Rapid Identification of Protein-Protein Interactions in Plants
Alisdair R. Fernie, Alisdair R. Fernie, Youjun Zhang, Youjun Zhang, Roberto Natale, Roberto Natale, Adilson Pereira Domingues Júnior, Adilson Pereira Domingues Júnior, Mitchell Rey Toleco, Mitchell Rey Toleco, Beata Siemiatkowska, Beata Siemiatkowska, Norma Fàbregas, Norma Fàbregas
Abstract
Enzyme-enzyme interactions can be discovered by affinity purification mass spectrometry (AP-MS) under in vivo conditions. Tagged enzymes can either be transiently transformed into plant leaves or stably transformed into plant cells prior to AP-MS. The success of AP-MS depends on the levels and stability of the bait protein, the stability of the protein-protein interactions, and the efficiency of trypsin digestion and recovery of tryptic peptides for MS analysis. Unlike in-gel-digestion AP-MS, in which the gel is cut into pieces for several independent trypsin digestions, we uses a proteomics-based in-solution digestion method to directly digest the proteins on the beads following affinity purification. Thus, a single replicate within an AP-MS experiment constitutes a single sample for LC-MS measurement. In subsequent data analysis, normalized signal intensities can be processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software. Following analysis of co-sublocalization of “bait” and “prey,” we suggest considering only the protein pairs for which the intensities were more than 2% compared with the bait, corresponding to FC-A values of at least four within-biological replicates, which we recommend as minimum. If the procedure is faithfully followed, experimental assessment of enzyme-enzyme interactions can be carried out in Arabidopsis within 3 weeks (transient expression) or 5 weeks (stable expression). © 2019 The Authors.
Basic Protocol 1 : Gene cloning to the destination vectors
Alternate Protocol : In-Fusion or Gibson gene cloning protocol
Basic Protocol 2 : Transformation of baits into the plant cell culture or plant leaf
Basic Protocol 3 : Affinity purification of protein complexes
Basic Protocol 4 : On-bead trypsin/LysC digestion and C18 column peptide desalting and concentration
Basic Protocol 5 : Data analysis and quality control
INTRODUCTION
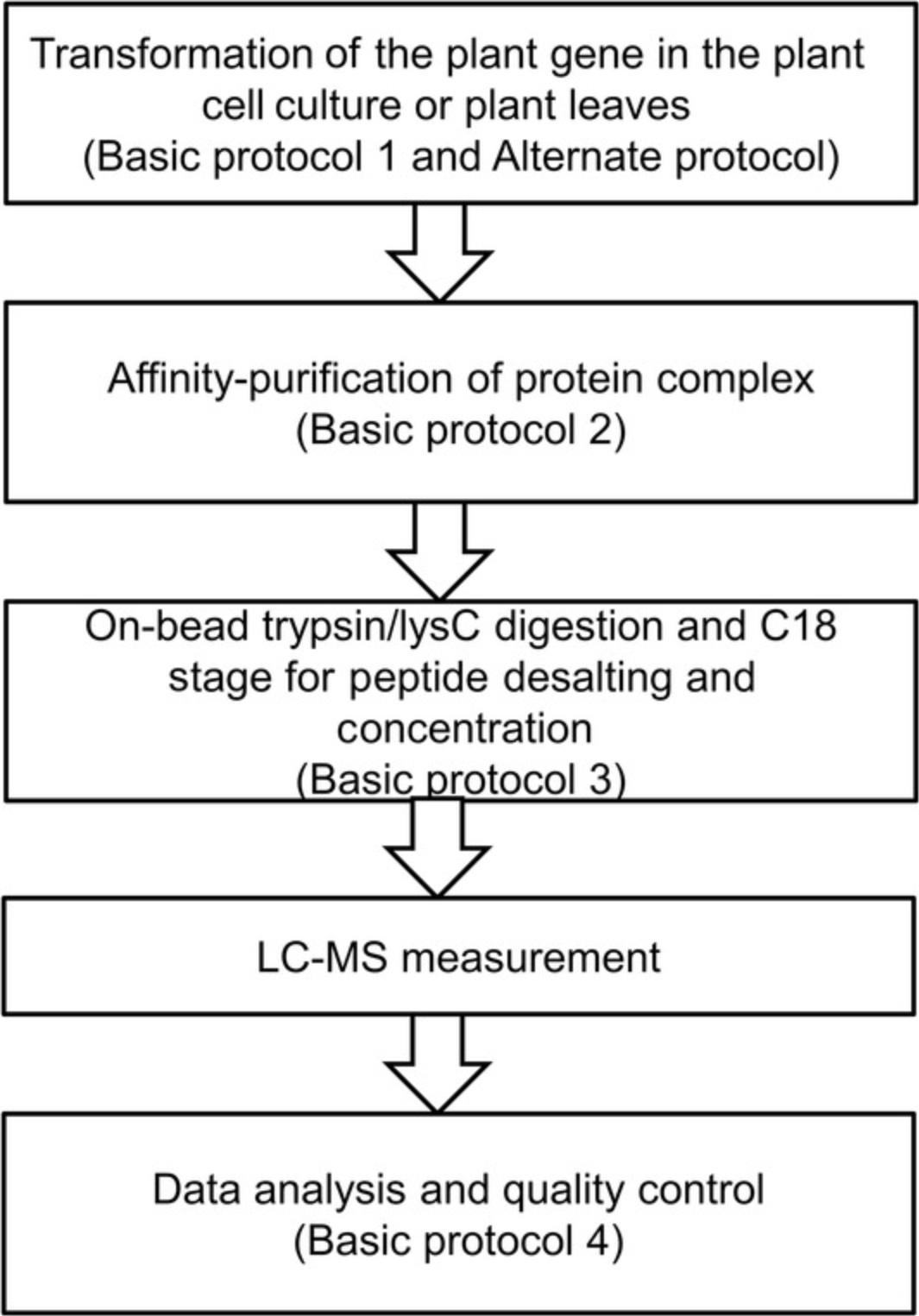
Affinity purification mass spectrometry is a highly effective method for isolating and identifying protein-binding partners of a target protein under in vivo conditions. Protein complexes can be captured by antibodies specific for the bait proteins or for tags fused to the bait proteins via recombinant DNA technologies. These complexes are thereby “pulled-down” onto immobilized-protein agarose beads via affinity purification, prior to their detection and identification via mass spectrometry. Given that AP-MS experiments have been widely used to generate meaningful interaction networks, it follows that they could also be used to produce information-rich data concerning extra-pathway protein-protein interactions (Bürckstümmer et al., 2006; Morris et al., 2014; Puig et al., 2001; Zhang, Beard, et al., 2017; Zhang, Sun, Zhang, Brasier, & Zhao, 2017; Zhang, Swart et al., 2018). Such interactions could aid in the characterization of the functions of the interacting proteins, provide detailed catalogs of proteins involved in protein complexes and biological processes, or reveal networks of biological processes on both the local and proteome-wide scale (Morris et al., 2014). In order to better understand these interactions, AP-MS can be readily performed in many plant species, with the main prerequisites being the ease of genetic transformation and availability of a sequenced reference genome. However, presently, these features apply to a multitude of plant species. The basic procedure can be divided into five stages: (i) gene cloning into the destination vectors (see Basic Protocol 1 and Alternate Protocol); (ii) plant cell culture transformation (see Basic Protocol 2); (iii) affinity purification of protein complexes (see Basic Protocol 3); (iv) on-beads trypsin/LysC digestion and C18 column–based peptide desalting and concentration (see Basic Protocol 4); and (v) data analysis and quality control (see Basic Protocol 5). The entire workflow is summarized in Figure 1.
STRATEGIC PLANNING
The Arabidopsis plant cell culture (PSBD, ABRC stock, CCL84840, Background: Ler, Landsberg erecta) can be obtained from the ABRC stock center and maintained according to a published protocol (Menges & Murray, 2002). Similarly, Arabidopsis seeds can be obtained from stock centers (ABRC), and Arabidopsis plants growth can be carried out as described in the literature (Sanchez-Serrano & Salinas, 2014; Zhang, Swart, et al., 2018).
Basic Protocol 1: GENE CLONING TO THE DESTINATION VECTORS
A variety of protocols have been described over the years in regard to gene cloning (Curtis & Grossniklaus, 2003; Katzen, 2007; Walhout et al., 2000). Described here is the gene cloning protocol used in our lab. A two-step polymerase chain reaction (PCR) is used to clone the genes of interest and link them to the donor vector using the Gateway BP reaction. As some genes cannot be linked to the donor vector by the Gateway BP reaction, we have alternatively used In-Fusion or Gibson assembly to sub-clone these genes (see Alternate Protocol). Next, the genes of interest are recombined into the destination vector by the Gateway LR reaction, under the control of the plant ubiquitin 10 promoter (Grefen et al., 2010).
Materials
-
Nuclease-free water
-
Phusion High-Fidelity DNA polymerase (ThermoFisher Scientific, F530L) and corresponding 5× buffer
-
dNTP mix (New England Biolabs, cat. no. N0447)
-
Template-specific primers: design the following template-specific primers; include 12 bases of the attB1 or attB2 site on the 5′ end of each primer, as appropriate:
- Gene forward: 5′-AAAAAAGCAGGCTCCACCNNNNNNNNN-
- Gene reverse: 5′-CAAGAAAGCTGGGTcatagccNNNNNNNN-
- NNNNNNNNNNNN represents the gene-specific primer with at least 20 bp; we strongly recommend using Primer, version 7 (https://www.primer-e.com/our-software/primer-version-7/) to evaluate the primers
-
Template DNA: cDNA from seedling of Arabidopsis (RevertAid First Strand cDNA Synthesis Kit, ThermoFisher Scientific, K1622)
-
Adapter primers: design the following adapter primers (required to install the complete attB sequences):
- attB1 adapter: 5′-G GGGACAAGTTTGTACAAAAAAGCAGGCTCCACC-3′
- attB2 adapter: 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTcatagcc-3′
- attB2St adapter: 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTcttagcc-3′ to amplified gene with stop code
-
1% agarose gel with RedSafe nucleic acid staining solution (Chembio Ltd., 21141; also see Current Protocols article: Voytas, 2000)
-
Nucleic acid gel extraction and purification kit (Qiagen, cat. no. 28704)
-
Donor vector: Gateway™ pDONR™221 vector (Thermo Fisher Scientific, 12536-017) or pDONR™207 vector (Thermo Fisher Scientific, 12213-013)
-
TE buffer, pH 8.0 (Current Protocols, 2001)
-
Gateway® BP Clonase® II enzyme mix (Thermo Fisher Scientific, 11789013, 11789020) including 2 µg/µl proteinase K solution
-
DH5α E. coli competent cells (Thermo Fisher Scientific,18265017)
-
Lysogeny broth (LB) medium (see recipe)
-
LB agar plates (see recipe) with 50 µg/ml kanamycin
-
M13F (GTAAAACGACGGCCAG) and M13R universal primers (CAGGAAACAGCTATGAC)
-
Gateway® LR Clonase® Enzyme Mix—to create a Gateway™ expression clone (Thermo Fisher Scientific, 11791019)
-
200-μl PCR tubes
-
Thermal cycler
-
42°C water bath for heat shock
-
37°C shaking incubator
-
Additional reagents and equipment for agarose gel electrophoresis (see Current Protocols article: Voytas, 2000) and DNA sequencing (see Current Protocols article: Shendure et al., 2011)
Gene cloning
1.Prepare gene-specific PCR mix (20 µl/reaction):
| Component | 20 µl reaction | Final concentration |
|---|---|---|
| Nuclease-free water | To 20 µl | |
| 5× Phusion HF buffera | 4 µl | 1× |
| 10 mM dNTPs | 0.4 µl | 200 µM |
| 10 µM gene-specific forward primer | 0.2 µl | 0.01 µM |
| 10 µM gene-specific reverse primer | 0.2 µl | 0.01 µM |
| Template DNA | 0.2 µl | <250 ng |
| DMSO (optional)b | (0.6 µl) | 3% |
| Phusion DNA polymerase | 0.2 µl | 0.02 U/µl |
-
aOptionally 5× Phusion GC buffer can be used.
-
bAddition of DMSO is recommended for GC-rich amplicons. DMSO is not recommended for amplicons with very low % GC or amplicons that are >20 kb.
2.Perform the first-step PCR in a thermal cycler using the following machine settings:
| Step | Time | Temperature | Cycles |
|---|---|---|---|
| Initial denaturation | 30 s | 98°C | 1× |
| Denaturation | 10 s | 98°C | 15× |
| Annealing | 20 s | 60° to 72°C | |
| Extension | 30 s/kb | 72°C |
3.Transfer 10 μl of the PCR reaction to a 40-μl PCR mixture containing 40 pmol each of the att B1 and att B2 adapter primers (note that attB1 and attB2St adapters are for the gene with stop code).
| Component | 40 µl reaction | Final concentration |
|---|---|---|
| Nuclease-free water | add to 40 µl | |
| 5× Phusion HF or GC buffer (see step 1) | 8 µl | 1× |
| 10 mM dNTPs | 0.8 µl | 200 µM |
| 10 µM adapter forward primer | 2 µl | 0.5 µM |
| 10 µM adapter reverse primer | 2 µl | 0.5 µM |
| DMSO (optional; see step 1) | (1.2 µl) | 3% |
| Phusion DNA polymerase | 0.4 µl | 0.02 U/µl |
4.Perform the second-step PCR in a thermal cycler using the following machine settings:
| Step | Time | Temperature | Cycles |
|---|---|---|---|
| Initial denaturation | 1 min | 98°C | 1× |
| Denaturation | 10 s | 98°C | 5× |
| Annealing | 20 s | 55°C | |
| Extension | 30 s/kb | 72°C | |
| Denaturation | 10 s | 98°C | 29× |
| Annealing | 20 s | 68°C | |
| Extension | 30 s/kb | 72°C | |
| Final extension | 5−10 min | 72°C | 1× |
5.Use agarose gel electrophoresis (see Current Protocols article: Voytas, 2000) to check quality and yield of the att B-PCR product, and then purify the PCR products for the BP reaction using a nucleic acid gel extraction and purification kit (e.g., Qiagen).
6.Perform a BP recombination reaction between an att B-flanked DNA fragment and an att P-containing donor vector (pDONR221 or pDONR207) to generate an entry clone.
- Add the following components to a 1.5-ml microcentrifuge tube at room temperature and mix::
| Clone (attB-PCR product, from step 5; ≥30 ng/μl; final amount up to 100−150 ng/μl) | 1−3.5 μl |
| pDONR™ vector (supercoiled, 150 ng/μl) | 0.5 μl |
| TE buffer, pH 8.0 | To 4.5 μl |
-
Vortex Gateway® BP Clonase® II enzyme mix briefly. Add 0.5 μl to the components above and mix well by vortexing briefly twice.
-
Incubate the reaction at 25°C for at least 1 hr (can be overnight).
-
Add 1 μl of 2 μg/μl proteinase K solution (included with the Clonase enzyme) and incubate at 37°C for 10 min.
E. coli transformation
7.Thaw 50 μl of chemically competent cells (DH5α or Top 10) for each transformation on ice. Add 5 μl of the BP recombination reaction to the competent cells and mix gently. Do not mix by pipetting up and down. Incubate the vial(s) on ice for 30 min. Heat-shock the cells for 45 s at 42°C without shaking. Remove the vial(s) from the 42°C bath and place them on ice for 2 min. Add 1 ml of room temperature LB medium to each vial. Cap the vial(s) tightly and put on a shaker (850 rpm) at 37°C for 1 hr. Microcentrifuge for 1 min at 14,000 × g , discard the supernatant, and resuspend the pellet by pipetting. Plate the bacteria onto the pre-warmed selective plate and incubate overnight at 37°C.
8.After sequencing by vector-specific primers (M13F and M13R for the pDONR221; see Current Protocols article: Shendure et al., 2011), perform an LR recombination reaction between an att L-flanked DNA fragment (produced before) and an att R-containing donor vector to generate a digestion vector:
- Add the following components to a 1.5-ml microcentrifuge tube at room temperature and mix (attL-Vector t or linearizedattL expression):
| Donor vector (≥ 30 ng/μl; final amount 100−150 ng) | 1–3.5 μl |
| Digestion vector (supercoiled, 150 ng/μl) | 0.5 μl |
| TE buffer, pH 8.0 | To 4.5 μl |
-
Vortex Gateway® LR Clonase® II enzyme mix briefly. Add 0.5 μl to the components above and mix well by vortexing briefly twice. Incubate the reaction at 25°C for at least 1 hr (can be overnight). Add 1 μl of 2 μg/μl Proteinase K solution and incubate at 37°C for 10 min. Transform competentE. coliand select for the appropriate antibiotic-resistant digestion vector following the method mentioned above.
Alternate Protocol: IN-FUSION OR GIBSON GENE CLONING
Although the BP reaction works for most genes, there are around 2% to 5% of genes that cannot be cloned by the BP reaction due to sequence-specific problems, among other reasons. Here, we provide an alternative protocol to sub-clone these genes into pDONR vector by In-Fusion and Gibson assembly.
Additional Materials (also see Basic Protocol 1)
- In-Fusion HD cloning (Takara, 639650)
- Gibson Assembly® Master Mix (NEB, E2611)
- pDONR-IF-f: GACCCAGCTTTCTTGTACAAAGT
- pDONR-IF-r: GGTGGAGCCTGCTTTTTTGT
1.Amplify the specific gene in a 50-µl PCR reaction as described in Basic Protocol 1, steps 1 and 2, using gene-specific primers.
2.Amplify the pDONR vectors using gene vector–specific primers (pDONR-IF-f/r) in a 50-µl PCR reaction at an annealing temperature of 58°C according to the manufacturer's instructions.
3.Use agarose gel electrophoresis (see Current Protocols article: Voytas, 2000) to check quality and yield of the products, then purify the PCR products using a nucleic acid gel extraction and purification kit.
4.Mix the two PCR products at a 1:1 ratio.
| PCR products (≥30 ng/μl; final amount, 100-150 ng) | 1-4 μl |
| pDONR vector (≥ 50 ng/μl; final amount, 100-150 ng) | 1-4 μl |
| TE buffer, pH 8.0 | To 8 μl |
5.Vortex the In-Fusion or Gibson enzyme mix briefly. Add 2 μl of the enzyme mix to the components above and mix well by vortexing briefly twice. Incubate the reaction at 50°C for 30 min to 1 hr.
6.Transform competent E. coli and select for the appropriate antibiotic-resistant pDONR vector as described in Basic Protocol 1.
Basic Protocol 2: TRANSFORMATION OF BAITS INTO THE PLANT CELL CULTURE OR PLANT LEAF
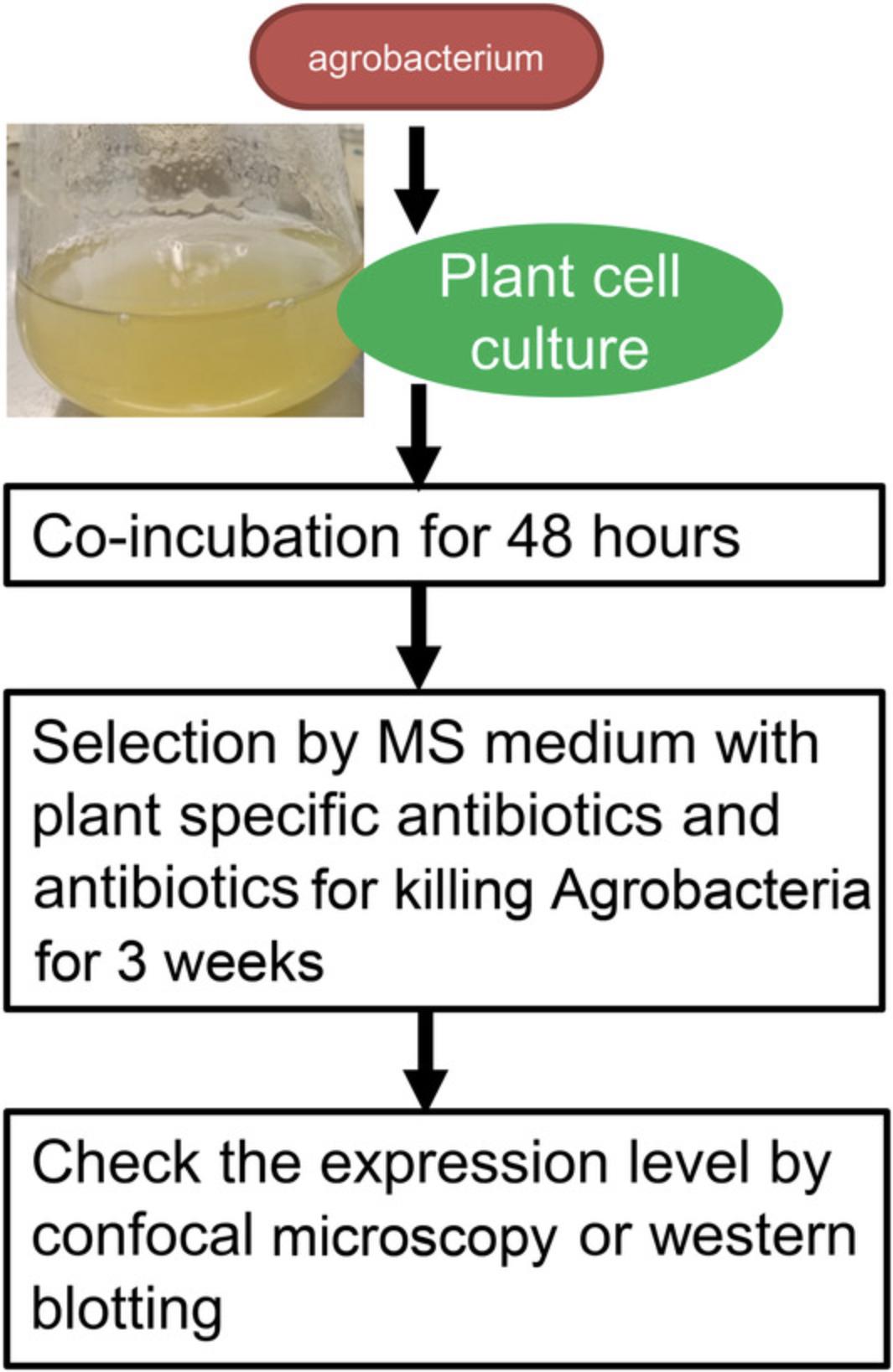
This protocol has been optimized for overexpressing the bait protein in the plant. The plant destination vectors containing plant promoter (such as ubquitin or actin) can be transformed into the plant cell culture within 1 month (Fig. 2). The cell culture could be harvested directly following transformation for the AP-MS.
Materials
-
Yeast extract beef (YEB) medium and agar plates (see recipe)
-
Appropriate antibiotics:
- Carbenicillin (Sigma, 4800-94-6)
- Rifampicin (Sigma, R3501)
- Ticarcellin clavulanic acid (Sigma, T5639)
- Vancomycin (Sigma, V1130)
- Kanamycin (Sigma, 60615)
- Vector-specific selection antibiotics
-
Agrobacterium tumefaciens AGL1 (Intact Genomics, 1283-12)
-
MSCC medium (see recipe)
-
0.1 M acetosyringone (Sigma, D134406) dissolved in dimethyl sulfoxide (DMSO)
-
2.5-ml culture tubes
-
2-ml microcentrifuge tubes
-
Electroporation apparatus and electroporation cuvettes
-
25° and 28°C shaking incubator
-
15-ml conical tubes (e.g., Corning Falcon)
-
50-ml and 100-ml Erlenmeyer flasks
-
Additional reagents and equipment for confocal microscopy (see Current Protocols article: Rajwa, 2005)
Agrobacterium tumefaciens transformation
1.Pour 20 ml YEB medium with carbenicillin (20 μg/L) and rifampicin (50 μg/L) but no gentamicin in a 2.5-ml liquid culture tube, add 200 μl from the frozen stock of Agrobacterium tumefaciens AGL1, and incubate the cultures overnight with shaking at 28°C.
2.Add 2 ml of the Agrobacterium overnight culture to a 2-ml tube and centrifuge for 30 s at 2000 × g , 4°C. Discard the supernatant, removing as much as the liquid as possible.
3.Put the sample on ice. Add 2 ml of ice-cold water, vortex, and centrifuge for 30 s at 14,000 × g , 4°C. Discard the supernatant. Repeat these steps with 1 ml, 500 μl, and 200 μl of ice-cold water. Do not discard the 200 μl water; resuspend the pellet in this volume and put on ice (these are the Agrobacterium competent cells).
4.Add 1 to 5 μl of the expression clone DNA sample into a 2-ml tube and place it on ice. Add 45 μl of Agrobacterium competent cell suspension from step 3 and incubate on ice 5 min. Put the solution into cold electroporation cuvettes and leave on ice.
5.Electroporate cells using an electroporation apparatus according to the manufacturer's instructions. Following electroporation, directly add 1 ml of YEB medium (with 20 µg/L carbenicillin and 50 µg/L rifampicin, but no gentamicin) and transfer the solution back into a new 2-ml tube.
6.Shake for 1 to 2 hr at 28°C.
7.Microcentrifuge for 1 min at 14,000 × g , discard the supernatant, and resuspend the pellet by pipetting up and down. Plate the bacteria on pre-warmed YEB plates (with 20 µg/L carbenicillin and 50 µg/L rifampicin, and the appropriate antibiotic for specific selection of your gene of interest, but no gentamicin) and incubate at 28°C for 2 to 3 days.
Cell culture transformation
8.DAY 1 (Wednesday) : Take one colony of the transformed Agrobacterium and plate it on a fresh YEB plate (with 20 µg/L carbenicillin, 50 µg/L rifampicin, and vector-specific antibiotics, but no gentamicin) with freshly grown Agrobacteria.
9.Incubate the YEB plate at 28°C for 2 days.
10.Dilute 10-ml 7-day-old Arabidopsis cell suspension cultures in 40 ml fresh MSCC (1/5 dilution).
11.DAY 3 (Friday) : Shave off Agrobacterium culture (see step 9) from plate, dissolve it in 2 ml MSCC medium in a 15-ml conical tube, and check the OD600. If OD600 is below 1.0, shave more Agrobacterium from plates and suspend in the same tube. Dilute the Agrobacteria to OD ∼1 using MSCC medium.
12.Co-cultivation/transformation : Take a 50-ml autoclaved Erlenmeyer flask and mix 6 ml 2-day-old Arabidopsis cell suspension culture with 12 µl 0.1 M acetosyringone. Combine 6 ml of Arabidopsis cell suspension/acetosyringone culture with 200 µl (300 µl and 400 µl for tests) of Agrobacterium culture from plate (OD ∼1.0). Close the flask and shake at 130 rpm for 72 hr at 25°C.
13.DAY 10 (Monday) : Add 20 ml MSCC plus 250 mg/L ticarcellin clavulanic acid (killing Agrobacteria), 250 mg/L vancomycin (killing Agrobacteria), and 25 mg/L kanamycin (plant cell culture selection) to a 50-ml flask and shake at 130 rpm 25°C for 5 days.
14.DAY 15 (Wednesday; only for direct transformation) : Transfer 10 ml (as much cells as possible) into a 100-ml flask containing 40 ml MSCC plus 250 mg/L ticarcellin clavulanic acid (killing Agrobacteria), 250 mg/L vancomycin (killing Agrobacteria) and 50 mg/L kanamycin (plant cell culture selection). Shake at 130 rpm at 25°C for 7 days.
15.DAY 22 (Wednesday; only for direct transformation : Transfer 10 ml of cells (after letting them sink down to the bottom) into 40 ml of MSCC plus 250 mg/L ticarcellin clavulanic acid (killing Agrobacteria), 250 mg/L vancomycin (killing Agrobacteria), and 50 mg/L kanamycin (plant cell culture selection) into a 100-ml flask. Shake at 130 rpm at 25°C for 7 days.
16.DAY 29 (Wednesday) only for direct transformation : Transfer as many cells as possible into 40 ml of MSCC with only 50 mg/L kanamycin (plant cell culture selection) in a 100-ml flask
17.Check the plants for protein expression via confocal microscopy (see Current Protocols article: Rajwa, 2005).
Basic Protocol 3: AFFINITY PURIFICATION OF PROTEIN COMPLEXES
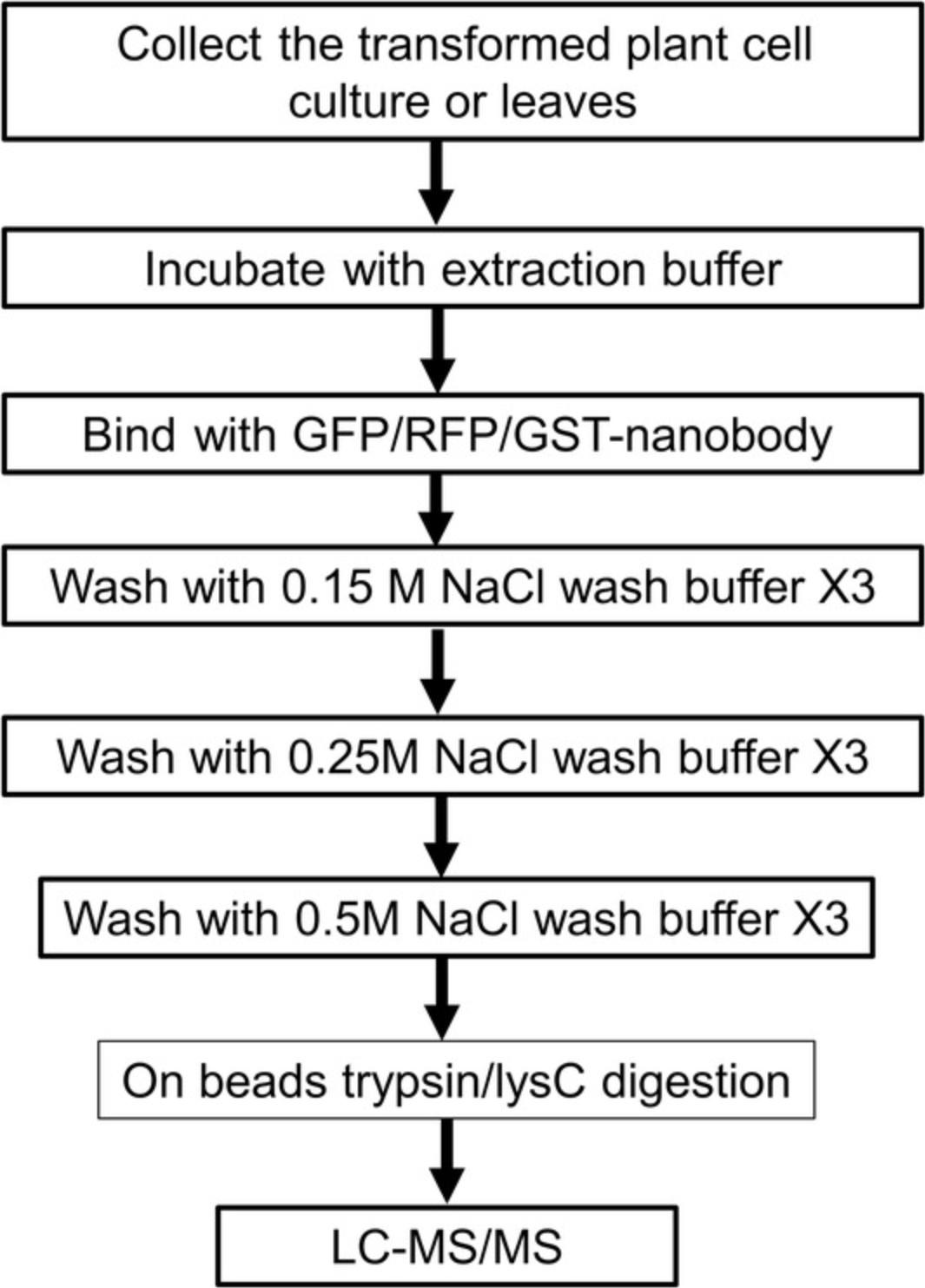
Protein complexes can be isolated through in vivo immunoprecipitation methods by using specific antibodies recognizing the bait protein. Given that it is incredibly laborious to directly use specific antibodies against plant proteins, an affinity tag protein (GFP or GS tag) fused to the protein of interest facilitates the development of a high-throughput affinity purification method for protein complexes (Van Leene et al., 2015). The target protein could be inserted into pUBC-GFP-Dest and pUBN-GFP-Dest (Grefen et al., 2010) and transformed into plant cell culture as mentioned above. Total protein extracts are collected from the transformed plant materials and incubated with affinity beads in order to purify the protein complexes (Fig. 3). The purified protein complexes are then measured by LC-MS.
Materials
-
Frozen plant cell power or plant tissue powder: grind plant cells or tissue into a fine powder with a mortar and pestle in the presence of liquid nitrogen; transfer powder to a tube and store in freezer
-
Extraction buffer (see recipe)
-
10× protease inhibitor cocktail (Sigma, P8340**)**.
-
ChromoTek GFP-Trap Nanobody beads (http://www.chromotek.com/products/nano-traps/)
-
Wash Buffer I (same as the extraction buffer)
-
Wash Buffer II (extraction buffer with 250 mM NaCl)
-
Wash Buffer III (extraction buffer with 500 mM NaCl)
-
15-ml conical polypropylene tubes (e.g., Corning Falcon)
-
2-ml microcentrifuge tubes
-
Refrigerated centrifuge
-
Spin Columns (ChromoTek, https://www.chromotek.com/products/detail/product-detail/spin-columns/)
-
Rotating shaker
1.Place ∼2 g of frozen plant cell powder or plant tissue powder in a 15-ml conical tube and add 2 ml extraction buffer. Use between three and four independent biological replicates.
2.Mix by vortexing for 1 min, incubate on ice for 5 min, add 100 µl 10× protease inhibitor, and repeat the vortexing three times.
3.Centrifuge 10 min at 3000 × g , 4°C, transfer 3 ml of the supernatant into 2-ml tubes, centrifuge at 20,000 × g at 4°C for 15 min, and transfer the supernatant to new 2-ml tubes. Repeat the centrifugation step and keep the supernatant for the following pull-down assays.
4.Wash 25 µl of GFP beads with 500 µl of extraction buffer three times in 2-ml tubes (remember to cut the ends of the pipette tips to widen the opening and mix well when taking the GFP beads), each time by centrifuging 1 min at 3000 × g , room temperature.
5.Transfer 2 ml of the supernatant from step 3 to a tube containing 25 µl of GFP beads, mix gently, and incubate at 4°C for 1 hr with rotation.
6.Centrifuge 1 min at 3000 × g , 4°C, and take out 1.6 ml of the supernatant. Using a cut-off pipet tip, transfer the rest of the supernatant and beads to a spin column. Centrifuge the columns 1 min at 3000 × g , 4°C.
7.Wash the spin columns containing the beads three times, each time for 1 min at 3000 × g , 4°C, using 500 µl of Wash buffer I.
8.Wash three times, each time for 1 min at 3000 × g , 4°C, using 500 µl of Wash buffer II.
9.Wash three times, each time for 1 min at 3000 × g , 4°C, using 500 µl of Wash buffer II.
Basic Protocol 4: ON-BEAD TRYPSIN/LysC DIGESTION AND C18 COLUMN PEPTIDE DESALTING AND CONCENTRATION
In-solution enzymatic protein digestion is a useful, and sometimes necessary, alternative to in-gel digestion. For samples of low content, or for samples not amenable to SDS-PAGE, in-solution digestion can be used and will provide similar results to in-gel digestion. However, protein folding can protect the amino acid chain from enzymatic cleavage, so denaturation is necessary for efficient cleavage. The conundrum with in-solution digestion is finding conditions to denature the sample without denaturing the protease. Detergents cannot be used in the denaturation process, since they will interfere with subsequent MS analysis. Common denaturants that we use in our laboratory for in-solution digestions include 8 M urea in 10 mM Tris·Cl (pH 8.0), 8 M guanidine HCl (pH 8.0), and 6 M urea/2 M thiourea in 10 mM Tris·Cl (pH 8.0). Unfortunately, trypsin, the most common protease for MS analysis, is not stable under any of these conditions, but fortunately another enzyme, LysC protease, is. LysC cleaves on the carboxyl side of lysine residues, while trypsin targets both lysine and arginine residues.
Materials
-
Protein sample of interest
-
6 M urea/2 M thiourea in 10 mM Tris·Cl, pH 8
-
Reduction buffer: 1 μg/μl (6.5 mM) dithiothreitol (DTT) in water
-
Alkylation buffer: 5 μg/μl (27 mM) iodoacetamide in water
-
10 mM Tris·Cl, pH 8
-
Trypsin/LysC proteases, modified sequencing grade (Promega): 0.4 µg/µl; i.e., in 50 µl, 20 µg)
-
100% methanol
-
80% (v/v) acetonitrile/0.1% trifluoroacetic acid (TFA) in distilled deionized water
-
0.1% and 2% (v/v) TFA in distilled deionized water
-
Resuspension solution: 0.2% (v/v) TFA/5% acetonitrile
-
Equilibration buffer A (100% H2O/0.2% TFA)
-
Elution buffer B (100% acetonitrile/0.2% TFA)
-
Bath sonicator
-
Refrigerated centrifuge
-
pH strips
-
C18 SepPak columns, 100 mg/ml
-
Visiprep™ 12- Port Vacuum Manifold (Sigma, 57044)
-
Vacuum pump
-
SpeedVac evaporator
-
Nano LC 1000 liquid chromatograph (ThermoFisher Scientific) with reversed-phase C18 column (Acclaim PepMap RSLC, 75 µm × 150 mm, C18, 2 µm, 100 Å; ThermoFisher Scientific)
In-solution digestion
1.Dissolve sample in a small volume of 6 M urea/2 M thiourea (pH 8.0). Use as low a volume as is compatible with your sample. Sonicate for 10 min to solubilize using a bath sonicator.
2.Centrifuge samples 10 min at 8000 × g , room temperature, to pellet any insoluble material.
3.Add 1 μl reduction buffer for every 50 μg of sample protein and incubate for 30 min at room temperature.
4.Add 1 μl alkylation buffer for every 50 μg sample protein and incubate for 20 min at room temperature in the dark.
5.Dilute sample with four volumes of 10 mM Tris·Cl, pH 8.
6.Add 1 μl of 0.4 µg/µl trypsin/LysC per 50 μg sample protein and incubate overnight at 37°C.
C18 Stage- SepPak for peptide desalting and concentration
7.Put the C18 Stage-SepPak columns in the Visiprep™ 12- Port Vacuum Manifolds and attach the vacuum pump.
8.Equilibrate the C18 SepPak columns with 1 ml 100% methanol and switch on the pump.
9.Equilibrate the C18 SepPak columns with 1 ml 80% acetonitrile/0.1% TFA in distilled, deionized water and switch on the pump.
10.Equilibrate with 1 ml of 0.1% TFA in distilled deionized water and switch on the pump. Repeat this step.
11.Dissolve samples in 0.1% TFA (add 1/10 volume of 2% TFA to reach pH 2.0). If the pH is too high, add 2% TFA, until it reaches a pH of 2.0.
12.Load the sample onto the SepPak columns, and switch on the pump.
13.Wash the tube that contained the digested sample with 200 µl 0.1% TFA, centrifuge 1 min at 1000 × g , and load this onto the column.
14.Wash the column with 1 ml of 0.1% TFA. Repeat this step.
15.Elute the peptides with 800 μl of 60% acetonitrile and 0.1% TFA into a new 1.5-ml microcentrifuge tube.
16.Dry the peptides in a SpeedVac evaporator.
17.Using a Nano LC 1000 liquid chromatograph with a reversed-phase C18 column (Acclaim PepMap RSLC, 75 µm × 150 mm, C18, 2 µm, 100 Å), perform mass spectrometric analysis as required by the experiment.
18.Add a final volume of 40 µl of resuspension solution (0.2% TFA/5% acetonitrile) to the sample and transfer it to a microtiter plate for mass spectrometric analysis.
Basic Protocol 5: DATA ANALYSIS AND QUALITY CONTROL
The ribosome protein and translation-related protein could be deleted at this step. The normalized signal intensities are processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software (Mellacheruvu et al., 2013). Compared with the GFP control, the background protein could be deleted at this step by FC-A values of at least four within at least three replicates (Morris et al., 2014). MS/MS information from the SUBA4 database (Hooper, Castleden, Tanz, Aryamanesh, & Millar, 2016) could give the subcellular localization of the bait and prey, and thus improve the reliably of the interactions. Finally, we consider only the protein pairs for which the intensities are in the top 2% compared with the bait intensity to represent positive interactions.
1.Following the LC-MS measurement, the ribosomal and translation-related proteins can be deleted unless there is a specific interest in translation-regulatory proteins.
2.The normalized intensity of all the bait and tag control lines can be analyzed by the CRAPome software (you need to follow the introduction to the software to prepare the interaction files (Fig. 4A).
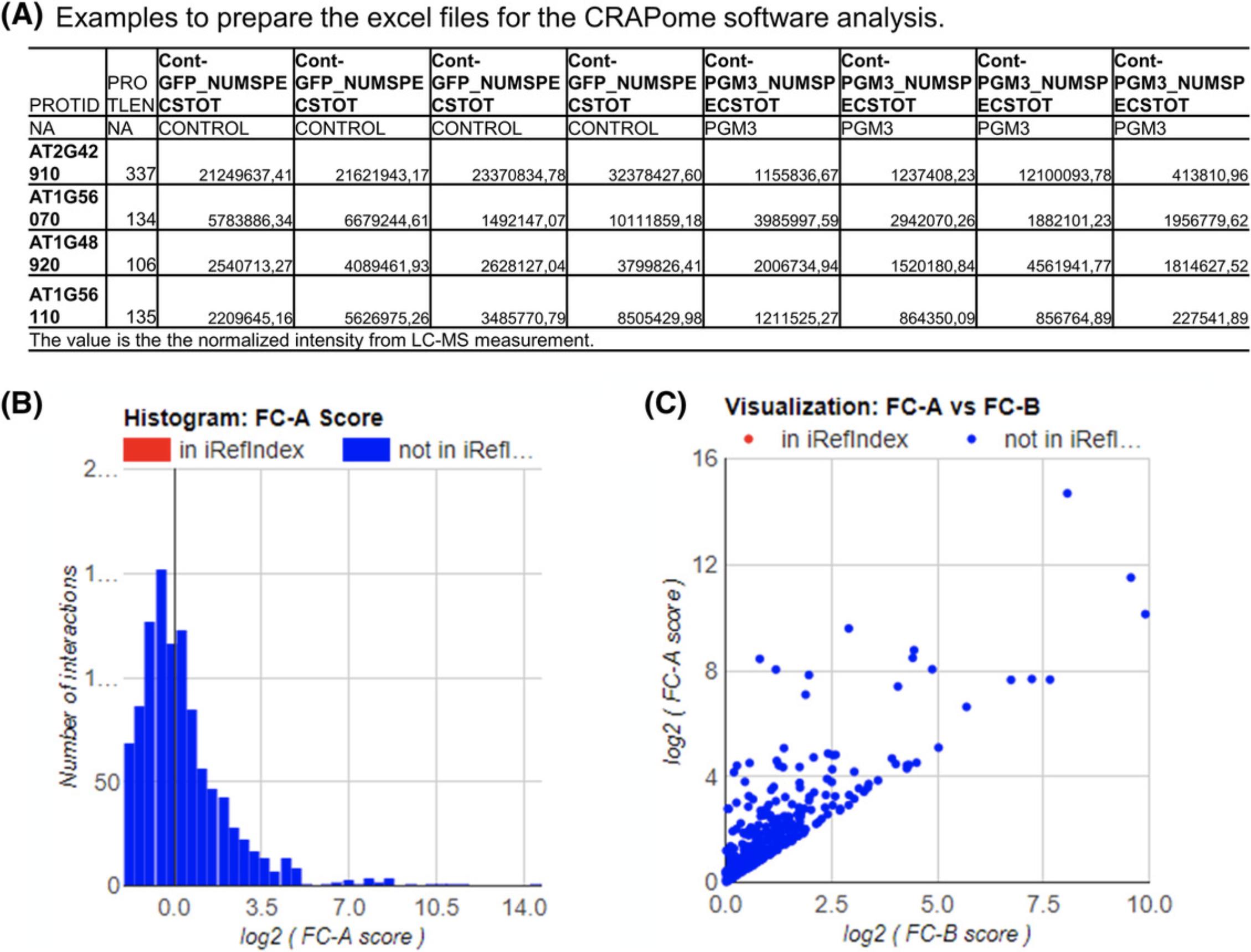
3.After getting the resulting lists of candidate interactors with the corresponding fold change (FC; Fig. 4B and C), interactions with FC greater than 4 can be selected as possible positive interactions. Only the intensities of interactors that have greater than 2% of the intensity of the GFP protein are selected.
4.The possible target prey proteins also need to be analyzed by the SUBA4 database to get the right subcellular localization with the bait protein.
5.The protein interaction network can be presented by Cytoscape (https://cytoscape.org/; Shannon et al., 2003).
REAGENTS AND SOLUTIONS
Extraction buffer
| Final concentration | For 10 ml | Stock | |
|---|---|---|---|
| Tris·Cl, pH 7.5 | 25 mM | 500 µl | 0.5 M |
| MgCl2 | 15 mM | 150 µl | 1 M |
| EGTA | 5 nM | 250 µl | 200 mM |
| DTT | 1 mM | 10 µl | 1 M |
| PMSF | 1 mM | 100 µl | 0.1 M |
| NaCl | 150 mM | 300 µl | 5 M |
| H2O | 8.69 ml |
- Store up to 6 months at 4°C
Lysogeny broth (LB) medium and agar plates
- 10 g/L tryptone
- 5 g/L yeast extract
- 10 g/L NaCl
- For LB agar plates, add 15 g agar per liter
- Store medium or plates up to 3 months at 4°C
MSCC medium
Prepare 4.43 g Murashige & Skoog salts with minimal organics (Sigma, M6899) with 30 g sucrose in a volume of 1 liter and adjust pH to 5.7 with KOH. Autoclave. Before use, add freshly prepared 50 µl kinetin [1 mg/ml kinetin (Sigma, K0753)/0.1 M NaOH] and 500 µl NAA [1 mg/ml α-naphtaleneacetic acid (NAA; Sigma, N0640)/0.1 M NaOH]. Also add the following antibiotics:
- 500 µl kanamycin (50 mg/ml, plant cell culture selection) or 500 µl hygromycin (10 mg/ml, plant cell culture selection)
- 2000 µl ticarcellin clavulanic acid, (125 mg/ml, killing Agrobacteria)
- 2500 µl vancomycin (100 mg/ml, killing Agrobacteria).
Yeast extract beef (YEB) medium and plates
- 1.0 g/L yeast extract
- 5.0 g/L beef extract
- 5.0 g/L peptone
- 5.0 g/L sucrose
- 20 g/L agarose (for plates)
- 1000 ml distilled water
- Autoclave
- Freshly add antibiotics:
- Rifampicin (50 µg/ml final concentration)
- Gentamicin (20 µg/ml final concentration)
- Vector-specific antibiotics
- Store up to 6 months at room temperature
COMMENTARY
Background Information
A protein complex is a group of two or more proteins associated by different or the same functional polypeptide chains by non-covalent interactions in vivo (Hartwell, Hopfield, Leibler, & Murray, 1999). They are usually organized into functional modules to play central roles in regulating DNA replication, transcription, translation, RNA splicing, protein secretion, cell cycle control, signal transduction, and intermediary metabolism (Bontinck et al., 2018). As they are the basis of many biological processes, studying these complexes and exposing their intricate interaction networks are thus of fundamental importance to understand not only basic cellular processes but also complex developmental programs. Several methods for analyzing protein-protein interactions (PPIs) are available, such as yeast two-hybrid (Y2H; Parrish, Gulyas, & Finley, 2006), co-immunoprecipitation followed by western blotting (co-IP; Antrobus & Borner, 2011), or protein-fragment complementation assays such as bimolecular fluorescence complementation (BiFC; Kerppola, 2008) and split luciferase (Chen et al., 2008; Fujikawa & Kato, 2007; Li, Bush, Xiong, Li, & McCormack, 2011). However, most of these methods allow testing PPIs only in a pairwise fashion or as three-protein interactions, and require prior knowledge to determine which combinations to test. Thus, a complementary method that is more suited to study co-complex memberships is AP-MS (Mellacheruvu et al., 2013). This method can affinity-purify protein complexes under near-physiological conditions to maintain PPIs intact, which is followed by their detection with mass spectrometry (Choi et al., 2011).
In AP-MS, PPIs can be captured by antibodies specific for the bait proteins or for tags that were introduced on the bait proteins and pulled down onto immobilized protein agarose beads or magnetic agarose beads (Zhang, Sun, et al., 2017). The affinity-purified protein complexes can be further digested into peptides by trypsin/LysC and identified by quantification of the resulting peptides via mass spectrometry. As the specific interactors are enriched in the bait sample, this method can produce a large amount of information-rich data that detail protein-protein interactions in different organisms and biological systems or different conditions and treatment (see Current Protocols article: Adelmant, Garg, Tavares, Card, & Marto, 2019). Using the putative interactions information, we can further confirm interactions by other binary interaction methods in order to characterize the functions of proteins and provide detailed catalogs of proteins involved in protein complexes and biological processes. These interactions could also reveal networks of biological processes at local and proteome-wide scales to further help us understand the genetic, epigenetic, and protein-based associations of these proteins. To establish a reliable protein interaction network, a well-established procedure is needed including sample preparation, diverse interaction scoring and clustering algorithms, methods for graph theory and data mining, and biological networks. In this article, we describe plant cell culture transformation, sample preparation, affinity purification, in-solution digestion, mass spectrometry detection, and, finally, data analysis required to produce meaningful networks.
In addition, the success of AP-MS depends on several factors, including high expression levels of bait protein, the extraction of protein complexes, the antibody to enrich the bait protein and preserve the protein complexes, the efficiency of trypsin digestion, and the recovery of the tryptic peptides for MS analysis (Oeffinger, 2012; Varjosalo et al., 2013). Here, we suggest using a GFP tag for the bait protein, which facilitates the detection of the protein localization by confocal microscopy (Dunham, Mullin, & Gingras, 2012; Keilhauer, Hein, & Mann, 2015). The efficiency of the trypsin digestion and the recovery of the resulting digested peptides for MS analysis are very important for the success of AP-MS (Zhang, Swart, et al., 2018). Instead of in-gel digestion for performing global proteomics profiling (Huang et al., 2016; Van Leene et al., 2015), several studies have used the improved efficiency of in-solution digestion on the beads, reducing time and steps (León, Schwämmle, Jensen, & Sprenger, 2013; Zhang, Sun, et al., 2017). These studies have shown that the choice of chaotropic agent, surfactant, or organic solvent has a significant impact on the efficiency, reproducibility, and completeness of trypsin digestion, and hence affects sequence coverage of protein identification by MS analysis. Here, we use the simplified method of digesting the protein in the urea/thiourea solution with both LysC and trypsin, and desalt the tryptic peptide in a C18 Stage-SepPak for the mass spectrometry.
Critical Parameters and Troubleshooting
The following troubleshooting guide does not include common issues that may arise when using reagents other than those recommended in the protocols (such as using anti-GFP-agarose and anti-RFP-agarose beads from suppliers other than ChromoTek), when a different tag is used for the bait protein (such as GS tag, GST tag), or when a different digestion method is used.
Protein expression in the plant cell culture
The protein expression level should be checked by confocal microscopy before starting the affinity purification. For subcellularly localized proteins (nuclear or membrane), the specific procedure for breaking the nucleus or membrane should be perfomed first and the protein expression level can be confirmed by western blotting before the AP-MS. Normally, the ubiquitin 10 promoter can express enough protein, while the 35S promoter can result in many false-positive interactions because of the higher expression. Plant cell cultures can be treated with different buffers or environments for different pull-down conditions.
Tag used for the affinity purification
As they enable protein expression level to be easily detected, GFP-, RFP-, or mCherry-tagged baits are suggested in this procedure. The GSrhino-TAP and glutathione S -transferase (GST) tags can also be used in affinity purification from plant, while the HA, FLAG, and His tags are not suggested for affinity purification from plant materials, since these three tags result in very strong background signals in plant.
Biological replicates and statistical analysis
In order to obtain a reliable protein interaction network, we recommend using more replicates (at least three) for the statistical analysis. Normalized intensities are used for the data analysis. For some poor reproducibility samples, six replicates are suggested. In addition, the proteins that only have intensity in bait samples are suggested as the candidate interactors.
Negative control
Given that the AP-MS produces large amounts of information-rich data, both the type of bead and the tag affect the affinity-purified interactors. Using the same subcellular localization of GFP as negative control, it can very importantly exclude the false-positive interactions. Proteins localized to subcellular compartments different from the bait protein are likely false positives and should be excluded. Here, we suggest using SUBA4 (http://suba.live/) to exclude subcellular localizations that are different from the bait protein. Given that large amounts of ribosomal proteins and proteins related to translation are detected from the AP-MS, these interactors should also be excluded. It is important to note that using a different plant material as control will result in lots of false-positive or false-negative interactors, e.g., if using transformed plant cell culture for the samples analysis while using the transformed seedling GFP lines as control.
Data analysis
In the data analysis, normalized signal intensities are processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software (Morris et al., 2014). Compared with intensity of bait, only the proteins for which the intensity score was more than 2%, corresponding to FC-A values of at least 4 within at least three replicates, should be regarded as positive interactions. Screening the SUBA4 database (Hooper et al., 2016), only pairs sharing the same subcellular localization are selected as positive interactors. In addition, transient interactions also play an important role within protein-protein interaction networks, especially post-translational modifications (Perkins, Diboun, Dessailly, Lees, & Orengo, 2010). The proteins that have low intensity in bait samples and no intensity in the negative control can be selected.
Understanding Results
The affinity purification protocol presented has been used by us and our collaborators to characterize binding partners of proteins spanning a variety of functional categories. This protocol should enable both novices and skilled biochemists alike to obtain valuable and meaningful information about interaction partners and help generate novel hypotheses.
TCA cycle interaction network
The 38 mitochondrial proteins of Arabidopsis thaliana were transformed into a PSB-D Arabidopsis plant cell culture, and a GFP-tag-based modified AP-MS procedure was implemented based on at least three biological replicates (Zhang, Beard, et al., 2017; Zhang, Swart, et al., 2018). Unlike normal AP-MS in which the gel is cut into pieces for several independent trypsin digestions (Morris et al., 2014), we used a proteomics-based in-solution digestion method to directly digest the proteins on the beads following affinity purification (Zhang, Sun, et al., 2017). Thus, an AP-MS experiment constitutes a single sample for the LC-MS measurement. In the subsequent data analysis, normalized signal intensities were processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software (Morris et al., 2014). A total of 3421 protein-protein interactions were obtained displaying in excess of four-fold changes in the five independent experiments. We considered only the protein pairs for which the protein intensity was in the top 2% compared with bait protein, corresponding to FC-A values of at least 4 within at least three of the replicates as positive interactions. A total of 449 potential positive protein-protein interactions were obtained according to these criteria, including those interactions with several ribosomal and protein-translation proteins.
As we are interested in mitochondrial interactions, only the mitochondrially targeted proteins were selected for network generation. It is, however, important to note that given that many of the enzymes of the TCA cycle have isoforms (exhibiting high identity), in more than one compartment the non-mitochondrial interactions, while not directly physiologically relevant, may well provide hints to interactions that do occur in vivo albeit extra-mitochondrially. Screening of the SUBA4 database (Hooper et al., 2016) revealed a total of 257 interactions between mitochondrially localized TCA cycle proteins and 37 of the proteins comprising the mitochondrial interaction network. Of these 257 interactions, 132 interactions between the enzymes of TCA cycle had already been reported (Zhang, Beard, et al., 2017), while there were 125 novel interactions between subunits of enzymes and other pathway enzymes or proteins (Zhang, Swart, et al., 2018).
Time Considerations
Identification of protein-protein interaction networks comprises multiple steps which can be accomplished within 1 to 2 months. The project is easily divided among the following independent stages: gene cloning will require 1 week, plant cell culture transformation will require 1 month, and finally the affinity purification with mass spectrometry and subsequent data analysis will require 2 to 4 weeks of work.
The time needed for processing of protein interaction networks depends on the (i) transformation of plant cell culture, (ii) transient expression of plant leaves, and (iii) mass spectrometry measurement. In our experiment, a full run that includes all the steps can be finished within 2 months.
Acknowledgment
This work was supported by funding from the Max Planck Society (Y.Z. and A.R.F.) and the European Union's Horizon 2020 research and innovation program, project PlantaSYST (Y.Z. and A.R.F.), and São Paulo Research Foundation (FAPESP, grant number 2018/20572-2 to A.P.D.-J).
Literature Cited
- Adelmant, G., Garg, B. K., Tavares, M., Card, J. D., & Marto, J. A. (2019). Tandem affinity purification and mass spectrometry (TAP-MS) for the analysis of protein complexes. Current Protocols in Protein Science , 96, e84. doi: 10.1002/cpps.84.
- Antrobus, R., & Borner, G. H. (2011). Improved elution conditions for native co-immunoprecipitation. PloS One , 6(3), e18218. doi: 10.1371/journal.pone.0018218.
- Bontinck, M., Van Leene, J., Gadeyne, A., De Rybel, B., Eeckhout, D., Nelissen, H., & De Jaeger, G. (2018). Recent trends in plant protein complex analysis in a developmental context. Frontiers in Plant Science , 9, 640. doi: 10.3389/fpls.2018.00640.
- Bürckstümmer, T., Bennett, K. L., Preradovic, A., Schütze, G., Hantschel, O., Superti-Furga, G., & Bauch, A. (2006). An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nature Methods , 3(12), 1013–1019. doi: 10.1038/nmeth968.
- Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., … Zhou, J.-M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiology , 146(2), 368–376. doi: 10.1104/pp.107.111740.
- Choi, H., Larsen, B., Lin, Z.-Y., Breitkreutz, A., Mellacheruvu, D., Fermin, D., … Nesvizhskii, A. I. (2011). SAINT: Probabilistic scoring of affinity purification—Mass spectrometry data. Nature Methods , 8(1), 70–73. doi: 10.1038/nmeth.1541.
- Current Protocols. (2001). Common stock solutions, buffer, and media. Current Protocols in Cytometry , 9, A.2A.1–A.2A.4. doi: 10.1002/0471142956.cya02as09.
- Curtis, M. D., & Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology , 133(2), 462–469. doi: 10.1104/pp.103.027979.
- Dunham, W. H., Mullin, M., & Gingras, A. C. (2012). Affinity-purification coupled to mass spectrometry: Basic principles and strategies. Proteomics , 12(10), 1576–1590. doi: 10.1002/pmic.201100523.
- Fujikawa, Y., & Kato, N. (2007). Technical advance: Split luciferase complementation assay to study protein interactions in Arabidopsis protoplasts. The Plant Journal , 52(1), 185–195. doi: 10.1111/j.1365-313X.2007.03214.x.
- Grefen, C., Donald, N., Hashimoto, K., Kudla, J., Schumacher, K., & Blatt, M. R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. The Plant Journal , 64(2), 355–365. doi: 10.1111/j.1365-313X.2010.04322.x.
- Hartwell, L. H., Hopfield, J. J., Leibler, S., & Murray, A. W. (1999). From molecular to modular cell biology. Nature , 402(6761 Suppl), C47–C52. doi: 10.1038/35011540.
- Hooper, C. M., Castleden, I. R., Tanz, S. K., Aryamanesh, N., & Millar, A. H. (2016). SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Research , 45(D1), D1064–D1074. doi: 10.1093/nar/gkw1041.
- Huang, H., Alvarez, S., Bindbeutel, R., Shen, Z., Naldrett, M. J., Evans, B. S., … Nusinow, D. A. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Molecular & Cellular Proteomics, 15(1), 201–217.
- Katzen, F. (2007). Gateway® recombinational cloning: A biological operating system. Expert Opinion on Drug Discovery , 2(4), 571–589. doi: 10.1517/17460441.2.4.571.
- Keilhauer, E. C., Hein, M. Y., & Mann, M. (2015). Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE-MS) rather than affinity purification mass spectrometry (AP-MS). Molecular & Cellular Proteomics, 14(1), 120–135.
- Kerppola, T. K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annual Review of Biophysics , 37, 465–487. doi: 10.1146/annurev.biophys.37.032807.125842.
- Lazo, G. R., Stein, P. A., & Ludwig, R. A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio/Technology , 9(10), 963–967. doi: 10.1038/nbt1091-963.
- León, I. R., Schwämmle, V., Jensen, O. N., & Sprenger, R. R. (2013). Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Molecular & Cellular Proteomics, 12(10), 2992–3005.
- Li, J.-F., Bush, J., Xiong, Y., Li, L., & McCormack, M. (2011). Large-scale protein-protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PloS One , 6(11), e27364. doi: 10.1371/journal.pone.0027364.
- Mellacheruvu, D., Wright, Z., Couzens, A. L., Lambert, J.-P., St-Denis, N. A., Li, T., … Low, T. Y. (2013). The CRAPome: A contaminant repository for affinity purification—Mass spectrometry data. Nature Methods , 10(8), 730–736. doi: 10.1038/nmeth.2557.
- Menges, M., & Murray, J. A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. The Plant Journal , 30(2), 203–212. doi: 10.1046/j.1365-313X.2002.01274.x.
- Morris, J. H., Knudsen, G. M., Verschueren, E., Johnson, J. R., Cimermancic, P., Greninger, A. L., & Pico, A. R. (2014). Affinity purification—Mass spectrometry and network analysis to understand protein-protein interactions. Nature Protocols , 9(11), 2539–2554. doi: 10.1038/nprot.2014.164.
- Oeffinger, M. (2012). Two steps forward—one step back: Advances in affinity purification mass spectrometry of macromolecular complexes. Proteomics , 12(10), 1591–1608. doi: 10.1002/pmic.201100509.
- Parrish, J. R., Gulyas, K. D., & Finley, Jr, R. L. (2006). Yeast two-hybrid contributions to interactome mapping. Current Opinion in Biotechnology , 17(4), 387–393. doi: 10.1016/j.copbio.2006.06.006.
- Perkins, J. R., Diboun, I., Dessailly, B. H., Lees, J. G., & Orengo, C. (2010). Transient protein-protein interactions: Structural, functional, and network properties. Structure , 18(10), 1233–1243. doi: 10.1016/j.str.2010.08.007.
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., … Séraphin, B. (2001). The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods , 24(3), 218–229. doi: 10.1006/meth.2001.1183.
- Rajwa, B. (2005). Modern confocal microscopy. Current Protocols in Cytometry , 31, 12.3.1–12.3.12. doi: 10.1002/0471142956.cy1203s31 .
- Sanchez-Serrano, J. J., & Salinas, J. (2014). Arabidopsis protocols. New York: Humana Press.
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., … Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research , 13(11), 2498–2504.doi: 10.1101/gr.1239303.
- Shendure, J. A., Porreca, G. J., Church, G. M., Gardner, A. F., Hendrickson, C. L., Kieleczawa, J., … Slatko, B.E. (2011). Overview of DNA sequencing strategies. Current Protocols in Molecular Biology , 96, 7.1.1–7.1.23. doi: 10.1002/0471142727.mb0701s96.
- Van Leene, J., Eeckhout, D., Cannoot, B., De Winne, N., Persiau, G., Van De Slijke, E., … Vandepoele, K. (2015). An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nature Protocols , 10(1), 169–187. doi: 10.1038/nprot.2014.199.
- Varjosalo, M., Sacco, R., Stukalov, A., Van Drogen, A., Planyavsky, M., Hauri, S., … Gstaiger, M. (2013). Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nature Methods , 10(4), 307–314. doi: 10.1038/nmeth.2400.
- Walhout, A. J., Temple, G. F., Brasch, M. A., Hartley, J. L., Lorson, M. A., van den Heuvel, S., & Vidal, M. (2000). GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods in Enzymology , 328, 575–592.
- Voytas, D. (2000). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51, 2.5A.1−2.5A.9. doi: 10.1002/0471142727.mb0205as51.
- Zhang, Y., Beard, K. F., Swart, C., Bergmann, S., Krahnert, I., Nikoloski, Z., … Fernie, A. R. (2017). Protein-protein interactions and metabolite channeling in the plant tricarboxylic acid cycle. Nature Communications , 8, 15212. doi: 10.1038/ncomms15212.
- Zhang, Y., Sun, H., Zhang, J., Brasier, A. R., & Zhao, Y. (2017). Quantitative assessment of the effects of trypsin digestion methods on affinity purification—Mass spectrometry-based protein-protein interaction analysis. Journal of Proteome Research , 16(8), 3068–3082. doi: 10.1021/acs.jproteome.7b00432.
- Zhang, Y., Swart, C., Alseekh, S., Scossa, F., Jiang, L., Obata, T., … Fernie, A. R. (2018). The extra-pathway interactome of the TCA cycle: Expected and unexpected metabolic interactions. Plant Physiology , 177(3), 966–979. doi: 10.1104/pp.17.01687.
Citing Literature
Number of times cited according to CrossRef: 24
- Youjun Zhang, Kezhen Qin, Alisdair R. Fernie, Plant Tissue Culture and Metabolite Profiling for High-Value Natural Product Synthesis, Plant Cell Culture Protocols, 10.1007/978-1-0716-3954-2_27, (405-416), (2024).
- I. A. Sedlov, I. A. Fesenko, Methods for Interactome Analysis of Microproteins Encoded by Small Open Reading Frames, Биоорганическая химия, 10.31857/S0132342323040395, 49 , 4, (333-347), (2023).
- I. A. Sedlov, I. A. Fesenko, Methods for Analysis of Interactome of Microproteins Encoded by Short Open Reading Frames, Russian Journal of Bioorganic Chemistry, 10.1134/S1068162023040179, 49 , 4, (717-730), (2023).
- Youjun Zhang, Moxian Chen, Tieyuan Liu, Kezhen Qin, Alisdair R. Fernie, Investigating the dynamics of protein–protein interactions in plants, The Plant Journal, 10.1111/tpj.16182, 114 , 4, (965-983), (2023).
- Ismail Dahmani, Kezhen Qin, Youjun Zhang, Alisdair R. Fernie, The formation and function of plant metabolons, The Plant Journal, 10.1111/tpj.16179, 114 , 5, (1080-1092), (2023).
- Haiji Qiu, Xiaoliang Zhang, Youjun Zhang, Xiaohui Jiang, Yujia Ren, Dawei Gao, Xiang Zhu, Björn Usadel, Alisdair R. Fernie, Weiwei Wen, Depicting the genetic and metabolic panorama of chemical diversity in the tea plant, Plant Biotechnology Journal, 10.1111/pbi.14241, 22 , 4, (1001-1016), (2023).
- Hongjian Wan, Youjun Zhang, Limin Wu, Guozhi Zhou, Luzhao Pan, Alisdair R Fernie, Yong-Ling Ruan, Evolution of cytosolic and organellar invertases empowered the colonization and thriving of land plants, Plant Physiology, 10.1093/plphys/kiad401, 193 , 2, (1227-1243), (2023).
- Youjun Zhang, Silvia Martínez Jaime, Mustafa Bulut, Alexander Graf, Alisdair R. Fernie, The conditional mitochondrial protein complexome in the Arabidopsis thaliana root and shoot, Plant Communications, 10.1016/j.xplc.2023.100635, 4 , 5, (100635), (2023).
- Youjun Zhang, Lorenz Wiese, Hao Fang, Saleh Alseekh, Leonardo Perez de Souza, Federico Scossa, John J. Molloy, Mathias Christmann, Alisdair R. Fernie, Synthetic biology identifies the minimal gene set required for paclitaxel biosynthesis in a plant chassis, Molecular Plant, 10.1016/j.molp.2023.10.016, 16 , 12, (1951-1961), (2023).
- Roberto Natale, Mariangela Coppola, Nunzio D'Agostino, Youjun Zhang, Alisdair Robert Fernie, Valeria Castaldi, Rosa Rao, In silico and in vitro approaches allow the identification of the Prosystemin molecular network, Computational and Structural Biotechnology Journal, 10.1016/j.csbj.2022.12.006, 21 , (212-223), (2023).
- Nur Hikmah Mostaffa, Ahmad Husaini Suhaimi, Aisyafaznim Al-Idrus, Interactomics in plant defence: progress and opportunities, Molecular Biology Reports, 10.1007/s11033-023-08345-0, 50 , 5, (4605-4618), (2023).
- Mst Hur Madina, Parthasarathy Santhanam, Yanick Asselin, Rajdeep Jaswal, Richard R. Bélanger, Progress and Challenges in Elucidating the Functional Role of Effectors in the Soybean-Phytophthora sojae Interaction, Journal of Fungi, 10.3390/jof9010012, 9 , 1, (12), (2022).
- Vivian Robin, Antoine Bodein, Marie-Pier Scott-Boyer, Mickaël Leclercq, Olivier Périn, Arnaud Droit, Overview of methods for characterization and visualization of a protein–protein interaction network in a multi-omics integration context, Frontiers in Molecular Biosciences, 10.3389/fmolb.2022.962799, 9 , (2022).
- Shijuan Yan, Ruchika Bhawal, Zhibin Yin, Theodore W. Thannhauser, Sheng Zhang, Recent advances in proteomics and metabolomics in plants, Molecular Horticulture, 10.1186/s43897-022-00038-9, 2 , 1, (2022).
- Youjun Zhang, Alisdair R. Fernie, Metabolite profiling of Arabidopsis mutants of lower glycolysis, Scientific Data, 10.1038/s41597-022-01673-z, 9 , 1, (2022).
- Sandra M. Kerbler, Roberto Natale, Alisdair R. Fernie, Youjun Zhang, From Affinity to Proximity Techniques to Investigate Protein Complexes in Plants, International Journal of Molecular Sciences, 10.3390/ijms22137101, 22 , 13, (7101), (2021).
- Jie Pan, Li-Ping Li, Chang-Qing Yu, Zhu-Hong You, Yong-Jian Guan, Zhong-Hao Ren, Sequence-Based Prediction of Plant Protein-Protein Interactions by Combining Discrete Sine Transformation With Rotation Forest, Evolutionary Bioinformatics, 10.1177/11769343211050067, 17 , (2021).
- Youjun Zhang, Alisdair R. Fernie, Stable and Temporary Enzyme Complexes and Metabolons Involved in Energy and Redox Metabolism, Antioxidants & Redox Signaling, 10.1089/ars.2019.7981, 35 , 10, (788-807), (2021).
- Youjun Zhang, Jonas Giese, Sandra M. Kerbler, Beata Siemiatkowska, Leonardo Perez de Souza, Jessica Alpers, David Barbosa Medeiros, Dirk K. Hincha, Danilo M. Daloso, Mark Stitt, Iris Finkemeier, Alisdair R. Fernie, Two mitochondrial phosphatases, PP2c63 and Sal2, are required for posttranslational regulation of the TCA cycle in Arabidopsis, Molecular Plant, 10.1016/j.molp.2021.03.023, 14 , 7, (1104-1118), (2021).
- Ramesh Namdeo Pudake, undefined Pallavi, Mrinalini Singh Pundir, Proteomics for Understanding the Interaction Between Plant and Rhizospheric Microflora, Omics Science for Rhizosphere Biology, 10.1007/978-981-16-0889-6_7, (113-129), (2021).
- Youjun Zhang, Alisdair Fernie, On the Detection and Functional Significance of the Protein–Protein Interactions of Mitochondrial Transport Proteins, Biomolecules, 10.3390/biom10081107, 10 , 8, (1107), (2020).
- Youjun Zhang, Arun Sampathkumar, Sandra Mae-Lin Kerber, Corné Swart, Carsten Hille, Kumar Seerangan, Alexander Graf, Lee Sweetlove, Alisdair R. Fernie, A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts, Nature Communications, 10.1038/s41467-020-18234-w, 11 , 1, (2020).
- Youjun Zhang, Alisdair R. Fernie, Metabolons, Enzyme–Enzyme Assemblies that Mediate Substrate Channeling, and Their Roles in Plant Metabolism, Plant Communications, 10.1016/j.xplc.2020.100081, (100081), (2020).
- Youjun Zhang, Moxian Chen, Beata Siemiatkowska, Mitchell Rey Toleco, Yue Jing, Vivien Strotmann, Jianghua Zhang, Yvonne Stahl, Alisdair R. Fernie, A Highly Efficient Agrobacterium-Mediated Method for Transient Gene Expression and Functional Studies in Multipe Plant Species, Plant Communications, 10.1016/j.xplc.2020.100028, (100028), (2020).

