Querying the NCBI database for GenomeTrakr data
Ruth Timme, Julie Haendiges, Maria Balkey, Candace Hope Bias
Disclaimer
Please note that this protocol is public domain, which supersedes the CC-BY license default used by protocols.io.
Abstract
This protocol describes methods to query GenomeTrakr sequencing records and metadata across multiple NCBI resources: BioSample, BioProject, Sequencing Read Archive, Pathogen Detection and Assembly databases.
Steps
NCBI Resources
Whole Genome Sequencing data submitted to NCBI is processed in multiple databases. If your laboratory or collaborator has submitted WGS data for foodborne pathogens to NCBI, you can locate the data at the NCBI resources: BioProject, BioSample, Sequencing Read Archive, Pathogen Detection and Assembly.
You can access NCBI databases at: https://www.ncbi.nlm.nih.gov/guide/all/
BioProject
BioProject is a collection of biological data related to a surveillance or research effort. Umbrella bioprojects contain several data-level projects. PRJNA593772 ( https://www.ncbi.nlm.nih.gov/bioproject/593772) comprises a set of umbrella bioprojects, each established for a pathogen being sequenced by the GenomeTrakr network. If you need to find a BioProjects for an specific organism processed by your lab, click at the corresponding organism umbrella Bioproject.
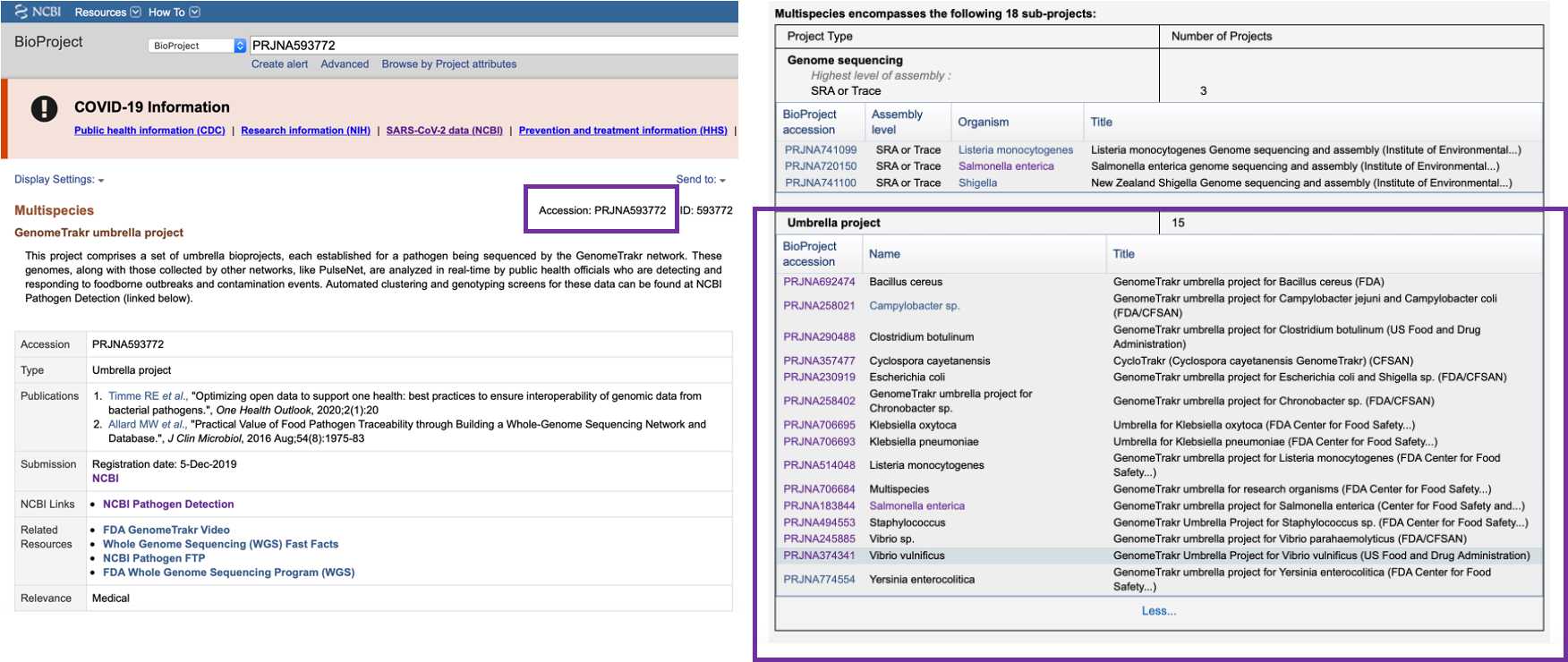
If you are interested in searching for specific type of data, you can click on Browse by Project attributes and narrow your search by using filters such as: Project, Data Type, Scope, Property , Kingdom, Group, Subgroup.
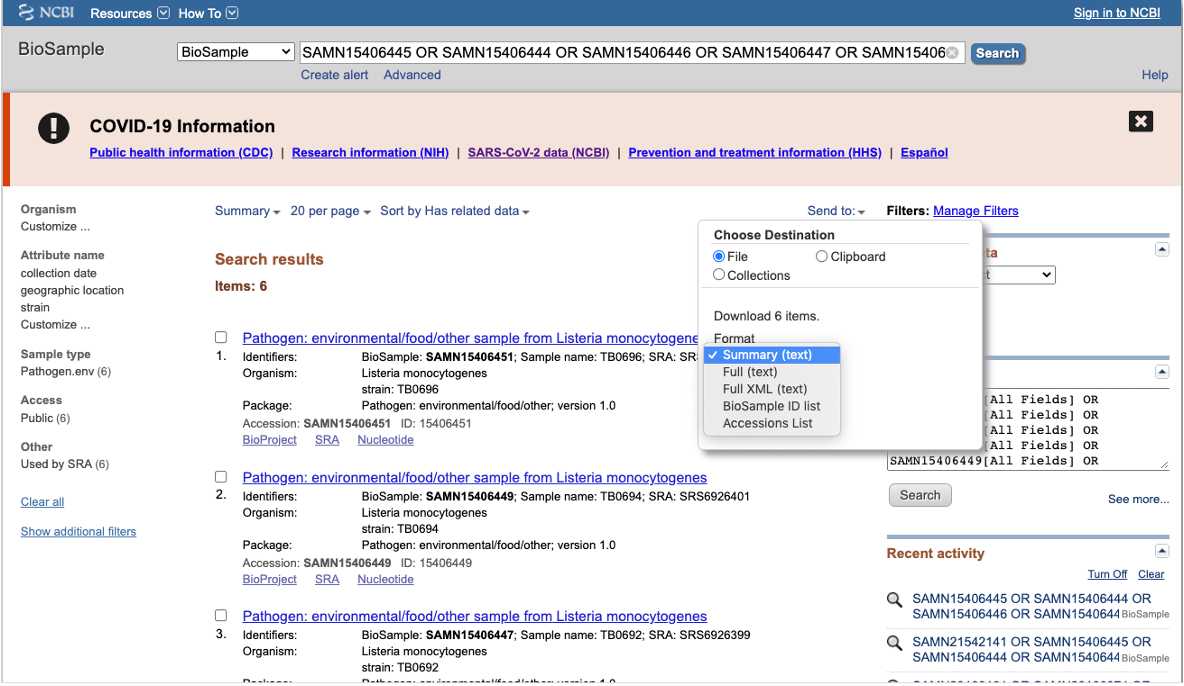
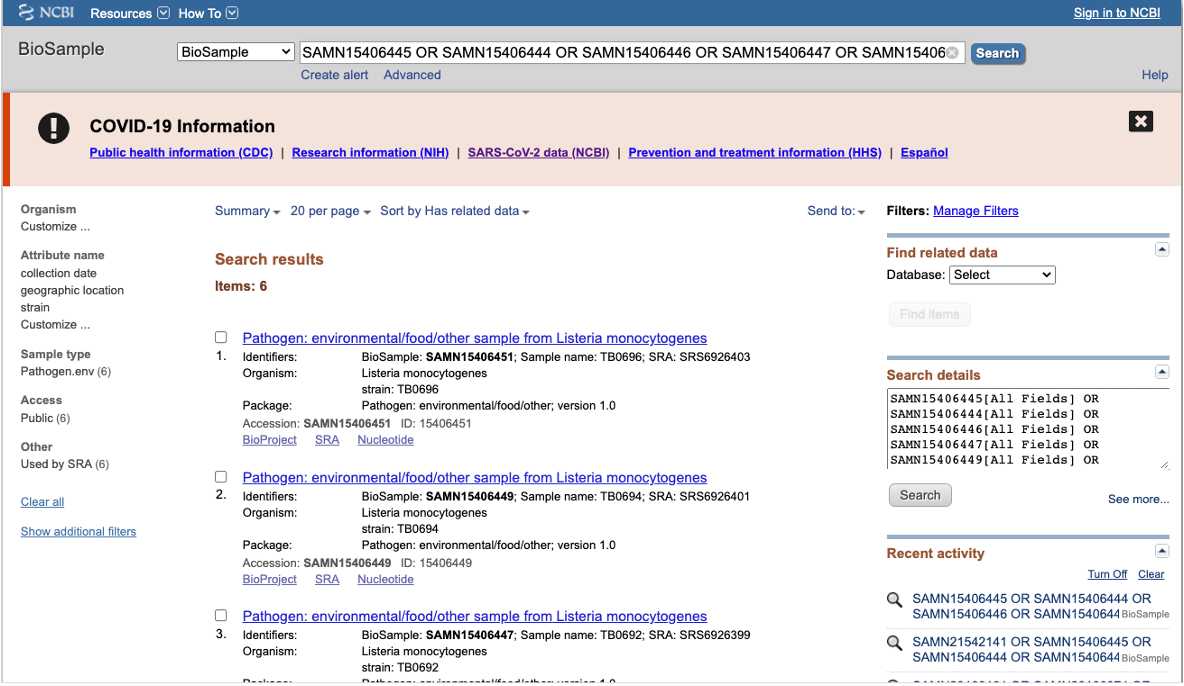
BioSample
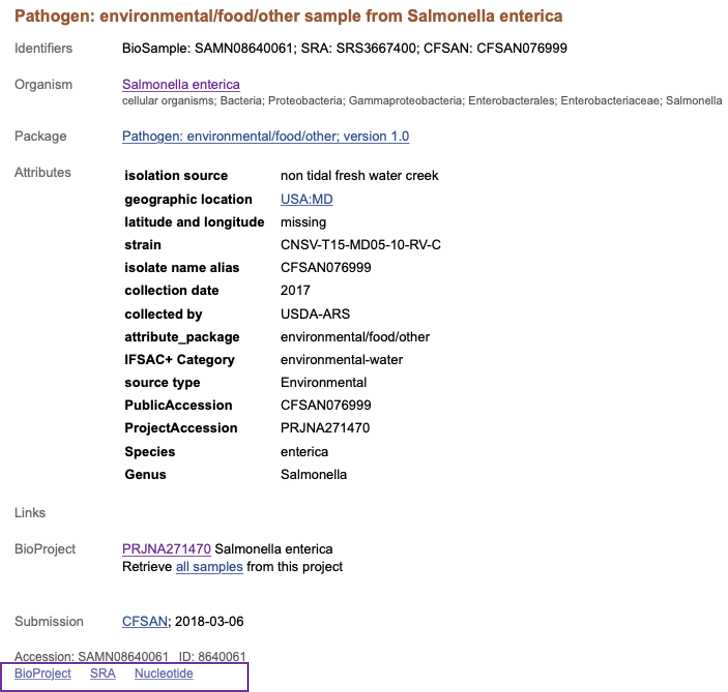
BioSample (https://www.ncbi.nlm.nih.gov/biosample/ ) is the database for the isolate or sample metadata. Users access biosample records at using the search box and typing laboratory identifiers ( strain, isolate name alias, FDA_Lab_ID, BioProjects) or specific attributes separated by " OR " e.g. "CFSAN0001 OR CFSAN0002 OR CFSAN0003" "Salmonella enterica".
Sequencing Read Archive
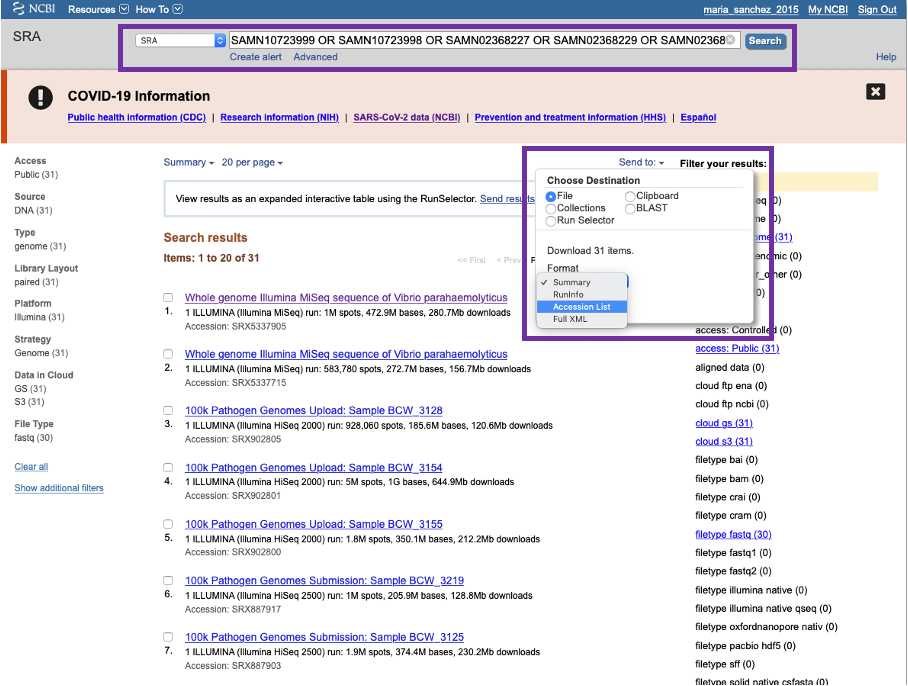
The Sequencing Read Archive (SRA) https://www.ncbi.nlm.nih.gov/sra/ is the primary repository of raw whole genome sequencing data. You can access records at SRA by typing laboratory identifiers ( strain, isolate name alias, FDA_Lab_ID, BioProjects) or specific attributes in the search box. Identifiers might need to be separated by " OR " e.g. -CFSAN0001 OR CFSAN0002 OR CFSAN0003.
You can export SRA accessions by clicking at the Send to bottom and choosing file and the accession list format.
You can download sequencing data files from NCBI using SRA Toolkit, Run Browser and the cloud.
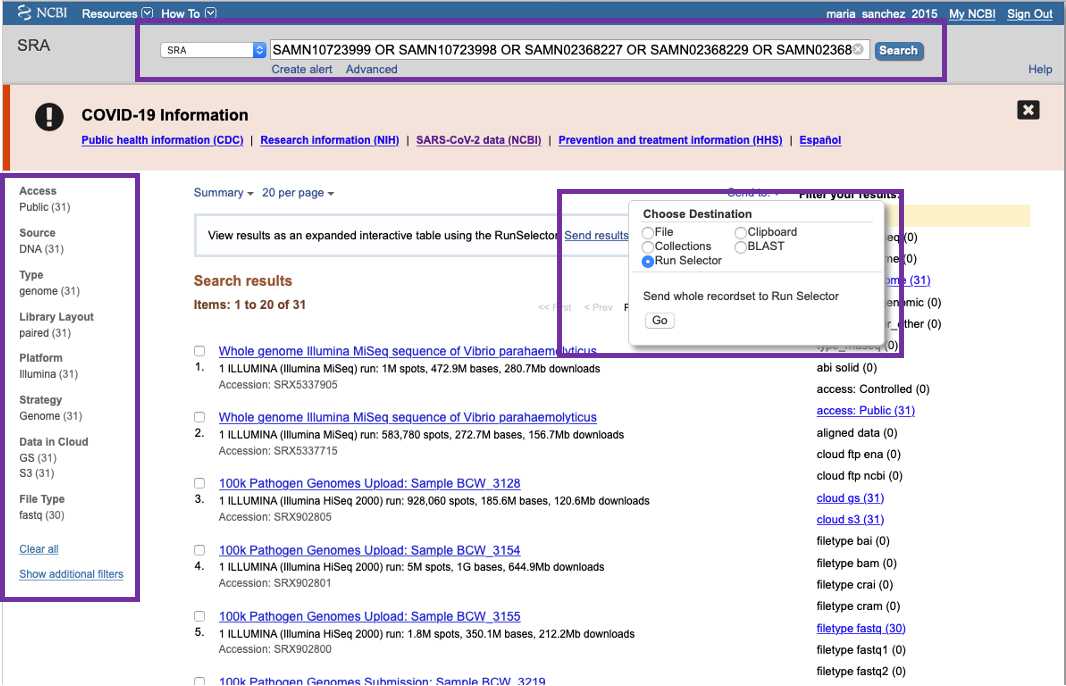
Run Selector
You can download a combination of sample/isolate and sequencing metadata in a tab-delimited file using Run Selector (https://www.ncbi.nlm.nih.gov/Traces/study/).
- Enter the accessions in the search box at the SRA browser, click Search , the output will include all the found records.
- Click Send to on the top of the SRA page, check the Run Selector radio button, and click the button Go .
- If necessary, refine your results by using various filters provided by the Run Selector 's interface.
- Click the Metadata button. This will generate a tabular file with metadata available for each Run.
Pathogen Detection
Visit the Pathogen Detection (https://www.ncbi.nlm.nih.gov/pathogens/) to access real-time analyses of isolates obtained from ongoing pathogen surveillance activities.
More details on how to navigate the Pathogen Detection can be found at: https://www.ncbi.nlm.nih.gov/pathogens/pathogens_help/
Resources from NCBI Pathogen Detection to address specific research questions.
- How To: Find an isolate you submitted(.pptx)
- How To: Find the latest Salmonella in the Isolates Browser(.pptx)
- How To: Download a list of human, clinical How To: Download a list of human, clinical E. faecailis isolates E. faecailis isolates (.pptx)
- How To: Identify isolates in the same SNP cluster that share a set of genes(.pptx)
- How To: Download a list of all carbapenem resistance genes and point mutations from the Reference Gene Catalog(.pptx)
- How To: Download all the reference sequences for a set of proteins(.pptx)
- How To: Find all the known resistance mechanisms to a given drug(.pptx)
- How To: Download the nucleotide sequence of all MCR-1 alleles(.pptx)
- How To: Identify all the contigs that share a set of genes(.pptx)
- How To: Identify isolates that have a pair of genes on the same contig(.pptx)
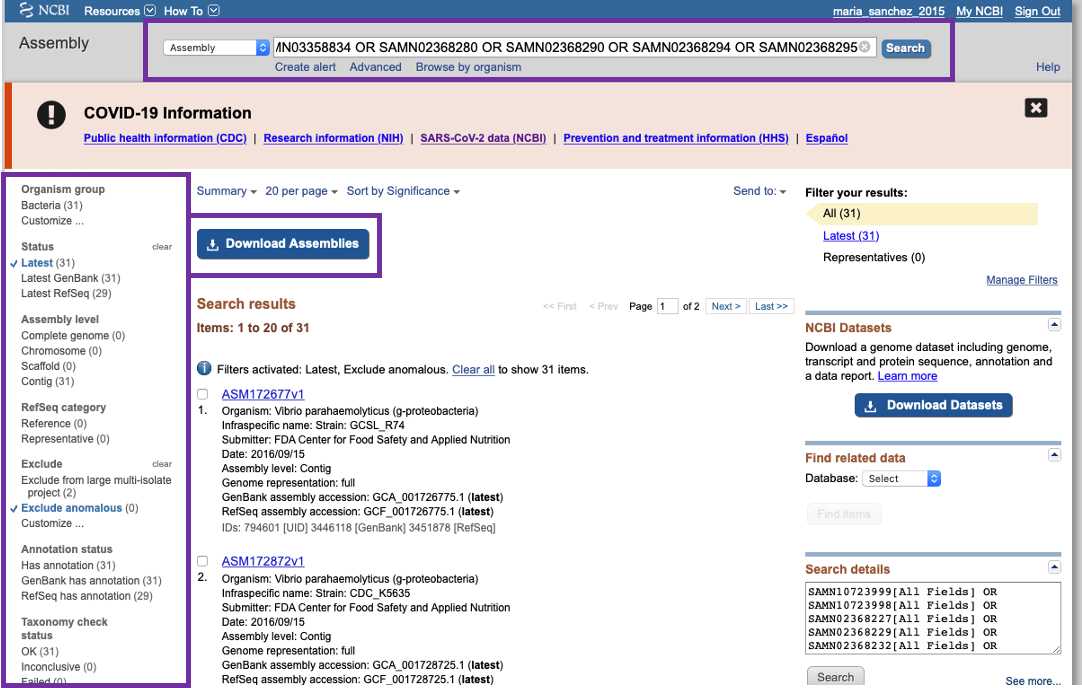
Assembly
If you need to download multiple assembled genomes, access the NCBI Assembly resource (https://www.ncbi.nlm.nih.gov/assembly/). Enter the identifiers in the search box and click in the "Download Assemblies" button. In the left side of this interface, you can refine your search by applying multiple filters. For more details on programatically download genomes from NCBI visit https://www.ncbi.nlm.nih.gov/genome/doc/ftpfaq/.
NCBI Insights
If you want to keep up with NCBI news, sign up for NCBI insights updates. The NCBI Insights Blog offers guidance on the latest NCBI resources.