Real-time quantitative polymerase chain reaction (RT-qPCR)
Qiaowa Gong, An.Huang
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Real-time quantitative polymerase chain reaction (RT-qPCR) helps determine the expression level of a certain gene by amplifying DNA according to the target mRNA template. This protocol describes the procedure of conducting RT-qPCR using Promega GoTaq® qPCR Master Mix A6001.
Steps
Thaw the GoTaq® Master Mix and Nuclease-Free Water. Thaw the cDNA templates and primer.
Vortex the GoTaq® Master Mix for 3–5 seconds to mix. Vortex at low speed to avoid aeration.
Determine the number of reactions to be set up, including negative control reactions. Add 1 or 2 reactions to this number to compensate for pipetting error.
Prepare the reaction mix (minus DNA template) by combining the GoTaq® qPCR Master Mix, PCR primers and Nuclease-Free Water as described below. The DNA template is added in Step 6. Vortex briefly to mix.
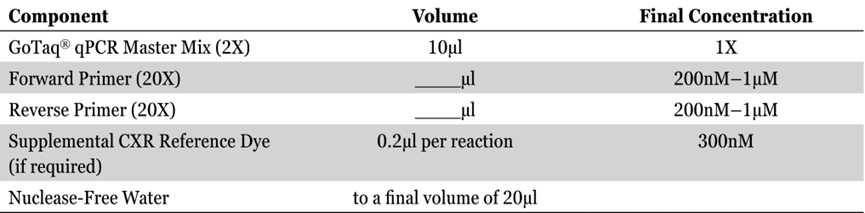
Add the DNA template (or water for the no-template control reactions) to the appropriate wells of the reaction plate.
Seal the tubes or optical plate, and centrifuge briefly to collect the contents of the wells at the bottom. The samples are ready for thermalcycling.
Thermalcycle in qPCR equipment. Realtime qPCR is performed with an initial denaturation of 3 min at 95°C, followed by 40 cycles of 20 s at 95°C, 20 s at 60°C, and 20s at 72°C.

