until they reach the coverslip.
13.Prepare 5 mg/ml collagen I solution, pH ∼7.4 (see recipe ).
14.Spin down the collagen solution for 10 s to remove air bubbles. Immediately place the solution back on ice. Take up the collagen solution in a prechilled 1-ml syringe with a plastic needle.
15.Immediately inject the collagen solution steadily into the microchannels via the uncut 10-μl pipet tip (inlet) until a meniscus forms on the cut 10-μl pipet tip (outlet). Add a 3-µl droplet of DPBS on top of the collagen I meniscus (outlet). Place the devices in the incubator at 37°C and 5% CO2 for 30 min to allow collagen polymerization.
16.Add 50 µl equilibrated EGM-2 on top of the polymerized collagen via the uncut 10-μl pipet tip (inlet). Place the devices in the incubator at 37°C with 5% CO2.
17.The next day, carefully remove the cut 10-μl pipet tip from the outlet port in a smooth twisting motion using blunt-tip forceps.
18.Insert a medium reservoir (cut 1000-μl pipet tip) into the outlet port, and add 20 µl of equilibrated EGM-2 to the medium reservoir using gel-load filter tips.
19.Remove the uncut 10-μl pipet tip from the inlet port in a smooth twisting motion using blunt-tip forceps.
20.Insert a medium reservoir into the inlet port and proceed to cell seeding.
Seeding cells in 3D lumen scaffolds (day 0)
In the following steps, the confluent monolayers of hiPSC-VSMCs (from Support Protocol 2; step 6) and hiPSC-ECs (from Support Protocol 2; step 12) are dissociated sequentially.
21.Prewarm B(P)EL and EC-SFM to room temperature. Place 5 ml EGM-2 in a slightly opened 15-ml tube in the incubator at 37°C with 5% CO2 and incubate at least 2 hr.
22.Dissociate the monolayer of hiPSC-VSMCs in 6-well plate as described in Basic Protocol 1, steps 39-42.
23.Resuspend the cell pellet in EGM-2 to obtain 2 × 106 cells/ml. Pipet the cell suspension up and down gently once or twice with a 200-µl pipet tip before injecting into each lumen.
24.Take up 10 µl of cell suspension in a 10-µl pipet tip. Insert the tip into the outlet medium reservoir with a light pressure.
25.Inject the cell suspension slowly and steadily until equal levels are reached across medium reservoirs and flow stops.
26.Place the devices on a cell culture rotator (channel longitudinal axis in line with rotating axis). Rotate the devices for 1 hr at 1 rpm in the incubator at 37°C with 5% CO2.
27.Add 25 µl EGM-2 to the outlet medium reservoir and 50 µl EGM-2 to the inlet medium reservoir.
28.Dissociate hiPSC-ECs in 6-well plates from Support Protocol 2, step 12.Aspirate EC-CGM and add 2 ml DPBS to each well.
29.Aspirate DPBS and add 0.5 ml TrypLE™ to each well. Incubate 3 min at room temperature.
30.Add 1.5 ml of EC-SFM to each well to dilute TrypLE™. Transfer cell suspension to a 15-ml tube using a 1000-µl pipet tip. Pipet up and down gently once or twice.
31.Count the cells and centrifuge 3 min at 1100 rpm (300 × g) at room temperature. Aspirate the supernatant.
32.Resuspend the cell pellet in EGM-2 supplemented with 50 ng/ml VEGF to reach 1 × 107 cells/ml. Pipet the cell suspension up and down gently once or twice with a 200-µl pipet tip before injecting into each lumen.
33.Remove any excess medium from medium reservoirs using a 10-µl pipet tip. Leave ∼10 µl medium in the reservoir to avoid injecting air bubbles.
34.Take up 5 µl of hiPSC-EC suspension in a 10-µl pipet tip. Insert the tip into the outlet medium reservoir with a light pressure.
35.Inject the cell suspension into microchannels as described in step 25.
36.Place the devices on a cell culture rotator (channel longitudinal axis in line with rotating axis). Rotate the devices for 4 hr at 1 rpm inside the 37°C, 5% CO2 incubator.
37.Add 25 µl EGM-2 supplemented with 50 ng/ml VEGF to the outlet medium reservoir and 50 µl to the inlet medium reservoir to create transient gravity-driven flow (day 0).
38.Remove 30 µl EGM-2 from each reservoir with 200-µl pipet tip. Add 25 µl EGM-2 to the outlet medium reservoir and 50 µl to the inlet medium reservoir to create transient gravity-driven flow (days 1-3).
Support Protocol 3: STRUCTURAL CHARACTERIZATION OF 3D VoC MODEL
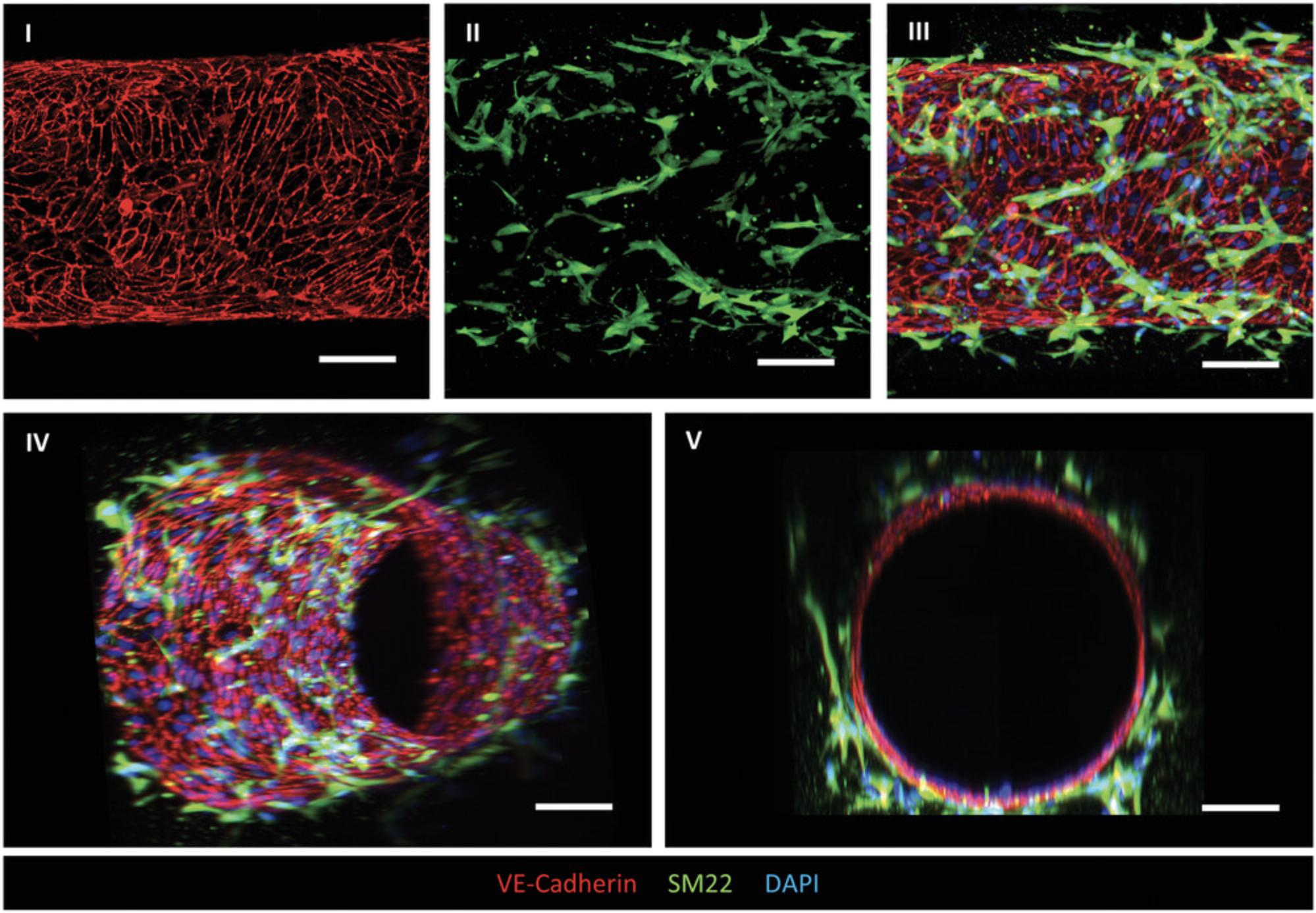
3D-engineered vessels generated following Basic Protocol 2 can be characterized on day 2 or 3.Here we describe the structural characterization of the 3D vessels using immunofluorescence staining. The fixed vessels are first permeabilized for intracellular staining followed by blocking for at least 1 hr to avoid nonspecific binding of the primary antibody. In this protocol, we use primary antibodies against VE-cadherin to visualize the EC cell junctions, and against contractile protein SM22 to visualize VSMC cell morphology. The secondary antibodies should be selected considering the host and target species of primary antibodies. The selection of specific fluorophores can vary between users and dilutions should be derived from manufacturer's recommendations. The stained 3D vessels are compatible with high-resolution imaging using advanced microscopy.
NOTE : Always follow the same sequence for adding solutions into the vessel lumens (Basic Protocol 2, step 38).
Materials
-
3D-engineered vessels (Basic Protocol 2)
-
4% (w/v) PFA (see recipe)
-
0.1% (v/v) Triton X-100 in DPBS (see recipe)
-
1% (w/v) BSA in DPBS (see recipe)
-
Dulbecco's phosphate-buffered saline (DPBS; Life Technologies, cat. no. 14190)
-
Primary antibodies:
- Anti-human VE-cadherin, Rabbit, 1:200 (Cell Signaling, cat. no. 2158S)
- Anti-human SM22 (TAGLIN/Transgelin), Rabbit, 1:200 (Abcam, cat. no. AB14106)
-
Parafilm sealing film, 10 cm × 38 m (Parafilm M, cat. no. 291-1213)
-
Leica SP8 microscope, Dragonfly® spinning disk (Andor)
-
Imaris 9.5 software (Bitplane, Oxford Instruments)
1.Aspirate EGM-2 from 3D vessels and add 4% (w/v) PFA to the medium reservoirs to fix the cells inside the lumens. Incubate 15 min at room temperature. Wash the lumens three times with DPBS, 10 min per wash, at room temperature.
2.Add 0.1% (v/v) Triton-X 100 in DPBS to each lumen to permeabilize the cells. Incubate 15 min at room temperature.
3.Add 1% (w/v) BSA in DPBS to each lumen to block nonspecific binding sites. Incubate 1 hr at room temperature.
4.Dilute the primary antibodies in 1% (w/v) BSA in DPBS and add it to the corresponding lumens. Incubate overnight at 4°C.
5.Wash the lumens three times with DPBS, 15 min per wash, at room temperature.
6.Dilute the secondary antibodies in 1% (w/v) BSA in DPBS and add to the corresponding lumens. Incubate 2 hr at room temperature.
7.Wash the lumens three times with DPBS, 15 min per wash, at room temperature. Store the devices at 4°C in a Parafilm-sealed Petri dish in the dark until microscopic imaging.
REAGENTS AND SOLUTIONS
L-Ascorbic acid 2-phosphate (AA2P), 5 mg ml–1
Add 250 mg AA2P (Sigma-Aldrich, cat. no. A8960) to 50 ml dH2O, nonsterile. Prepare aliquots and store up to 1 year at −20°C.
Basic fibroblast growth factor (FGF2) stock solution, 10 μg ml–1
Reconstitute at 100 μg ml–1 in distilled water (dH2O; Gibco™; Thermo Fisher Scientific, cat. no. 15230089). Further dilute to 10 μg ml–1 in 0.1% (w/v) BSA in DPBS (see recipe). Prepare aliquots and store up to 1 year at –80°C. Avoid repeated freeze-thaw cycles.
B(P)EL
Prepare 250 ml of B(P)EL using the volumes listed below. Sterilize medium using a Stericup filter and store up to 3 weeks at 4°C.
| Composition | Volume | Final concentration |
|---|---|---|
| IMDM | 107.63 ml | |
| F12 nut. mix | 113.88 ml | |
| PFHM-II | 12.5 ml | 5% |
| 10% (w/v) BSA/IMDM (see recipe) | 6.25 ml | 0.25% |
| Lipids (100×) | 2.5 ml | 1× |
| ITS-X (100×) | 250 μl | 0.1× |
| αMTG (1.3%; see recipe) | 750 μl | 450 μM |
| AA2P (5 mg/ml; see recipe) | 2.5 ml | 0.05 m/ml |
| GlutaMAX (200 mM) | 2.5 ml | 2 mM |
| Pen/Strep (5000 U/ml) | 1.25 ml | 0.5% |
IMDM, Iscove's modified Dulbecco's medium, no phenol red (Gibco™; Thermo Fisher Scientific, cat. no. 21056023); F12 nut. mix, Ham's F-12 nutrient mix, GlutaMAX supplement (Gibco™; Thermo Fisher Scientific, cat. no. 31765027); PFHM-II, Protein-Free Hybridoma Medium, 1× (Gibco™; Thermo Fisher Scientific, cat. no. 12040077); Lipids, chemically defined lipid concentrate (Gibco™; Thermo Fisher Scientific, cat. no. 11905031); ITS-X, insulin-transferrin-selenium-ethanolamine (100×; Gibco™, Thermo Fisher Scientific, cat. no. 51500056); GlutaMAX, GlutaMAX-1 supplement (Gibco™; Thermo Fisher Scientific, cat. no. 35050038); Pen/Strep, Penicillin-Streptomycin (5000 U/ml; Gibco™; Thermo Fisher Scientific, cat. no. 15070063).
BSA, 1% (w/v) in DPBS
Dissolve 0.5 g BSA (BovoStar BSA, Bovogen Biologicals, cat. no. BSAS 0.1) in 50 ml of DPBS on a bench shaker at cold room. Sterilize the solution by filtration using 0.22-µm-pore-size membrane filter and store up to 4 weeks at 4°C.
BSA, 10% (w/v) in IMDM
Add 5 g of BSA (BovoStar) to 40 ml of IMDM in a 50-ml tube. Leave the tube for a few hours on a roller bank at 4°C until the BSA is dissolved completely. Add 10 ml IMDM to reach 10% (w/v) BSA final. Sterilize the solution by filtration with a 0.22-μm-pore-size membrane filter and store for up to 4 weeks at 4°C.
CHIR99021 (CHIR), 4 mM
Reconstitute CHIR99021 (Tocris Bioscience, cat. no. 4423) at 4 mM in DMSO. Prepare aliquots and store up to 1 year at −20°C.
Citric acid, 5 mM (pH ∼3)
Add 960.6 mg citric acid (Sigma-Aldrich, cat. no. 27487) to 1 liter dH2O. Sterilize the solution by filtration with a 0.22-μm-pore-size membrane filter and store up to 3 months at 4°C.
Collagen I stock solution, 10 mg/ml
Prepare 200-μl aliquots of 10 mg/ml collagen I from rat tail collagen stock solution (Ibidi, cat. no. 50204). Store the aliquots according to the manufacturer's instructions.
Collagen I solution, 5 mg/ml (pH ∼7.4)
| Mixing order | Component | Volume (μl) |
|---|---|---|
| i | 10× M199 | 42.7 |
| ii | dH2O | 142.8 |
| iii | 7.5% (w/v) NaHCO3 | 12.5 |
| iv | 1 M NaOH | 2 |
| v | 10 mg/ml collagen I stock (see recipe) | Weight (mg) 200 |
10× M199 and 7.5% sodium bicarbonate (NaHCO3) solution are from Gibco™ (Thermo Fisher Scientific, cat. nos. 11825015 and 25080060) and 1 M sodium hydroxide (NaOH) from Merck (cat. no. 1064981000).
Cut 10-μl pipet tips, 7 mm (for VFP)
Cut off the tip of 10-μl pipet tips (Greiner Bio-One, cat. no. 741015) 7 mm from the tip using a razor blade. Autoclave the cut pipet tips at least 3 days before experiment.
Endothelial Cell Complete Growth Medium (EC-CGM)
Prepare EC-CGM using the volumes listed below. Sterilize the medium with a Stericup filter and store up to 2 weeks at 4°C.
| Composition | Volume | Final concentration |
|---|---|---|
| EC-SFM | 247.5 ml | |
| hPPS | 2.5 ml | 1% |
| VEGF, 50 μg/ml | 150 µl | 30 ng/ml |
| bFGF, 100 μg/ml | 50 µl | 20 ng/ml |
EC-SFM, Invitrogen, cat. no. 11111-044); hPPS, human serum from platelet-poor plasma (hPPS; Sigma-Aldrich, cat. no. P2918); VEGF, human VEGF (165) IS, premium grade (Miltenyi Biotec, cat. no. 130-109-386); bFGF, human bFGF, premium grade (Miltenyi Biotec, cat. no. 130-093-842).
EDTA, 0.5 M (pH 8.0)
Add 186.1 g EDTA disodium salt (Sigma-Aldrich, cat. no. E5134) to 800 ml dH2O. Adjust the pH to 8.0 with NaOH and add dH2O to a final volume of 1 liter. Filter the solution through a 0.22-μm-pore-size membrane filter and sterilize by autoclaving. Store up to 1 year at room temperature.
EGM-2
Prepare a 500-ml bottle of EGM-2 as described by the manufacturer, adding all supplements from the kit to the basal medium. Additionally, add 2.5 ml Penicillin-Streptomycin (5000 U/ml; Gibco™; Thermo Fisher Scientific, cat. no. 15070063). Store the medium up to 6 weeks at 4°C.
Fibronectin-coated 96-well plate
Dilute fibronectin (Sigma-Aldrich, cat. no. F1141) in DPBS to final concentration of 5 μg/ml. Add 100 μl to each well of 96-well plate. Incubate at room temperature for at least 1 hr before use. Stored up to 2 weeks at 4°C.
Fluorescence-activated cell sorting buffer (FACSB)
Dissolve 1.25 g BSA in 250 ml DPBS and add 1 ml of 0.5 M EDTA (pH 8.0; see recipe). Sterilize using a Stericup filter (0.22 μm pore size) and store up to 4 weeks at 4°C.
Gelatin-coated 6-well plate
Add 0.1% (w/v) gelatin solution (see recipe) to a culture plate or flask. Ensure that the gelatin covers the whole surface. Use 1 ml gelatin solution per well of a 6-well plate. Leave the plate with gelatin for 1 hr at 37°C before use. The coated plates can be stored up to 2 weeks at 4°C.
Gelatin solution, 0.1% (w/v)
Add 1 g gelatin (from porcine skin, type A; Sigma-Aldrich, cat. no. G1890) to 100 ml of dH2O to prepare a 1% (w/v) gelatin solution. Autoclave the solution and store 10-ml aliquots up to 1 year at −20°C.
For 0.1% (w/v) gelatin working solution, dilute stock solution 10× with Dulbecco's phosphate-buffered saline (DPBS; Life Technologies, cat. no. 14190). Sterilize the solution by filtration with a 0.22-μm-pore-size membrane filter and store up to 1 year at room temperature.
Matrigel-coated 6-well plate
Thaw 0.5 mg Matrigel (Growth Factor Reduced; Corning, cat. no. 354230) on ice for each 6-well culture plate. Aliquot 6 ml cold (4°C) Dulbecco's modified Eagle's medium/nutrient mixture F-12 (DMEM/F12) medium into a 50-ml tube. Cool a pipet tip by pipetting up and down the cold DMEM/F12 medium several times and use it to transfer thawed Matrigel into the medium. Mix with a cold (4°C) 5-ml pipet and add 1 ml to each well. Leave the plate at room temperature for 1 hr. Use the plate right away or seal with Parafilm and store up to 2 weeks at 4°C.
Medium reservoirs for microchannels (cut 1000-μl pipet tips)
Cut 1000-μl pipet tips (Greiner Bio-One, cat. no. 740295) ∼20 mm from the tip using razor blade. Autoclave the cut pipet tips at least 3 days before experiment.
α-Monothioglycerol (αMTG) solution, 1.3% (v/v)
Add 130 μl mono-thio glycerol (α-MTG; Sigma-Aldrich, cat. no. M6145-25 ml) to 9.87 ml Iscove's modified Dulbecco's medium (IMDM) and store up to 1 year at 4°C protected from light.
Neural crest (NC) differentiation medium
Prepare neural crest differentiation medium using the volumes listed below. Always prepare fresh.
| Composition | Volume | Final concentration |
|---|---|---|
| B(P)EL (see recipe) | 4 ml | |
| SB431542 (20 nM; see recipe) | 2 μl | 10 µM |
| CHIR99021 (4 mM; see recipe) | 1 μl | 1 µM |
| FGF2 (10 μg/ml; see recipe) | 4 μl | 10 ng/ml |
PDGF-BB stock solution, 20 μg/ml
Reconstitute PDGF-BB (PeproTech, cat no. 100-14B) at 20 μg/ml in 0.1% (w/v) BSA in DPBS. Prepare aliquots and store at –20°C or below for up to 1 year. Avoid repeated freeze-thaw cycles.
PFA, 4% (w/v)
Mix 1 vol of 8% (w/v) PFA (see recipe) with 1 vol of 0.2 M phosphate buffer (pH 7.4; see recipe). Keep for up to 2 weeks stored at 4°C.
PFA, 8% (w/v)
Add 40 g paraformaldehyde (PFA; Merck, cat. no. 1.04005.1000) to 400 ml Milli-Q water. Heat to 60°C and stir it at medium speed. After a few minutes, add ∼10 drops of 1 N NaOH to dissolve the PFA granules. Eventually, the solution will become translucent. Let the solution cool down and add Milli-Q water to a total volume of 500 ml. Store the solution up to 2 months at 4°C.
Phosphate buffer, 0.2 M (pH 7.4)
To prepare solution 1, dissolve 8.28 g sodium dihydrogen phosphate dihydrate (NaH2PO4·2H2O; J.T. Baker, cat. no. 0326) in 300 ml Milli-Q ultrapurified water (e.g., from a Millipore system). To prepare solution 2, dissolve 10.78 g disodium hydrogen phosphate monohydrate (NaH2PO4·1H2O; Merck, cat. no. 1.06346.1000) in 300 ml Milli-Q water. Add solution 1 to solution 2 until a pH of 7.4 is obtained, to make 0.2 M phosphate buffer (pH 7.4). This buffer can be stored at room temperature indefinitely (no expiration date).
Polydopamine (PDA), 2 mg/ml (w/v)
Dissolve dopamine hydrochloride (Merck, cat. no. H8502-5G) in 10 mM Tris·Cl buffer (pH 8.5; see recipe) to obtain 2 mg/ml (w/v) polydopamine (PDA) solution. Filter the buffer through a 0.22-μm-pore-size membrane filter. Always prepare fresh.
SB43152 solution, 20 mM
Reconstitute SB43152 (Tocris Bioscience, cat. no. 1614) at 20 mM in DMSO. Prepare aliquots and store up to 1 year at −20°C.
Transforming growth factor beta-3 (TGF-β3) stock solution, 5 μg/ml
Reconstitute TGF-β3 (PeproTech, cat. no. 100-36E) at 100 μg/ml in 5 mM citric acid (see recipe). Further dilute to 5 μg/ml in 0.1% (w/v) BSA in DPBS (see recipe). Prepare aliquots and store up to 1 year at −20°C.
Tris·Cl buffer (pH 8.5), 10 mM
Dissolve 1.2 g/liter Tris (Trizma®) base (Tris(hydroxymethyl)aminomethane; Sigma-Aldrich, cat. no. T6066-1KG) and 1.0 g/liter sodium azide (Sigma-Aldrich, cat. no. S8032-25G) in ∼90% final volume with cold dH2O. Add cold dH2O to obtain final volume. Adjust to pH 8.5 with 5 M HCl or 5 M NaOH as needed. Filter the buffer through a 0.22-μm-pore-size membrane filter. Store up to 4 years at room temperature.
Triton X-100, 0.1% (v/v) in DPBS
Add 250 µl Triton X-100 (Sigma-Aldrich, cat. no. T8787) to 50 ml DPBS. Sterilize the solution by filtration using a 0.22-µm-pore-size membrane filter. Store up to 1 year at room temperature.
Vascular endothelial growth factor (VEGF) stock solution, 50 μg/ml
Reconstitute human VEGF (165) IS, premium grade (Miltenyi Biotec, cat. no. 130-109-386) at 50 μg ml–1 in 0.1% (w/v) BSA in DPBS. Prepare aliquots and store up to 1 year at −80°C.
Vitronectin-coated 6-well plates
Prewarm Cell Adhere dilution buffer (Stemcell Technologies, cat. no. 07183) to room temperature. For each well of a 6-well plate, thaw 40 μl VitronectinXF (Stemcell Technologies, cat. no. 07180) at room temperature and add to 1.21 ml dilution buffer. Mix well and add 1.25 ml per well to the wells of a 6-well cell suspension plate. Distribute vitronectin to cover the whole well and incubate at room temperature for 1 hr. Use the plate right away or seal with Parafilm and store up to 2 weeks at 4°C.
Vascular smooth muscle cell (VSMC) differentiation medium
Prepare VSMC differentiation medium using the volumes listed below. Always prepare fresh.
| Composition | Volume | Final concentration |
|---|---|---|
| B(P)EL (see recipe) | 25 ml | |
| TGFβ3 (5 µg/ml; see recipe) | 10 μl | 2 ng/ml |
| PDGFBB (20 µg/ml; see recipe) | 12.5 μl | 10 ng/ml |
COMMENTARY
Background Information
Since the initial discovery of hiPSCs, directed differentiation protocols to generate vascular cell types under defined conditions have significantly improved. The potential applications of hiPSC-derived vascular cells are broad and include engineering of human blood vessels and the study of vascular-disease-related abnormalities. Developmentally, VSMCs originate from multiple lineages including mesoderm and NC, the main source of VSMCs in the cerebral vasculature (Majesky, 2007). Several protocols have reported methods to induce NC formation in hiPSC using different chemical cocktails based on the factors that target bone morphogenic protein, WNT, and fibroblast growth factor signaling. Our protocol described here induces NC formation by combining an inhibitor of glycogen synthase kinase 3, CHIR99021, to activate canonical WNT signaling together with basic fibroblast growth factor and the activin/nodal inhibitor SB431542. After 12 days of culture, more than half of the cells become CD271+ and can then be cryopreserved and thawed for further differentiation and expansion. At P3, hiPSC-NCCs derived from this protocol show a high total CD271+ NCC yield and high reproducibility across different hiPSC lines and batches. Importantly, these hiPSC-NCCs preserve their phenotype up to P7, which provides a distinct advantage for the generation of large cell batches for disease modelling in vitro. To induce VSMC differentiation from hiPSC-NCCs, we used PDGF-BB and TGF-β3 for another 12 days, based on previously described protocols (Cheung et al., 2012; Dash et al., 2016; Granata et al., 2016; Halaidych et al., 2019a; Wanjare et al., 2013, 2014). hiPSC-VSMCs can be cryopreserved on day 8 and later thawed for further differentiation until day 12. At this stage, hiPSC-VSMCs exhibit increased expression of the contractile proteins calponin 1, α-smooth muscle actin (αSMA), and SM22, as previously described (Halaidych et al., 2019a). This protocol has the particular advantage that it successfully induces hiPSC-VSMC differentiation and upregulation of contractile proteins and is based on serum-free (defined) medium. To assess hiPSC-VSMC functionality, we developed an automated pipeline that includes the determination of intracellular Ca2+ release and contraction in response to vasoconstrictors. Specifically, this defines a robust approach for quantification and specification of VSMC function by providing multiple parameters to accurately assess phenotype in the overall cell population across hiPSC-VSMCs differentiated from different hiPSC lines (Halaidych et al., 2019b).
Additionally, we previously described a protocol to efficiently induce EC differentiation from hiPSCs through mesoderm specification under defined culture conditions (Orlova et al., 2014b). We carried out extensive functional characterization of these hiPSC-ECs using multiple assays, including a comparative assessment of barrier function and inflammatory responses upon treatment with various proinflammatory agents. We further demonstrated the capacity of stimulated hiPSC-EC monolayers to recruit cells of the human monocytic cell line THP1 (Halaidych et al., 2018), blood monocytes, and hiPSC-derived monocytes (Cao et al., 2019, 2020) by using 2D microfluidic flow assays. Overall, we can robustly produce large quantity of hiPSC-derived VSMCs and ECs that are fully functional after cryopreservation. Recently, we demonstrated the potential utility of these hiPSC-derived ECs and VSMCs by developing a robust multicellular 3D VoC model that supports the formation of a functional self-assembled microvasculature network within fibrin hydrogel inside a microfluidic chip (Vila Cuenca et al., 2021).
Current VoC models based on the self-organization of vascular cells into microvasculature networks are of great value for studying physiological aspects of vascular development in vitro. However, the complex and intrinsically variable architecture makes it challenging to precisely control fluid flow and the wall shear stress exposed to the vascular barrier during dynamic flow assays. To address this issue, alternative VoC platforms are being developed to fabricate controlled and defined vascular architectures. One widely used approach is the insertion of removable templates (e.g., a small-diameter needle or silicon rod) inside an unpolymerized hydrogel to create 3D lumen scaffolds that are subsequently used for cell culture (Hasan, Paul, Memic, & Khademhosseini, 2015; Jiménez-Torres et al., 2015). Although it is straightforward, this method carries certain limitations: it is laborious, time consuming, and challenging to scale up. In 2012, Bischel et al. reported a practical approach for generating lumen scaffolds inside microfluidic channels by pipetting only fluids. The method exploits the viscosity difference between two fluids: less viscous fluid (cell culture medium) flows through and displaces more viscous fluid (unpolymerized hydrogel), leaving a hollow structure upon polymerization (Bischel, Lee, & Beebe, 2012). The method was applied to pattern a variety of controlled geometries, including multiple parallel lumens, to study vascular barrier function and angiogenesis (Bischel, Young, Mader, & Beebe, 2013). The lumen structures supported the multicellular culture of primary ECs, pericytes, and astrocytes with the aim of modeling blood-brain barrier (BBB) in vitro (Herland et al., 2016).
In our recent work, we implemented the VFP technique and optimized it further to improve the reproducibility of patterning uniform lumen scaffolds in collagen gel. We extended the patterning pathway using a cut 10-μl pipet tip on the outlet port to create a constant driving force for patterning. An extension length of 7 mm was found to be optimal to achieve a uniform intra- and interlumen diameter profile, giving reproducible results in the context of dynamic functional studies (de Graaf et al., 2019). Before collagen patterning, surface functionalization of PDMS is required to promote the strength of adhesion to collagen I hydrogel. In this protocol, we use a single-step functionalization technique based on polydopamine (PDA; Park, Georgescu, Oh, Kwon, & Huh, 2019). Alternative methods for surface functionalization have been reported previously that can be implemented prior to collagen patterning using the VFP protocol (de Graaf et al., 2019; Polacheck, Kutys, Tefft, & Chen, 2019; Yadav, Sriram, Carter, & Miller, 2014). Our VFP protocol has particular advantages compared to other approaches, including its robustness, high reproducibility between experiments, low cost, and potential for scale-up without the need for specialized equipment, as dozens of lumen scaffolds can be efficiently patterned in a few minutes.
Here, we also describe the successful integration of hiPSC-VSMCs and hiPSC-ECs into patterned collagen lumen scaffolds in a sequential manner. 3D vessel structures are efficiently formed in a couple of days that reveal close interactions of hiPSC-ECs and hiPSC-VSMCs, which is crucial for recapitulating physiological EC-mural cell crosstalk found in vivo (Gaengel et al., 2009). In our experience, 3D vessels can be stable for up to 7 days; however, long-term culture is challenging and is impractical for dynamic studies. The cells remodel collagen gel and over time the lumen scaffold becomes deformed over, mostly because of the high proliferation of hiPSC-VSMCs. Another limitation is that the model is not optimal for high-throughput studies such as drug screening, as the time required for manual fabrication of PDMS devices limits truly mass production. Overall, 3D multi-cell-type VoC model generated using this protocol can potentially be used in many applications integrating hemodynamic and mechanical parameters for physiological vessel function. Using hiPSCs derived from patients with specific disease genotypes presents opportunities for applications that include, but are not limited to, cardiovascular and brain vascular disease modeling.
Critical Parameters
hiPSCs used for differentiation in this protocol are expanded on vitronectin-coated plates in defined TeSR-E8 medium. It is important to limit the spontaneous differentiation of hiPSC colonies in culture for a robust differentiation. To successfully maintain the pluripotent state, the colonies should be passaged regularly every week. Before seeding, large colonies should be fragmented into small aggregates (100-200 µm) by pipetting to increase their adherence and survival. Compact colonies with >90% confluency should be achieved after 1 week of passaging. Initial seeding density of hiPSCs is a crucial step for efficient NC differentiation. The recommended split ratio is 1:30 or 1:40 from a well that is >90% confluent; this will result in 20-30 hiPSC colonies attached the next day. However, the split ratio may need further optimization, depending on the line, to achieve similar results. On day 0, before NC induction, the colonies should be 200-400 µm in size. As differentiated hiPSC-NCCs start migrating from the colony edges on day 5-7, mechanical elimination of the cells in the colony centers becomes essential for efficient differentiation (Fig. 1A; day 8a and day 8b). hiPSC-NCCs should be further differentiated and expanded to P3 before VSMC induction. The initial density of hiPSC-NCCs is crucial for efficient VSMC differentiation (day 0, Fig. 1C). It is best to cryopreserve hiPSC-VSMCs on day 8 and to prepare a batch of cells from the same differentiation that is sufficient for multiple experiments.
The generation of 3D-engineered vessels consists of three consecutive parts: fabricating and assembling the PDMS devices, patterning the collagen I lumen scaffold, and integrating the hiPSC-VSMCs and hiPSC-ECs in a sequential manner. Microfluidic devices are fabricated by casting PDMS on SU-8 master mold. The inlet and outlet ports should be opened by using a biopsy puncher with a diameter of 1.2 mm, which is an optimal size for inserting pipet tips, reservoirs, and tubing of external microfluidic pumps. PDMS devices can be stored in a dry, dust-free place and should be cleaned with Scotch tape before assembly. Air-plasma treatment provides a transient change in the property of the surface by rendering it hydrophilic for a short time, and therefore treated surfaces should be contact bound immediately. Pressing gently after bonding ensures that there is no air trapped in between surfaces. In this protocol, we use PDA for surface functionalization, which does not require freshly plasma-treated surfaces. One possible drawback is the slight darkening of PDMS if high concentrations of PDA are used or in the case of a prolonged incubation time. The final concentration of 2 mg/ml PDA with a 1-hr incubation hr gives efficient results and no interference with the optical transparency of PDMS. Before patterning, the channels should be dried completely for efficient collagen attachment.
The collagen gel preparation contains critical steps concerning the final concentration, pH, and temperature, which particularly affect the success rate and the lumen diameter. In this method, we recommend using highly concentrated collagen I derived from rat tail. The specifications of the stock collagen I solution can vary between different batches and manufacturers. The calculations should be empirically determined following the manufacturer's instructions for each different batch. To improve the reproducibility in this step, we verify the exact quantity of each collagen I aliquot by weight. The volumes of each reagent should be adjusted according to the stock concentration and the final weight of the collagen aliquot. The mechanical properties of collagen fibers re greatly influenced by the final pH and polymerization temperature. The pH of the collagen solution should be adjusted to ∼7.4; acidic or alkaline collagen solutions should not be used for patterning as that will dramatically affect the success rate and lumen diameter. All reagents should be always kept ice cold to delay the polymerization of collagen fibers. Patterning fails dramatically when partially polymerized collagen gel is used. Therefore, we recommend limiting the procedure time to 5 min per batch of mixed collagen. Consistency is the key to generating reproducible lumen scaffolds that create constant physiological conditions for culturing vascular cells. A uniform lumen diameter profile between experiments is achieved by creating a constant driving force for patterning. For this, we insert the 10-μl pipet tips of consistent 7-mm length into the outlet ports. After patterning, the pipet tips need to be removed by twisting to allow access to the lumen scaffolds for cell seeding.
Air bubbles are the most common reason for lumen deformation during the cell seeding step. However, this can be avoided if the cell seeding step is carried out correctly (Fig. 3B and 3I). We illustrate the potential ways in which air bubbles may be injected in Figure 3B, III and IV; we recommend checking each port for air bubbles and removing it if needed (Fig. 3B, II) before proceeding with cell seeding. Equilibrating cell culture medium helps to reduce the emergence of air bubbles over time due to the temperature differences. We found that injection of small air bubbles during cell seeding can enlarge lumen scaffolds. To eliminate variations in diameter, cells should be injected slowly and steadily. The initial cell seeding density is absolutely crucial for the formation of a confluent 3D vessel barrier (Fig. 2D). Low seeding densities of hiPSC-ECs fail to form a confluent endothelial barrier (Fig. 3C, II and IV). High seeding densities of hiPSC-VSMCs can induce deformation of collagen scaffold in time due to the cells’ high proliferation. Inconsistency in seeding densities between channels can be avoided by keeping the cell suspension homogeneous. After cell seeding, the liquid levels in opposite reservoirs should be equilibrated to stop the flow of cells. Rotating the devices promotes even distribution of the cells along the collagen lumen surface. Compared to the protocol published previously (de Graaf et al., 2019), we reduced the time for which the devices need to be rotated after hiPSC-EC seeding to 4 hr, which is optimal for hiPSC-EC attachment to lumen scaffolds. The rotation time might need to be optimized for ECs and mural cells from other sources. The 3D vessels should be refreshed every day due to the small volume of cell culture medium, a total of 75 µl per channel. Severe evaporation of the medium can be prevented by placing water in a dish next to the devices. Care should be taken to avoid injecting air bubbles when refreshing the vessels as discussed before for cell seeding. A transient flow is induced during daily refreshments by creating a hydrostatic pressure difference across medium reservoirs. This should be consistent for each refreshment or washing step.
Troubleshooting
Detailed troubleshooting guidelines can be found in Table 1.
| Step(s) | Problem(s) | Possible cause(s) | Solution(s) |
|---|---|---|---|
| Basic Protocol 1, step 12 or 16 |
Low efficiency of NC differentiation; low % CD271+SOX2– and high % CD271–SOX2+ |
Starting seeding density of hiPSCs was not optimal | Ensure that 20-30 hiPSC colonies 200-400 µm in size are present on day 0, before start of differentiation. |
| Colony centers were not eliminated sufficiently | Mechanically eliminate all colony centers on day 8 or 9 (Fig. 1A; days 8a and 8b). | ||
| Basic Protocol 1, step 34 | Low efficiency of VSMC differentiation | Initial seeding density of hiPSC-NCCs was not optimal | Seed ∼2.5 × 105 hiPSC-NCCs in each well of a 6-well plate. |
| Basic Protocol 2, steps 9-10 | Dark PDMS channels after surface functionalization | Final concentration of PDA solution was too high, or incubation duration was too long. | Reduce the final concentration of PDA solution or the duration of incubation. |
| Washing after PDA incubation was insufficient | Wash the microchannels thoroughly with dH2O. | ||
| Basic Protocol 2, step 13 | Inconsistent pH values between batches for the final collagen solution | Calculations for the preparation of the final collagen solution were wrong | Weigh the stock collagen I aliquots to verify the quantity. Update the volume of each reagent according to the quantity and concentration of stock aliquots. |
| Basic Protocol 2, steps 13-15 | Inconsistent lumen diameter and uneven geometry; partially patterned lumens | Driving force created for patterning was inconsistent | Ensure that all cut 10-μl pipet tips are 7 mm long. Use a cutting guide to improve consistency. Introduce adequate volume of driving fluid (3 µl of DPBS) on top of the cut pipet tip to create stable driving force. |
| Preparation of collagen solution was inconsistent | Check the calculations for collagen final solution. Weigh the collagen to verify quantity. Ensure the final pH is ∼7.4 and the solution is pink in color. | ||
| Partially polymerized collagen solution was used for patterning | Keep all reagents on ice during collagen preparation. Use the neutralized collagen solution within 5 min. Immediately place the patterned channels in the incubator at 37°C. | ||
| Collagen stock is old | Store high-concentration collagen I stock solution as recommended and ≤4 months. | ||
| Basic Protocol 2, step 9 | Delamination of collagen lumen scaffold during cell seeding or culture | Surface functionalization was inefficient | Optimize concentration and incubation time for PDA treatment. Thoroughly the PDMS channels and dry completely with pressured air. |
| Basic Protocol 2, step 25 | Air bubbles are injected into the lumens | Equilibrate the cell culture medium; inspect the channel ports before the injection step and remove any air bubbles (Fig. 3B, II). | |
| Basic Protocol 2, steps 23-25 and 32 | Inconsistent cell seeding density | Cell suspension was not mixed well before seeding | Pipet the cell suspension up and down before injecting into each lumen. |
| Seeded cells in the lumens are flushed out due to gravity-driven flow | Equilibrate the medium levels across reservoirs to stop the flow of cells in the lumens. |
Understanding Results
Initial seeding density of hiPSCs is a crucial step for efficient NC differentiation. In our experience, low-density cultures yield higher differentiation efficiency. A yield of 20-30 attached hiPSC colonies is expected the next day. These colonies expand continuously from day –2 to day 4, reaching 200-400 µm in size on day 0 of NC induction. Differentiated hiPSC-NCCs should start migrating outside the colonies from around day 5-7, and their number should grow continuously until day 12. On day 12 (P0), >50% of the cells are expected to be CD271+ hiPSC-NCCs. A low percentage of CD271+ cells indicates inefficient differentiation or mechanical elimination of colony centers (Fig. 1A; day 8a and day 8b). An enriched cell population with >90% CD271+ cells should be obtained for hiPSC-NCCs at P3 (Fig. 1B). In our experience, the phenotype is maintained up to P7. Both fresh or cryopreserved hiPSC-NCCs (P3) can be used to differentiate VSMCs. To induce VSMC differentiation, ∼2.5 × 105 hiPSC-NCCs should be seeded to each well of the 6-well plate (day –1). The next day, hiPSC-NCCs should have a density as shown in Figure 1C before the start of VSMC differentiation (day 0). Until day 8, cells will expand and acquire a bigger and spindle-like morphology (Fig. 1C). A confluent monolayer should be observed before passaging (day 4) and cryopreservation (day 8). Thawed hiPSC-VSMCs should continue further differentiation and reach >90% confluency 4 days after thawing (day 12, Fig. 2A). hiPSC-VSMCs should express the contractile markers SM22, calponin 1 (CNN1), and αSMA (ACTA2) on day 12 (Fig. 1D). Optionally, functional assays can be performed to further characterize the hiPSC-VSMCs (Halaidych et al., 2019a, 2019b).
Microfluidic devices are fabricated by casting PDMS and assembled with PDMS-coated cover glasses creating watertight microchannels. The efficiency of the contact bonding can be checked by slightly lifting the device or injecting a solution into the microchannels. Surface functionalization with PDA can cause slight darkening of PDMS, but this is negligible under the conditions we use in this protocol. Inefficient functionalization can cause the detachment of collagen I scaffold from the PDMS surfaces. Sterilization of devices with UV and ozone is effective against contamination and does not interfere with PDA treatment. Before patterning, a stock collagen I aliquot (10 mg/ml) with a defined weight is diluted to 5 mg/ml final following the recipe in Reagents and Solutions. The pH is estimated by the color of the phenol red from the 10× M199 medium, which is expected to be pink, indicating a pH of ∼7.4 (Fig. 2C and 2I). Acidic gel solutions (yellow) can be adjusted by adding NaOH dropwise until the color changes to pink. Alkaline solutions (purple) should be discarded. The patterning is initiated by introducing a droplet of DPBS (3 µl) into the cut 10-μl pipet tip (outlet), which immediately flows through the collagen gel and disappears. Any DPBS remaining on the cut 10-μl pipet tip is an indication of failed patterning. The VFP protocol can be further scaled up using a syringe to inject collagen I and a multi-dispenser pipet to add DPBS droplet. In this way, 24 channels can be easily patterned within 5 min. After removal of the pipet tips next day, the patterned lumens are expected to be perfusable and accessible via the ports. The structure of 3D lumens and collagen fibers can be visualized using the two-photon second-harmonic generation (2P-SHG) technique without the need to stain the collagen fibers (Fig. 2C, III and IV). Additionally, perfusability of the lumens can be quickly assessed by flowing fluorescent beads.
Thawed hiPSC-VSMCs and hiPSC-ECs are prepared for VoC culture in 4 days (Fig. 2A). Representative images of sequential cell-seeding steps with optimal cell densities, using genetically targeted hiPSC-VSMCs (ACTB; green) and hiPSC-ECs (mCherry; red), are shown in Figure 2D. First, hiPSC-VSMC suspension is injected in the lumens via the inlet medium reservoir (Fig. 2D and 2I). The collagen lumen scaffold is evenly covered with attached hiPSC-VSMC and the devices are rotated on a cell culture rotator for 1 hr (Fig. 2D, II). Next, hiPSC-EC suspension is injected into the lumens (Fig. 2D, III) and the devices are rotated for 4 hr. The next day (day 1), a confluent hiPSC-EC layer is formed, with surrounding hiPSC-VSMCs spreading and covering the hiPSC-EC layer in the following days. The hiPSC-EC layer remains stable, while hiPSC-VSMCs continue proliferating and maintain cell-cell interaction with hiPSC-ECs. Success rate of vessel formation in lumen scaffolds is >90%, and the resulting diameter is consistent between experiments, averaging 298 ± 17.82 μm (n = 12; N = 4) on day 3. In our experience, wider lumen diameters are expected after cell seeding as the lumen scaffolds can easily be deformed by high pressures or air bubbles during cell seeding (de Graaf et al., 2019). 3D-engineered vessels can be used for functional and dynamic assays on day 2 or 3 under physiologically relevant wall shear stress. Using genetically targeted hiPSC lines, the cell-cell interactions can be tracked in real time. The structure of 3D vessels can be visualized by immunofluorescence analysis (Support Protocol 3) and cell-cell interaction can be assessed using high-resolution fluorescence microscopy. 3D reconstruction reveals a continues hiPSC-EC barrier with integral VE-Cadherin junctions (red) and closely interacting SM22 (green) positive hiPSC-VSMCs (Fig. 4).
Time Considerations
Basic Protocol 1
Differentiation of NCCs from hiPSCs takes 12 days in total:
Steps 1-8: ∼20 min to passage hiPSCs for maintenance;
Steps 9-13: ∼20 min to passage hiPSCs to a Matrigel-coated plate for NC differentiation;
Step 15: ∼15 min every 2 days to refresh the cells;
Steps 16-17: ∼15 min to detach the centers of colonies and refresh each well.
Differentiation and expansion of hiPSC-NCCs to P3 take 8-11 days in total:
Steps 19-25: ∼20 min to passage hiPSC-NCCs for expansion;
Steps 27-30: ∼30 min to cryopreserve hiPSC-NCCs.
Differentiation of VSMCs from hiPSC-NCCs takes 12 days:
Steps 31-34: ∼20 min to thaw cryopreserved hiPSC-NCCs;
Steps 36-37: ∼15 min every 2 days to change the medium;
Steps 38-44: ∼20 min to passage cells;
Steps 45 and 47: ∼15 min to refresh the cells;
Steps 48-51: ∼30 min to cryopreserve hiPSC-VSMCs.
Support Protocol 1
Analysis of hiPSC-NCCs by flow cytometry takes 1 day:
Steps 1-11:∼2.5 hr to perform surface staining followed by intracellular staining of hiPSC-NCCs;
Step 12: ∼30 min are needed to run the samples by flow cytometry.
Immunofluorescence staining of hiPSC-VSMCs takes 2 days:
Steps 13-15: ∼20 min to dissociate and seed hiPSC-VSMCs followed by 24 hr culture;
Steps 16-19: ∼1.5 hr to fix, wash, permeabilize, and block the cells;
Step 20: ∼ 15 min to dilute primary antibodies and add to them the wells;
Steps 21-22:∼45 min to wash the cells and add diluted secondary antibodies;
Step 23: ∼45 min to wash the cells.
Support Protocol 2
Further differentiation of hiPSC-VSMCs takes 4 days:
Steps 1-5: ∼20 min to thaw cryopreserved cells and seed into the wells;
Step 6: ∼15 min to refresh on day 10.
Expansion of hiPSC-ECs takes 4 days:
Steps 7-11: ∼20 min to thaw cryopreserved cells and seed into the wells;
Step 12: ∼15 min to change the medium on day 3.
Basic Protocol 2
The fabrication of PDMS devices takes 2 days:
Steps 1-3: ∼20 min to prepare PDMS followed by overnight curing;
Step 4: ∼20 min to coat the cover glasses with PDMS followed by overnight curing;
Steps 5-8: ∼30 min to cut and assemble PDMS devices;
Step 9: ∼1.5 hr min to functionalize PDMS surface, including 1 hr PDA incubation;
Step 10: ∼1 hr to run UV + ozone sterilization.
Patterning of collagen I lumen scaffolds takes 2 days:
Step 12: ∼5 min to insert pipet tips into the inlet and outlet ports;
Steps 13-14: ∼5 min to prepare the collagen gel;
Step 15: ∼5 min to pattern the lumens followed by 30 min for collagen polymerization;
Step 16: ∼5 min to add medium on polymerized collagen gel followed by overnight incubation.
Cell seeding takes 1 day, with 2 hr total active work and 5 hr for rotation of the devices:
Steps 17-20: ∼10 min to replace pipet tips with medium reservoirs;
Steps 21-26: ∼30 min to dissociate hiPSC-VSMCs, count and seed into the lumens followed by 1 hr rotation;
Step 27: ∼5 min to add medium into the lumens;
Steps 28-36: ∼30 min to dissociate hiPSC-Ecs and count and seed them into the lumens followed by 5 hr rotation;
Steps 37-38: ∼10 min to refresh the lumens every day.
Support Protocol 3
Immunofluorescence staining of vessels takes 2 days:
Steps 1-3: ∼1.5 hr min to fix, permeabilize, and block the cells;
Step 4: ∼15 min to dilute primary antibodies and add to the lumens;
Steps 5-6: ∼45 min to wash the lumens and add diluted secondary antibodies followed by 2 hr incubation;
Step 7: ∼45 min to wash the lumens.
Acknowledgments
The work is supported by the European Union's Horizon 2020 research and innovation program under grant agreement No. 812954 and the Netherlands Organ-on-Chip Initiative, which is an NWO Gravitation project (024.003.001) funded by the Ministry of Education, Culture and Science of the government of the Netherlands. The Allen Cell Collection, available from Coriell Institute for Medical Research, provided materials. Images were generated using Biorender.com.
Author Contributions
Merve Bulut : Writing─original draft; Marc Vila Cuenca : Writing─original draft, writing─review & editing; Mees de Graaf : Methodology; Francijna E. van den Hil : Writing─original draft, writing─review & editing; Christine L. Mummery : Funding acquisition, writing─review & editing; Valeria V. Orlova : Conceptualization, funding acquisition, supervision, writing─review & editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data, tools, and materials (or their sources) that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Bischel, L. L., Lee, S. H., & Beebe, D. J. (2012). A practical method for patterning lumens through ECM hydrogels via viscous finger patterning. Journal of Laboratory Automation , 17(2), 96–103. doi: 10.1177/2211068211426694
- Bischel, L. L., Young, E. W. K., Mader, B. R., & Beebe, D. J. (2013). Tubeless microfluidic angiogenesis assay with three-dimensional endothelial-lined microvessels. Biomaterials , 34(5), 1471. doi: 10.1016/J.BIOMATERIALS.2012.11.005
- Cao, X., van den Hil, F. E., Mummery, C. L., & Orlova, V. v. (2020). Generation and functional characterization of monocytes and macrophages derived from human induced pluripotent stem cells. Current Protocols in Stem Cell Biology , 52(1), e108. doi: 10.1002/cpsc.108
- Cao, X., Yakala, G. K., van den Hil, F. E., Cochrane, A., Mummery, C. L., & Orlova, V. v. (2019). Differentiation and functional comparison of monocytes and macrophages from hiPSCs with peripheral blood derivatives. Stem Cell Reports , 12(6), 1282–1297. doi: 10.1016/j.stemcr.2019.05.003
- Cheung, C., Bernardo, A. S., Trotter, M. W. B., Pedersen, R. A., & Sinha, S. (2012). Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nature Biotechnology , 30(2), 165. doi: 10.1038/NBT.2107
- Cochrane, A., Albers, H. J., Passier, R., Mummery, C. L., van den Berg, A., Orlova, V. v., & van der Meer, A. D. (2019). Advanced in vitro models of vascular biology: Human induced pluripotent stem cells and organ-on-chip technology. Advanced Drug Delivery Reviews , 140, 68–77. doi: 10.1016/J.ADDR.2018.06.007
- Dash, B. C., Levi, K., Schwan, J., Luo, J., Bartulos, O., Wu, H., … Qyang, Y. (2016). Tissue-engineered vascular rings from human iPSC-derived smooth muscle cells. Stem Cell Reports , 7(1), 19. doi: 10.1016/J.STEMCR.2016.05.004
- de Graaf, M. N. S., Cochrane, A., van den Hil, F. E., Buijsman, W., van der Meer, A. D., van den Berg, A., … Orlova, V. v. (2019). Scalable microphysiological system to model three-dimensional blood vessels. APL Bioengineering , 3(2), 026105. doi: 10.1063/1.5090986
- Gaengel, K., Genové, G., Armulik, A., & Betsholtz, C. (2009). Endothelial-mural cell signaling in vascular development and angiogenesis. In Arteriosclerosis, thrombosis, and vascular biology (Vol. 29, Issue 5, pp. 630–638). Lippincott Williams & Wilkins. doi: 10.1161/ATVBAHA.107.161521
- Granata, A., Serrano, F., Bernard, W. G., McNamara, M., Low, L., Sastry, P., & Sinha, S. (2016). An iPSC-derived vascular model of Marfan syndrome identifies key mediators of smooth muscle cell death. Nature Genetics 2017 , 49(1), 97–109. doi: 10.1038/ng.3723
- Halaidych, O. V., Freund, C., van den Hil, F., DCF, S., M, R., CL, M., & VV, O. (2018). Inflammatory responses and barrier function of endothelial cells derived from human induced pluripotent stem cells. Stem Cell Reports , 10(5), 1642–1656. doi: 10.1016/J.STEMCR.2018.03.012
- Halaidych, O. V., Cochrane, A., van den Hil, F. E., Mummery, C. L., & Orlova, V. v. (2019a). Quantitative analysis of intracellular Ca2+ release and contraction in hiPSC-derived vascular smooth muscle cells. Stem Cell Reports , 12(4), 647–656. doi: 10.1016/J.STEMCR.2019.02.003
- Halaidych, O. v., Mummery, C. L., & Orlova, V. v. (2019b). Quantifying Ca2+ signaling and contraction in vascular pericytes and smooth muscle cells. Biochemical and Biophysical Research Communications , 513(1), 112–118. doi: 10.1016/J.BBRC.2019.03.143
- Hasan, A., Paul, A., Memic, A., & Khademhosseini, A. (2015). A multilayered microfluidic blood vessel-like structure. Biomedical Microdevices , 17(5), 88. doi: 10.1007/S10544-015-9993-2
- Herland, A., van der Meer, A. D., FitzGerald, E. A., Park, T. E., Sleeboom, J. J. F., & Ingber, D. E. (2016). Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood-brain barrier on a chip. PLOS ONE , 11(3), e0150360. doi: 10.1371/JOURNAL.PONE.0150360
- Jiménez-Torres, J. A., Peery, S. L., Sung, K. E., Beebe, D. J., Jiménez-Torres, J. A., Peery, S. L., … Beebe, D. J. (2015). LumeNEXT: A practical method to pattern luminal structures in ECM gels. Advanced Healthcare Materials , 5(2), 198–204. doi: 10.1002/adhm.201500608
- Majesky, M. W. (2007). Developmental basis of vascular smooth muscle diversity. Arteriosclerosis, Thrombosis, and Vascular Biology , 27(6), 1248–1258. doi: 10.1161/ATVBAHA.107.141069
- Orlova, V. V., Drabsch, Y., Freund, C., Petrus-Reurer, S., van den Hil, F. E., Muenthaisong, S., … Mummery, C. L. (2014a). Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arteriosclerosis, Thrombosis, and Vascular Biology , 34(1), 177–186. doi: 10.1161/ATVBAHA.113.302598
- Orlova, V. v., van den Hil, F. E., Petrus-Reurer, S., Drabsch, Y., ten Dijke, P., & Mummery, C. L. (2014b). Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nature Protocols , 9(6), 1514–1531. doi: 10.1038/nprot.2014.102
- Owens, G. K., Kumar, M. S., & Wamhoff, B. R. (2004). Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiological Reviews , 84(3), 767–801. doi: 10.1152/PHYSREV.00041.2003/ASSET/IMAGES/LARGE/Z9J0030403131003.JPEG
- Park, S. E., Georgescu, A., Oh, J. M., Kwon, K. W., & Huh, D. (2019). Polydopamine-based interfacial engineering of extracellular matrix hydrogels for the construction and long-term maintenance of living three-dimensional tissues. ACS Applied Materials & Interfaces, 11(27), 23919–23925. doi: 10.1021/ACSAMI.9B07912
- Passier, R., Orlova, V., & Mummery, C. (2016). Complex tissue and disease modeling using hiPSCs. Cell Stem Cell , 18(3), 309–321. doi: 10.1016/j.stem.2016.02.011
- Patsch, C., Challet-Meylan, L., Thoma, E. C., Urich, E., Heckel, T., Sullivan, J. F. O., … Cowan, C. A. (2015). Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nature Cell Biology , 17(8), 994–1003. doi: 10.1038/ncb3205
- Polacheck, W. J., Kutys, M. L., Tefft, J. B., & Chen, C. S. (2019). Microfabricated blood vessels for modeling the vascular transport barrier. Nature Protocols , 14(5), 1425–1454. doi: 10.1038/s41596-019-0144-8
- Roberts, B., Haupt, A., Tucker, A., Grancharova, T., Arakaki, J., Fuqua, M. A., … Gunawardane, R. N. (2017). Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Molecular Biology of the Cell , 28, 2854–2874.
- Rufaihah, A. J., Huang, N. F., Kim, J., Herold, J., Volz, K. S., Park, T. S., … Cooke, J. P. (2013). Human induced pluripotent stem cell-derived endothelial cells exhibit functional heterogeneity. American Journal of Translational Research , 5(1), 21.
- Samuel, R., Duda, D. G., Fukumura, D., & Jain, R. K. (2015). Vascular diseases await translation of blood vessels engineered from stem cells. Science Translational Medicine , 7(309), 309rv6. doi: 10.1126/SCITRANSLMED.AAA1805
- Shi, Y., Inoue, H., Wu, J. C., & Yamanaka, S. (2016). Induced pluripotent stem cell technology: A decade of progress. Nature Reviews Drug Discovery , 16(2), 115–130. doi: 10.1038/nrd.2016.245
- Sweeney, M., & Foldes, G. (2018). It takes two: Endothelial-perivascular cell cross-talk in vascular development and disease. Frontiers in Cardiovascular Medicine , 5, 154. doi: 10.3389/FCVM.2018.00154
- Vila Cuenca, M., Cochrane, A., van den Hil, F. E., de Vries, A. A. F., Lesnik Oberstein, S. A. J., Mummery, C. L., & Orlova, V. v. (2021). Engineered 3D vessel-on-chip using hiPSC-derived endothelial- and vascular smooth muscle cells. Stem Cell Reports , 16(9), 2159–2168. doi: 10.1016/J.STEMCR.2021.08.003
- Wanjare, M., Kuo, F., & Gerecht, S. (2013). Derivation and maturation of synthetic and contractile vascular smooth muscle cells from human pluripotent stem cells. Cardiovascular Research , 97(2), 321. doi: 10.1093/CVR/CVS315
- Wanjare, M., Kusuma, S., & Gerecht, S. (2014). Defining differences among perivascular cells derived from human pluripotent stem cells. Stem Cell Reports , 2(5), 561. doi: 10.1016/J.STEMCR.2014.03.004
- Yadav, A. R., Sriram, R., Carter, J. A., & Miller, B. L. (2014). Comparative study of solution-phase and vapor-phase deposition of aminosilanes on silicon dioxide surfaces. Materials Science & Engineering. C, Materials for Biological Applications, 35(1), 283–290. doi: 10.1016/J.MSEC.2013.11.017
- Zhang, J., Chu, L.-F., Hou, Z., Schwartz, M. P., Hacker, T., Vickerman, V., … Thomson, J. A. (2017). Functional characterization of human pluripotent stem cell-derived arterial endothelial cells. Proceedings of the National Academy of Sciences of the United States of America , 114, E6072–E6078.
Citing Literature
Number of times cited according to CrossRef: 1
- Ungsig Nam, Jaesang Kim, Hee‐gyeong Yi, Jessie S. Jeon, Investigation of the Dysfunction Caused by High Glucose, Advanced Glycation End Products, and Interleukin‐1 Beta and the Effects of Therapeutic Agents on the Microphysiological Artery Model, Advanced Healthcare Materials, 10.1002/adhm.202302682, 13 , 21, (2024).

