Synthetic DNA Assembly Using Golden Gate Cloning and the Hierarchical Modular Cloning Pipeline
Sylvestre Marillonnet, Sylvestre Marillonnet, Ramona Grützner, Ramona Grützner
DNA assembly
DNA construction
Golden Gate cloning
MoClo
modular cloning
multigene constructs
synthetic biology
Abstract
Methods that enable the construction of recombinant DNA molecules are essential tools for biological research and biotechnology. Golden Gate cloning is used for assembly of multiple DNA fragments in a defined linear order in a recipient vector using a one-pot assembly procedure. Golden Gate cloning is based on the use of a type IIS restriction enzyme for digestion of the DNA fragments and vector. Because restriction sites for the type IIS enzyme used for assembly must be present at the ends of the DNA fragments and vector but absent from all internal sequences, special care must be taken to prepare DNA fragments and the recipient vector with a structure suitable for assembly by Golden Gate cloning. In this article, protocols are presented for preparation of DNA fragments, modules, and vectors suitable for Golden Gate assembly cloning. Additional protocols are presented for assembly of defined parts in a transcription unit, as well as the stitching together of multiple transcription units into multigene constructs by the modular cloning (MoClo) pipeline. © 2020 The Authors.
Basic Protocol 1 : Performing a typical Golden Gate cloning reaction
Basic Protocol 2 : Accommodating a vector to Golden Gate cloning
Basic Protocol 3 : Accommodating an insert to Golden Gate cloning
Basic Protocol 4 : Generating small standardized parts compatible with hierarchical modular cloning (MoClo) using level 0 vectors
Alternate Protocol : Generating large standardized parts compatible with hierarchical modular cloning (MoClo) using level –1 vectors
Basic Protocol 5 : Assembling transcription units and multigene constructs using level 1, M, and P MoClo vectors
INTRODUCTION
DNA cloning has traditionally been done by digesting a source plasmid or DNA fragment and a recipient vector with restriction enzymes, extracting the digested fragments from a gel, and ligating the purified fragments using DNA ligase (see Current Protocols article Struhl, 1991). This approach is suitable for ligating one or two DNA fragments in a vector, but does not work well for ligating multiple fragments in a vector in a single step. Fortunately, new cloning methods are available that allow assembly of several fragments in a vector in a single step, including homology-based cloning methods (e.g., Gibson assembly) and methods relying on type IIS restriction enzymes, such as Golden Gate cloning (named in reference to Gateway cloning, but also as word play with the name of the bridge). Golden Gate is performed using a restriction-ligation reaction in a thermal cycler. Parameters for setting up and performing a standard Golden Gate assembly reaction are described in Basic Protocol 1 and outlined in Figure 1. Golden Gate cloning requires that the vector and inserts have a specific structure regarding the presence or absence of specific restriction sites. A method for converting a regular cloning vector into a Golden Gate–compatible cloning vector is provided in Basic Protocol 2. An example for accommodating an insert to Golden Gate cloning is provided in Basic Protocol 3, which explains how to design primers for amplification and cloning of a gene of interest into a Golden Gate vector.
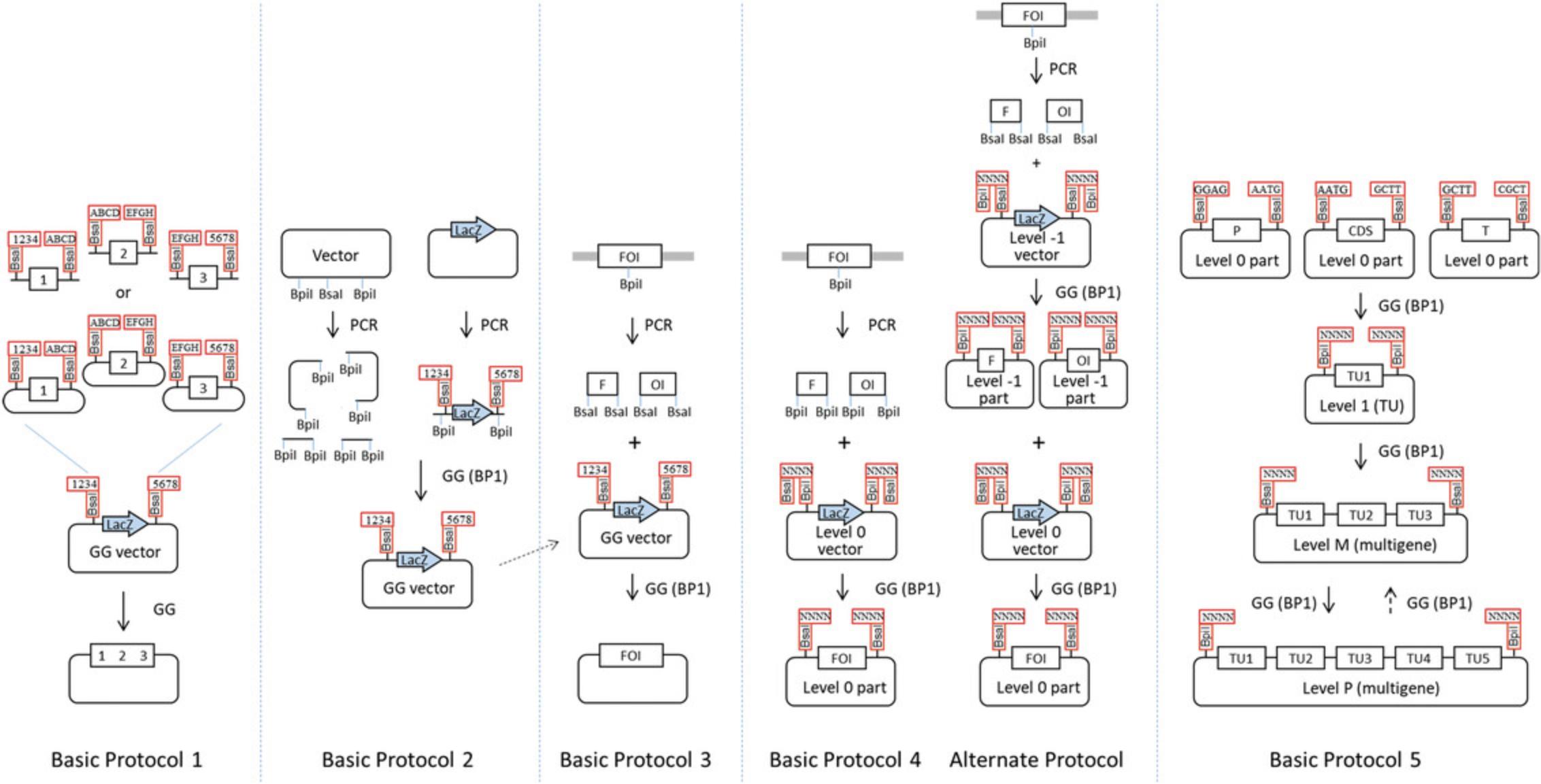
Generating defined transcription units and subsequent assembly of transcription units into multigene constructs using traditional cloning methods is not a simple task. This is due not only to the DNA assembly itself, but also because of the difficulty of designing cloning strategies for large constructs. One reason for this limitation is that recognition sites for the restriction enzymes used at each cloning step are usually still present in the constructs after ligation. Therefore, different restriction enzymes have to be used for each successive step, rendering the planning of a cloning strategy extremely difficult or impossible when working with a large number of genes. A solution to this problem is provided by standardizing the parts and the assembly strategy, combined with the use of Golden Gate cloning for DNA assembly at each cloning step. This strategy is employed by the modular cloning system MoClo (Engler et al., 2014; Weber, Engler, Gruetzner, Werner, & Marillonnet, 2011; Werner, Engler, Weber, Gruetzner, & Marillonnet, 2012). The MoClo system is based on the use of standard parts that are made by PCR, cloned into a vector backbone, and sequenced. Standard parts contain the DNA sequence of a basic genetic element, such as a promoter, coding sequence, or terminator. Basic Protocol 4 provides a method for cloning parts from PCR products in a single step (Fig. 1), and an Alternate Protocol provides a method for cloning larger parts in two successive steps. The standardized structure of the parts allows them to be re-used in many different constructs without the need for PCR amplification. It also allows standardization of the assembly procedure. Multigene constructs are assembled from basic parts using a series of one-pot assembly steps. The cloning strategy and the different cloning steps used with the MoClo system are described in Basic Protocol 5.
Basic Protocol 1: PERFORMING A TYPICAL GOLDEN GATE CLONING REACTION
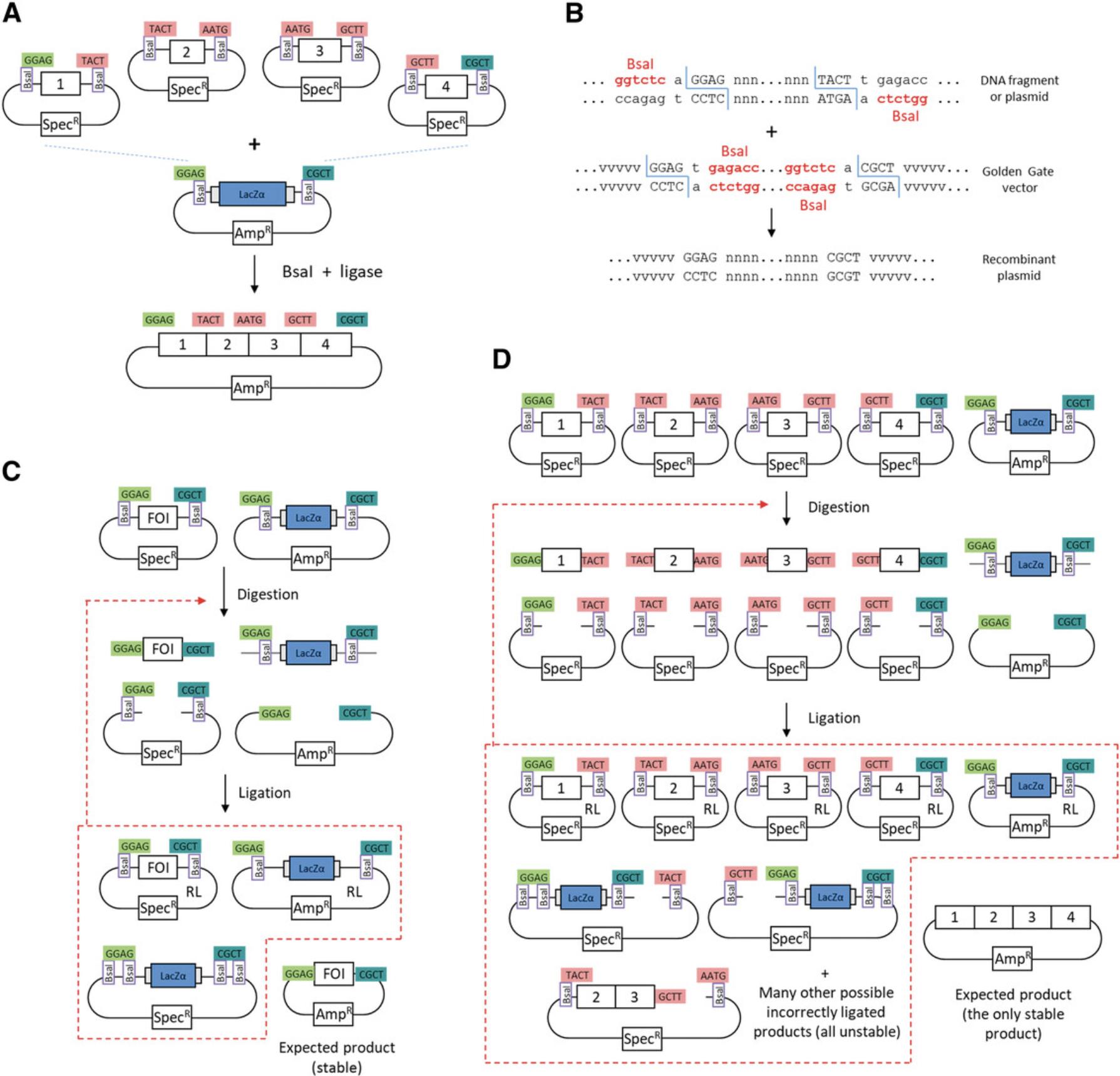
The principle of Golden Gate cloning consists of using a type IIS restriction enzyme and ligase in a restriction-ligation to assemble several DNA fragments in a defined linear order in a vector in a single step (Fig. 2A). Type IIS restriction enzymes cleave DNA outside of their DNA recognition site sequence. For example, the restriction site of Bsa I (GGTCTC N ↓N1N2N3N4) consists of a DNA recognition site sequence (GGTCTC) and a DNA cleavage site (↓) leading to a 4-nt single-stranded DNA overhang (N1N2N3N4) after digestion. To be suitable for Golden Gate cloning, all DNA fragments must be flanked by restriction sites for a type IIS restriction enzyme, with an inward orientation such that the DNA recognition site sequences are cleaved from the DNA fragments during the digestion step (Fig. 2B). The destination vector must also contain two restriction sites for the same type IIS enzyme, but with an outward orientation such that the DNA recognition sites are also cleaved from the vector by the digestion step. Finally, all single-stranded 4-nt overhangs (also called fusion sites) at the ends of the DNA fragments and vector must be unique and complementary to allow annealing to the end of the next fragment or the vector. Because the expected circular recombinant DNA molecule does not contain restriction sites for the enzyme used, it cannot be re-digested, allowing restriction and ligation to be performed in the same reaction mix. This allows DNA fragments that were ligated back in their initial plasmid backbone to be subject to more cycles of digestion and ligation until they finally become incorporated into the desired recombinant product (Fig. 2C). This is useful for transfer of a fragment from one vector to the next, but is increasingly important when several fragments need to be cloned in a single step. This is because the number of possible ligation events increases exponentially with the number of plasmids in the ligation mix, while the chance of getting a correctly ligated product in a single step becomes increasingly unlikely (Fig. 2D). The restriction-ligation leads to a high cloning efficiency and allows up to 24 fragments to be ligated in a single cloning reaction (Potapov et al., 2018).
The DNA fragments used can be linear fragments obtained by PCR, but can also be sequences cloned in a circular plasmid. This protocol describes how to perform a Golden Gate cloning reaction when both DNA parts and the destination vector are available as plasmids.
Materials
-
Purified plasmid DNA:
- Golden Gate destination vector (see Basic Protocol 2 or available from Addgene or other researchers)
- Inserts (plasmid DNA)
-
3 U/μl T4 DNA ligase and 10× ligation buffer (Promega, cat. no. M1804)
-
Type IIS restriction enzyme:
- 10 U/µl Bsa I (New England Biolabs, cat. no. R0535L)
- 10 U/µl Bpi I (Thermo Fisher Scientific, cat. no. ER1012)
- 10 U/µl Bsm BI (New England Biolabs, cat. no. R0580s)
-
Chemically or electrocompetent Escherichia coli cells (e.g., DH10B) (see Current Protocols article Seidman, 1997)
-
LB liquid medium (e.g., LB Broth High Salt, Duchefa, cat. no. L1704)
-
LB plates (LB Agar High Salt, Duchefa, cat. no. L1706; see Current Protocols article Elbing & Brent, 2018) containing appropriate antibiotic (kanamycin, spectinomycin, or carbenicillin) and X-gal
-
DNA miniprep kit (e.g., NucleoSpin Plasmid EasyPure, Macherey Nagel, cat. no. 740727.250)
-
1× TAE (see Current Protocols article Moore, 1996)
-
Agarose (Biozym cat. no. 840006)
-
Spectrophotometer (e.g., Nanodrop 1000, Peqlab)
-
0.2-ml PCR plates (Thermo Fisher Scientific, cat. no. AB0600)
-
Adhesive PCR plate seals (Thermo Fisher Scientific, cat. no. AB0588)
-
Thermal cycler (e.g., C1000 Touch, BioRad)
-
1.5-ml microcentrifuge tubes
-
37° and 42°C block incubator (e.g., Eppendorf ThermoMixer)
-
37°C incubator and shaker incubator
-
Culture flasks or tubes
-
DNA analysis software (e.g., Vector NTI, Thermo Fisher Scientific)
-
Additional reagents and equipment for agarose gel electrophoresis and sequencing
NOTE : Type IIS enzymes that generate a 3-nt overhang upon cleavage, such as Lgu I (Thermo Fisher Scientific, cat. no. ER1931), can also be used.
Perform restriction-ligation
1.Measure the DNA concentration of insert and vector plasmids using a spectrophotometer.
2.Add equal amounts of each insert and vector to the reaction mix. Use 20 fmol of each plasmid in a total reaction volume of 15 or 25 µl as follows:
For one to three fragments :
- x 1 µl vector DNA
- x 2 µl insert 1
- x 3 µl insert 2
- x 4 µl insert 3
- 1.0 µl T4 DNA ligase
- 0.5 µl type IIS enzyme
- 1.5 µl 10× ligation buffer
- H2O to 15 µl
For more than three fragments :
- x 1 µl vector
- x 2 µl insert 1
- x 3 µl insert 2
- … …
- xn µl insert n
- 1.5 µl T4 DNA ligase
- 1.0 µl type IIS enzyme
- 2.5 µl 10× ligation buffer
- H2O to 25 µl
3.Incubate the mix in a thermal cycler with the following conditions:
For one to three fragments :
- 37°C for 2-4 hr
- 50°C for 5 min
- 80°C for 10 min
- 16°C
For more than three fragments when using BsaI or BsmBI :
- 50 cycles: 37°C for 2 min, 16°C for 5 min
- 50°C for 5 min
- 80°C for 10 min
- 16°C
For more than three fragments when using BpiI :
- 50 cycles: 37°C for 2 min, 16°C for 5 min
- 37°C for 5 min
- 80°C for 10 min
Transform product into E. coli
4.Add the entire ligation mix (15 or 25 µl) to 50 µl competent E. coli cells and incubate on ice for 30 min.
5.Heat shock at 42°C for 90 s. Let cells recover on ice for 2 min.
6.Add 500 µl LB or SOC medium and shake cells for 45 min at 37°C in a ThermoMixer.
7.Plate on LB plates containing the appropriate antibiotic and X-gal. Incubate overnight at 37°C.
Pick colonies and assess clones
8.Pick individual colonies and inoculate 5 ml LB medium. Incubate overnight at 37°C in a shaker incubator.
9.Prepare DNA using a miniprep kit, eluting in a volume of 50 μl.
10.Digest 1 µl eluate in a 10-µl reaction using an appropriate enzyme (often Bsa I or Bpi I when using the MoClo system; see Basic Protocol 4) to confirm correct assembly by DNA fingerprinting.
11.If PCR amplification was used to prepare the DNA fragments, sequence the plasmid.
Basic Protocol 2: ACCOMMODATING A VECTOR TO GOLDEN GATE CLONING
Golden Gate cloning requires that the DNA fragments and recipient vector conform to specific requirements, such as the presence of type IIS enzyme restriction sites at the ends of the fragments and vector, and a lack of the same restriction sites in internal sequences of the fragments and vector. Many vectors suitable for Golden Gate cloning are available from several research groups, and the number of vectors available will likely grow in the future. Nevertheless, converting a standard cloning vector into a Golden Gate cloning vector is sometimes required for a user's specific needs. There are many strategies to do that, including using Gibson DNA assembly or Golden Gate cloning.
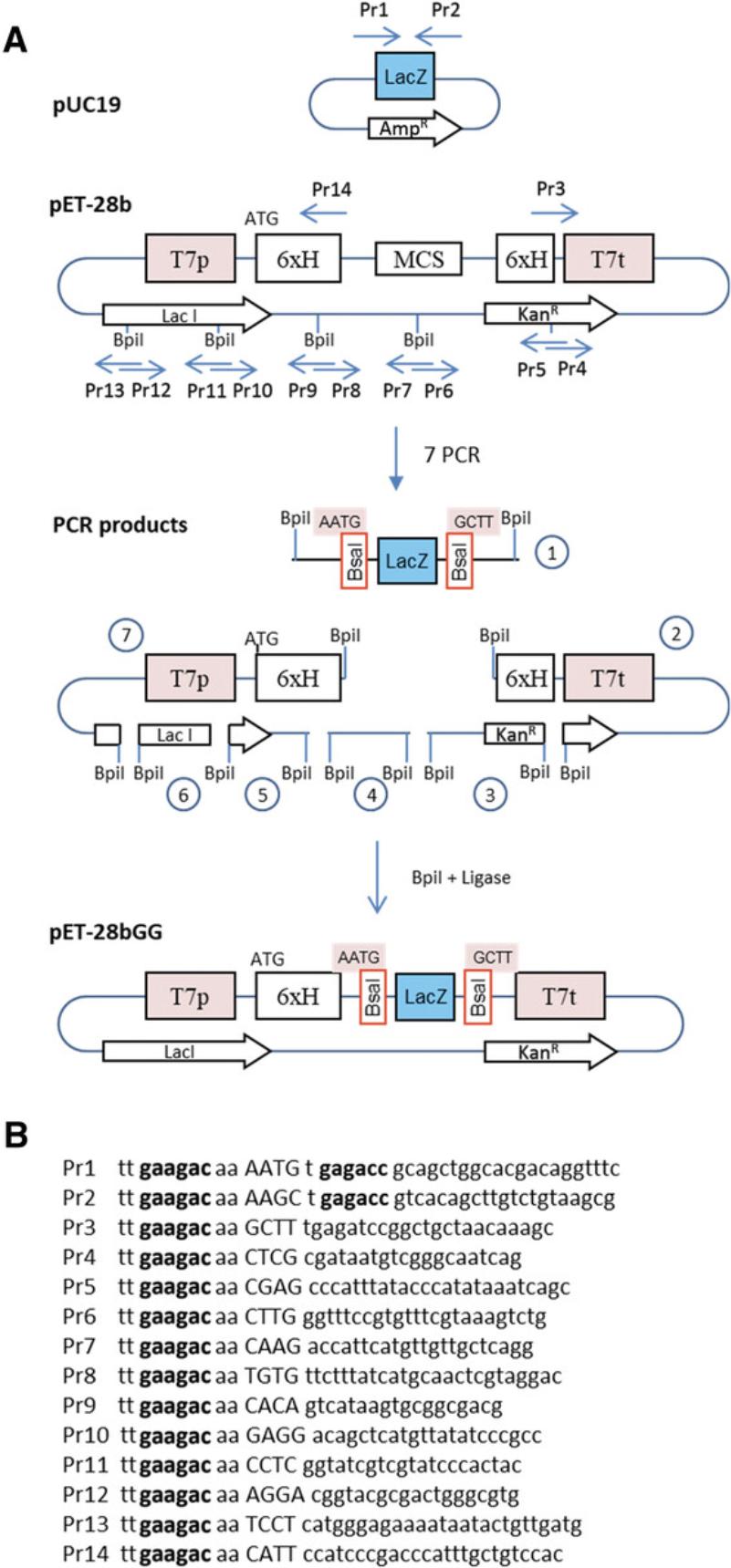
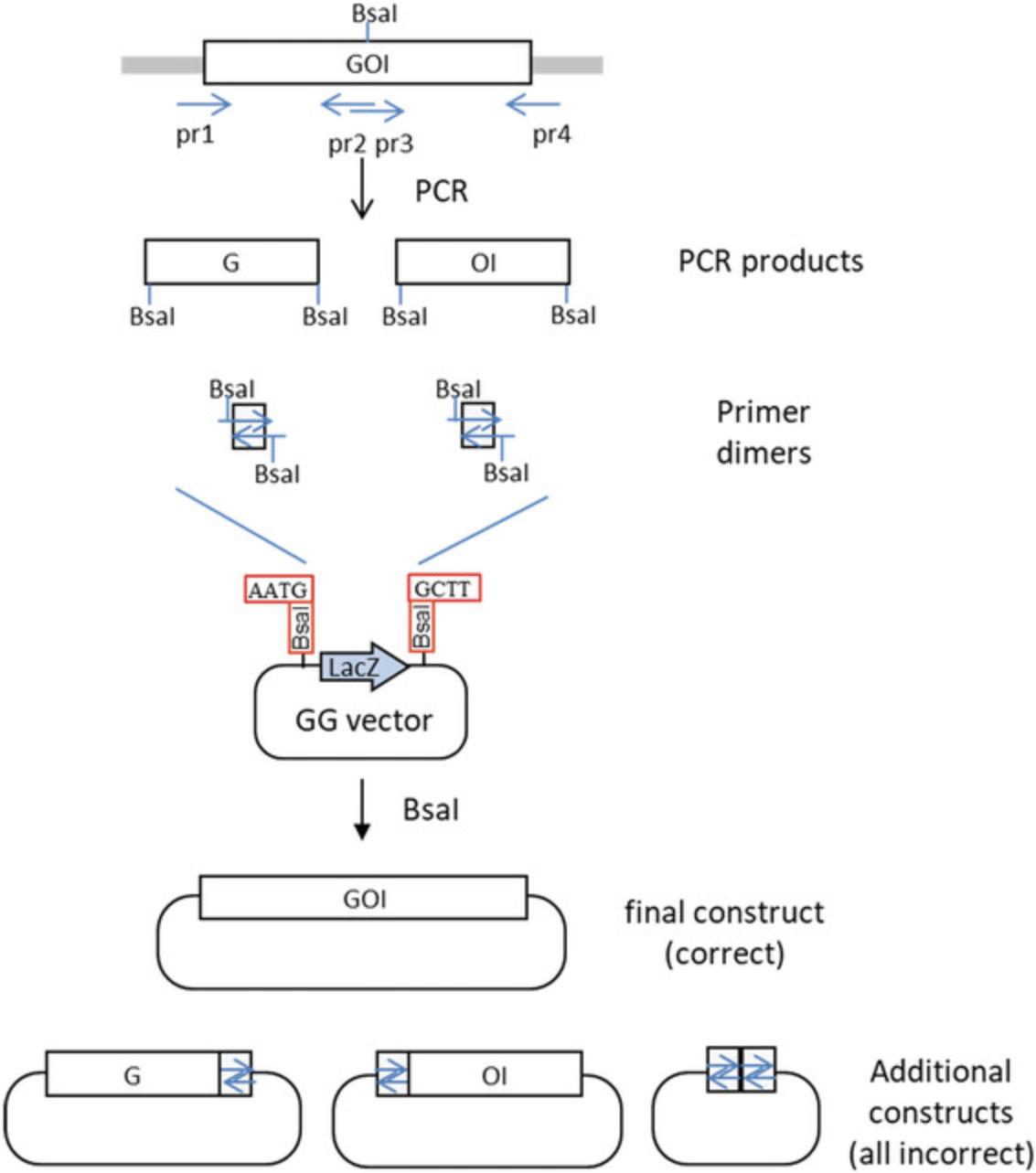
An example is provided here for converting the pET-28b (+) vector for use with Golden Gate cloning. In this example, a LacZ alpha fragment flanked by two Bsa I sites is inserted in the vector, replacing the multicloning site (Fig. 3A). The LacZ fragment will be used later for blue-white screening. Here, the two fusion sites flanking the LacZ fragment are AATG and GCTT, corresponding to the MoClo standard for cloning of coding sequences (see Basic Protocol 4). However, they can be any other sequences that a user might want, as long as they are different from each other and non-palindromic. Indeed, overhangs consisting of palindromic sequences can anneal to the same overhang of an additional copy of the same fragment in the other orientation, leading to stable ligation products that are nevertheless incorrect and resulting in a significant reduction of the cloning efficiency. In the example provided here, the ATG that is part of the first fusion site corresponds to a methionine start codon, which is cloned in-frame with a His tag present in the upstream vector sequences. The vector and the LacZ fragment are obtained by PCR, and assembly is performed using a second type IIS enzyme, Bpi I. Since there are four Bpi I sites in the vector, they are removed using primers designed to insert single-nucleotide mutations in the DNA recognition sequences.
Additional Materials (also see Basic Protocol 1)
- Gene-specific primers from commercial vendor (e.g., Eurofins Genomics)
- Plasmid template for amplification of LacZ cassette (e.g., pUC19, New England Biolabs, cat. no. N3041S)
- Vector plasmid DNA (e.g., pET-28b (+), Novagen, Merck Millipore, cat. no. 69418)
- High-fidelity PCR amplification kit (e.g., KOD Hot Start DNA Polymerase, Millipore Sigma, VWR, cat. no. 71086-3)
- PCR fragment purification kit (e.g., NucleoSpin Gel and PCR Clean-up, Macherey Nagel, cat. no. 740609.250)
1.Design primers to amplify LacZ fragment.
Primer locations and sequences for this example are shown in Figure 3.The first two primers, Pr1 and Pr2, are designed to amplify the LacZ fragment from pUC19.pUC19 is selected as a template because it has a different antibiotic resistance gene (AmpR) from the recipient vector (KanR). This reduces the probability of obtaining incorrect constructs by carryover of the undigested template vector when screening clones after transformation. The amplified fragment contains a functional unit for the LacZ marker for blue-white screening. Primers Pr1 and Pr2 have Bpi I restriction sites (gaagac in Fig. 3B) with 4-nt fusion sites corresponding to sequences AATG and GCTT, respectively. The fusion sites in the two primers are followed by Bsa I restriction sites in opposite orientation (reverse complement of ggtctc = gagacc in Fig. 3B). The Bsa I sites will become part of the resulting vector and will be used for Golden Gate cloning once the vector is made.
The pET-28b vector contains four Bpi I restriction sites in internal sequences, which need to be removed by making single-nucleotide substitutions (a process called domestication). Eight primers are needed for this purpose (Pr3 and Pr6-14). Silent mutations are made for restriction sites located in coding sequences (in this case, the Lac I repressor, which has two Bpi I sites). A final pair of primers (Pr4 and Pr5) is made to avoid having to amplify fragments larger than 2 kb; this pair of primers is optional. Primers Pr3-14 all have a Bpi I site with a fusion site selected to be compatible with the fusion site of the fragment to which it will be ligated.
2.Perform amplification.
-
Set up one 50-μl PCR reaction with primer pair Pr1-2 using pUC19 as a template.
-
Set up six 50-μl reactions with primers pairs Pr3-4 to Pr13-14 using pET-28b (+) as a template.
3.Run 5 μl from each reaction on a gel to see if the PCR was successful.
4.Purify the remaining 45 μl from each reaction using a column-based kit (i.e., Macherey Nagel NucleoSpin DNA Clean-up kit).
This will separate the PCR product from any remaining DNA polymerase and dNTPs, as well as primer dimers that are produced at various levels in most PCR reactions.
5.Quantify purified PCR products using a spectrometer such as a Nanodrop.
6.Perform the Golden Gate cloning reaction (see Basic Protocol 1) using the enzyme Bpi I. Transform the product into E. coli (see Basic Protocol 1) and select blue colonies.
In this particular example, transformants should be plated on LB containing kanamycin and X-gal. Correct colonies should not be white, but blue.
7.Prepare DNA using a miniprep kit.
8.Digest 1 µl with Bsa I in a 10-µl reaction.
Two fragments of 5.3 kb and 506 bp are expected for this particular construct.
9.Sequence the plasmid.
Sequencing the entire plasmid with primers designed at various positions within the plasmid is desirable because PCR was used for amplification of the entire vector. However, since some regions required for replication or expression of the LacZ marker should be functional if the plasmid replicates and the colonies are blue, the entire plasmid may not need to be sequenced. At a minimum, all elements that will later be required for expression of the gene should be sequenced, including the region extending from the T7 promoter to the terminator. Sequencing of the region containing the two introduced BsaI sites is essential to make sure that the sequence of the two fusion sites is as expected.
Basic Protocol 3: ACCOMMODATING AN INSERT TO GOLDEN GATE CLONING
This protocol explains how to design primers to accommodate a sequence of interest to Golden Gate cloning and then clone the amplified fragments in a Golden Gate destination vector. An example is given for cloning the coding sequence of an Arabidopsis thaliana isoflavone reductase amplified from an Arabidopsis cDNA template (Genbank accession NM_106183).
Additional Materials (also see Basic Protocols 1 and 2)
- cDNA or DNA template for PCR (here, cDNA from Arabidopsis)
- Plasmid DNA of Golden Gate destination vector
1.Design silent mutations in the open reading frame of the sequence of interest in silico using cloning software (e.g., Vector NTI).
In this example, the coding sequence contains one Bsa I site (Fig. 4A). It is removed in silico by a single-nucleotide substitution, resulting in a silent mutation (shown in red in Fig. 4B).
2.Add to the coding sequence two flanking fusion sites for compatibility with the vector.
In this case, add one A before the start codon to give AATG, and add GCTT after the stop codon.
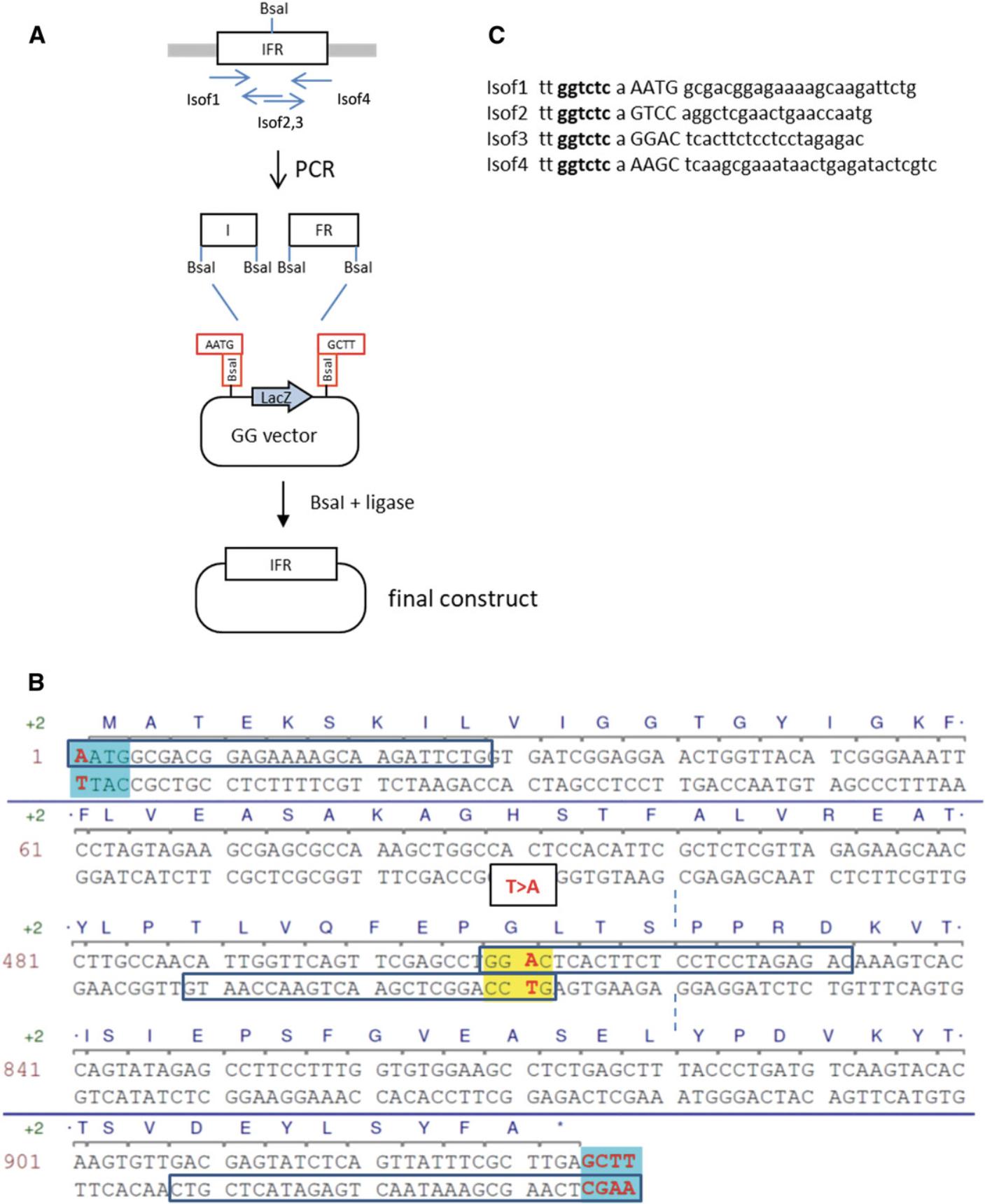
3.Select a fusion site at the site of the mutation.
Two PCR fragments will be amplified, one on each side of the mutation (I and FR in Fig. 4A), and then assembled via a fusion site that overlaps or is near the mutated nucleotide. The fusion site needs to be different from the other fusion sites (AATG and GCTT) and should not be palindromic. It should contain at least one G or C (more, if possible), as sites containing only As or Ts may work less well for DNA assembly (Potapov et al., 2018). When many fusion sites are needed, the Ligase Fidelity Viewer program (New England Biolabs, http://ggtools.neb.com/viewset/run.cgi) can be used to check the suitability of all sites. In the present example, a single sequence, GGAC, is selected (shown in yellow in Fig. 4B).
4.Design primers starting at all fusion sites. Select two primers in opposite orientation for each mutated site (in this case, only one site). Make the primers long enough to give an appropriate melting temperature for PCR amplification.
5.Add the sequence of the Bsa I recognition site (tt ggtctc a) to the beginning of all primers.
6.Amplify the two PCR fragments using the appropriate primers pairs (Isof1-2 and Isof3-4; Fig. 4A) from the Arabidopsis cDNA template in a 50-µl reaction. Run 5 µl on a gel to check that the correct fragment was amplified. Purify the remaining 45 µl using a column-based kit according to manufacturer's instructions.
7.Set up and perform the assembly reaction as described (see Basic Protocol 1).
Because this example describes a simple cloning with only two fragments cloned into the vector, a simple incubation at 37°C for 2 hr, 50°C for 5 min, and 80°C for 10 min should be sufficient.
8.Transform the ligation mix into competent E. coli , plate on LB containing kanamycin and X-gal, and screen the resulting colonies.
Minipreps are digested with two enzymes with restriction sites flanking the insert, or with enzymes that have sites in the vector in insert. Because PCR was used for cloning, the cloned insert must be sequenced using primers for sequences in the vector flanking the insert.
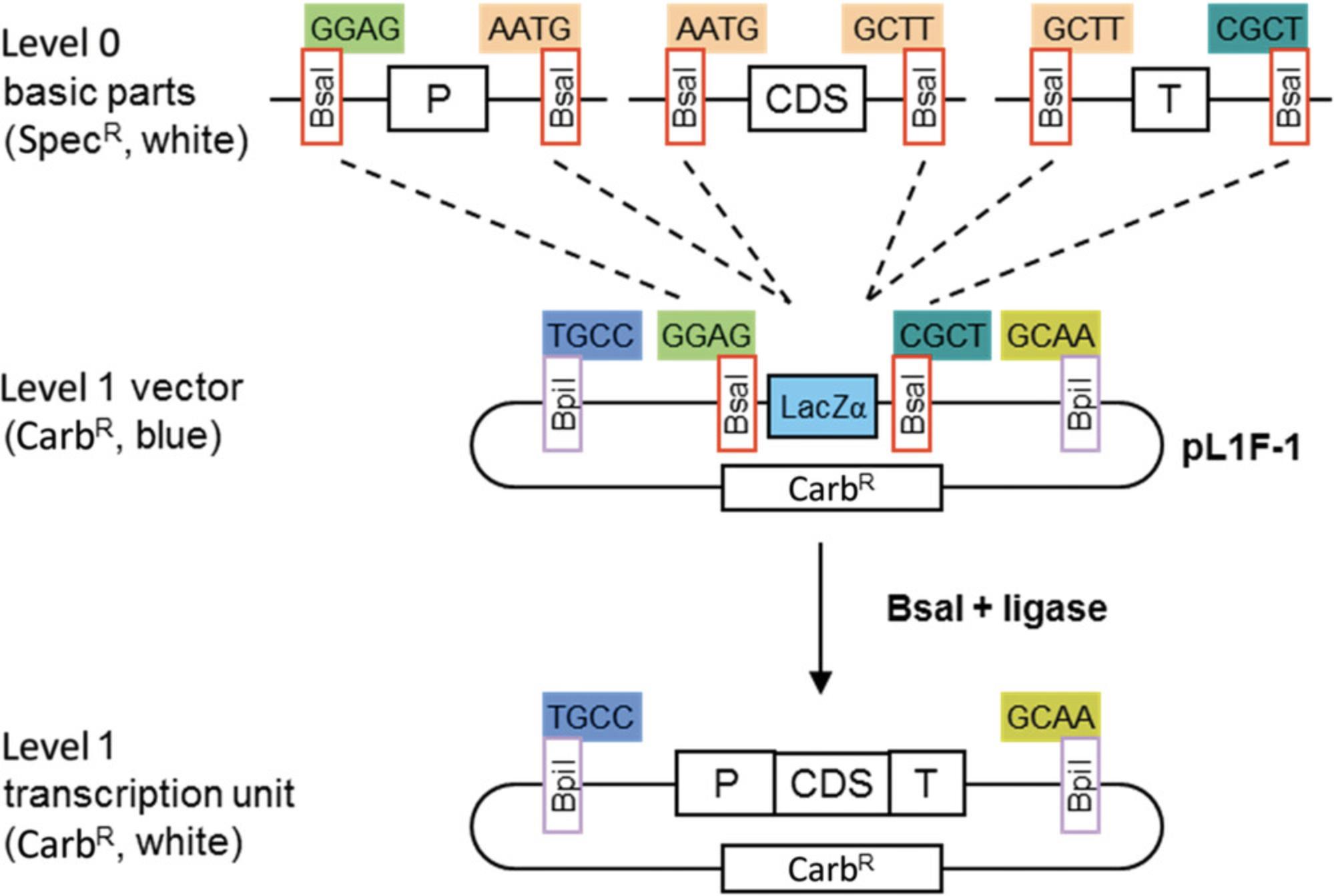
Basic Protocol 4: GENERATING SMALL STANDARDIZED PARTS COMPATIBLE WITH HIERARCHICAL MODULAR CLONING (MoClo) USING LEVEL 0 VECTORS
Assembly of DNA fragments from plasmids using Golden Gate cloning can be done quickly and efficiently. In contrast, assembly from PCR products requires more work, because PCR products often contain primer dimers formed by mis-annealing of primers during PCR amplification. Because primer dimers are flanked by the same type IIS restriction sites as the PCR products, they can be cloned, resulting in incorrect constructs and lowering the cloning efficiency (Fig. 5). Even if no primer dimers are made, the cloned constructs need to be sequenced to make sure that they do not contain PCR-induced mutations. Therefore, it makes a lot of sense to have a system where cloned and sequenced DNA parts can be reused in different constructs. For example, parts that contain regulatory sequences such as promoters and terminators can be reused in many different constructs.
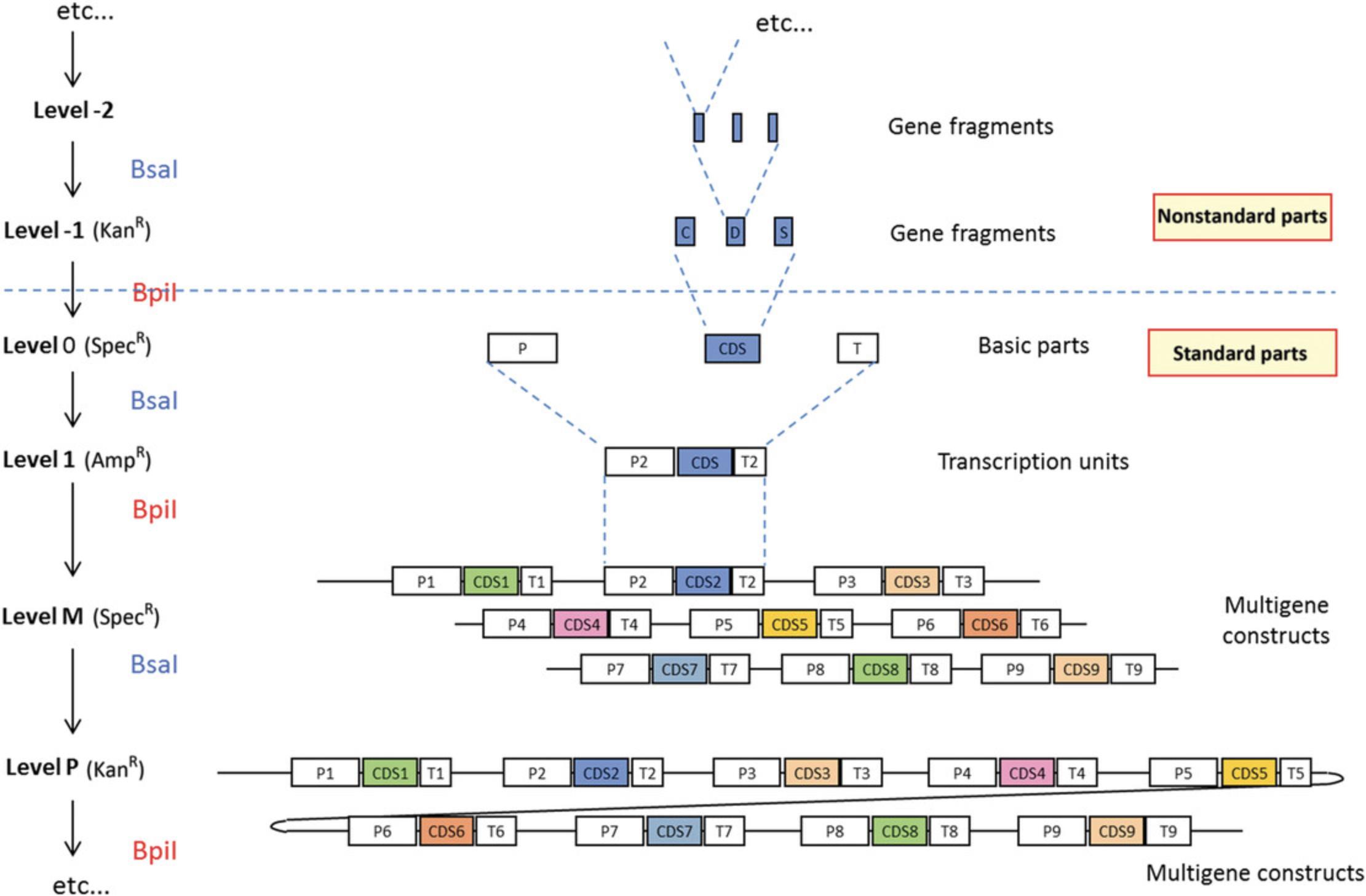
The modular cloning system MoClo was designed to facilitate the reuse of cloned parts in different constructs (Fig. 6). It is based on the use of basic parts that are put together using Golden Gate cloning. The basic parts are cloned as level 0 modules and assembled in level 1 cloning vectors to make transcription units. The level 1 constructs are then assembled to make level M and P multigene constructs. Basic parts contain the coding sequence of a basic genetic element flanked by two Bsa I restriction sites in opposite orientation (Fig. 7). Cloning of the basic parts requires amplification from DNA or cDNA as a template. The amplified product is cloned in a recipient vector using Golden Gate cloning. The cloning steps consist of defining the part type, design primers containing Bsa I restriction sites at the ends of the fragments, removing sites from internal sequences, and cloning the amplified fragments in a vector.
Additional Materials (also see Basic Protocols 1 and 2)
- cDNA or DNA template for PCR (here, cDNA from Arabidopsis)
- Universal level 0 cloning vector pAGM9121 (Addgene, plasmid #51833)
1.Define the type of level 0 module (part).
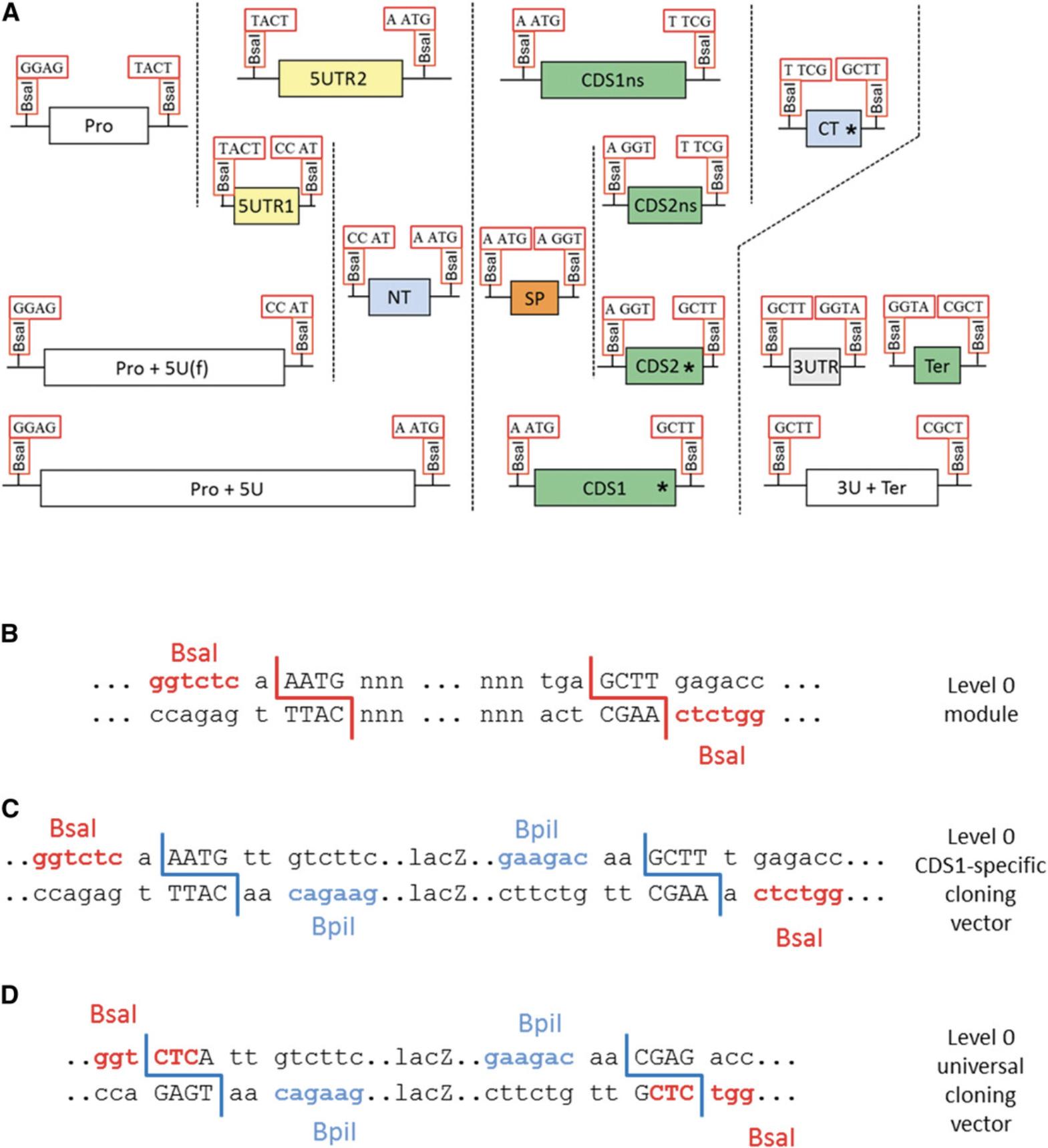
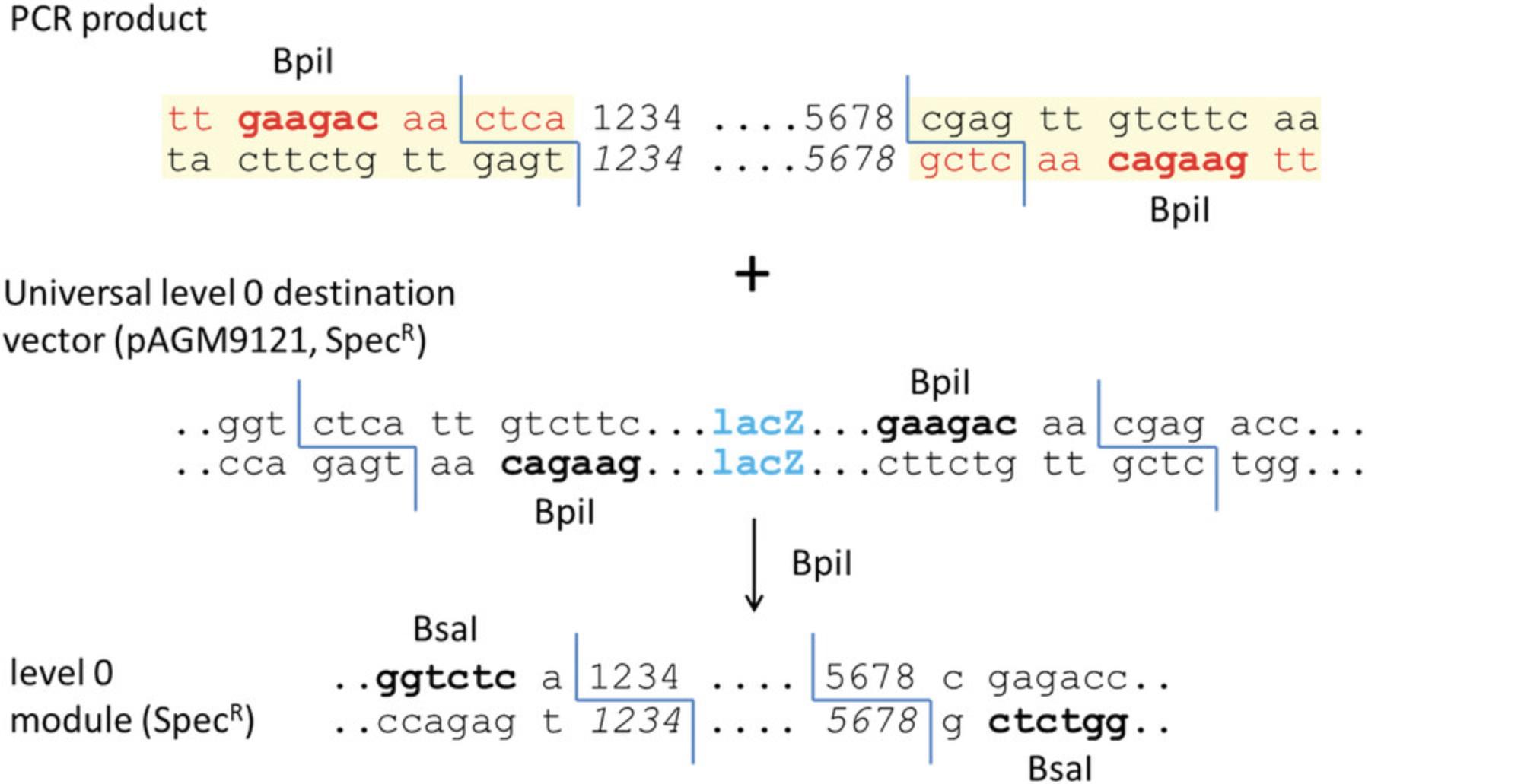
Parts can consist of promoters, coding sequences with or without a stop codon, C- or N-terminal fusion tags, and terminators (Fig. 7A). Each part is flanked by two Bsa I sites in opposite orientation (Fig. 7B). The 4-bp sequences of the Bsa I cleavage sites (fusion sites) are specific for each type of module and define which parts can be fused together. For example, a promoter module with TACT as the 3′ fusion site (Pro in Fig. 7A) can be fused only to an untranslated leader sequence with the same fusion site at the 5′ end (5UTR2 in Fig. 7A). For each part type, a specific cloning vector is available with compatible fusion sites. Alternatively, all part types can be cloned into the universal level 0 cloning vector pAGM9121 (Figs. 7D and 8) as described in this protocol. Level 0 modules may also be composites of several basic parts. For example, constructs containing a complete transcription unit can be cloned directly as level 0 modules between fusion sites GGAG and CGCT. Other types of genetic elements, such as a phage recombination site or a matrix attachment region, can also be cloned as level 0 modules between fusion sites GGAG and CGCT.
2.Introduce silent mutations in the gene sequence in silico.
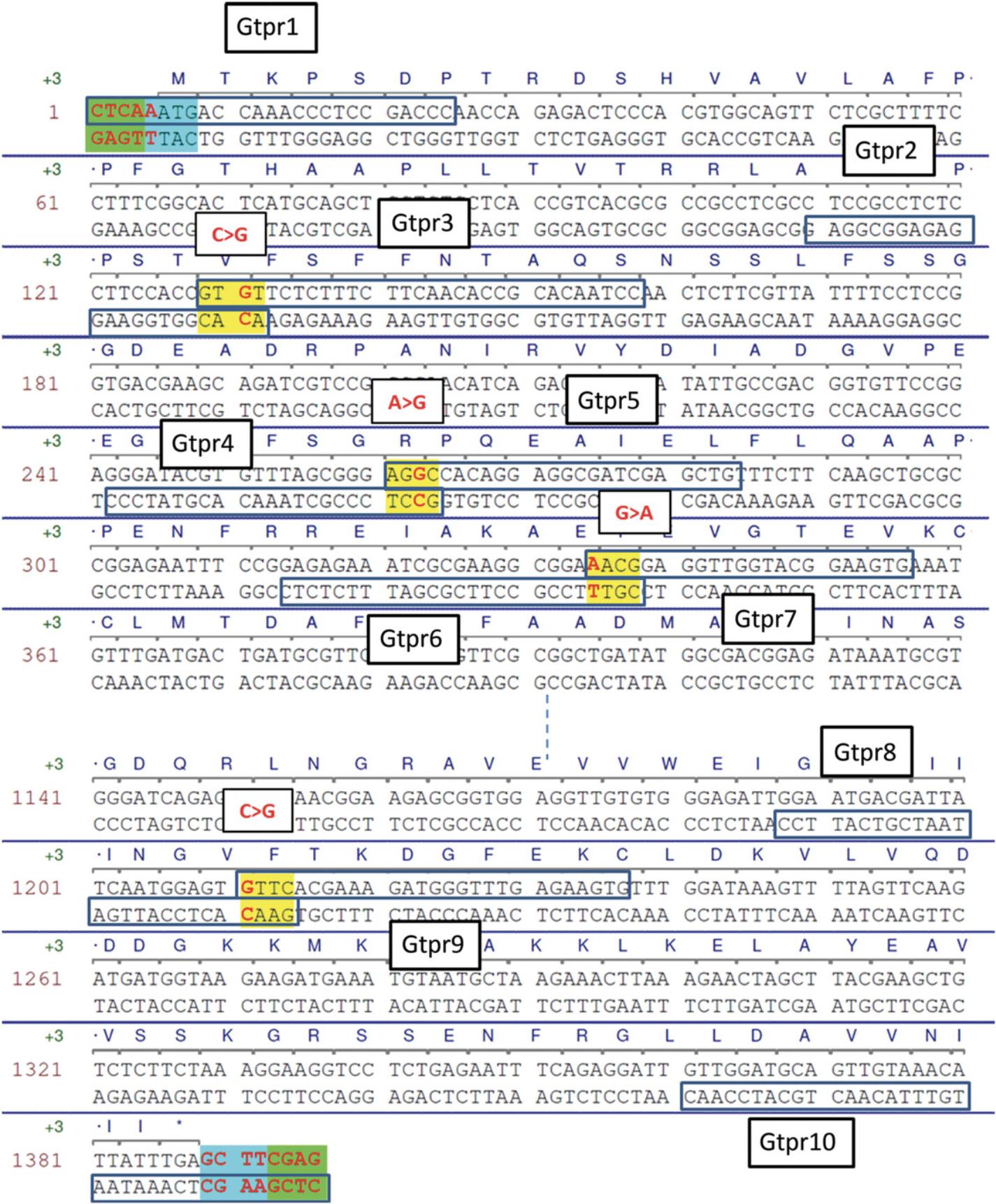
Basic parts should not contain restriction sites for the type IIS enzymes that are used with the MoClo system (Bsa I, Bpi I, and optionally Bsm BI). Restriction sites are removed by introducing single-nucleotide mutations, which is first performed in silico. Removal of internal sites in coding sequences can always be done with a silent mutation. Removal of sequences in promoter regions is more difficult, because sequences important for promoter function are not always known. Therefore, after domesticating promoter sequences, it will be necessary to validate experimentally in the host organism that the promoter still works as intended. Domestication of the Arabidopsis UGT78D2 gene (GenBank accession NM_121711) is presented here as an example. This gene contains one Bsa I site, two Bpi I sites, and one Bsm BI site (Fig. 9A), which are removed by introducing four silent mutations (red in Fig. 10).
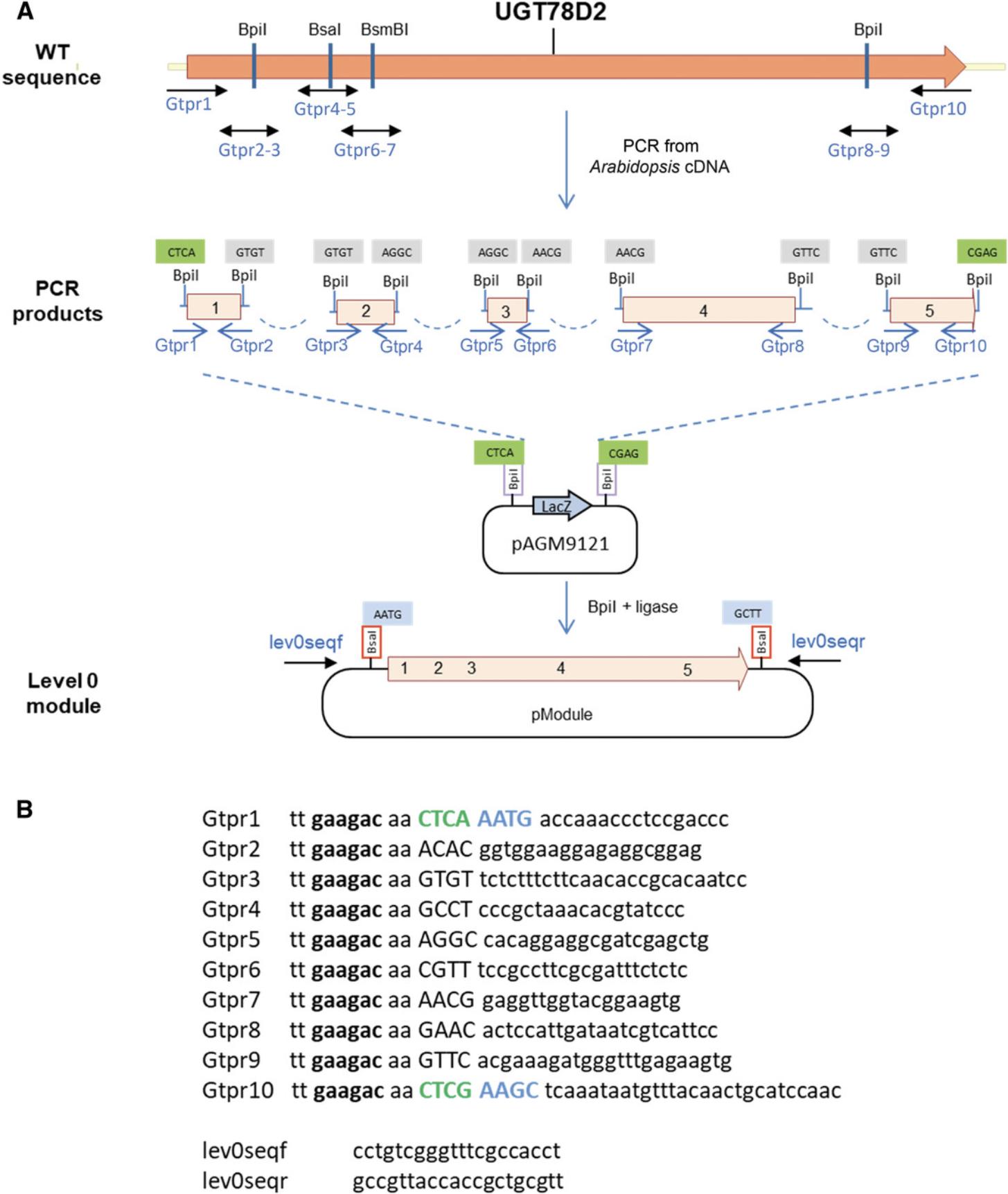
3.Add the two standard fusion sites flanking the part.
Because this module will be a coding sequence with a stop codon (CDS1), one A is added before the start codon to give AATG, and the sequence GCTT added after the stop codon (both fusion sites are highlighted in blue in Fig. 10).
4.Add sequences for compatibility with the universal cloning vector.
In this example, the part will be cloned into the universal level 0 cloning vector pAGM9121.Therefore, CTCA and CGAG are added at the beginning and end of the fragment to provide the necessary compatibility (highlighted in green in Fig. 10). These sequences will become part the Bsa I restriction sites that will flank the final level 0 module.
5.Select the nonstandard fusion sites that will be used for PCR fragment assembly.
These are the 4-nt sequences that will be used to assemble the PCR products during cloning. They need to be different from each other and from the fusion sites that will be used in the vector (CTCA and CTCG for the universal cloning vector, or AATG and GCTT for the CDS1-specific cloning vector). They also need to be different from the reverse complement of any of the selected fusion sites to avoid inappropriate fragment ligation. The selected sequences are highlighted in yellow in Figure 10.
6.Design primers starting at all fusion sites, standard and nonstandard.
For fusion sites at sites mutated to remove restriction sites, primers are designed in both orientations. The primers should be long enough to have an appropriate melting temperature. This can be calculated using any of the many programs available online or can be done manually.
7.Add the type IIS enzyme recognition sequence.
Because Bpi I is used for assembly, the sequence tt gaagac aa is added to all primers. Primer sequences and locations are shown in Figure 9.
8.PCR amplify the fragments.
Amplify five fragments from Arabidopsis cDNA in separate 50-µl reactions using primer pairs Gtpr1-2 to Gtpr9-10 and a proofreading polymerase. Run 5 µl on a gel to check that each fragment was amplified as expected.
9.Column purify the fragments.
The amplified fragments need to be purified to remove remaining polymerase, primers, and dNTPs, and most of the primer dimers that are often present in the reaction. If a single fragment was amplified, using a DNA purification kit (e.g., Macherey Nagel DNA purification kit) is sufficient. Gel extraction of the PCR fragments of the correct size may be necessary if several DNA fragments were amplified.
10.Set up cloning in the universal level 0 vector pAGM9121 as described (see Basic Protocol 1).
11.Transform in E. coli and pick two white colonies. More colonies can be picked later if sequencing of these clones does not show the correct sequence.
12.Screen colonies by digesting miniprep DNA with Bsa I.
13.Sequence the clones using the vector primers lev0seqf and lev0seqr (Fig. 9B).
Alternate Protocol: GENERATING LARGE STANDARDIZED PARTS COMPATIBLE WITH HIERARCHICAL MODULAR CLONING (MoClo) USING LEVEL –1 VECTORS
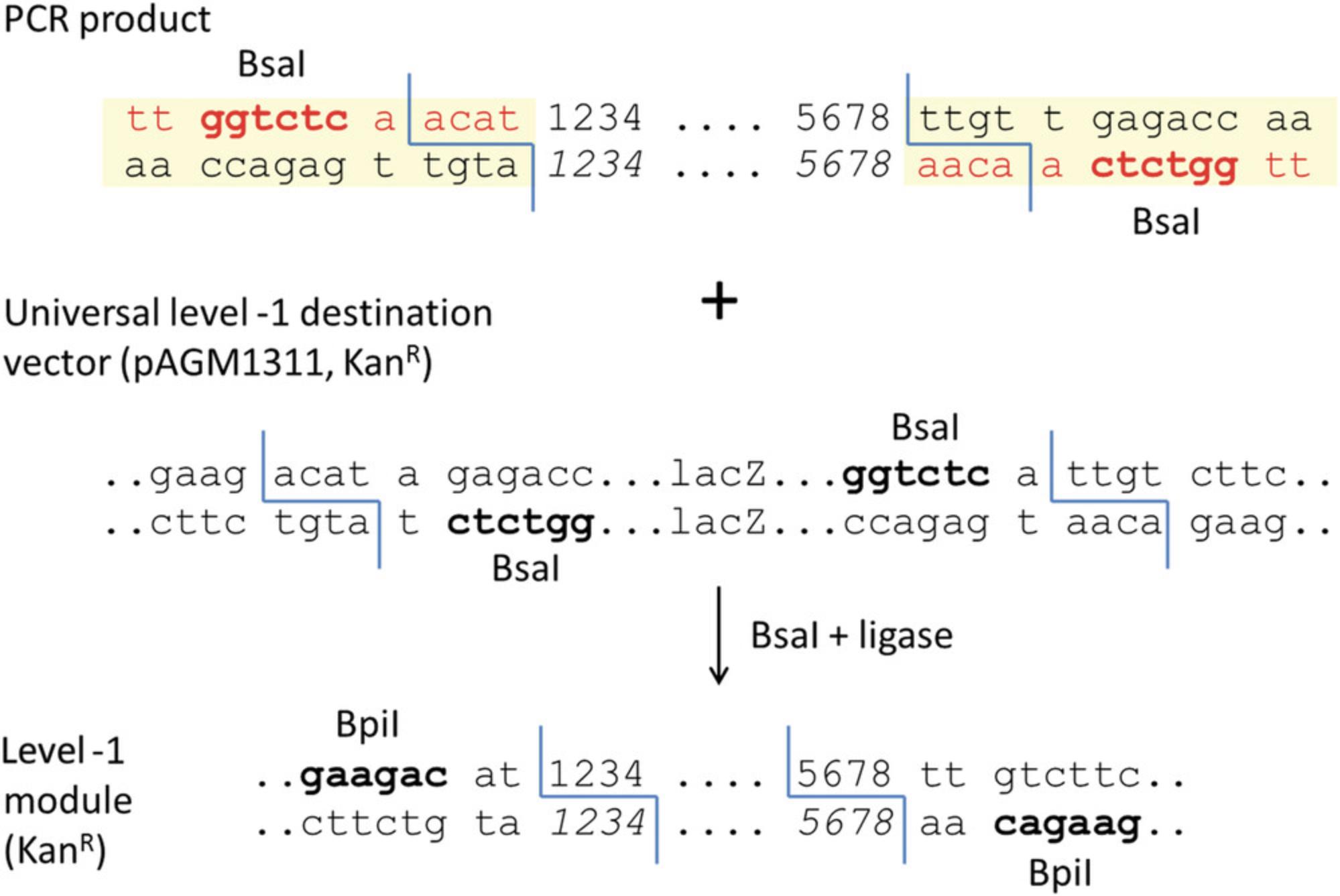
While small parts (<1.2 kb) can be sequenced using standard primers located in flanking vector sequences, large parts cannot be sequenced entirely from vector primers. Additional sequencing primers have to be designed within the part to sequence its middle section. To facilitate sequencing without the need for custom sequencing primers for each part, an alternative strategy consists of cloning part fragments (sub-parts) that are no more than 1-1.2 kb long, sequencing the sub-parts using vector primers, and then assembling the sequenced sub-parts using a second assembly reaction to make the final level 0 module (Fig. 11). Sub-parts are cloned in a universal level –1 cloning vector (pAGM1311, Fig. 12).
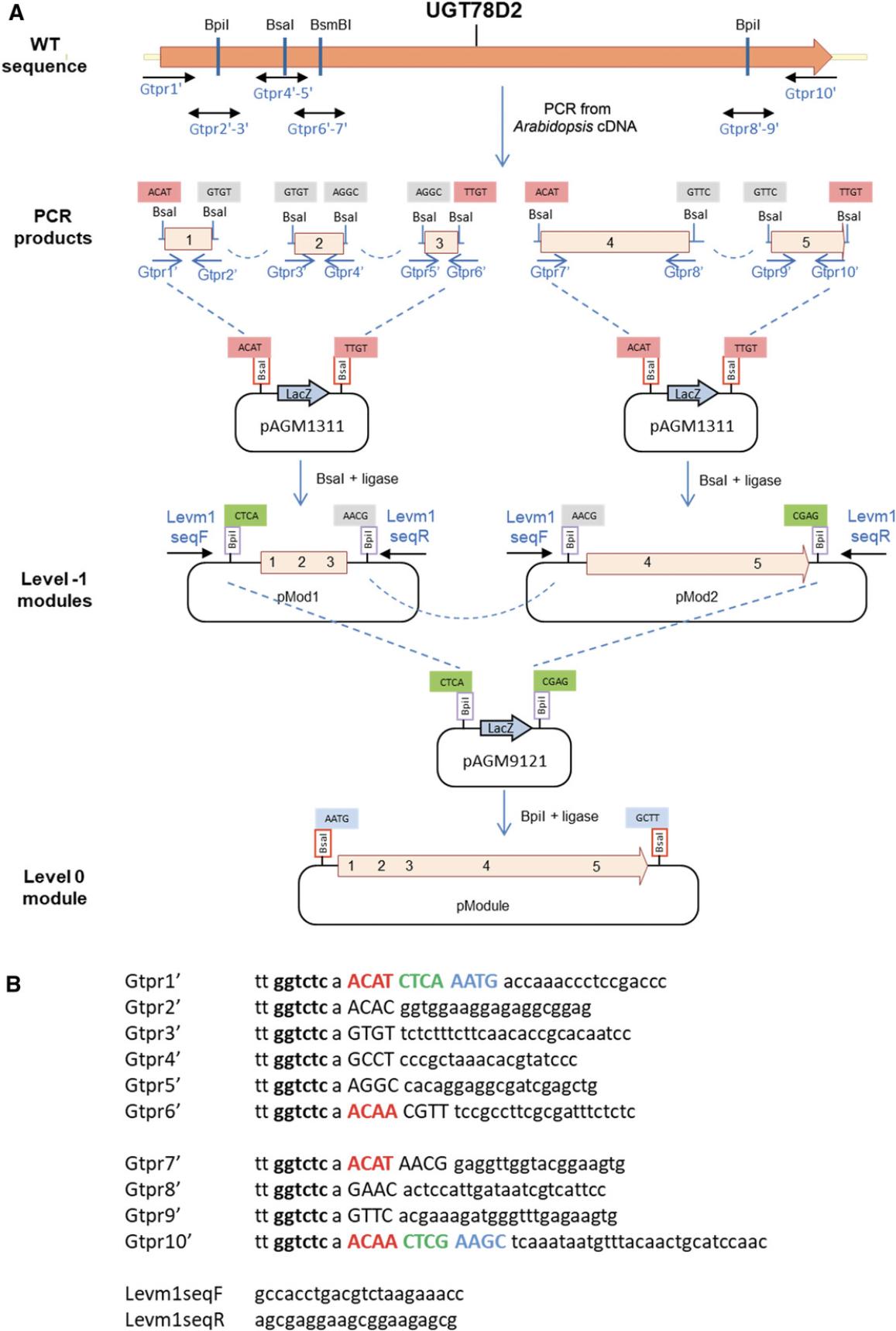
Additional Materials (also see Basic Protocol 4)
- Universal level –1 pAGM1311 cloning vector (Addgene, plasmid #47983)
1.Define the part type, introduce silent mutations, add the standard fusion sites, and add sequences for compatibility with the universal cloning vector as described (see Basic Protocol 4, steps 1-4).
2.Select the nonstandard fusion sites that will be used for PCR fragment assembly.
These are the 4-nt sequences that will be used to assemble the PCR products during cloning. They need to be different from each other and from the fusion sites that will be used in the vector (in this case, ACAT and TTGT for the universal level –1 vector; see step 5 below). They also need to be different from the reverse complement of any of the selected fusion sites to avoid inappropriate fragment ligation. The selected sequences are the same as those in Basic Protocol 4 (highlighted in yellow in Fig. 10).
3.Design primers starting at all fusion sites, standard and nonstandard, as described (see Basic Protocol 4, step 6).
4.Add the type IIS enzyme recognition sequence.
For cloning in level –1 vectors, Bsa I is used for assembly, and thus the sequence tt ggtctc a is added to the 5′ end of all primers.
5.Add sequences for compatibility with the universal level –1 cloning vector.
The two fusion sites of the universal cloning vector are ACAT and TTGT, and they contain part of a Bpi I restriction site that will flank the sub-part after cloning (Fig. 12). Therefore, the sequences ACAT and ACAA (reverse complement of TTGT) must be added to the two primers that delimit the group of PCR fragments that will be cloned together. These sequences are added immediately 3′ of the Bsa I site and before the universal level 0 vector fusion site (CTCA or CTCG). In this example, the first three PCR products will be cloned together in the universal level –1 vector pAGM1311, and the fourth and fifth products will also be cloned together in pAGM1311.Primer sequences and locations for the first level –1 construct (Gtpr1′ to 6′) and second level –1 construct (Gtpr7′ to 10′) are shown in Figure 11.
6.PCR amplify the fragments (see Basic Protocol 4, step 8) using the appropriate primer pairs (Gtpr1′-2′ to Gtpr9′-10′).
7.Column purify the fragments (Basic Protocol 4, step 9).
8.Set up two Golden Gate cloning reactions as described (see Basic Protocol 1) using the universal level –1 vector pAGM1311.Clone the first three products together in one reaction and the fourth and fifth products in the second reaction (level –1 modules in Fig. 11A).
9.Transform in E. coli and pick two white colonies. More colonies can be picked later if sequencing of these clones does not provide the correct sequence.
10.Screen colonies by digesting miniprep DNA with Bpi I.
11.Sequence clones using the vector primers Levm1seqF and Levm1seqR (Fig. 11B).
12.Assemble level 0 modules from the level –1 sub-parts. Set up a Golden Gate cloning reaction to assemble the two selected level –1 clones in the universal level 0 cloning vector pAGM9121 (see Basic Protocol 1).
13.Transform in E. coli cells.
14.Screen colonies by digesting miniprep DNA with Bsa I.
Basic Protocol 5: ASSEMBLING TRANSCRIPTION UNITS AND MULTIGENE CONSTRUCTS USING LEVEL 1, M, AND P MoClo VECTORS
Assembly of multigene constructs using the MoClo system is done hierarchically (Fig. 6). The first step is assembly of level 0 parts in level 1 destination vectors to make constructs that usually contain a transcription unit. The second is assembly of multigene constructs which is done in alternating stages using level M and P vectors. Up to six transcription units can be assembled in a level M cloning vector to make a level M multigene construct. Then, up to six level M constructs can be further assembled into a level P vector to make a level P construct containing up to 36 transcription units. Level P constructs can themselves be further assembled into level M vectors to make even larger constructs. This process can be repeated indefinitely, until constructs become too large to be cloned as plasmids in E. coli. Level M and P assembly use Bsa I and Bpi I in an alternating fashion.
Additional Materials (also see Basic Protocol 1)
- Level 0 parts
- Level 1, M, and P destination vectors from MoClo Toolkit (Addgene, kit #1000000044)
Assemble transcription units in level 1 vectors
1.Select standard level 0 parts.
The first step for planning the assembly of a multigene construct is to identify all basic parts required for each transcription unit. For example, to make a construct containing four transcription units, each composed of three parts—including a promoter with 5′-UTR (pro + 5U), a coding sequence (CDS1), and a 3′-UTR with terminator (3U + Ter)—a total of twelve parts is required.
A list of all part types is shown in Figure 7.Parts that are not already available are cloned as described (see Basic Protocol 4). Some are already available for plants (MoClo Plant Parts Kit, Addgene, kit #1000000047), Chlamydomonas (https://www.chlamycollection.org/product/moclo-toolkit/; Crozet et al., 2018), and cyanobacteria (http://www.addgene.org/browse/article/28196941/; Vasudevan et al., 2019).
2.Select level 1 destination vectors.
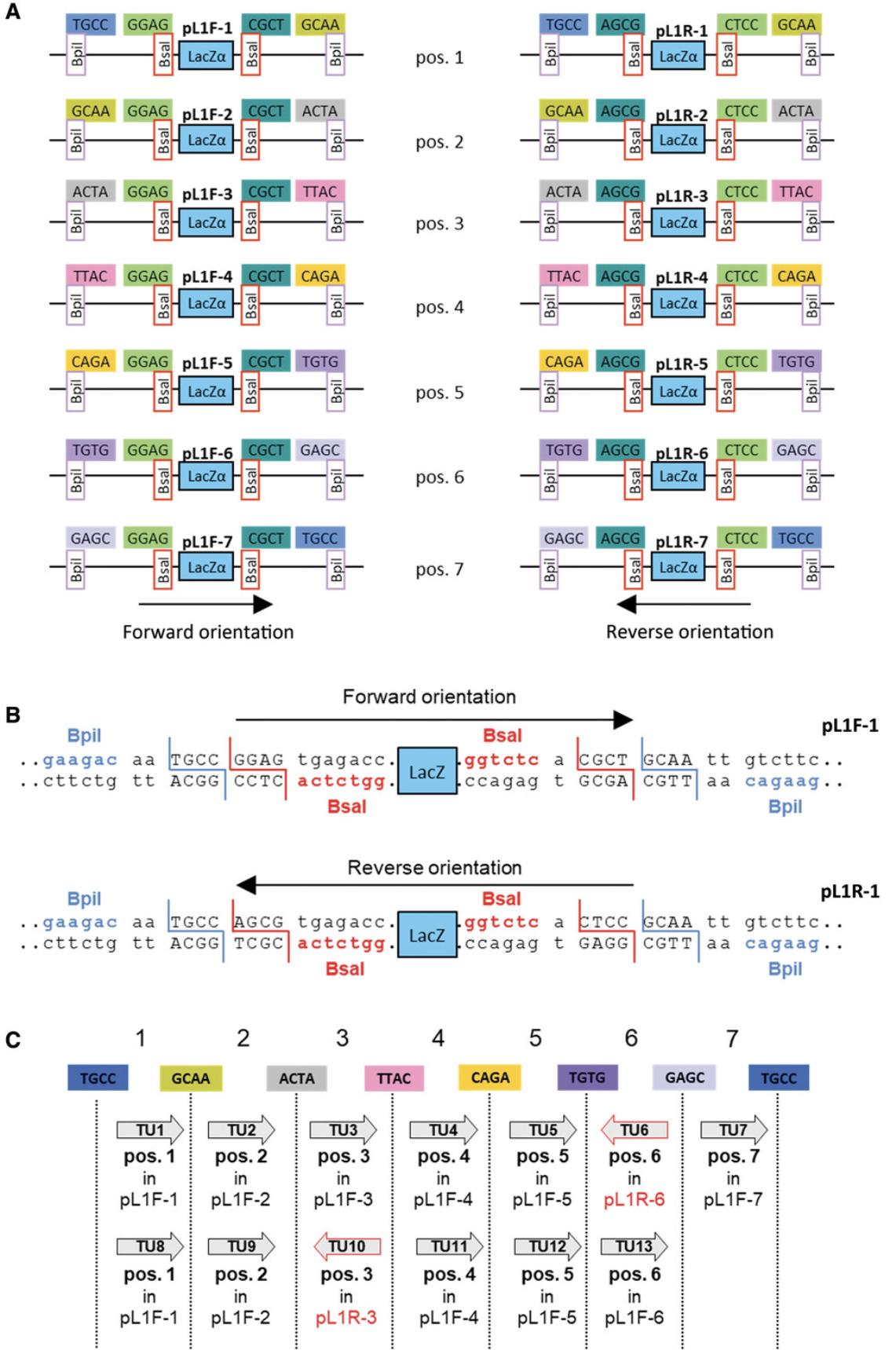
Fourteen level 1 vectors are available: seven for cloning a transcription unit in the forward orientation in seven different positions in the final construct, and seven for the reverse orientation (Fig. 13A). All contain two Bsa I sites and the fusion sites GGAG and CGCT to provide compatibility with level 0 modules. They also contain two Bpi I sites for excision of the assembled transcription unit in the next step of assembly. The Bpi I fusion sites are designed to provide compatibility from one vector to the next. In an example where four transcription units are cloned in the forward orientation, vectors for positions 1 to 4 for the forward orientation (pL1F-1, pL1F-2, pL1F-3, and pL1F-4) are selected.
The Bpi I fusion site at the end of vector 7 (TGCC) is the same as the site at the beginning of vector 1, allowing one to clone more than seven genes in a multigene construct by reusing the same seven vectors indefinitely. Thus, an eighth transcription unit is cloned using vector 1, a ninth transcription unit is cloned using vector 2, and so on (Fig. 13C).
In the example in Figure 13C, two transcription units are cloned in reverse orientation, and therefore vectors for cloning in the reverse orientation are selected. The choice of whether a transcription unit should be cloned in one orientation or the other is decided solely by the user needs.
3.Assemble level 1 constructs.
Constructs are assembled using four Golden Gate cloning reactions, each containing the three selected basic parts and level 1 vector. An example for cloning the first transcription unit is shown in Figure 14.The cloning reaction is performed as described (see Basic Protocol 1). Because level 1 vectors contain a LacZ fragment for blue-white screening, two white colonies are picked. As assembled transcription units are flanked by Bpi I sites, miniprep DNA is checked by Bpi I digestion.
Assemble multigene constructs in level M vectors
4.Design level M constructs.
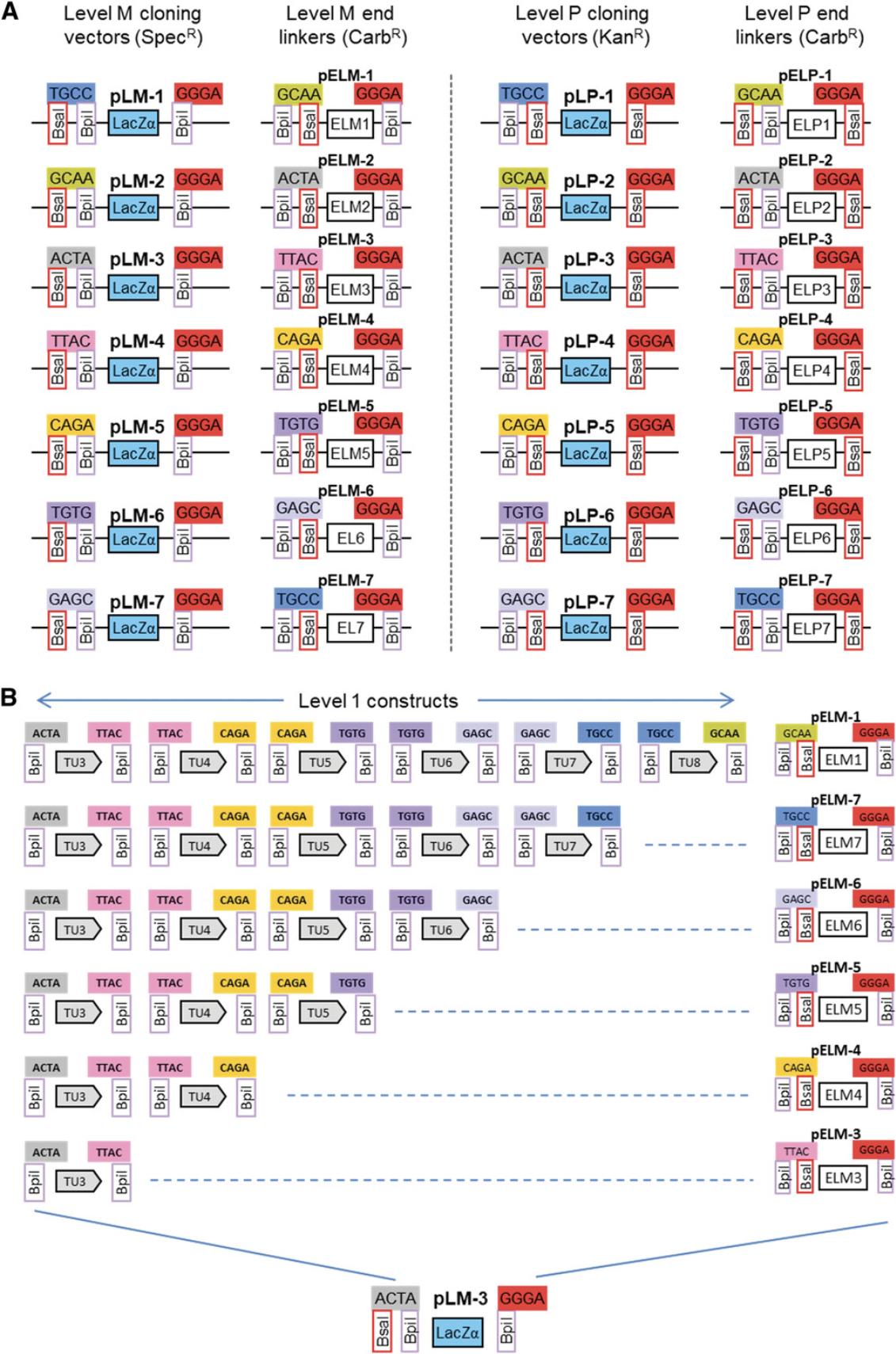
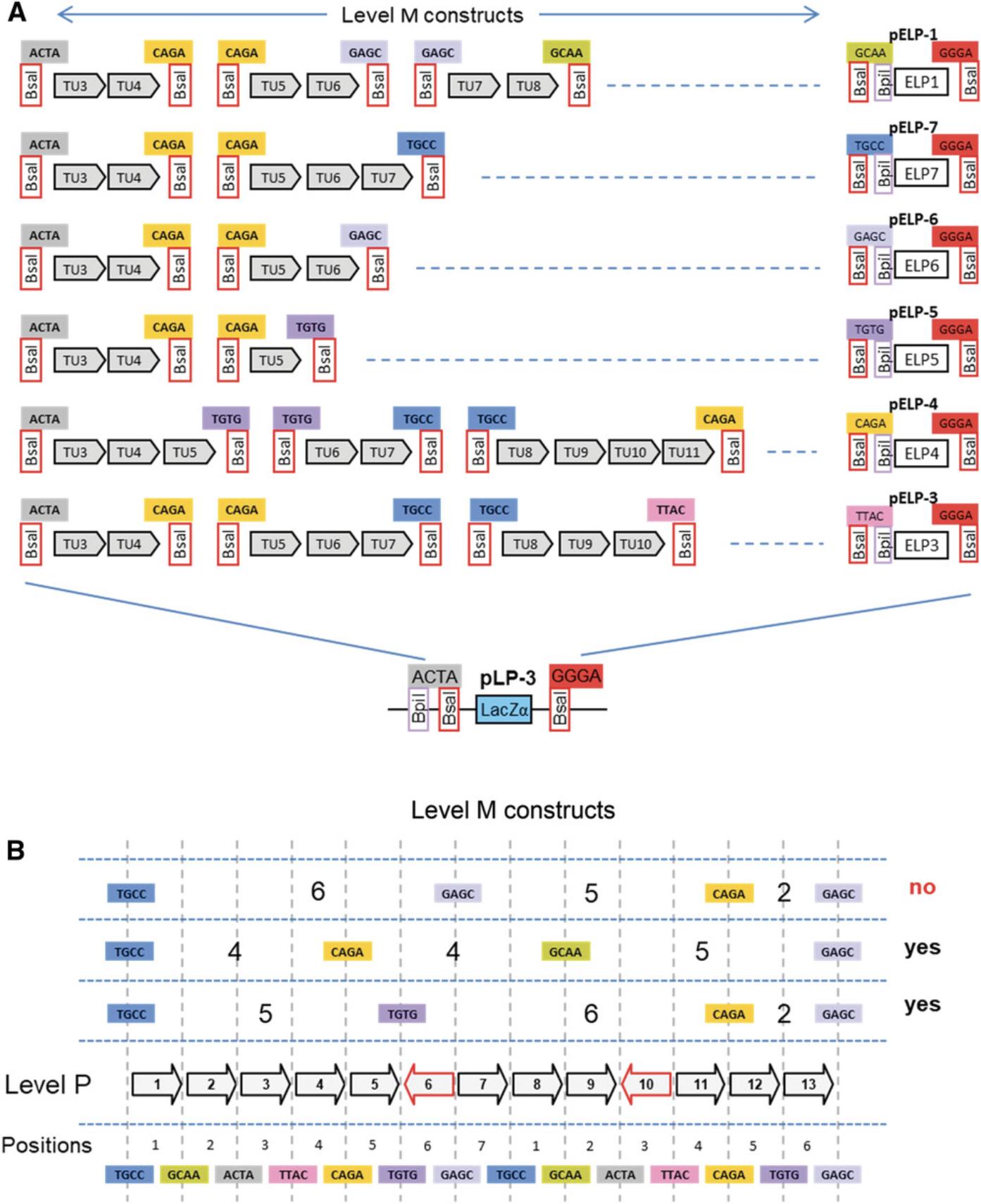
Seven level M vectors and seven level M end-linkers are available (Fig. 15A). The vectors contain two Bpi I restriction sites in opposite orientation for insertion of level 1 modules. The upstream fusion site defined by the first Bpi I site (e.g., ACTA for pLM-3) is compatible with fusion sites flanking the upstream fusion sites of level 1 vectors of the corresponding position (Fig. 15B). The downstream fusion site has of a universal sequence (GGGA). The end of the last transcription unit is fused to the vector using the compatible end-linker (Fig. 15B). This system of vectors and end-linkers provides the possibility to clone up to six transcription units in the same vector, avoiding the need for a considerably larger number of level M vectors for all possible cloning combinations. A maximum of six transcription units can be assembled in a level M construct in one step because the last fusion site of level 1 vector 7 (TGCC) is the same as the first fusion site of the first level 1 vector (Fig. 13A).
Using more genes in this step would lead to incorrect clones, because the same fusion site would be present on different modules. Therefore, cloning than six transcription units requires making at least two level M constructs. For example, to clone thirteen transcription units, at least three level M constructs must be made. In such a case, it is important to plan the cloning strategy carefully, because not all combinations are compatible with level P cloning. For example, one option would be to clone six, five, and two transcription units into vectors pLM-1, pLM-7, and pLM-5, respectively (Fig. 16B). The fusion sites that flank a level M construct are the same as the fusion sites that flank the first and last assembled transcription units. Therefore, fusion sites flanking a level M construct position 1 containing six transcription units are TGCC and GAGC. Fusion site pairs flanking the next two level M constructs of five and two transcription unit are GAGC/CAGA and CAGA/GAGC (Fig. 16B). This combination is not viable, because GAGC would used twice in the level P assembly. Alternative strategies using groups of four, four, and five or five, six, and two would be compatible with level P cloning (Fig. 16B).
In the present example, the four transcription units can easily be assembled in one step in level M vector pLM-1 using the end-linker pELM-4. This would be the final construction step. However, as an example of the use of level P constructs, the four transcription units can be cloned in groups of two in two level Mvectors:
- TU1 and TU2 in vector pLM-1 with end-linker pELM-2 (Fig. 17A)
- TU3 and TU4 in vector pLM-3 with end-linker pELM-4 (Fig. 17B)
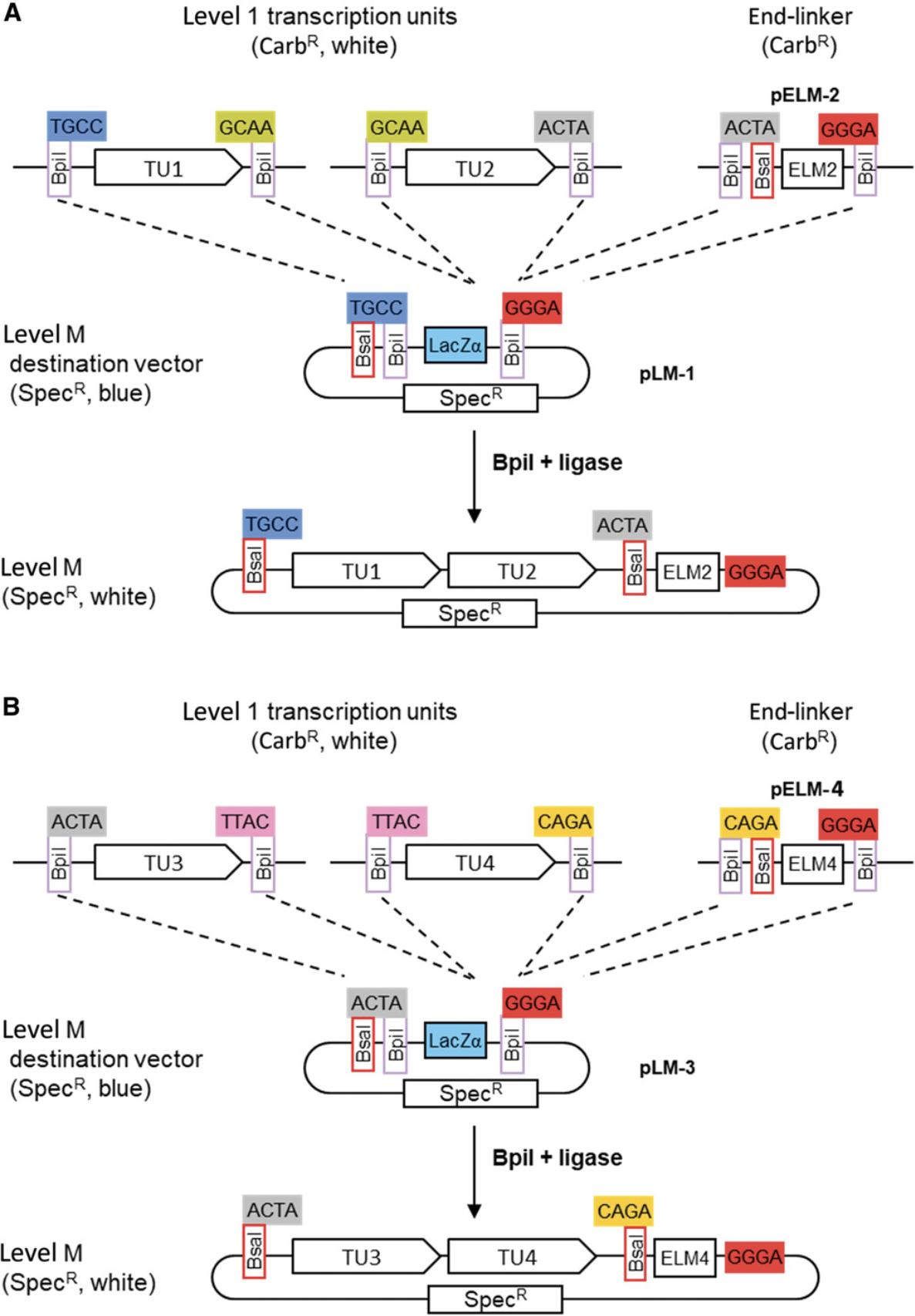
5.Assemble level M constructs.
The two level M constructs are assembled in separate Golden Gate reactions using Bpi I and ligase as described (see Basic Protocol 1). Because level M vectors contain a LacZ fragment for blue-white selection and spectinomycin resistance, cells are grown on spectinomycin-containing X-gal plates and two white colonies are picked for each construct. As assembled transcription units are flanked by Bsa I sites, miniprep DNA is checked with a Bsa I digest.
Assemble larger constructs in level P vectors
6.Select level P constructs.
Seven level P vectors and seven level P end-linkers are available (Fig. 15A). Level P vectors and end-linkers are similar to level M, except that all Bpi I sites and Bsa I sites are reciprocally exchanged, and level P vectors have a kanamycin resistance gene instead of a spectinomycin resistance gene. As with level M constructs, cloning level P constructs requires selection of suitable vectors and end-linkers. In the example, where our current example with two level M constructs containing two transcription units each, vector pLP-1 and end-linker pELP-4 are used (Fig. 18).
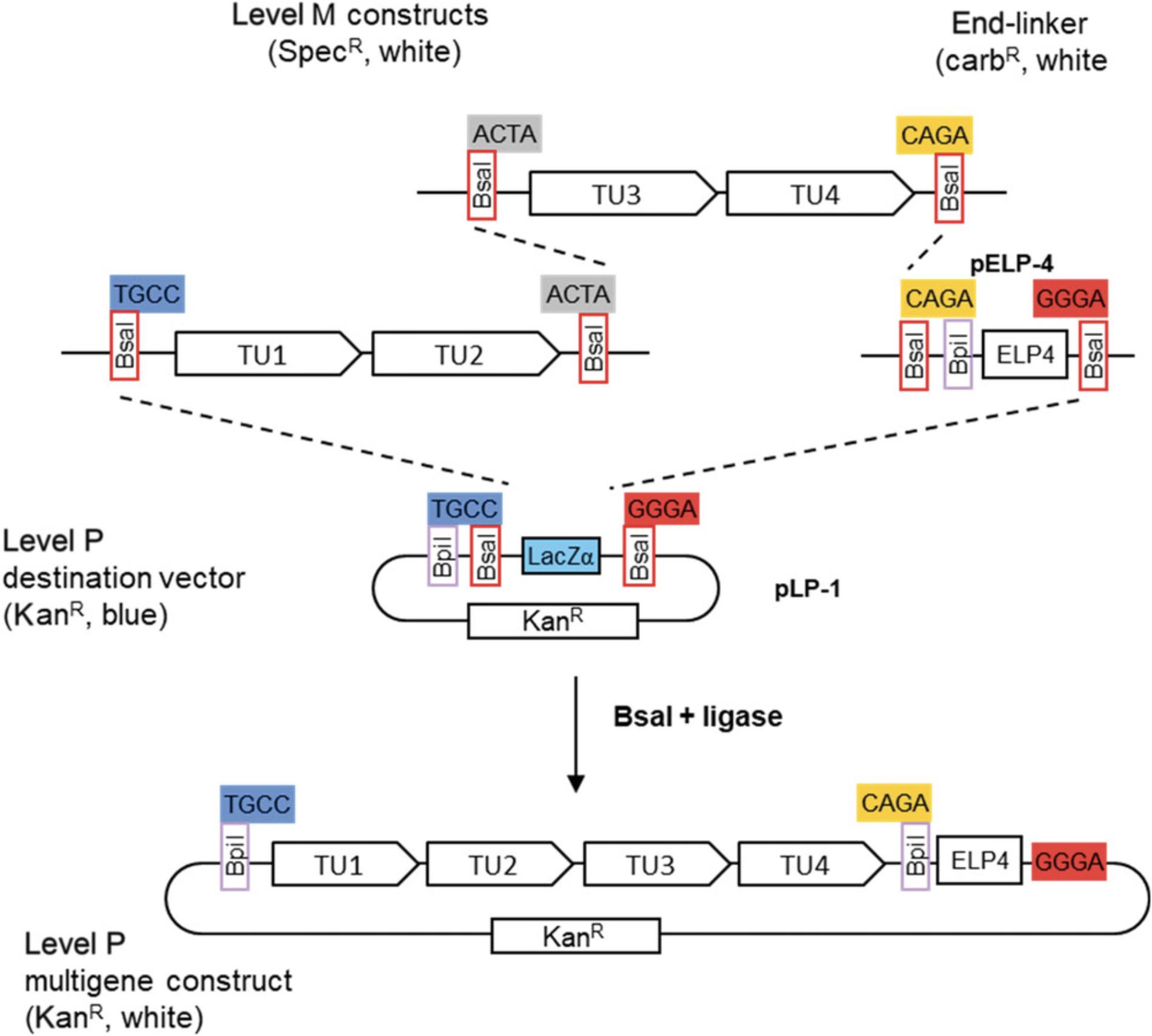
7.Assemble level P constructs.
The reaction is set up with Bsa I and ligase as described (see Basic Protocol 1). Because level P vectors contain a LacZ fragment for blue-white selection and kanamycin resistance, cells are grown on kanamycin-containing X-gal plates and two white colonies are picked. As assembled transcription units are flanked by Bpi I sites, miniprep DNA is checked with a Bpi I digest.
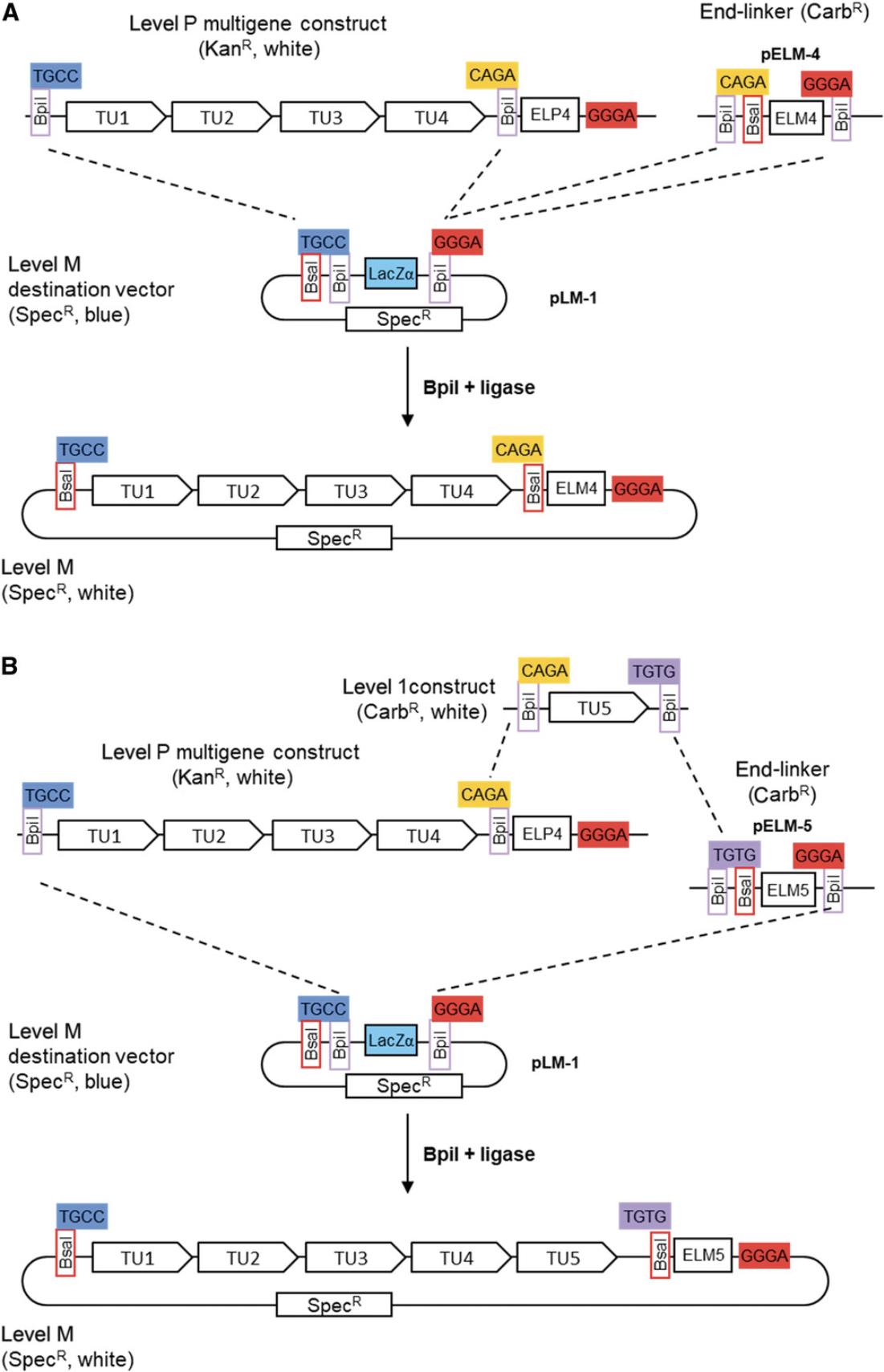
Level P constructs are flanked by two Bpi I restriction sites and thus have a similar structure to level 1 constructs (except for the selectable marker). Like level 1 constructs, several level P constructs can be assembled into level M vectors using Bpi I. As an example, the level P construct above can be subcloned in a level M vector using a suitable level M end-linker (Fig. 19A). Level P and compatible level 1 constructs can also be assembled in a level M vector (Fig. 19B). As discussed for level M constructs, it is important to be sure that all level P vectors are compatible with respect to their fusion sites. The suitability of the fusion sites has to be determined at the beginning of the project, before starting to make level M constructs.
COMMENTARY
Background Information
Type IIS restriction enzymes have been known for a long time, since the discovery of the first type IIS enzyme, Hga I, in 1974, and the identification of the recognition sites of type IIS enzymes Hph I and Mbo II in 1976 (reviewed in Szybalski; Szybalski, Kim, Hasan, & Podhajska, 1991). The first use of a type IIS enzyme, Hga I, was published in 1979 (Moses & Horiuchi, 1979). In this work, six fragments obtained by digestion of bacteriophage F1 DNA with Hga I were re-ligated in vitro to reconstitute infectious phage DNA molecules. This work took advantage of the fact that the overhangs produced by a type IIS enzyme are variable and depend on the specific sequence of a given restriction site. This feature could be used to reassemble the six DNA fragments, as the sequences of all overhangs were different. In 1991, using the same principle, type IIS enzymes were used to assemble a gene lacking introns by PCR-amplifying exon sequences from genomic DNA and ligating the resulting fragments in a recipient vector (Lebedenko, Birikh, Plutalov, & Berlin Yu, 1991). In 1992, type IIS enzymes were used to introduce mutations and even small insert sequences in a plasmid by amplifying the entire plasmid with primers flanked by type IIS enzymes and re-ligating the digested linear fragment (Stemmer & Morris, 1992). In 1996, a DNA insert and recipient vector amplified with primers containing type IIS restriction sites were first digested with the type IIS enzyme and then ligated in the presence of the enzyme (Padgett & Sorge, 1996). This led to a low amount of incorrect constructs, because unwanted ligation products were re-digested by the enzyme. The same year, Rebatchouk et al. proposed a system for modular plasmid construction using two type IIS enzymes (Rebatchouk, Daraselia, & Narita, 1996). With this system, modules were inserted in a recipient plasmid, one a time, in successive cloning steps. This system did not, however, allow seamless assembly of DNA fragments and did not allow assembly of multiple fragments in a single step. A decade later, Fromme & Klingenspor (2007) used a type IIS enzyme in combination with a type II enzyme to transfer a DNA fragment from one vector to another.
Despite this early work, type IIS enzymes were relatively rarely used until recently, with researchers relying mostly on the use of standard type II restriction enzymes for cloning work, or on the Gateway recombination system (Invitrogen), which allows transfer of a DNA fragment from an entry plasmid to a destination vector using the phage lambda recombination system. Golden Gate cloning, first published in 2008 (Engler, Kandzia, & Marillonnet, 2008), combines the use of a type IIS restriction enzyme with a restriction-ligation reaction, but also uses strategies to optimize cloning efficiency, such as donor and recipient vectors with different antibiotic selection markers. The use of a visual marker (or a counter-selectable marker such as ccdB) in the recipient vector also permits the identification of colonies that contain the correct constructs and not the original vector. Finally, to obtain maximal cloning efficiency, the cloning reaction is performed in a thermal cycler by repeated cycles of two successive incubation steps optimized for digestion and ligation (Engler, Gruetzner, Kandzia, & Marillonnet, 2009). This results in a strategy that enables the transfer of a DNA fragment from one vector to the next using a simple one-pot assembly with a very high efficiency. It also enables the transfer and seamless assembly of several fragments from different source plasmids into a recipient vector with very high efficiency. Type IIS enzymes can be used for assembly of PCR products in a restriction-ligation, but the most important benefit of Golden Gate cloning is the ability to transfer and assemble cloned and sequenced DNA fragments from one or several source plasmids to a recipient vector. One application of Golden Gate cloning is the assembly of designer transcription activator-like (TAL) effector and TAL nuclease constructs, which require assembly of 18 DNA repeat modules selected from libraries of modules with different specificities (Cermak et al., 2011; Geissler et al., 2011; Weber, Gruetzner, Werner, Engler, & Marillonnet, 2011). Golden Gate cloning is also frequently used for assembly of CRISPR gRNA expression arrays for site-specific mutagenesis in a variety of organisms (McCarty, Shaw, Ellis, & Ledesma-Amaro, 2019; Vad-Nielsen, Lin, Bolund, Nielsen, & Luo, 2016). Finally, Golden Gate cloning has led to the development of the modular cloning system MoClo (Engler et al., 2014; Kowarschik, Hoehenwarter, Marillonnet, & Trujillo, 2018; Weber, Engler, et al., 2011; Werner et al., 2012) and of several other related modular cloning systems (Agmon et al., 2015; Andreou & Nakayama, 2018; Binder et al., 2014; Larroude et al., 2019; Lee, DeLoache, Cervantes, & Dueber, 2015; Martella, Matjusaitis, Auxillos, Pollard, & Cai, 2017; Moore et al., 2016; Occhialini et al., 2019; Pollak et al., 2019; Prielhofer et al., 2017; Sarrion-Perdigones et al., 2011, 2013).
Critical Parameters and Troubleshooting
A successful Golden Gate cloning reaction normally results in a large number of white colonies and very few blue colonies. Therefore, a transformation that produces a few white colonies and a high blue-to-white colony ratio suggests that something may be wrong with the cloning reaction. If this is the case, screening as many white colonies as possible may not necessarily produce the correct construct. There are several potential reasons for the failure of a Golden Gate cloning reaction, and these are listed below.
An insert or the vector is degraded or in low concentration
One common reason for low cloning efficiency is that one of the modules has degraded. This is not necessarily visible when DNA concentration is evaluated using a Nanodrop, but will be visible on a gel. Thus, the first thing that should be done when a Golden Gate cloning reaction is not working is to run all DNA components on a gel, including all donor plasmids and the vector. They should be run in the same amount as used in the ligation mix, or in a higher amount but with the same relative proportions as used in the ligation. The DNAs can be run uncut, but running DNA digested with the type IIS enzyme used in the ligation can provide additional information. The solution to a degraded plasmid module is simply to prepare a fresh miniprep and repeat the cloning reaction.
A second problem may be that one of the plasmids used in the ligation is present in a much lower (or sometimes higher) amount than expected. In that case, the ligation should be repeated by visually estimating how much is present on the gel and estimating how much more or less should be used for this module.
A third problem is that one of the modules of the cloning reaction is not the correct one. The pattern of the restriction digest on the gel should detect this problem.
Incorrect design of a fusion site
Another possibility for a failed cloning reaction may be a mistake in the design of a fusion site when making a level –1 or 0 construct. To avoid this problem, all selected fusions sites may be checked in an online tool from New England Biolabs for the design of fusion sites (http://ggtools.neb.com/viewset/run.cgi). In contrast with construction of level −1 or 0 modules, there cannot be a mistake in the design of fusion sites for level 1, M, or P constructs, as all fusion sites are already part of the vectors and are standard, and thus do not need to be designed by the user.
Incorrect design of cloning strategy
Another possibility may be a mistake in the design of the cloning strategy, such as incorrect design of a primer used for amplification or incorrect choice of a cloning vector or end-linker. A solution to this problem is to generate all constructs in silico using software for plasmid assembly (vector NTi or Geneious, among others) to make sure that the entire cloning strategy is viable. It is preferable to do this before ordering primers and before starting work at the bench.
Another solution is to use software designed to facilitate the design of primers for Golden Gate cloning. For example, open source software is available (https://msbi.ipb-halle.de/GoldenMutagenesisWeb/) for designing Golden Gate primers and/or introducing site-specific mutations (Pullmann et al., 2019). Software from commercial companies, such as Geneious (Biomatters, Inc.), is also available.
Pipetting errors
Another possibility for a failed cloning reaction may be a pipetting mistake for one of the cloning vectors or inserts. When making a construct containing three transcription units, for instance, three level 1 constructs are cloned using three level 1 vectors for as a first step. If the wrong level 1 vector is added to a reaction (e.g., vector position 3 instead of vector position 2), the cloning of all three level 1 constructs will seem correct, with the exact restriction digest pattern for all three constructs, but the assembly of the level M vectors will fail. The only way to figure out what went wrong is to sequence all three level 1 constructs, making such mistakes very difficult to elucidate. The only solution to avoid such problems is to be extremely careful when pipetting all vectors and modules.
Presence of an unknown type IIS restriction site in amplified sequences
Basic parts can be made from PCR fragments amplified from a sequence cloned in a plasmid. However, more often, PCR fragments are amplified from DNA or cDNA prepared from an organism of choice. The PCR fragments should not contain restriction sites for the type IIS enzyme used for cloning, as primers are designed to remove all of these. Nevertheless, it sometimes happens that an unknown DNA polymorphism present in the host genome (a single-nucleotide substitution is sufficient) may lead to the presence of a restriction site in amplified sequences. This will not be seen when the PCR products are run on a gel, as they will have the expected size. The presence of this site will nevertheless significantly reduce the cloning efficiency, as the correct construct will be re-digested at the end of the restriction-ligation. If few white colonies are present and none contain a correct construct, the same PCR products as those added to the ligation can be digested with the type IIS enzyme and run on a gel. Mutations that are not too close to the primers can be identified this way. Alternatively, the PCR product can be directly sequenced with the same primers used for amplification. This will help identify restriction sites (at least those that are not very close to the sequencing primers). If a restriction site is identified, a new pair of primers can be designed to inactivate it. PCR amplification and cloning is then repeated with the new set of primers. Alternatively, the part can be ordered from a gene synthesis provider.
The gene used as a PCR template is part of multigene family
When a gene is part of a gene family, it sometimes happens that primer pairs designed to amplify the gene (or fragments of the gene) also amplify DNA fragments from several homologs. When a basic part is assembled from several fragments, it may consist of chimeric sequences derived from different homologs. A solution for this problem is to clone all amplified PCR fragments individually in the universal level –1 vector. Sequencing of several level –1 clones for each fragment will identify clones with the correct sequence. The final level 0 module is then assembled from the selected level –1 clones. Alternatively, the part can be ordered from a gene synthesis provider.
Assembly of large constructs
When very large constructs need to be made, preparing miniprep DNA using column-based kits can be problematic, as DNAs >50 kb can be sheared on the column. In this case, miniprep DNA should be prepared using another method, such as the standard alkaline lysis method. Alternatively, the NucleoBond PC from Macherey-Nagel can be used to make miniprep DNA for constructs up to 300 kb.
In addition, large constructs can become very unstable in E. coli (and probably other hosts) if they contain repeats. Therefore, it is useful to avoid any repeat sequence when planning large constructs. Another element that may help the stability of large constructs is the use of origins of replication for low copy or even single copy number in E. coli and, in some cases, the final host organism.
Ordering genes from a gene synthesis company
Ordering genes or gene fragments from a gene synthesis company is an alternative to cloning of level 0 basic parts from PCR products. This solution will become increasingly attractive as the cost of gene synthesis continues to decrease. Using gene synthesis also has other benefits. For example, it allows the preparation of parts for which no DNA template is available for PCR amplification, and allows codon optimization and removal of restriction sites at the same time. When ordering DNA from a gene synthesis company, it is important to consider which vector will be used by the company. The plasmid backbone should have an antibiotic resistance gene different from the vector in which the part is designed to be assembled. When ordering linear DNA fragments rather than a sequence cloned in a plasmid, the gene fragment should be designed with restriction sites directly compatible for Golden Gate assembly with the vector that will be used, such as the universal level –1 or level 0 cloning vectors for the MoClo system.
Understanding the Results
Determining whether the correct constructs are obtained can be achieved by DNA sequencing for basic parts or by DNA fingerprinting for all other constructs. Large constructs require careful analysis, as the restriction pattern of digested DNA on a gel can be complex and the copy number of the plasmid is usually low.
An important element in determining whether a cloning reaction has worked as planned is observing the blue-to-white colony ratio. A high blue-to-white colony ratio is a sign that something may have gone wrong. In addition, plasmids can be unstable, in particular if they contain repeats, and some sequences may be toxic in E. coli. The size of the colonies obtained is also a meaningful criterion, as large colonies may contain incorrect clones. Therefore, when a cloning experiment results in both small (slowly growing) and large (quickly growing) colonies, it is important to analyze some of the small colonies, as these may contain the correct constructs, especially when making large constructs.
Time Considerations
Making constructs that require PCR: Cloning level 0 modules (Basic Protocol 4) or level –1 sub-parts (first part of Alternate Protocol) or domesticating a vector (Basic Protocol 2)
In advance, 2-3 days should be allowed for ordering and receiving primers. From receipt of primers, a total of 3-4 days are needed to obtain clones and send them for sequencing. Days 1-2 are needed for PCR amplification, gel electrophoresis, and column purification; setting up and performing the cloning reaction; and transforming the mixture into E. coli. Colonies are picked the next day (day 2 or 3) and grown overnight. On day 3 or 4, miniprep DNA is made, analyzed, by digestion and electrophoresis, and sent for sequencing. It should take 1-3 days to receive sequencing results.
Making constructs that do not require PCR: Cloning level 0 constructs from sequenced level –1 constructs (part of Alternate Protocol), cloning level 1 constructs (Basic Protocol 5), or cloning level M and P constructs (Basic Protocol 6)
Clones can be obtained in 3 days. On day 1, the cloning reactions are set up and carried out and products are transformed into E. coli. On day 2, colonies are picked and inoculated for overnight culture. On day 3, miniprep DNA is made, digested, and run on an agarose gel. For constructs that require the assembly of large numbers of parts, the assembly reaction is done overnight. Therefore, obtaining the construct requires one more day (total of 4 days).
Acknowledgments
Preparation of this manuscript was funded by the Leibniz Institute of Plant Biochemistry, Germany.
Literature Cited
- Agmon, N., Mitchell, L. A., Cai, Y., Ikushima, S., Chuang, J., Zheng, A., … Boeke, J. D. (2015). Yeast Golden Gate (yGG) for the efficient assembly of S. cerevisiae transcription units. ACS Synthetic Biology , 4(7), 853–859. doi: 10.1021/sb500372z.
- Andreou, A. I., & Nakayama, N. (2018). Mobius assembly: A versatile Golden-Gate framework towards universal DNA assembly. PLoS One , 13(1), e0189892. doi: 10.1371/journal.pone.0189892.
- Binder, A., Lambert, J., Morbitzer, R., Popp, C., Ott, T., Lahaye, T., & Parniske, M. (2014). A modular plasmid assembly kit for multigene expression, gene silencing and silencing rescue in plants. PLoS One , 9(2), e88218. doi: 10.1371/journal.pone.0088218.
- Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., … Voytas, D. F. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research , 39(12), e82. doi: 10.1093/nar/gkr218.
- Crozet, P., Navarro, F. J., Willmund, F., Mehrshahi, P., Bakowski, K., Lauersen, K. J., … Lemaire, S. D. (2018). Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synthetic Biology , 7(9), 2074–2086. doi: 10.1021/acssynbio.8b00251.
- Elbing, K. L., & Brent, R. (2018). Recipes and tools for culture of Escherichia coli. Current Protocols in Molecular Biology , 125, e83. doi: 10.1002/cpmb.83.
- Engler, C., Gruetzner, R., Kandzia, R., & Marillonnet, S. (2009). Golden Gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One , 4(5), e5553. doi: 10.1371/journal.pone.0005553.
- Engler, C., Kandzia, R., & Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One , 3(11), e3647. doi: 10.1371/journal.pone.0003647.
- Engler, C., Youles, M., Gruetzner, R., Ehnert, T. M., Werner, S., Jones, J. D., … Marillonnet, S. (2014). A Golden Gate modular cloning toolbox for plants. ACS Synthetic Biology , 3(11), 839–843. doi: 10.1021/sb4001504.
- Fromme, T., & Klingenspor, M. (2007). Rapid single step subcloning procedure by combined action of type II and type IIs endonucleases with ligase. Journal of Biological Engineering , 1, 7. doi: 10.1186/1754-1611-1-7.
- Geissler, R., Scholze, H., Hahn, S., Streubel, J., Bonas, U., Behrens, S. E., & Boch, J. (2011). Transcriptional activators of human genes with programmable DNA-specificity. PLoS One , 6(5), e19509. doi: 10.1371/journal.pone.0019509.
- Kowarschik, K., Hoehenwarter, W., Marillonnet, S., & Trujillo, M. (2018). UbiGate: A synthetic biology toolbox to analyse ubiquitination. The New Phytologist , 217(4), 1749–1763. doi: 10.1111/nph.14900.
- Larroude, M., Park, Y. K., Soudier, P., Kubiak, M., Nicaud, J. M., & Rossignol, T. (2019). A modular Golden Gate toolkit for Yarrowia lipolytica synthetic biology. Microbial Biotechnology , 12(6), 1249–1259. doi: 10.1111/1751-7915.13427.
- Lebedenko, E. N., Birikh, K. R., Plutalov, O. V., & Berlin Yu, A. (1991). Method of artificial DNA splicing by directed ligation (SDL). Nucleic Acids Research , 19(24), 6757–6761. doi: 10.1093/nar/19.24.6757.
- Lee, M. E., DeLoache, W. C., Cervantes, B., & Dueber, J. E. (2015). A highly characterized yeast toolkit for modular, multipart assembly. ACS Synthetic Biology , 4(9), 975–986. doi: 10.1021/sb500366v.
- Martella, A., Matjusaitis, M., Auxillos, J., Pollard, S. M., & Cai, Y. (2017). EMMA: An extensible mammalian modular assembly toolkit for the rapid design and production of diverse expression vectors. ACS Synthetic Biology , 6(7), 1380–1392. doi: 10.1021/acssynbio.7b00016.
- McCarty, N. S., Shaw, W. M., Ellis, T., & Ledesma-Amaro, R. (2019). Rapid assembly of gRNA arrays via modular cloning in yeast. ACS Synthetic Biology , 8(4), 906–910. doi: 10.1021/acssynbio.9b00041.
- Moore, D. (1996). Commonly used reagents and equipment. Current Protocols in Molecular Biology , 35, A.2.1–A.2.8. doi: 10.1002/0471142727.mba02s35.
- Moore, S. J., Lai, H. E., Kelwick, R. J., Chee, S. M., Bell, D. J., Polizzi, K. M., & Freemont, P. S. (2016). EcoFlex: A multifunctional MoClo kit for E. coli synthetic biology. ACS Synthetic Biology , 5(10), 1059–1069. doi: 10.1021/acssynbio.6b00031.
- Moses, P. B., & Horiuchi, K. (1979). Specific recombination in vitro promoted by the restriction endonuclease Hga I. Journal of Molecular Biology , 135(2), 517–524. doi: 10.1016/0022-2836(79)90452-2.
- Occhialini, A., Piatek, A. A., Pfotenhauer, A. C., Frazier, T. P., Stewart, C. N., Jr., & Lenaghan, S. C. (2019). MoChlo: A versatile, modular cloning toolbox for chloroplast biotechnology. Plant Physiology , 179(3), 943–957. doi: 10.1104/pp.18.01220.
- Padgett, K. A., & Sorge, J. A. (1996). Creating seamless junctions independent of restriction sites in PCR cloning. Gene , 168(1), 31–35. doi: 10.1016/0378-1119(95)00731-8.
- Pollak, B., Cerda, A., Delmans, M., Alamos, S., Moyano, T., West, A., … Haseloff, J. (2019). Loop assembly: A simple and open system for recursive fabrication of DNA circuits. The New Phytologist , 222(1), 628–640. doi: 10.1111/nph.15625.
- Potapov, V., Ong, J. L., Kucera, R. B., Langhorst, B. W., Bilotti, K., Pryor, J. M., … Lohman, G. J. S. (2018). Comprehensive profiling of four base overhang ligation fidelity by T4 DNA ligase and application to DNA assembly. ACS Synthetic Biology , 7(11), 2665–2674. doi: 10.1021/acssynbio.8b00333.
- Prielhofer, R., Barrero, J. J., Steuer, S., Gassler, T., Zahrl, R., Baumann, K., … Marx, H. (2017). GoldenPiCS: A Golden Gate–derived modular cloning system for applied synthetic biology in the yeast Pichia pastoris. BMC Systems Biology , 11(1), 123. doi: 10.1186/s12918-017-0492-3.
- Pullmann, P., Ulpinnis, C., Marillonnet, S., Gruetzner, R., Neumann, S., & Weissenborn, M. J. (2019). Golden mutagenesis: An efficient multi-site-saturation mutagenesis approach by Golden Gate cloning with automated primer design. Scientific Reports , 9(1), 10932. doi: 10.1038/s41598-019-47376-1.
- Rebatchouk, D., Daraselia, N., & Narita, J. O. (1996). NOMAD: A versatile strategy for in vitro DNA manipulation applied to promoter analysis and vector design. Proceedings of the National Academy of Sciences of the United States of America , 93(20), 10891–10896. doi: 10.1073/pnas.93.20.10891.
- Sarrion-Perdigones, A., Falconi, E. E., Zandalinas, S. I., Juarez, P., Fernandez-del-Carmen, A., Granell, A., & Orzaez, D. (2011). GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS One , 6(7), e21622. doi: 10.1371/journal.pone.0021622.
- Sarrion-Perdigones, A., Vazquez-Vilar, M., Palaci, J., Castelijns, B., Forment, J., Ziarsolo, P., … Orzaez, D. (2013). GoldenBraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiology , 162(3), 1618–1631. doi: 10.1104/pp.113.217661.
- Seidman, C. E., Struhl, K., Sheen, J., & Jessen, T. (1997). Introduction of plasmid DNA into cells. Current Protocols in Molecular Biology , 37, 1.8.1–1.8.10. doi: 10.1002/0471142727.mb0108s37.
- Stemmer, W. P., & Morris, S. K. (1992). Enzymatic inverse PCR: A restriction site independent, single-fragment method for high-efficiency, site-directed mutagenesis. Biotechniques , 13(2), 214–220.
- Struhl, K. (1991). Subcloning of DNA fragments. Current Protocols in Molecular Biology , 13, 3.16.1–3.16.2. doi: 10.1002/0471142727.mb0316s13.
- Szybalski, W., Kim, S. C., Hasan, N., & Podhajska, A. J. (1991). Class-IIS restriction enzymes—A review. Gene , 100, 13–26. doi: 10.1016/0378-1119(91)90345-c.
- Vad-Nielsen, J., Lin, L., Bolund, L., Nielsen, A. L., & Luo, Y. (2016). Golden Gate assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes. Cellular and Molecular Life Sciences: CMLS , 73(22), 4315–4325. doi: 10.1007/s00018-016-2271-5.
- Vasudevan, R., Gale, G. A. R., Schiavon, A. A., Puzorjov, A., Malin, J., Gillespie, M. D., … McCormick, A. J. (2019). CyanoGate: A modular cloning suite for engineering cyanobacteria based on the plant MoClo syntax. Plant Physiology , 180(1), 39–55. doi: 10.1104/pp.18.01401.
- Weber, E., Engler, C., Gruetzner, R., Werner, S., & Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One , 6(2), e16765. doi: 10.1371/journal.pone.0016765.
- Weber, E., Gruetzner, R., Werner, S., Engler, C., & Marillonnet, S. (2011). Assembly of designer TAL effectors by Golden Gate cloning. PLoS One , 6(5), e19722. doi: 10.1371/journal.pone.0019722.
- Werner, S., Engler, C., Weber, E., Gruetzner, R., & Marillonnet, S. (2012). Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioengineered Bugs , 3(1), 38–43. doi: 10.1371/journal.pone.001676510.4161/bbug.3.1.18223.
Citing Literature
Number of times cited according to CrossRef: 60
- Cody Kamoku, Cheyanna Cooper, Ashley Straub, Nathan Miller, David R. Nielsen, Delivery of Novel Replicating Vectors to Synechococcus sp. PCC 7002 Via Natural Transformation of Plasmid Multimers, Synthetic Biology and Engineering, 10.35534/sbe.2024.10014, 2 , 3, (10014-10014), (2024).
- Vamsi Krishna Gali, Kang Lan Tee, Tuck Seng Wong, Crafting Genetic Diversity: Unlocking the Potential of Protein Evolution, SynBio, 10.3390/synbio2020009, 2 , 2, (142-173), (2024).
- Hyo Lee, Sangkyu Park, Saet Buyl Lee, Jaeeun Song, Tae-Hwan Kim, Beom-Gi Kim, Tailored biosynthesis of diosmin through reconstitution of the flavonoid pathway in Nicotiana benthamiana, Frontiers in Plant Science, 10.3389/fpls.2024.1464877, 15 , (2024).
- Nan-Sun Kim, Kyeong-Ryeol Lee, Jihyea Lee, Eui-Joon Kil, Juho Lee, Seon-Kyeong Lee, High production of recombinant protein using geminivirus-based deconstructed vectors in Nicotiana benthamiana, Frontiers in Plant Science, 10.3389/fpls.2024.1407240, 15 , (2024).
- Luke J. Webster, Denys Villa-Gomez, Reuben Brown, William Clarke, Peer M. Schenk, A synthetic biology approach for the treatment of pollutants with microalgae, Frontiers in Bioengineering and Biotechnology, 10.3389/fbioe.2024.1379301, 12 , (2024).
- Cintia H. D. Sagawa, Geoffrey Thomson, Benoit Mermaz, Corina Vernon, Siqi Liu, Yannick Jacob, Vivian F. Irish, An efficient multiplex approach to CRISPR/Cas9 gene editing in citrus, Plant Methods, 10.1186/s13007-024-01274-4, 20 , 1, (2024).
- Carolina Cano-Prieto, Agustina Undabarrena, Ana Calheiros de Carvalho, Jay D. Keasling, Pablo Cruz-Morales, Triumphs and Challenges of Natural Product Discovery in the Postgenomic Era, Annual Review of Biochemistry, 10.1146/annurev-biochem-032620-104731, 93 , 1, (411-445), (2024).
- Patryk Strzelecki, Nicolas Joly, Pascal Hébraud, Elise Hoffmann, Grzegorz M Cech, Anna Kloska, Florent Busi, Wilfried Grange, Enhanced Golden Gate Assembly: evaluating overhang strength for improved ligation efficiency, Nucleic Acids Research, 10.1093/nar/gkae809, (2024).
- Bhanu Prakash Potlapalli, Jörg Fuchs, Twan Rutten, Armin Meister, Andreas Houben, The potential of ALFA-tag and tyramide-based fluorescence signal amplification to expand the CRISPR-based DNA imaging toolkit, Journal of Experimental Botany, 10.1093/jxb/erae341, (2024).
- Sunny Ahmar, Babar Usman, Goetz Hensel, Ki-Hong Jung, Damian Gruszka, CRISPR enables sustainable cereal production for a greener future, Trends in Plant Science, 10.1016/j.tplants.2023.10.016, 29 , 2, (179-195), (2024).
- Matteo Gionfriddo, Timothy Rhodes, Spencer M. Whitney, Perspectives on improving crop Rubisco by directed evolution, Seminars in Cell & Developmental Biology, 10.1016/j.semcdb.2023.04.003, 155 , (37-47), (2024).
- Patryk Strzelecki, Anastasiia Shpiruk, Grzegorz M. Cech, Anna Kloska, Pascal Hébraud, Nicolas Beyer, Florent Busi, Wilfried Grange, Robust, high-yield, rapid fabrication of DNA constructs for Magnetic Tweezers, Biochemical and Biophysical Research Communications, 10.1016/j.bbrc.2024.150370, 731 , (150370), (2024).
- Jiazhang Lian, DNA assembly techniques for the reconstitution of plant natural product biosynthetic pathways in Saccharomyces cerevisiae, Engineering Biology for Microbial Biosynthesis of Plant-Derived Bioactive Compounds, 10.1016/B978-0-443-15558-1.00015-1, (33-46), (2024).
- Aparna Tiwari, Siddhesh B. Ghag, Base editing and prime editing in horticulture crops: Potential applications, challenges, and prospects, CRISPRized Horticulture Crops, 10.1016/B978-0-443-13229-2.00002-8, (105-126), (2024).
- Gregory J. S. Lohman, Selection of Fusion-Site Overhang Sets for High-Fidelity and High-Complexity Golden Gate Assembly, Golden Gate Cloning, 10.1007/978-1-0716-4220-7_3, (41-60), (2024).
- Jesús Laborda-Mansilla, Eva García-Ruiz, Advancements in Golden Gate Cloning: A Comprehensive Review, Golden Gate Cloning, 10.1007/978-1-0716-4220-7_27, (481-500), (2024).
- Peter Emelin, Sarah Abdul-Mawla, Felix Willmund, Golden Gate Cloning for the Standardized Assembly of Gene Elements with Modular Cloning in Chlamydomonas, Golden Gate Cloning, 10.1007/978-1-0716-4220-7_25, (451-465), (2024).
- Gregory Foran, Ryan D Hallam, Marvel Megaly, Anel Turgambayeva, Aleksandar Necakov, Playback Cloning: Simple, Reversible, Cost-effective Cloning for the Combinatorial Assembly of Complex Expression Constructs, BioTechniques, 10.2144/btn-2023-0042, 75 , 4, (168-178), (2023).
- Asma Rafique, Amber Afroz, Nadia Zeeshan, Umer Rashid, Muhammad Azmat Ullah Khan, Muhammad Irfan, Waheed Chatha, Muhammad Ramzan Khan, Nazia Rehman, Production of Sitobion avenae-resistant Triticum aestivum cvs using laccase as RNAi target and its systemic movement in wheat post dsRNA spray, PLOS ONE, 10.1371/journal.pone.0284888, 18 , 5, (e0284888), (2023).
- Sofia Markakiou, Ana Rute Neves, Ahmad A. Zeidan, Paula Gaspar, Development of a Tetracycline-Inducible System for Conditional Gene Expression in Lactococcus lactis and Streptococcus thermophilus, Microbiology Spectrum, 10.1128/spectrum.00668-23, 11 , 3, (2023).
- Norman Adlung, Silvan Scheller, Application of the Fluorescence-Activating and Absorption-Shifting Tag (FAST) for Flow Cytometry in Methanogenic Archaea, Applied and Environmental Microbiology, 10.1128/aem.01786-22, 89 , 4, (2023).
- Yongyu Huang, Roop Kamal, Nandhakumar Shanmugaraj, Twan Rutten, Venkatasubbu Thirulogachandar, Shuangshuang Zhao, Iris Hoffie, Goetz Hensel, Jeyaraman Rajaraman, Yudelsy Antonia Tandron Moya, Mohammad-Reza Hajirezaei, Axel Himmelbach, Naser Poursarebani, Udda Lundqvist, Jochen Kumlehn, Nils Stein, Nicolaus von Wirén, Martin Mascher, Michael Melzer, Thorsten Schnurbusch, A molecular framework for grain number determination in barley, Science Advances, 10.1126/sciadv.add0324, 9 , 9, (2023).
- Camilo Calvache, Marta Vazquez‐Vilar, Elena Moreno‐Giménez, Diego Orzaez, A quantitative autonomous bioluminescence reporter system with a wide dynamic range for Plant Synthetic Biology, Plant Biotechnology Journal, 10.1111/pbi.14146, 22 , 1, (37-47), (2023).
- Bangmian Du, Mengjuan Sun, Wenyang Hui, Chengjia Xie, Xian Xu, Recent Advances on Key Enzymes of Microbial Origin in the Lycopene Biosynthesis Pathway, Journal of Agricultural and Food Chemistry, 10.1021/acs.jafc.3c03942, 71 , 35, (12927-12942), (2023).
- Maliha Tasnim, T. Jacob Selph, Jason Olcott, Jonathon T. Hill, The type IIS restriction enzyme MmeI can cut across a double-strand break, Molecular Biology Reports, 10.1007/s11033-023-08375-8, 50 , 6, (5495-5499), (2023).
- Tom Lawrenson, Mark Youles, Monika Chhetry, Martha Clarke, Wendy Harwood, Penny Hundleby, Efficient Targeted Mutagenesis in Brassica Crops Using CRISPR/Cas Systems, Plant Genome Engineering, 10.1007/978-1-0716-3131-7_16, (253-271), (2023).
- Andrew P. Sikkema, S. Kasra Tabatabaei, Yan‐Jiun Lee, Sean Lund, Gregory J. S. Lohman, High‐Complexity One‐Pot Golden Gate Assembly, Current Protocols, 10.1002/cpz1.882, 3 , 9, (2023).
- Yanglei Huang, Ziling Ye, Xiukun Wan, Ge Yao, Jingyu Duan, Jiajia Liu, Mingdong Yao, Xiang Sun, Zixin Deng, Kun Shen, Hui Jiang, Tiangang Liu, Systematic Mining and Evaluation of the Sesquiterpene Skeletons as High Energy Aviation Fuel Molecules, Advanced Science, 10.1002/advs.202300889, 10 , 23, (2023).
- Shawn P Shortill, Mia S Frier, Ponthakorn Wongsangaroonsri, Michael Davey, Elizabeth Conibear, The VINE complex is an endosomal VPS9-domain GEF and SNX-BAR coat, eLife, 10.7554/eLife.77035, 11 , (2022).
- Ryan R. Cochrane, Arina Shrestha, Mariana M. Severo de Almeida, Michelle Agyare-Tabbi, Stephanie L. Brumwell, Samir Hamadache, Jordyn S. Meaney, Daniel P. Nucifora, Henry Heng Say, Jehoshua Sharma, Maximillian P. M. Soltysiak, Cheryl Tong, Katherine Van Belois, Emma J. L. Walker, Marc-André Lachance, Gregory B. Gloor, David R. Edgell, Rebecca S. Shapiro, Bogumil J. Karas, Superior Conjugative Plasmids Delivered by Bacteria to Diverse Fungi, BioDesign Research, 10.34133/2022/9802168, 2022 , (2022).
- M. Moniruzzaman, Yun Zhong, Zhifeng Huang, Guangyan Zhong, Having a Same Type IIS Enzyme’s Restriction Site on Guide RNA Sequence Does Not Affect Golden Gate (GG) Cloning and Subsequent CRISPR/Cas Mutagenesis, International Journal of Molecular Sciences, 10.3390/ijms23094889, 23 , 9, (4889), (2022).
- Taehoon Kim, Fábio Ometto Dias, Agustina Gentile, Marcelo Menossi, Kevin Begcy, ScRpb4, Encoding an RNA Polymerase Subunit from Sugarcane, Is Ubiquitously Expressed and Resilient to Changes in Response to Stress Conditions, Agriculture, 10.3390/agriculture12010081, 12 , 1, (81), (2022).
- Soyoung Park, Vimalraj Mani, Jin A. Kim, Soo In Lee, Kijong Lee, Combinatorial transient gene expression strategies to enhance terpenoid production in plants, Frontiers in Plant Science, 10.3389/fpls.2022.1034893, 13 , (2022).
- Yuwei Mao, Ella Catherall, Aranzazú Díaz-Ramos, George R L Greiff, Stavros Azinas, Laura Gunn, Alistair J McCormick, The small subunit of Rubisco and its potential as an engineering target, Journal of Experimental Botany, 10.1093/jxb/erac309, 74 , 2, (543-561), (2022).
- Joaquín Gutiérrez Mena, Sant Kumar, Mustafa Khammash, Dynamic cybergenetic control of bacterial co-culture composition via optogenetic feedback, Nature Communications, 10.1038/s41467-022-32392-z, 13 , 1, (2022).
- Jasmine E. Bird, Jon Marles-Wright, Andrea Giachino, A User’s Guide to Golden Gate Cloning Methods and Standards, ACS Synthetic Biology, 10.1021/acssynbio.2c00355, 11 , 11, (3551-3563), (2022).
- Jonas De Saeger, Mattias Vermeersch, Christophe Gaillochet, Thomas B. Jacobs, Simple and Efficient Modification of Golden Gate Design Standards and Parts Using Oligo Stitching, ACS Synthetic Biology, 10.1021/acssynbio.2c00072, 11 , 6, (2214-2220), (2022).
- Kairen Tian, Xia Hong, Manman Guo, Yanni Li, Hao Wu, Qinggele Caiyin, Jianjun Qiao, Development of Base Editors for Simultaneously Editing Multiple Loci in Lactococcus lactis , ACS Synthetic Biology, 10.1021/acssynbio.1c00561, 11 , 11, (3644-3656), (2022).
- John M. Pryor, Vladimir Potapov, Katharina Bilotti, Nilisha Pokhrel, Gregory J. S. Lohman, Rapid 40 kb Genome Construction from 52 Parts through Data-optimized Assembly Design, ACS Synthetic Biology, 10.1021/acssynbio.1c00525, 11 , 6, (2036-2042), (2022).
- Koray Malcı, Emma Watts, Tania Michelle Roberts, Jamie Yam Auxillos, Behnaz Nowrouzi, Heloísa Oss Boll, Cibele Zolnier Sousa do Nascimento, Andreas Andreou, Peter Vegh, Sophie Donovan, Rennos Fragkoudis, Sven Panke, Edward Wallace, Alistair Elfick, Leonardo Rios-Solis, Standardization of Synthetic Biology Tools and Assembly Methods for Saccharomyces cerevisiae and Emerging Yeast Species , ACS Synthetic Biology, 10.1021/acssynbio.1c00442, 11 , 8, (2527-2547), (2022).
- Fabio Pasin, Assembly of plant virus agroinfectious clones using biological material or DNA synthesis, STAR Protocols, 10.1016/j.xpro.2022.101716, 3 , 4, (101716), (2022).
- Michael Cohen, David E. Randolph, Maria E. Lozano, Paul W. Anderson, John Crissman, Franz J. Triana, Thomas Cujec, A fully automated high-throughput plasmid purification workstation for the generation of mammalian cell expression-quality DNA, SLAS Technology, 10.1016/j.slast.2022.01.005, (2022).
- Zihe Liu, Junyang Wang, Jens Nielsen, Yeast synthetic biology advances biofuel production, Current Opinion in Microbiology, 10.1016/j.mib.2021.10.010, 65 , (33-39), (2022).
- Reena Sharma, Scott C. Lenaghan, Duckweed: a potential phytosensor for heavy metals, Plant Cell Reports, 10.1007/s00299-022-02913-7, 41 , 12, (2231-2243), (2022).
- Samuel Mortensen, Lauren F. Cole, Diana Bernal-Franco, Suphinya Sathitloetsakun, Erin J. Cram, Carolyn W. T. Lee-Parsons, EASI Transformation Protocol: An Agrobacterium-Mediated Transient Transformation Protocol for Catharanthus roseus Seedlings, Catharanthus roseus, 10.1007/978-1-0716-2349-7_18, (249-262), (2022).
- Beatriz González, Marta Vazquez-Vilar, Javier Sánchez-Vicente, Diego Orzáez, Optimization of Vectors and Targeting Strategies Including GoldenBraid and Genome Editing Tools: GoldenBraid Assembly of Multiplex CRISPR/Cas12a Guide RNAs for Gene Editing in Nicotiana benthamiana, Recombinant Proteins in Plants, 10.1007/978-1-0716-2241-4_12, (193-214), (2022).
- Lee Ling Tan, Elena Heng, Nadiah Zulkarnain, Wan-Chi Hsiao, Fong Tian Wong, Mingzi M. Zhang, CRISPR/Cas-Mediated Genome Editing of Streptomyces, Recombineering, 10.1007/978-1-0716-2233-9_14, (207-225), (2022).
- Sheng-An Yang, Jose L. Salazar, David Li-Kroeger, Shinya Yamamoto, Functional Studies of Genetic Variants Associated with Human Diseases in Notch Signaling-Related Genes Using Drosophila, Notch Signaling Research, 10.1007/978-1-0716-2201-8_19, (235-276), (2022).
- Verónica Aragonés, Flavio Aliaga, Fabio Pasin, José‐Antonio Daròs, Simplifying plant gene silencing and genome editing logistics by a one‐Agrobacterium system for simultaneous delivery of multipartite virus vectors, Biotechnology Journal, 10.1002/biot.202100504, 17 , 7, (2022).
- Ramona Grützner, Ramona Schubert, Claudia Horn, Changqing Yang, Thomas Vogt, Sylvestre Marillonnet, Engineering Betalain Biosynthesis in Tomato for High Level Betanin Production in Fruits, Frontiers in Plant Science, 10.3389/fpls.2021.682443, 12 , (2021).
- Wenfang Wang, Guosong Zheng, Yinhua Lu, Recent Advances in Strategies for the Cloning of Natural Product Biosynthetic Gene Clusters, Frontiers in Bioengineering and Biotechnology, 10.3389/fbioe.2021.692797, 9 , (2021).
- Suyu Jiang, Marie-Françoise Jardinaud, Jinpeng Gao, Yann Pecrix, Jiangqi Wen, Kirankumar Mysore, Ping Xu, Carmen Sanchez-Canizares, Yiting Ruan, Qiujiu Li, Meijun Zhu, Fuyu Li, Ertao Wang, Phillip S. Poole, Pascal Gamas, Jeremy D. Murray, NIN-like protein transcription factors regulate leghemoglobin genes in legume nodules, Science, 10.1126/science.abg5945, 374 , 6567, (625-628), (2021).
- Ramona Grützner, Patrick Martin, Claudia Horn, Samuel Mortensen, Erin J. Cram, Carolyn W.T. Lee-Parsons, Johannes Stuttmann, Sylvestre Marillonnet, High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns, Plant Communications, 10.1016/j.xplc.2020.100135, 2 , 2, (100135), (2021).
- Subha Sankar Paul, Heykel Trabelsi, Yazen Yaseen, Upasana Basu, Hiyam Adil Altaii, Debarun Dhali, Advances in long DNA synthesis, Microbial Cell Factories Engineering for Production of Biomolecules, 10.1016/B978-0-12-821477-0.00014-3, (21-36), (2021).
- Amber Afroz, Safeena Aslam, Umer Rashid, Muhammad Faheem Malik, Nadia Zeeshan, Muhammad Ramzan Khan, Muhammad Qasim Shahzad But, Sabaz Ali Khan, Intron-containing hairpin RNA interference vector for OBP8 show promising mortality in peach potato aphid, Plant Cell, Tissue and Organ Culture (PCTOC), 10.1007/s11240-021-02174-4, 148 , 1, (155-166), (2021).
- Maren Schubert, Manfred Nimtz, Federico Bertoglio, Stefan Schmelz, Peer Lukat, Joop van den Heuvel, Reproducible and Easy Production of Mammalian Proteins by Transient Gene Expression in High Five Insect Cells, Structural Proteomics, 10.1007/978-1-0716-1406-8_6, (129-140), (2021).
- Fabio Pasin, Oligonucleotide abundance biases aid design of a type IIS synthetic genomics framework with plant virome capacity, Biotechnology Journal, 10.1002/biot.202000354, 16 , 5, (2021).
- Guoliang Yuan, Md. Mahmudul Hassan, Degao Liu, Sung Don Lim, Won Cheol Yim, John C. Cushman, Kasey Markel, Patrick M. Shih, Haiwei Lu, David J. Weston, Jin-Gui Chen, Timothy J. Tschaplinski, Gerald A. Tuskan, Xiaohan Yang, Biosystems Design to Accelerate C 3 -to-CAM Progression , BioDesign Research, 10.34133/2020/3686791, 2020 , (2020).
- John M. Pryor, Vladimir Potapov, Rebecca B. Kucera, Katharina Bilotti, Eric J. Cantor, Gregory J. S. Lohman, Enabling one-pot Golden Gate assemblies of unprecedented complexity using data-optimized assembly design, PLOS ONE, 10.1371/journal.pone.0238592, 15 , 9, (e0238592), (2020).
- Alejandro Sarrion‐Perdigones, Yezabel Gonzalez, Koen J.T. Venken, Rapid and Efficient Synthetic Assembly of Multiplex Luciferase Reporter Plasmids for the Simultaneous Monitoring of Up to Six Cellular Signaling Pathways, Current Protocols in Molecular Biology, 10.1002/cpmb.121, 131 , 1, (2020).

