Protocol of the Double-Click Seq Method to Evaluate the Deposition Bias of Chromatin Proteins
Petra E. van der Wouden, Petra E. van der Wouden, Martijn R. H. Zwinderman, Martijn R. H. Zwinderman, Deng Chen, Deng Chen, Michela Borghesan, Michela Borghesan, Frank J. Dekker, Frank J. Dekker
chromatin proteins
DNA replication
double-click seq
Ethynyl-2′-deoxyuridine (EdU)
histone deposition
l-Azidohomoalanine (AHA)
Abstract
Symmetrical deposition of parental and newly synthesized chromatin proteins over both sister chromatids is important for the maintenance of epigenetic integrity. However, the mechanisms to maintain equal distribution of parental and newly synthesized chromatid proteins over sister chromatids remains largely unknown. Here, we describe the protocol for the recently developed double-click seq method that enables mapping of asymmetry in the deposition of parental and newly synthesized chromatin proteins on both sister chromatids in DNA replication. The method involved metabolic labeling of new chromatin proteins with l-Azidohomoalanine (AHA) and newly synthesized DNA with Ethynyl-2′-deoxyuridine (EdU) followed by two subsequent click reactions for biotinylation and subsequently by corresponding separation steps. This enables isolation of parental DNA that was bound to nucleosomes containing new chromatin proteins. Sequencing of these DNA samples and mapping around origins of replication in the cellular DNA enables estimation of the asymmetry in deposition of chromatin proteins over the leading and lagging strand in DNA replication. Altogether, this method contributes to the toolbox to understand histone deposition in DNA replication. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Metabolic labeling with AHA and EdU and isolation of nuclei
Basic Protocol 2 : First click reaction, MNase digestion and streptavidin enrichment of labeled nucleosomes
Basic Protocol 3 : Second click reaction, Replication-Enriched Nucleosome Sequencing (RENS) Protocol
INTRODUCTION
Post-translational modifications of histone (PTMs) are crucial for regulating gene transcription and nuclear organization (Allis & Jenuwein, 2016). Some modifications are stable and can be inherited during DNA replication (Audergon et al., 2015; Ragunathan et al., 2015), but they differ between parental and newly synthesized histones (Reverón-Gómez et al., 2018), making it important to maintain balanced deposition during replication to ensure similar chromatin states for the sister chromatids after cell division. The preservation of repressed chromatin domains by the redeposition of parental histones highlights the functional importance of this process (Escobar et al., 2019). Accurate mechanisms for deposition of old and new histones are therefore critical (Alabert et al., 2015).
Recent studies have shed light on how histones are deposited during DNA replication by using immunoprecipitation of PTMs that are characteristic of either parental histones (H4K20me2) or new histones (H4K5ac) (Petryk et al., 2018; Yu et al., 2018). When this approach was coupled with the labeling of new DNA using the thymidine analog 5-ethynyl-2′deoxyuridine (Edu) to separate parental from newly synthesized DNA, a slight deposition bias of new histones to the lagging strand was revealed in mouse embryonic stem cells (Petryk et al., 2018) and a slight bias of new histones deposition onto the leading strand in budding yeast cells treated with hydroxyureas (HU) (Yu et al., 2018). Of note, both studies concluded that the structural integrity of the replisome is essential to maintain near-symmetrical histone inheritance. Expanding further the technology to differentiate parental versus new chromatin proteins would be certainly of benefit for the research field.
Despite recent advances in understanding how histones are deposited during DNA replication, there are several limitations to existing approaches. Firstly, the current methods rely on specific post-translational modifications (PTMs) of histones, such as H4K20me2, H5K5ac, and H4K5ac, to distinguish between parental and newly synthesized histones. However, these PTMs may not be entirely reliable markers, as they can also be present in other contexts or exhibit dynamic changes during replication or other cellular processes. Secondly, histone deposition during DNA replication may vary depending on the cell type, tissue context, and disease state, and it is important to consider these factors in future research. Thirdly, while the current approaches have provided insights into the biased deposition of new histones to the leading or lagging strand during replication, the underlying molecular mechanisms are not fully understood. The role of replisome integrity in maintaining near-symmetrical histone inheritance needs further investigation, as well as the potential involvement of other factors or regulatory pathways.
Despite these limitations, improving our understanding of histone deposition during DNA replication could greatly advance our knowledge of epigenetic regulation and nuclear organization, and have significant implications for biology and disease research. For example, a better understanding of how histones are accurately deposited during replication could shed light on how epigenetic information is faithfully transmitted during cell division and inherited across generations. Additionally, dysregulation of histone deposition has been implicated in various diseases, including cancer and neurodegenerative disorders, highlighting the importance of studying this process for therapeutic development. Therefore, further advancements in technology and approaches to accurately differentiate parental from new chromatin proteins could provide valuable insights into the mechanisms underlying histone deposition during DNA replication and its functional significance in biology.
Here, we present the double-click-seq method, which is a novel approach complementary to previously established immunoprecipitation-based methods for enriching parental DNA from new chromatin proteins. The double-click-seq technique utilizes stable labeling of de novo synthesized chromatin proteins by co-translational incorporation of the methionine surrogate azidohomoalanine (AHA), which can be covalently attached to a biotin affinity tag via copper-catalyzed alkyne to azide cycloaddition reaction, commonly referred to as the "click" reaction (Kolb et al., 2001). This enables enrichment of nucleosomes containing new histones by conducting a click reaction with a biotin affinity tag. The incorporation of this technology into the double-click-seq protocol offers a powerful method to investigate the distribution of parental versus new histones over the leading and lagging strand during DNA replication. The double-click-seq method provides several advantages compared to other methods for enriching parental DNA from newly synthesized chromatin proteins:
- 1.Complementary approach: The double-click-seq method complements existing immunoprecipitation-based methods, offering an additional tool for investigating chromatin proteins and allowing for a more comprehensive understanding of parental versus new histone distribution during DNA replication.
- 2.Stable labeling: The double-click-seq method utilizes stable labeling of de novo synthesized chromatin proteins with azidohomoalanine (AHA) during translation, ensuring reliable capture and enrichment of labeled chromatin proteins for accurate assessment.
- 3.Covalent attachment with biotin affinity tag: The double-click-seq method employs a specific and stable covalent attachment of a biotin affinity tag to AHA-labeled chromatin proteins using a "click" reaction, minimizing nonspecific binding and background noise.
- 4.High sensitivity and specificity: The use of biotin affinity tag in the double-click-seq method enables efficient and specific enrichment of nucleosomes containing new histones, resulting in a highly sensitive and specific approach for studying chromatin protein dynamics during DNA replication.
- 5.Powerful and versatile: The incorporation of double-click-seq technology into the protocol provides a powerful and versatile method for investigating the distribution of parental versus new histones in various biological contexts, offering insights into chromatin dynamics and epigenetic regulation.
The double-click-seq method involves three basic protocols (Fig. 1). In Basic Protocol 1, newly synthesized proteins in the cell are labeled with AHA, and newly synthesized DNA is labeled with 5-ethynyl-2′deoxyuridine (EdU). After cell lysis and isolation of the nuclei, Basic Protocol 2 involves subjecting the isolated nuclei to the first click reaction using alkyne-labeled biotin to biotinylate AHA-labeled proteins. It is important to carefully design and optimize the click reactions to minimize any potential cross-reactivity or competition between different components. In Basic Protocol 2, where the first click reaction is performed to biotinylate AHA-labeled proteins using alkyne-labeled biotin, it is crucial to ensure that the click reaction is selective and specific for the intended targets (AHA-labeled proteins) and does not cross-react with other components, such as the DNA alkyne or azide groups.
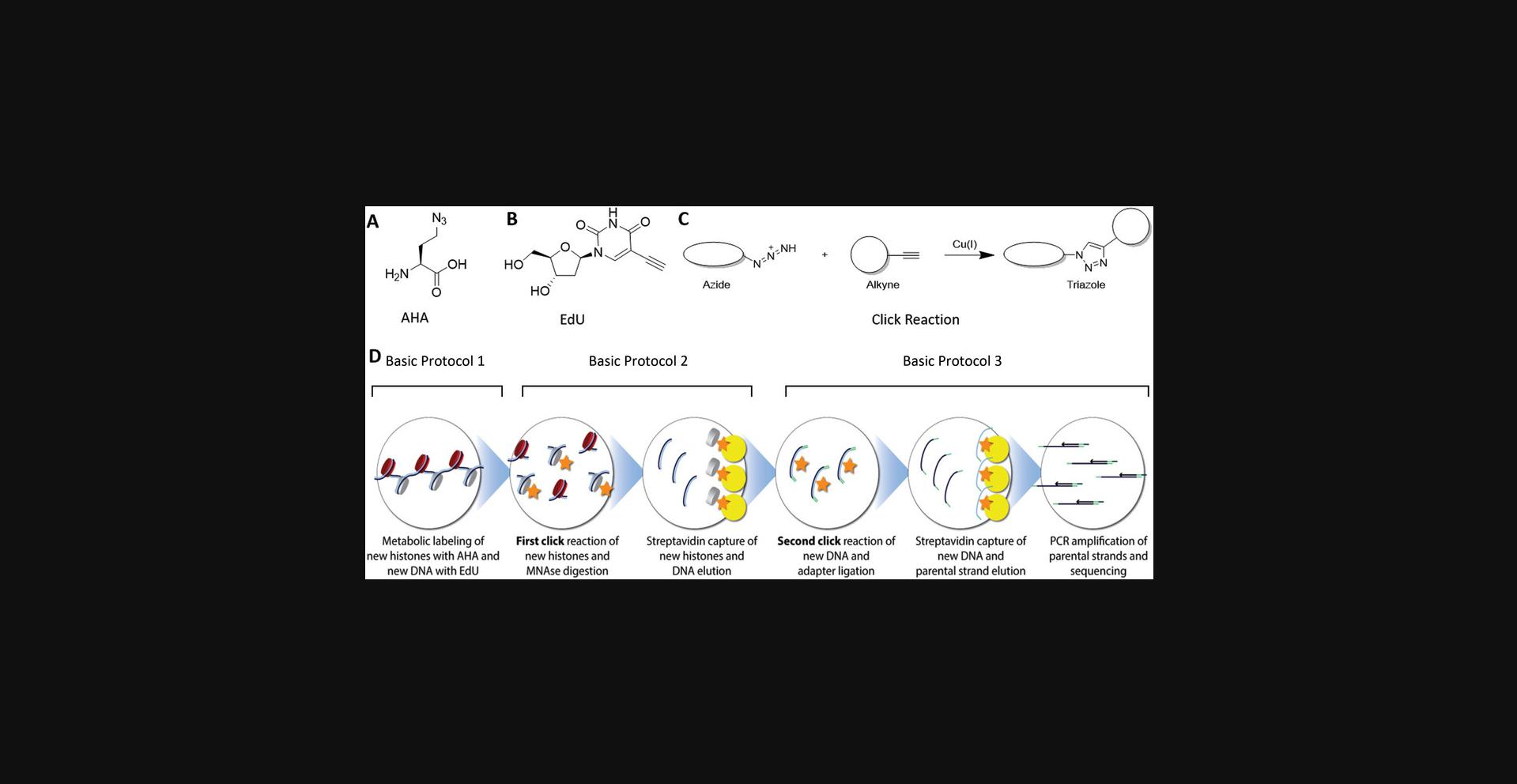
To minimize the potential for cross-reactivity between the DNA alkyne and the protein azide, it is important to choose appropriate click reaction conditions, such as optimizing the concentration and ratio of the reactants, reaction time, and temperature. Additionally, using highly selective click chemistry reactions, such as copper-catalyzed azide-alkyne cycloaddition (CuAAC) or strain-promoted azide-alkyne cycloaddition (SPAAC), can help minimize cross-reactivity and maximize specificity.
It is recommended to carefully validate and optimize the click reactions in your specific experimental setup to ensure that the biotinylation of AHA-labeled proteins is efficient and specific, while minimizing any potential cross-reactivity with DNA alkyne or azide groups. This may involve conducting control experiments and adjusting reaction conditions as needed to achieve the desired specificity and efficiency in the click reaction.
Mono-nucleosomes are then prepared by DNA digestion with MNase treatment, and biotinylated nucleosomes are isolated using streptavidin-coated magnetic beads. The double-stranded DNA is eluted from the beads by Proteinase to digest the nucleosomes. In Basic Protocol 3, the second click reaction is then performed using azide-labeled biotin to biotinylate Edu that is incorporated into newly synthesized DNA. This step is also used for aptamer ligation. Subsequently, the biotinylated DNA is captured using streptavidin-coated magnetic beads, and the complementary strand is eluted to provide the parental DNA strand. Finally, the resulting DNA library is amplified by PCR and prepared for next-generation sequencing (NGS).
Basic Protocol 1: METABOLIC LABELING WITH AHA AND EdU AND ISOLATION OF NUCLEI
This protocol describes the method for de novo labeling of newly synthesized histones and DNA. Cells are grown in appropriate medium containing the methionine analog azidohomoalanine (AHA) and 5-ethynyl-2′deoxyuridine (EdU). Dividing cells incorporate AHA in their newly synthesized proteins including histones. EdU will be incorporated in newly synthesized DNA during the S-phase. In our method we used the hTERT RPE-1 cell line. This cell line was chosen because it is an immortalized nearly diploid cell line from human retinal pigment epithelial cell with an average chromosome number of 46.This feature makes sequencing analysis feasible. The nuclei are being isolated through a simple and fast method using 0.1% of NP-40 in PBS and a few short centrifugation steps.
Materials
-
RPE-1 cells (hTERT RPE-1, ATCC®, CRL-4000™)
-
Standard medium (see recipe)
-
GlutaMAX (Gibco, cat. no. 35050-038)
-
Penicillin-streptomycin (10,000 U penicillin and 10 mg streptomycin/ml) (Gibco, cat. no. 11548876)
-
Fetal bovine serum (Serana, cat. no. S-FBS-SA-0150)
-
AHA (Azidohomoalanine) medium
-
5-ethynyl-2′deoxyuridine (EdU) (stock solution of 67 mM in DMSO, stored at −20°C) (Bio-Connect cat. no. CLK-N001)
-
Phosphate-buffered saline (PBS; Gibco, cat. no. 10010056)
-
Trypsin-EDTA (Gibco, cat. no. 15400-054)
-
0.1% NP-40 in PBS with protease inhibitor (see recipe)
-
175-cm2 cell culture flask (Greiner, cat. no. 660175)
-
Inverted microscope (Leica)
-
50-ml centrifuge tubes (Sarstedt, cat. no. 62547254)
-
Swing-out centrifuge (Eppendorf 5810)
-
ULT Freezer
-
Benchtop centrifuge (Eppendorf 5415R)
-
1.5-ml centrifuge tubes (Sarstedt, cat. no. 72690001)
-
Vortex mixer
Harvest cells
1.Seed four 175-cm2 flasks each with 3 × 106 RPE-1 cells per flask (12 million cells in total) in appropriate (standard) medium. Incubate overnight at 37°C with 5% CO2.
2.Prepare 95% AHA medium by mixing 28.5 ml AHA medium with 1.5 ml standard medium per 175-cm2 flask.
3.Aspirate the medium from the cells.
4.Add 9 µl EdU (67 mM stock) to 30 ml of the 95% AHA medium (this results in 20 µM final concentration of EdU) to the cells and incubate for 24 hr at 37°C with 5% CO2.
5.Incubate the cells for 24 hr at 37°C with 5% CO2.
6.Aspirate the medium and wash the cells twice, each time with 10 ml PBS.
7.Add 3 ml Trypsin-EDTA and incubate at 37°C until all the cells are detached from the bottom of the flask.
8.Quench the trypsin by adding 3 ml standard medium with FBS.
9.Collect the cells in 50-ml tubes.
10.Centrifuge the cells for 5 min at 200 × g at room temperature.
11.Wash the cells with 5 ml ice-cold PBS and centrifuge again for 5 min at 200 × g at room temperature.
12.Remove the supernatant and store the pellets at −80°C until further use or continue directly to step 13 to extract the nuclei.
Nuclei extraction
13.To extract the nuclei, the thawed pellet is resuspended in 1 ml of ice-cold PBS containing 0.1% NP-40 supplemented with 1× protease inhibitor cocktail.
14.Transfer the cell suspension into a 1.5-ml centrifuge tube.
15.Vortex the cells suspension for 5 sec at maximum speed.
16.Centrifuge the lysed cell suspension for 5-10 sec at 9000 × g , 4°C, in a benchtop centrifuge.
17.Discard the supernatant.
18.Resuspend the nuclei in 1 ml ice-cold PBS containing 0.1% NP-40 and supplemented with 1× protease inhibitor cocktail.
19.Centrifuge the washed nuclei for 5-10 sec at 9000 × g , 4°C, in a benchtop centrifuge and discard the supernatant.
20.Store the nuclei at −80°C until further use.
Basic Protocol 2: FIRST CLICK REACTION, MNASE DIGESTION AND STREPTAVIDIN ENRICHMENT OF LABELED NUCLEOSOMES
In this second protocol we describe the first click reaction performed on isolated nucleosomes. During the incubation with AHA in Basic Protocol 1, newly synthesized proteins have incorporated the unnatural methionine analog. This molecule contains an azide moiety, which is employed in a click reaction to covalently link to a biotinylated alkyne. After the click reaction, the protocol proceeds with an MNase digestion of the nuclei to obtain mononucleosomes. It is important to optimize the MNase concentration per cell type and per cell density in order to avoid over digestion or under digestion (Fig. 2). After digestion, the mono-nucleosomes can diffuse out of the nuclei where they are captured in a streptavidin pull-down with magnetic beads, which enables enrichment of intact newly synthesized nucleosomes.
Materials
-
Isolated nuclei (Basic Protocol 1)
-
Nuclei buffer (see recipe)
-
100 mM Alkyne-PEG-Biotin in Methanol (Merck, cat. no. 764213)
-
CuSO4 (50 mM)
-
THPTA (Merck, cat. no. 762342)
-
Sodium ascorbate (100 mM)
-
MNase enzyme (New England Biolabs cat. no. M0247S) containing:
-
MNase reaction buffer
-
Bovine serum albumin (BSA; 20 mg/ml) (New England Biolabs cat. no. B9000) (provided with the MNase)
-
0.5 M EDTA (Gibco, cat. no. 15575-038)
-
Wash buffer (see recipe)
-
Dynabeads™ MyOne™ Streptavidin C1 (Invitrogen™, cat. no. 65001)
-
Phosphate-buffered saline (PBS; Gibco, cat. no. 10010056)
-
Proteinase K (Fisher Scientific, cat. no. 10181030)
-
NaCl (5 M)
-
Ampure XP beads (Beckman, cat. no. A63881)
-
Nuclease-free water
-
Generuler 1 kb+ marker (Thermo Scientific cat. no. SM1333)
-
End-over-end rotator (VWR, cat. no. 444-500)
-
1.5-ml centrifuge tubes (Sarstedt, cat. no. 72690001)
-
Heated water bath (Grant Sub 28 or similar)
-
Benchtop centrifuge (Eppendorf 5415R)
-
Magnet rack (Dynal MPC-S or similar)
First click reaction: Biotinylation of proteins metabolically labeled with AHA
1.Resuspend the nuclei in 250 µl nuclei buffer.
2.Add 10 µl of 100 mM alkyne-PEG-biotin to click histones.
3.Pre-mix 5 µl 50 mM CuSO4 with 5 µl 250 mM THPTA and 8 µl of this mix to the sample.
4.Add 10 µl freshly prepared 100 mM sodium ascorbate.
5.Place the nuclei in an end-over-end rotator for 30 min at room temperature.
6.Repeat the cycloaddition reaction with freshly prepared ingredients (steps 2 to 5).
7.Pellet the nuclei for 5-10 sec at 10,000 rpm.
8.Discard the supernatant.
MNase digestion to obtain mono-nucleosomes
9.Resuspend the nuclei in 100 µl MNase buffer + BSA (100 µg/ml final concentration) and warm in a water bath for to 37°C for about 5 min.
10.Add 2 µl MNase enzyme to 398 µl MNase buffer to obtain a concentration of 10 gel Units of enzyme per µl in MNase buffer. Store on ice.
11.Add 60 µl of the diluted MNase to the nuclei and incubate for 5 to 10 min at 37°C.
12.Add 10 µl 100 mM EDTA and cool the sample on ice.
13.Add 100 µl wash buffer.
14.Spin down 5-10 sec at 10,000 rpm to pellet the nuclei and store the supernatant in a new 1.5-ml microtube. Discard the pellet.
First streptavidin pull-down: Enrichment of AHA labelled nucleosomes
15.Resuspend the tube containing the Dynabeads™ MyOne™ Streptavidin C1 for at least 30 s.
16.Transfer 30 µl of the resuspended Dynabeads™ MyOne™ Streptavidin C1 to a new 1.5 ml centrifuge tube.
17.Add 1 ml PBS.
18.Place the tube on a magnet rack for at least 1 min and discard the supernatant.
19.Remove the tube containing the beads from the magnet rack and resuspend the washed beads in 30 µl PBS.
20.Repeat the washings for a total of 3 washes (3 × 30 µl).
Capturing and washing of biotinylated mononucleosomes and release of associated DNA
21.Add the mononucleosomes from step 14 to the Dynabeads™ MyOne™ Streptavidin C1 and incubate for 1 hr at room temperature in an end-over-end rotator.
22.Place the sample on a magnet for 3-4 min and remove the supernatant.
23.Wash the beads five times, each time with 200 µl wash buffer.
24.Add 100 µl PBS containing 1 mg/ml proteinase K and incubate for 1 hr at 50°C.
25.Add 100 µl 5 M NaCl and incubate for 15 min on an end-over-end rotator.
26.Transfer 200 µl of elution sample (containing DNA) into a new 1.5-ml tube.
27.Add 360 µl (1.8× sample volume) Ampure beads and purify the DNA according to the manual.
28.Elute the sample in 80 µl nuclease-free water.
Basic Protocol 3: REPLICATION-ENRICHED NUCLEOSOME SEQUENCING (RENS) PROTOCOL
This protocol describes all the steps necessary to obtain a library from parental DNA originating from newly synthesized histones.
Materials
-
Sample: Double-stranded DNA (Basic Protocol 2)
-
100 mM Azide-PEG3-Biotin (in methanol) (Merck, cat. no. 762024)
-
50 mM CuSO4
-
THPTA (Merck, cat. no. 762342)
-
Sodium Ascorbate (100 mM)
-
Ampure beads (Beckman)
-
Nuclease-free water
-
NEB next UltraTM DNA library kit for Illumina® (New England Biolabs, cat. no. E7645)
-
DynabeadsTM MyOneTM Streptavidin T1 (C1 (Invitrogen™, cat. no. 65601)
-
2× B&W buffer (10 mM Tris·Cl, pH 7.5, 1 mM EDTA, 2 M NaCl) (see recipe)
-
1× B&W buffer (diluted 2× B&W buffer with an equal amount of water)
-
1× B&WT buffer (see recipe)
-
Alkaline buffer (see recipe)
-
Acetic acid
-
20 mM EDTA, pH 8.0 (Gibco)
-
NEBNext® multiplex oligos for Illumina® index primer set 1 or 2 (New England Biolabs, cat. no. E7335 or cat. no. E7500)
-
Qubit™ dsDNA high-sensitivity assay kit (Thermo Scientific, cat. no. Q32854)
-
End-over-end rotator
-
Magnet rack for 1.5-mltubes
-
1.5-ml centrifuge tubes
-
Vivaspin® 2 5000 Da MW cut-off centrifugal concentrator
-
Benchtop centrifuge
-
Swing bucket centrifuge for 15-ml centrifuge tubes
-
Thermal cycler
-
PCR tubes (strips or single tubes)
-
Vortex mixer
-
Qubit Fluorometer Tapestation System (Agilent)
-
High-Sensitivity D1000 ScreenTape (Agilent, cat. no. 5067-5584)
Second click reaction: Biotinylation of DNA metabolically labeled with EdU
1.Add 1 µl 100 mM Azide-PEG3-Biotin to the eluted DNA from the previous step.
2.Pre-mix 5 µl of 50 mM CuSO4 with 5 µl mM of 250 mM THPTA and add 8 µl of this mix to the sample.
3.Add 10 µl freshly prepared 100 mM sodium ascorbate to the sample.
4.Place the sample on an end-over-end rotator for 45 min at room temperature.
5.Add 0.7× sample volume of Ampure beads and incubate for 5 min at room temperature.
6.Place the sample on a magnet rack and wait until the supernatant is clear.
7.Discard the beads and pipet the supernatant into a new 1.5-ml microcentrifuge tube.
8.Add 1.8× sample volume Ampure beads to the sample and perform the clean-up according to the manufacture's protocol.
9.Elute the sample in 55.5 µl nuclease-free water.
End prep and adaptor ligation
10.Perform end prep and adaptor ligation according to the manual of the NEB next UltraTM DNA library kit for Illumina®.
11.Elute the DNA in 50 µl nuclease-free water.
12.Add 25 µl DynabeadsTM MyOneTM Streptavidin T1 into a 1.5-ml centrifuge tube.
13.Place on a magnet until the supernatant is clear.
14.Remove the supernatant.
15.Add 1 ml of 1× B&W buffer and resuspend the beads.
16.Place the tube on the magnet rack for about 1 min and discard the supernatant.
17.Repeat the washing steps for a total of three washes.
18.Add the sample obtained in step 11 to 25 µl washed DynabeadsTM MyOneTM Streptavidin T1 magnetic beads.
Isolation of replicated double-stranded DNA fragments
19.Incubate the sample with the beads for 15 min at room temperature in an end-over-end rotator.
20.Place the tube with the beads on the magnet rack and wait until the supernatant is clear.
21.Wash the beads four times, each time with 200 µl of 1× B&WT buffer.
22.Wash the beads once with 200 µl of 2× concentrated B&W buffer.
Isolation of replicated parental strands
23.Add 100 µl alkaline buffer to the beads and incubate for 1 min at room temperature to elute the parental strand.
24.Place the tube with the beads on a magnet and wait until the supernatant is clear.
25.Collect the supernatant and pipet into a clean 1.5-ml centrifuge tube.
26.Repeat the elution for a total of three times. Collect the supernatant in the same tube. The final volume will be 300 µl.
27.Neutralize the combined sample to a final concentration of 0.1 M acetic acid and 2 mM EDTA pH = 8.0.
28.Dilute the sample to a final volume of 1 ml with nuclease-free water.
29.Add the sample to a Vivaspin® 2 (5 kDa cut-off) centrifugal concentrator and concentrate to approximately 20 µl final volume by centrifuging for 15 min at 4000 × g , room temperature, in a swinging bucket.
30.Perform a reversed spin according to the manufacturer's instruction. Centrifuge for 2 min at 3000 × g , room temperature, in a swinging bucket to collect the sample.
Library amplification
31.Amplify libraries using the NEBNext Ultra DNA library prep kit for Illumina using 13 PCR cycles.
| Adapter ligated DNA Fragments | 15 µl |
| NEBNext Q5 Hot Start HiFi PCR Master Mix | 25 µl |
| Index primer/i7 primer | 5 µl |
| Universal PCR primer/i5 primer | 5 µl |
| Total volume | 50 µl |
| Cycle step | Temperature °C | Time | Cycles |
| Initial denaturation | 98°C | 30 sec | 1 |
| Denaturation | 98°C | 10 sec | 13 |
| Annealing/extension | 65°C | 75 sec | |
| Final extension | 65°C | 5 min | 1 |
| Hold | 4°C | Indefinitely |
32.Purify the libraries according to the manual with 0.9× Ampure Beads (45 µl).
33.Elute the sample in 30 µl nuclease-free water.
34.Check the quantity and the quality of the DNA library using Qubit and Tapestation analysis.
REAGENTS AND SOLUTIONS
Alkaline buffer
- 0.1 M NaOH (Acros Organics, cat. no. 424330010)
- 0.05% (v/v) Tween 20 (Promega, cat. no. H5151)
- Store up to 1 month at 4°C
Azidohomoalanine (AHA) medium
- DMEM high glucose, no glutamine, no methionine, no cysteine (Gibco, cat. no. 11570546)
- 10% Fetal bovine serum (FBS; Serana, cat. no. S-FBS-SA-0150)
- Penicillin-streptomycin (10,000 units penicillin and 10 mg streptomycin/ml) (Gibco, cat. no. 11548876)
- 4 mM GlutaMAX (Gibco, cat. no. 35050-038)
- 0.2 mM l Cystine 2HCl (Sigma-Aldrich, cat. no. C8755)
- 1 mM Sodium pyruvate (Gibco cat. no. 11360070)
- 0.2 mM AHA (Santa Cruz Biotechnology, cat. no. sc285947)
- 20 µM EdU (Bio-Connect, cat. no. CLK-N001)
- Store up to 2 months at 4°C
B&W buffer, 2×
- 10 mM Tris·Cl, pH 7.5 (Merck, cat. no. T5941)
- 1 mM EDTA (Fisher Scientific, cat. no. 15575-020)
- 2 M NaCl (Duchefa Biochemie, cat. no. S0520)
- Store up to 1 month at 4°C
B&WT buffer, 1×
- 5 mM Tris·Cl, pH 7.5 (Merck, cat. no. T5941)
- 0.5 mM EDTA (Fisher Scientific, cat. no. 15575-020)
- 1 M NaCl (Duchefa Biochemie, cat. no. S0520)
- 0.05% (v/v) Tween 20 (Promega, cat. no. H5151)
- Store up to 1 month at 4°C
NP-40 in PBS with protease inhibitor, 0.1%
- Phosphate-buffered saline (PBS; Fisher Scientific, cat. no. 11540486)
- 0.1% (v/v) NP-40 (Sigma, cat. no. 74385)
- Protease inhibitor cocktail (cOmplete™, EDTA-free Protease Inhibitor Cocktail; Roche, cat. no. 11873580001)
- NP-40 in PBS can be stored up to 1 month at 4°C. Add Protease inhibitor cocktail just before use.
Nuclei buffer
- 320 mM Sucrose (Duchefa Biochemie, cat. no. S0809.0025)
- 5 mM MgCl2 (Merck, cat. no. 1.05833.1000)
- 10 mM HEPES, pH 7.4 (Duchefa Biochemie, cat. no. H1504)
- 1× cOmplete™, EDTA-free Protease Inhibitor Cocktail (Roche, cat. no. 11873580001)
- Prepare a 50× concentrated stock solution of the Protease inhibitor cocktail (store at −20°C)
- Add just before use
- Store up to 1 month at 4°C
Standard medium
- DMEM, high glucose, pyruvate, GlutaMAX™ (Gibco, cat. no. 31966047)
- 10% Fetal bovine serum (FBS; Serana, cat. no. S-FBS-SA-0150)
- 1% Penicillin-streptomycin (10,000 units penicillin and 10 mg streptomycin/ml; Gibco, cat. no. 11548876)
- Store up to 6 months at 4°C
Wash buffer
- Phosphate-buffered saline (PBS; Fisher Scientific, cat. no. 11540486)
- 0.1% (w/v) bovine serum albumin (BSA; VWR, cat. no. 1.12018.0100)
- 0.01% Tween 20 (Promega, cat. no. H5151)
- Store up to 1 month at 4°C
COMMENTARY
Background Information
The advancement in the study of histone deposition during DNA replication has been achieved through the application of immunoprecipitation techniques on histone post-translational modifications (PTMs) like H4K20me2 and H4K5ac (Petryk et al., 2018; Yu et al., 2018). Research in mouse embryonic stem cells and budding yeast cells using labelling of new DNA with a thymidine analogue (EdU) in the presence of hydroxyurea (HU) showed a slight bias of new histone deposition on the lagging strand in mouse cells and leading strand in yeast cells. Several methods are currently considered state of the art in unravelling the deposition bias in histones in DNA replication.
The sequencing-based techniques, such as ChOR-seq, SCAR-seq, and eSPAN, enable the investigation of various chromatin components during replication in a time-sensitive and locus-specific manner. These methods use labeling of newly synthesized DNA strands with various nucleotide analogs (EdU, CldU, IdU), followed by chromatin immunoprecipitation (ChIP) of histones and DNA sequencing. By analyzing the distribution of labeled DNA strands in relation to histone modifications, the directionality and magnitude of histone deposition bias during DNA replication can be revealed. Specifically, these techniques have demonstrated that histones H3-H4 are symmetrically recycled to both daughter strands during replication. Replication factors with chaperone activity, including MCM2, POLE3/4, and POLA1, have been found to recycle H3-H4 to either the leading or the lagging strand (Petryk et al., 2018; Yu et al., 2018). In a recent study (Flury et al., 2023) the ChOR-seq and SCAR-seq methods were used to track the occupancy of H2AK119ub1, H2BK120ub1, and H2A.Z on newly replicated DNA in mouse embryonic stem cells. Based on these technologies it was found that the histones H2A-H2B are efficiently and accurately recycled during replication, through a distinct mechanism independent of H3-H4 recycling, involving the lagging strand polymerase POLA1. Furthermore, the H2A-H2B PTMs landscapes were found to be rapidly restored post-replication, with H2AK119ub1 guiding the timely and accurate restoration of H3K27me3, which plays an important role in propagating the epigenome.
Other advanced techniques are available to analyze the epigenome landscape, including labeling DNA with nucleotide analogues in pulse-chase experiments, followed by CHIP-seq of histones and DNA sequencing; amino acid labeling in cell culture using isotopes couple with mass spectrometry (pSILAC) (Nakamura et al., 2022), and DNA labeling and isolation by 5-ethynyl-deoxyuridine for mass spectrometry (iDEMS) (Stewart-Morgan et al., 2023). In addition to histone deposition bias, these methods reveal details about the regulation of PTMs during replication, such as the dynamics of H3K56 acetylation, H4K20 methylation, and hydroxymethylation.
Generally the methods described above all rely on histone PTMs for separation of parental versus newly synthesized histones. Application of a metabolic labelling provides a complementary method that does not rely on histone PTMs but provides an unambiguous labelling of newly synthesized proteins. Concomitantly, the metabolic labelling method does not unambiguously allow for the isolation of only histones and might also include other tightly associated chromatin proteins. Ideally, metabolic labelling methods can be combined with a histone chromatin immunoprecipitation followed by separation of the parental and novel DNA strand by EdU labelling. However, such a triple-separation protocol would not deliver sufficient sample material for amplification and sequencing with the current state of technology.
The double-click-seq protocol recently developed by us (Zwinderman et al., 2021) is a complementary method that enriches parental DNA from new histones. It uses stable labelling of newly synthesized histones through co-translational incorporation of azidohomoalanine (AHA). A copper catalyzed reaction, known as the "click" reaction, is used to attach a biotin affinity tag to the labelled histones. This allows for enrichment of nucleosomes containing new histones for analysis. This protocol provides a novel approach for understanding the distribution of parental versus new histones on the leading and lagging strands in DNA replication, without the risk of ambiguous results due to the dynamic nature of PTMs.
It is important to consider the potential limitations and caveats of using azidohomoalanine (AHA) or ethynyluridine as labeling methods in the study of histone deposition during DNA replication.
AHA and ethynyluridine are analogs that are incorporated into newly synthesized proteins or DNA, respectively, during replication. However, these analogs may not perfectly mimic the native molecules, and their incorporation could potentially disrupt normal biological interactions. For example, AHA incorporation could affect protein folding, stability, or interactions with other cellular components, which may in turn affect histone deposition or other cellular processes. The "click" reaction used in the AHA labeling protocol involves copper-catalyzed chemistry, which may have potential limitations. Copper ions can generate reactive oxygen species that could potentially cause oxidative damage to cellular components, including DNA and proteins. Additionally, copper ions can also interact with other cellular components, potentially leading to off-target effects or interference with normal cellular processes.
The AHA and ethynyluridine labeling methods may require specialized techniques and expertise for their implementation, as such it is important to be aware of their potential limitations and consider appropriate controls and validation measures to ensure the accuracy and reliability of the results.
Critical Parameters
Cell culture conditions
The exact number of cells needed for optimal amounts of DNA to use for library amplification and sequencing steps will depend on several factors, such as the type of sequencing technology, the complexity of the genome, and the desired depth of sequencing. As a general rule, most next-generation sequencing platforms require between 1 and 10 nanograms (ng) of DNA for library preparation. However, this amount can vary widely depending on the platform and the sample type.
To obtain sufficient amounts of DNA for library amplification and sequencing, it is recommended to start with a concentration of cells above 20 million, although this can vary depending on the type of cell and the experimental conditions. It is important to note that the yield and quality of DNA obtained from cells will depend on the extraction method used, the age and health of the cells, and the presence of contaminants in the samples. It is important to routinely test cell cultures once a month for mycoplasma contamination to ensure that samples are suitable for use in experiments.
To ensure optimal yields and quality of DNA, it is recommended to culture cells under appropriate conditions. For example, cells should be dividing at the time of harvesting, as this ensures that DNA replication has occurred, and there is sufficient genomic material for extraction. Additionally, the confluency of the cells in culture should not exceed 70%, as overcrowding can lead to stress and changes in gene expression that may affect the yield and quality of DNA. Finally, the cell's culture medium should contain 3% to 5% methionine to allow proper cell growth and AHA incorporation.
MNase enzyme
The concentration of the MNase enzyme should be optimized according to the number of cells seeded. The MNase should be promptly aliquoted upon arrival to avoid freeze-thaw cycles.
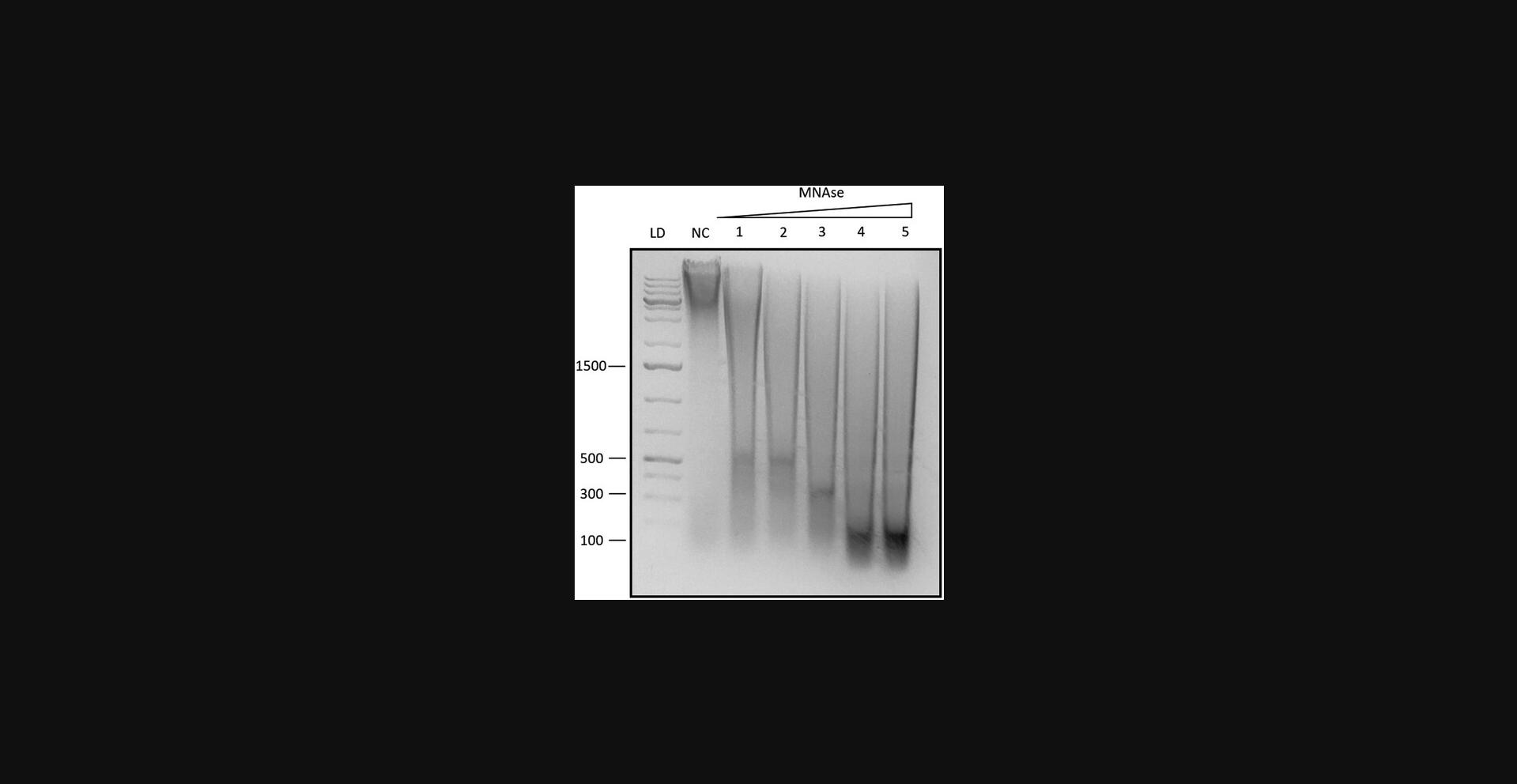
To ensure that the amount of MNase is optimal, we recommend performing a titration experiment to test the range of enzyme concentrations to determine the optimal amount required for efficient chromatin digestion. In the titration experiment, fixed chromatin samples must be incubated with different concentrations of MNase (e.g., 0.5U, 1U, 2U, 5U and 10U of enzyme per microgram of chromatin) and then check the chromatin digestion efficiency by running the samples on a 2% agarose gel. The optimal concentration of MNase is the one that generates a chromatin digestion pattern with a range of DNA fragments from mononucleosomes to higher-order oligonucleosomes, without over-digestion or under-digestion. Over-digestion may lead to the loss of some regions of interest, while under-digestion may result in incomplete chromatin fragmentation.
DNA measurements
To determine the amount of DNA obtained for sequencing and downstream analysis it is recommended to measure the absorbance of the DNA samples at 260 nm by using a spectrophotometer or a Qubit fluorometer (ThermoFisher Scientific).
The absorbance value or the fluorescence signal emitted by the samples is then used to calculate the concentration of DNA. One advantage of using the fluorometry method over traditional spectrophotometric methods is the ability to selectively detect only the target molecule (DNA in this case) and no other contaminants, such as salts or organic compounds that may be present in the sample. For the click-seq protocol a minimum of 5-10 ng/µl of DNA is required for sequencing applications.
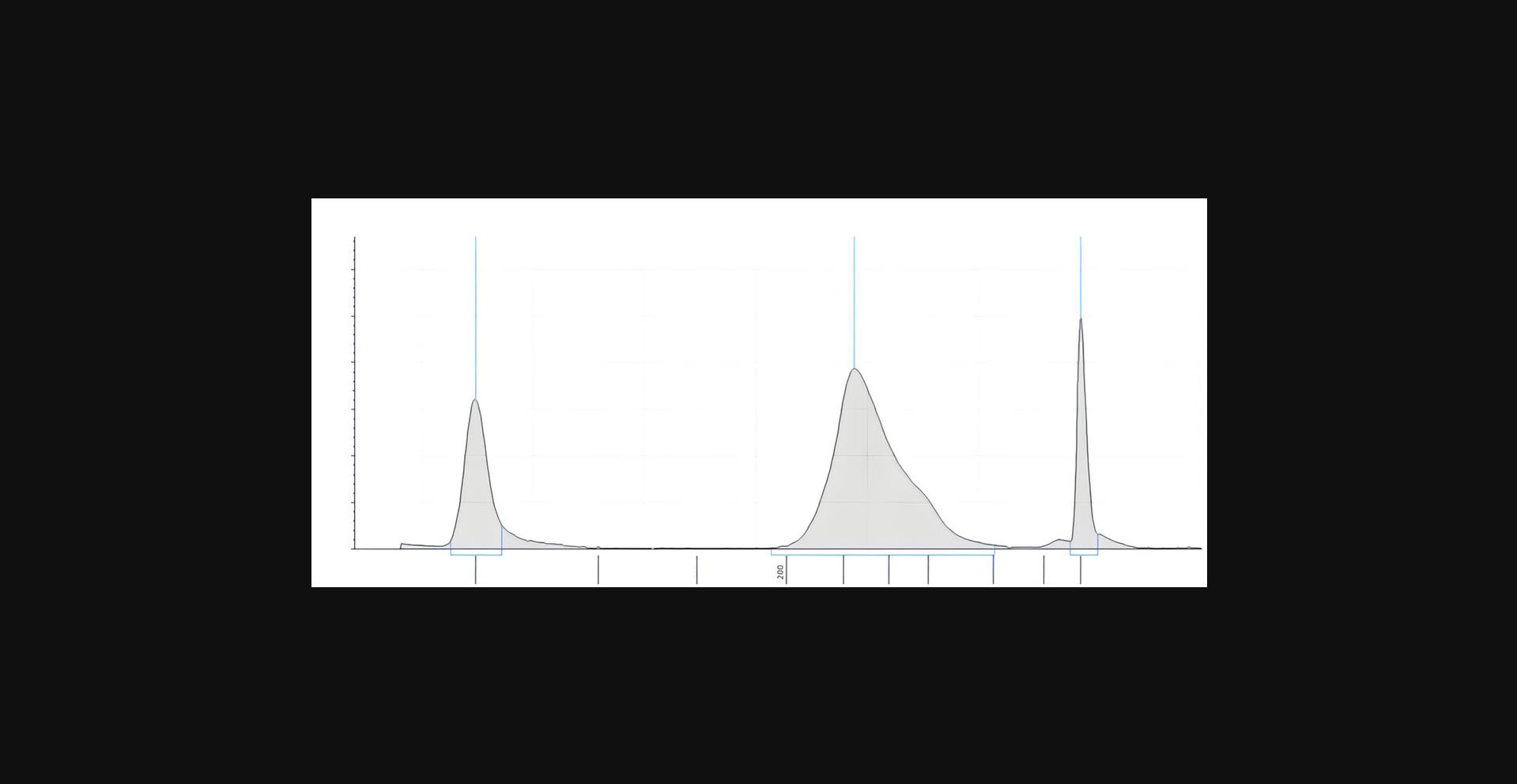
The TapeStation (Agilent) electropherogram can be used to determine the quality and the quantity of the sample, as well as to assess the degree of sample fragmentation, identify the presence of contaminants, and confirm the success of a particular step in the library preparation for sequencing. The electropherogram displays peaks representing the DNA fragments of a specific size, the height of each peak represents the relative concentration of the corresponding fragment. Multiple peaks can be indicative of different issues including: adapter dimers due to insufficient removal of unligated adaptor (peak at 60 bp), insufficient fragmentation, overloaded sample and degraded DNA.
Adapter dimers can be removed by using gel purification, while to improve fragmentation it is possible to increase the MNase shearing time and or conditions. To prevent DNA degradation it is recommended to store the samples at −20°C and to use fresh reagents. Multiple peaks can also be indicative of low library complexity, which can result from inefficient adapter ligation or low input DNA. To improve library complexity, it is recommended to optimize the adapter ligation conditions and increasing the amount of input DNA.
Troubleshooting
The frequently encountered problems and their possible causes and solutions are listed in Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| Low DNA yield | Insufficient amount of starting material | Increase amount of cells. Start with ≥12 million cells per experiment. |
| Incomplete MNase digestion | Optimize MNase digestion/check activity of MNase | |
| Dried/temperature Ampure Beads | Before elution Ampure Beads should not be overdried. The beads should be at room temperature before using. | |
| Multiple peaks at Tapestation | Insufficient removal of unligated adaptors (peak at 60 bp) |
Perform an extra clean-up step using Ampure Beads Use recommended amount of DNA input in library preparation |
| Bubble product (extra peak at the double size as expected bp) | Libraries with a bubble product can be sequenced without any problem |
Understanding Results
MNase enzyme
MNase (Micrococcal Nuclease) is an enzyme commonly used in chromatin research to digest chromatin into mono-nucleosomes, di-nucleosomes, and higher-order nucleosome fragments. It cleaves the linker DNA regions between nucleosomes, resulting in characteristic banding patterns on agarose gels that can be used to assess the efficiency of chromatin digestion. In this protocol, the optimal concentration of MNase enzyme was determined by testing different concentrations and analyzing the resulting nucleosome fragments on an agarose gel. Figure 2 shows a representative example of the gel with increasing concentrations of MNase enzyme and the corresponding banding pattern. Mono-nucleosomes (150 bp), di-nucleosomes (300 bp), and tri-nucleosomes (450 bp) bands are clearly observed. The concentration of MNase enzyme that provides the highest yield of mono-nucleosomes is considered optimal for further experiments (Fig. 2, lane 5).
In our experiments we achieved a final concentration of DNA between 5 and 100 ng/µl measured by Qubit in a total volume of 30 µl. After DNA quality control analysis that was performed with the TapeStation system (Agilent) a peak of approximately 320 bp was observed (Fig. 3). The expected peak size is around 270 bp (150 bp DNA from mononucleosomes and two adapters, which are each 60 bp in size). The higher average peak size of 320 bp can be explained by the fact that the DNA wrapped around the histones is flanked by linker DNA.
Time Considerations
The complete double-click seq protocol, from cell to DNA libraries can be estimated to take 5-6 days. It is advised to have all the buffers and reagents prepared before the start of the protocols. Basic Protocol 1: the seeding, treatment, and harvesting of the cells may take 3 days up to a few weeks in order to obtain the optimal cell amount of 20 million. It is important to note that this is an estimate, and the actual time required may vary depending on the specific experimental condition. It is always recommended to monitor cell growth carefully, and to ensure that cells are healthy before harvesting. For example, some cell lines may double in number every 24 hr under optimal condition, while others may take several days or longer to reach the same number of cells. The amount of time it takes to reach 1 million cells can vary widely depending on several factors, including the type of cell, the growth condition, and the start density. The preparation of nuclei can be performed in as little time as 15 min. At this point it is safe to store the cell pellets and/or nuclei in the freezer at −80°C until further processing. It is possible to perform Basic Protocol 2 (5 hr) and Basic Protocol 3 (3 hr) without stopping. However, if necessary, it is possible to incorporate a stop after Basic Protocol 2. The sample can be stored safely overnight at −20°C. Basic Protocol 2 and Basic Protocol 3 can be performed in approximately 8 hr, although the time needed is dependent on the amount of samples that are being processed.
Acknowledgments
F.J.D. was funded by a starting grant (Nr 309782) from the European Research Council and a VIDI grant (Nr 639.033.907) from the Netherlands Organization of Scientific Research. D.C. is supported with grant no. 201907720019 from the China Scholarship Council (CSC).
Author Contributions
Petra van der Wouden did the: formal analysis, investigation, methodology development, validation, visualization and writing of the original draft. Martijn Zwinderman did the: conceptualization, formal analysis, investigation, methodology development, validation and visualization. Deng Chen did the: formal analysis, investigation, methodology development, validation and visualization. Michaela Borghesan did the: writing, review & editing. Frank Dekker did: conceptualization, funding acquisition, project administration, supervision, writing, review & editing.
Conflict of Interest
The authors declare no conflict of interest. The funding sources had no involvement in collection, analysis and interpretation of the data.
Open Research
Data Availability Statement
As described in the main paper on the development of the double-click seq method (cite: ACS Chem. Biol. 2021, 16, 2193−2201) the data sets generated during this study are available at EBI ArrayExpress under accession number E-MTAB-8624. The code generated during this study is available at GitHub; https://github.com/thamarlobo/histone_deposition_analysis. Further permission related to the material excerpted should be directed to the ACS.
Literature Cited
- Alabert, C., Barth, T. K., Reverón-Gómez, N., Sidoli, S., Schmidt, A., Jensen, O. N., Imhof, A., & Groth, A. (2015). Two distinct modes for propagation of histone PTMs across the cell cycle. Genes & Development, 29(6), 585–590. https://doi.org/10.1101/gad.256354.114
- Allis, C. D., & Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. In Nature reviews genetics (Vol. 17, Issue 8, pp. 487–500). Nature Publishing Group. https://doi.org/10.1038/nrg.2016.59
- Audergon, P. N. C. B., Catania, S., Kagansky, A., Tong, P., Shukla, M., Pidoux, A. L., & Allshire, R. C. (2015). Restricted epigenetic inheritance of H3K9 methylation. Science , 348(6230), 132–135. https://doi.org/10.1126/science.1260638
- Escobar, T. M., Oksuz, O., Saldaña-Meyer, R., Descostes, N., Bonasio, R., & Reinberg, D. (2019). Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA replication. Cell , 179(4), 953–963.e11. https://doi.org/10.1016/j.cell.2019.10.009
- Flury, V., Reveron-Gomez, N., Alcaraz, Stewart-Morgan, K. R., Wenger, A., Klose, R. J., & Groth, A. (2023). Recycling of modified H2A-HEB provides short-term memory of chromatin states. Cell , 186, 1050–1065. https://doi.org/10.1016/j.cell.2023.01.007
- Nakamura, K., Groth, A., & Alabert, C. (2022). Investigating mitotic inheritance of histone posttranslational modifications by triple pSILAC coupled to nascent chromatin capture. Methods in Molecular Biology , 2529, 407–417. https://doi.org/10.1007/978-1-0716-2481-4_17
- Petryk, N., Dalby, M., Wenger, A., Stromme, C. B., Strandsby, A., Andersson, R., & Groth, A. (2018). MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science , 361(6409), 1389–1392. https://doi.org/10.1126/science.aau0294
- Ragunathan, K., Jih, G., & Moazed, D. (2015). Epigenetic inheritance uncoupled from sequence-specific recruitment. Science , 348(6230), https://doi.org/10.1126/science.1258699
- Reverón-Gómez, N., González-Aguilera, C., Stewart-Morgan, K. R., Petryk, N., Flury, V., Graziano, S., Johansen, J. V., Jakobsen, J. S., Alabert, C., & Groth, A. (2018). Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Molecular Cell , 72(2), 239–249.e5. https://doi.org/10.1016/j.molcel.2018.08.010
- Kolb, H. C., Finn, M. G., & Sharpless, K. B. (2001). Click chemistry: Diverse chemical function from a few good reactions. Angewandte Chemie (International ed in English) , 40(11), 2004–2021. https://doi.org/10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5
- Stewart-Morgan, K. R., Requena, E. C., Flury, V., Du, Q., Heckhausen, Z., Hajkova, P., & Groth, A. (2023). Quantifying propagation of DNA methylation and hydroxymethylation with iDEMS. Nature Cell Biology , 25(1), 183–193.
- Yu, C., Gan, H., Serra-Cardona, A., Zhang, L., Gan, S., Sharma, S., Johansson, E., Chabes, A., Xu, R.-M., & Zhang, Z. (2018). A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science , 361, 1386–1389. https://www.science.org
- Zwinderman, M. R. H., Jessurun Lobo, T., van der Wouden, P. E., Spierings, D. C. J., van Vugt, M. A. T. M., Lansdorp, P. M., Guryev, V., & Dekker, F. J. (2021). Deposition bias of chromatin proteins inverts under DNA replication stress conditions. ACS Chemical Biology , 16(11), 2193–2201. https://doi.org/10.1021/acschembio.1c00321
Key References
- Petryk, N., Dalby, M., Wenger, A., Stromme, C. B., Strandsby, A., Andersson, R., & Groth, A. (2018). See above.
Study on histone deposition biased using antibody based methods in eukaryotic cells.
- Yu, C., Gan, H., Serra-Cardona, A., Zhang, L., Gan, S., Sharma, S., Johansson, E., Chabes, A., Xu, R.-M., & Zhang, Z. (2018). See above.
Study on histone deposition biased using antibody based histone separation in yeast.
- Zwinderman, M. R. H., Jessurun Lobo, T., van der Wouden, P. E., Spierings, D. C. J., van Vugt, M. A. T. M., Lansdorp, P. M., Guryev, V., & Dekker, F. J. (2021). Deposition bias of chromatin proteins inverts under DNA replication stress conditions. ACS Chemical Biology , 16(11), 2193–2201. See above.
Original publication of the double-click seq method in which this protocol is used.

