SCRMshaw: supervised cis-regulatory module prediction for insect genomes
Hasiba Asma, Luna Liu, Marc S. Halfon
Abstract
As the number of sequenced insect genomes continues to grow, there is a pressing need for rapid and accurate annotation of their regulatory component. SCRMshaw is a computational tool designed to predict cis -regulatory modules (CRMs) in the genomes of various insect species (Kantorovitz 2009; Kazemian 2011; Asma and Halfon 2019; Asma 2024).
One of the key advantages of SCRMshaw is its accessibility. It requires minimal resources—just a genome sequence and training data from known Drosophila CRMs, which are readily available for download. Even users with modest computational skills can run SCRMshaw on a desktop computer for basic applications, although an HPC cluster is recommended for optimal results. SCRMshaw can be tailored to specific needs. Users can employ a single set of training data to predict CRMs associated with a particular gene expression pattern, or utilize multiple sets to provide a rough regulatory annotation for a newly-sequenced genome.
This protocol provides an update to the protocol in Kazemian and Halfon (2019) and incorporates modifications introduced in Asma and Halfon (2019) and Asma et al. (2024).
Before start
Software and Dependencies:
Ensure that the following software is installed :
- Python: Python3
Pybedtools
Statistics
Pandas
Numpy
Biopython (for “extract_unannoatatedScaffolds.py”)
- Perl: Perl
Bioperl
- Other: MACS2
Download - https://pypi.org/project/MACS2/
NCBI Download Utility (optional)
Download - https://www.ncbi.nlm.nih.gov/home/tools/
- Optional (for Orthology assignment steps): Docker
Download - https://www.docker.com/products/docker-desktop/
Orthologer Software
Requirements: Docker on Mac or Linux
Download - https://hub.docker.com/r/ezlabgva/orthologer (ver2.4.3)
Documentation - https://hub.docker.com/r/ezlabgva/orthologer
- Optional (for final merging of the SCRMshaw results) BEDTools (https://github.com/arq5x/bedtools2/releases))
Steps
Create Project directory
Create a Project Directory and subdirectories
# “create project directory”
mkdir project1
Under the project directory create subdirectories for saving input and output files respectively, for training data, and for scripts
#“create subdirectories”
mkdir GenomeFiles HD_output TrainingSets Scripts

Install SCRMshaw
From the project directory, clone the SCRMshaw_HD software from
https://github.com/HalfonLab .
#“Download SCRMshaw”
git clone https://github.com/HalfonLab/SCRMshaw_HD.git
If necessary, decompress the tar archive (e.g., tar xvf SCRMshaw_HD.tar). This will create a single “SCRMshaw_HD” directory with the following subdirectories:
bin/
src/
code/
include/
example/
Navigate to the SCRMshaw directory
#“Navigating to the SCRMshaw directory"
cd SCRMshaw_HD
Run “make.” Detailed information on how to install and run the software is provided in the “README.txt” file located within the software package.
#“Executing the “make” command”
make
Obtain training set sequences
Obtain up-to-date training data from
https://github.com/HalfonLab/Training_sets.
Find details on how to construct custom training sets in Kazemian and Halfon (2019). Place training sets into the “TrainingSets” directory.
Download required genome files
Download the genome files from NCBI Datasets
(https://www.ncbi.nlm.nih.gov/datasets/)) and place them in the GenomeFiles directory. Necessary files are:
- Genome sequences (FASTA)
- Annotation features (GFF)
- Protein (FASTA)
We recommend using the RefSeq builds if available.
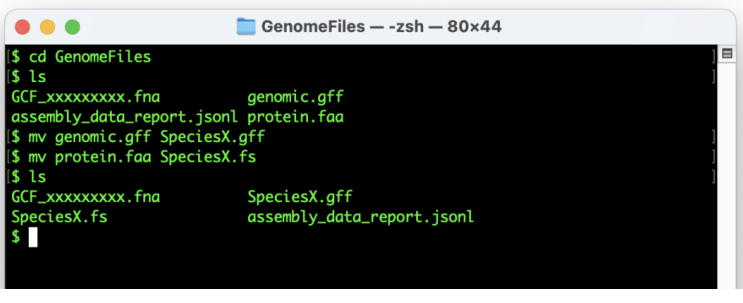
Rename the “genomic.gff” and “protein.faa” files to “SpeciesX.gff” and “SpeciesX_protein.fs”, respectively (where “X” is the name of your species). This avoids future confusion due to the generic “genome” and “protein” designations, and provides compatibility for downstream steps.
#“Rename the “genomic.gff”
mv genomic.gff SpeciesX.gff
#“Rename the “protein.faa”
mv protein.faa SpeciesX.fs
The GenomeFiles directory should now look similar to this:
Assessing genome files using preflight
Assess genome and annotation files for proper content and formatting using our preflight script, which can be downloaded from
https://github.com/HalfonLab/UtilityPrograms. We recommend placing this into the “Scripts” directory.
#“running preflight”
perl ../Scripts/preflight.pl -gff genome.gff -fasta GCF_xxxxxxxxx.fna
Preflight validates the formats of these files and produces a comprehensive log file that highlights any issues along with basic information such as the number of chromosomes/scaffolds and their sizes, data types present in the annotation (e.g., ‘gene’, ‘exon’, ‘ncRNA’, etc.), and average intergenic distances.
Preflight also provides a sample output of the SCRMshaw-generated ‘gene’ and ‘exon’ files. This feature allows users to identify any discrepancies or errors stemming from the input files and to reformat these files as needed before running SCRMshaw. Be sure to check the following:
- In the first section, no GFF3 errors are reported
- Gene and Exon data appear in the following sections
- The ‘FASTA’ section reports “all sequences have proper characters”
- The following section reports that “All seqids in GFF are also in FASTA”
If any of these conditions is not met, correct the input files before moving on to run SCRMshaw, or SCRMshaw will fail.
Removing unannotated genes
Preflight will also indicate whether there are sequence scaffolds that do not contain any annotated genes (reported as “seqids not in the GFF file.”) Any such sequence scaffolds should be removed before passing the genome sequence to the main SCRMshaw program. Perform this using the “extract_unannotatedScaffolds.py” script (from
https://github.com/HalfonLab/UtilityPrograms)) that takes the preflight output file and FASTA sequence file as input and generates a fasta file, “mappedOnly_GCF_xxxxx.fna”, that contains only the desired sequences.
#“extracting unannotated scaffolds”
python ../Scripts/extract_unannotatedScaffolds.py -po preflight_output_file.txt -fasta GCF_xxxxxxxxx.fna
Masking Tandem Repeats
Download the current version of TRF for your computer architecture from
https://tandem.bu.edu/trf/home (e.g., trf409.linux64).
#“example download command for TRF”
curl -JLO ../Scripts/ https://github.com/Benson-Genomics-Lab/TRF/releases/download/v4.09.1/trf409.linux64
Change the downloaded TRF file to an executable form:
#“changing to executable”
chmod +x ../Scripts/trf409.linux64
Run TRF on your FASTA files (e.g., mappedOnly_GCF_xxxxx.fna) using parameters as shown:
#“running TRF”
../Scripts/trf409.linux64 mappedOnly_GCF_xxxxx.fna 2 7 7 80 10 50 500 –m –h
- The resulting output file will be named “mappedOnly_GCF_xxxxx.fna.2.7.7.80.10.50.500.mask”
Running SCRMshaw
SCRMshaw requires the following files and directories:
- A genome file (e.g., “genome.fa”), a multi-FASTA file including all of the genomic sequences needing to be scored. This is typically a downloaded genome file that has been masked for tandem repeats, and is passed to the program using the “--genome" option.
- One or more dataset directories each containing two files, “crms.fasta” and “neg.fasta.” These files are respectively the positive and negative training sequences, both in FASTA file format. We highly recommend tandem repeat masking of genome.fa, crms.fasta, and neg.fasta.
- A text file containing a list of the dataset directories (e.g., “trainingSet.lst”; see below step 17). This is specified using the “--traindirlst” option.
Navigate back to the project directory.
#“move back to the project directory”
cd ..
Under the project directory, create a list file “trainingSet.lst” that contains the path to the training datasets you wish to use (e.g., project1/TrainingSets/dataset1/), one line per dataset. You can create this file using a simple text editor or using the following “echo” commands (adjust path as necessary):
#“creating file for dataset1”
echo “~/project1/TrainingSets/dataset1/” >> trainingSet.lst
#“creating file for dataset2”
echo “~/project1/TrainingSets/dataset2/” >> trainingSet.lst
#“creating file for dataset3”
echo “~/project1/TrainingSets/dataset3/” >> trainingSet.lst
At the conclusion of these steps, your directory should resemble Figure 3 below.
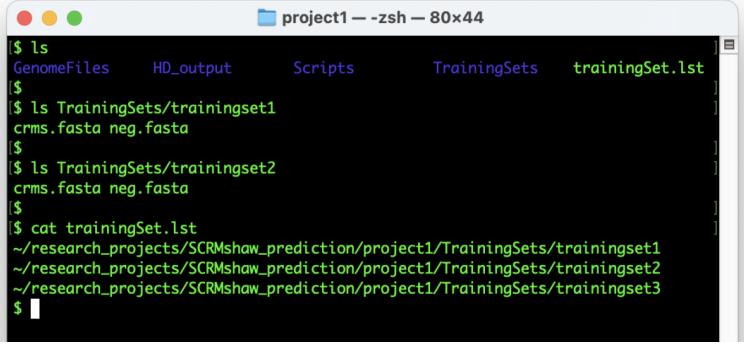
Run SCRMshaw by providing the full paths to the genome file, gene annotation file, and training set list after the --genome, --gff, and --traindirlst flags, respectively (e.g. GenomeFiles/SpeciesX.gff), and providing appropriate values for --lb and --outdir (see Note).
#“Running SCRMshaw”
perl SCRMshaw_HD/code/scrm.pl --thitw 5000 --gff GenomeFiles/SpeciesX.gff --genomeGenomeFiles/mappedOnly_GCF_xxxxx.fna.2.7.7.80.10.50.500.mask --traindirlst trainingSet.lst --imm --
We recommend that the base directory for --outdir be set to ~/project1/HD_output for consistency with the examples in this protocol.
The output of SCRMshaw for the 25 offset instances will be saved in subdirectories, such as 'task_offset_0_1', 'task_offset_10_2', 'task_offset_20_3', and so on up to 'task_offset_240_25', as specified by the '--outdir' parameter in the Slurm script. These directories will be created automatically if they don't already exist. If Slurm is not utilized, it is crucial to ensure that the output directory and file names adhere to the specified format (e.g., 'task_offset_0_1' to 'task_offset_240_25') to prevent any downstream disruptions.
The SCRMshaw command line options include:
Use the name of scoring method(s) for prediction (“--imm” for IMM, “--hexmcd” for HexMCD, and “--pac” for PAC): required. Select any individual or combination of methods to run (e.g. “--imm --pac” to run IMM and PAC). For details about the individual scoring methods see (Kantorovitz et al. 2009, Kazemian et al. 2011).
The gene annotation file of the genomic regions to be scored in GFF3 format (e.g., “--gff SpeciesX.gff”): optional but strongly recommended.
- Alternatively, separate “gene” and “exon” files can be used. It is also possible to use SCRMshaw without providing any annotation, but this will not allow for exclusion of coding sequences or calculation of “local rank” (see below).
The number of top scoring regions to be reported as CRM predictions (--thitw N): optional, default = 2000.
The offset parameter --lb: optional, default = 0. The --lb tells the algorithm to ignore the base pairs at the beginning of a chromosome/scaffold before this point, i.e., to start creating the analysis windows from this point in the genome. The Slurm script described above sets --lb from 0 to 240 with a step size of 10 (i.e 10, 20, 30,..., 240) for the 25 instances being run for each training set.
Which steps of the SCRMshaw pipeline to run (--step 123): optional, default = 123. Step 1 processes the genome and genome annotation, step 2 processes the training data, and step 3 scores the genome.
Detailed information on how to run SCRMshaw from the command line and additional available options can be obtained by the following command: “perl SCRMshaw_HD/code/scrm.pl” or from the README document accompanying the SCRMshaw distribution.
- An example benchmark data set along with how to run and interpret its results is provided in the example directory within the software package.
Post-processing
The post-processing steps merge the data from the 25 SCRMshaw instances and select the top-scoring regions as CRM predictions. To run the post-processing steps, download the following scripts from GitHub and place in the
“Scripts” directory:
post_processing_complete.sh (https://github.com/HalfonLab/UtilityPrograms))
Generate_top_N_SCRMhits.pl (https://github.com/HalfonLab/UtilityPrograms))
postProcessingScrmshawPipeline.py
(https://github.com/HalfonLab/post_processing_SCRMshaw_pipeline))
Execute the shell script post_processing_complete.sh from the project directory. Use the path to the GFF annotation file for SCRMshaw and enter as a command line argument:
#“post-processing”
./Scripts/post_processing_complete.sh GenomeFiles/SpeciesX.gff
The post-processing procedure will create the following files:
scrmshawOutput_offset_0to240.bed: this is the concatenated results of the top 5000 results from each of the individual SCRMshaw instances
scrmshawOutput_peaksCalled_[hexmcd/imm/pac]: BED-formatted files with the MACS-generated peaks with the results for each training set and SCRMshaw method
peaks_AllSets.bed: the concatenated final results from the three separate peaks files in BED format
log_flankingMoreThanOneGenesFromAllSets.txt: a log file documenting cases of SCRMshaw predictions where there is more than a single flanking gene either up- or downstream of the prediction (e.g., where genes overlap or are nested).
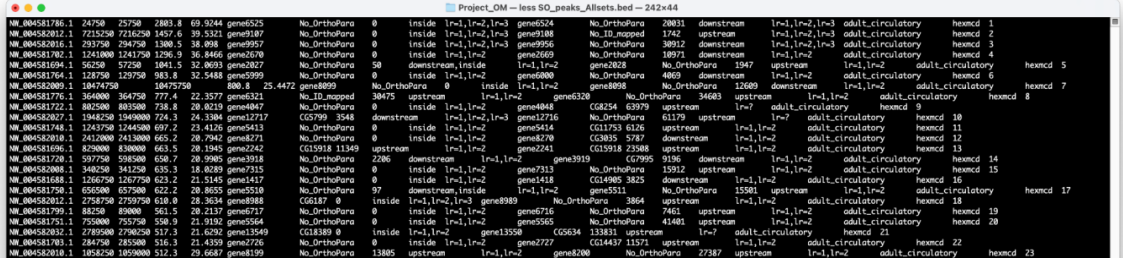
It is possible to finish your SCRMshaw processing at this step, and use the information from the peaks_AllSets.bed file for any desired downstream analysis.
- However, we recommend continuing with the Orthology Mapping steps below, as it makes the output more understandable by providing recognizable gene names to what otherwise would be arbitrary ID numbers.
- If you choose to end your SCRMshaw processing here, we recommend you still follow the “cleanup” procedures in the final section of this protocol, “Cleaning Up.”
Running Orthologer
Before running Orthologer, create a directory titled Project_OM.
#“Make Project_OM directory for running Orthologer”
mkdir Project_OM
Navigate to the Project_OM directory
#“Navigate to the Project_OM directory”
cd Project_OM
Obtain the Orthologer Docker image (see note above about Docker and HPC platforms).
#“Pulling Orthologer from Docker”
docker pull ezlabgva/orthologer:v3.2.2
Create a subdirectory of Project_OM titled “protein_files”.
#“Create protein_files directory”
mkdir Project_OM/protein_files
Copy the protein FASTA files for the current species as well as for Drosophila melanogaster into the protein_files directory.
#“Copy protein FASTA files”
cp GenomeFiles/SpeciesX_protein.fs path/DMEL_protein.fs Project_OM/protein_files
Create a text file specifying the FASTA files to be used, as follows:
#“Create ‘mydata.txt’ file”
for x in $(ls files/*.fs); do echo "+$(basename $x .fs) $x"; done > mydata.txt
Import the specified proteomes:
#“import proteomes”
docker run -u $(id -u) -v $(pwd):/odbwork ezlabgva/orthologer:v3.2.2 ./orthologer.sh manage -f mydata.txt
Run the Orthologer container
#“Running Orthologer”
docker run -u $(id -u) -v $(pwd):/odbwork ezlabgva/orthologer:v3.2.2 ./orthologer.sh -t todo/mydata.todo - r all
Saving Orthologer outputs for orthology mapping
Navigate back to the main project directory
#“Navigate back to the main project directory”
cd ..
Create a directory “orthologer_output”
#“create “orthologer_output” directory
mkdir orthologer_output
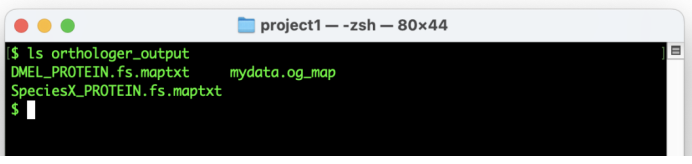
Copy the desired Orthologer output files to the orthologer_output directory
#“copy files”
cp Project_OM/Cluster/mydata.og_map Project_OM/Rawdata/SpeciesX_protein.fs.maptxt Project_OM/Rawdata/DMEL_protein.fs.maptxt orthologer_output/
Mapping DMEL orthologs to SpeciesX SCRMshaw predictions
Generate a gene ID ⇔ protein ID mapping file
#“Sample code for generating a gene⇔protein mapping file (Using Perl)”
perl -F'\t' -ane 'next if $_ =~ /#/; if ($F[2] =~ /CDS/){$F[8] =~ /gene=(.*?);.*protein_id=(.*)/; print "$1\t$2\n";}' speciesX.gff | sort -u > speciesX_A.map
#“Sample code for generating a gene⇔protein mapping file (Using sed)”
sed -n '/#/d; /CDS/ {s/.*gene=\([^;]*\);.*protein_id=\([^;]*\).*/\1\t\2/; p; }' speciesX.gff | sort -u > speciesX_A.map
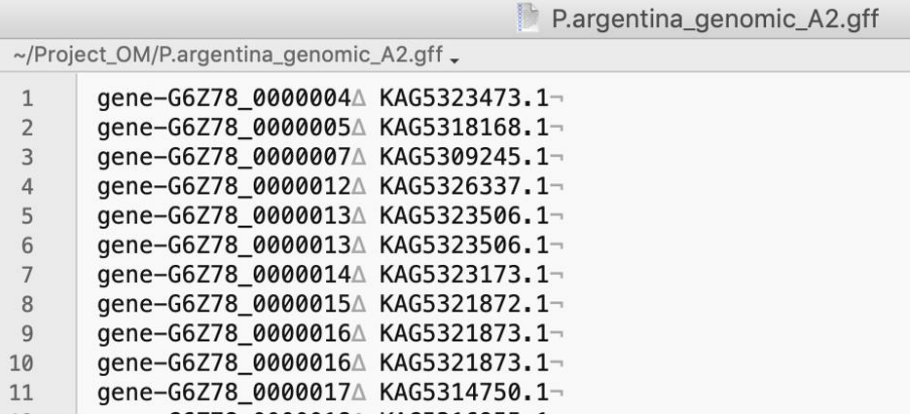
Inspect the speciesX_A.map file and check that the first column of this file has the gene IDs formatted the same as that in column 6 of the SCRMshaw output file, and that the protein IDs in the second column match those in the Orthologer-generated “mydata.og_map” file. If necessary, edit these files for appropriate formatting. The following figures provide an example:
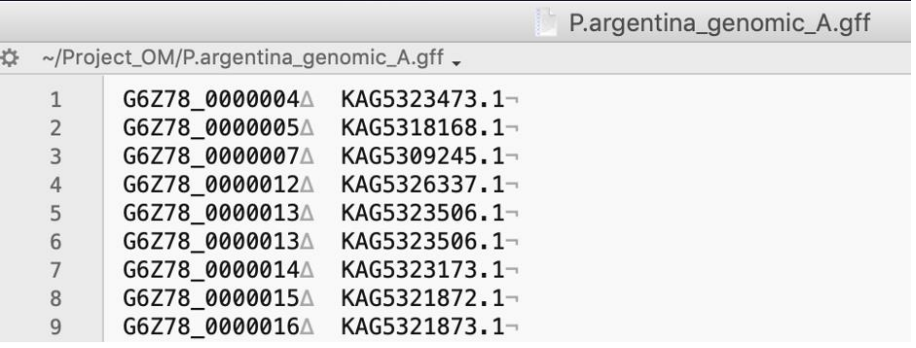
Looking at this file we can see that protein ID (2nd) column matches the format of the Orthologer output, but the gene ID (1st) column is formatted slightly differently than SCRMshaw output (the latter have ‘gene-’ at the beginning of each ID). We therefore edit the file as follows:
#“editing P.argentina_A.map”
sed ‘s/^/gene-/’ P.argentina_A.map > P.argentina_Aedited.map
Obtain the necessary scripts and files from the Halfon Lab GitHub
#“Clone the GitHub repository”
git clone https://github.com/HalfonLab/Mapping-D.mel-Orthologs.git
Run the script “OM_mappingFlyOrthologsToSCRMshawPredictions.py”.
You will need to supply the following command line options:
- –ft: A table of D. melanogaster annotation features (included in the Mapping-D.mel-Orthologs repository)
- –mD: D. melanogaster Orthologer “maptxt” output file (included in the Mapping-D.mel-Orthologs repository or from your output in the Orthologer step)
- –mX: The SpeciesX Orthologer “maptxt” file generated in the Orthologer step
- –og: The “mydata.og_map Orthologer output file generated in the Orthologer step
- –sp1id: The appropriately edited SpeciesX.map file from step 40
- –so: The SCRMshaw final output “peaks_Allsets.bed” file
#command title = “Running the final orthology mapping”
python Mapping-D.mel Orthologs/OM_mappingFlyOrthologsToSCRMshawPredictions.py -ft Mapping-D.mel Orthologs/GCF_000001215.4_Release_6_plus_ISO1_MT_feature_table.txt -mD Mapping-D.mel-Orthologs/DMEL_PROTEIN.fs.maptxt -mX orthologer_output/SpeciesX_protein.fs.maptxt -og orthologer_output/mydata.og_map -sp1id SpeciesX_Aedited.map -so peaks_Allsets.bed
Interpreting the output
The output file will have the same file name as SCRMshaw’s output file with an additional “SO_” prefix. As noted above, this file may have duplicate and/or overlapping predictions, e.g. predict the same or similar sequences by more than one training set, or more than one method. This can be resolved using the “sort” and “merge” commands in BEDTools (Quinlan and Hall, 2010) as follows:
#“BEDTools Sort and Merge”
bedtools sort -i SO_peaks_Allsets.bed | bedtools merge -c 4,5,6,7,8,9,10,11,12,13,14,15,16,17,18 -o max,max,distinct,distinct,distinct,distinct,distinct,distinct,distinct,distinct,distinctdistinct,distinct,distinct,min
Cleaning up
Download the “cleanup_fastaAndScoredirectory_SCRMshawHD.sh” from
(https://github.com/HalfonLab/UtilityPrograms)) and place into the “Scripts” directory.
From the Project1 directory, run the cleanup script.
#“cleanup directories”
./Scripts/cleanup_fastaAndScoredirectory_SCRMshawHD.sh