Protocol 2 - MRI data protocol
Sophie Adler, Mathilde Ripart, Meld Project, Konrad Wagstyl
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
The MELD Project is an international collaboration aiming to create open-access, robust and generalisable tools for FCD detection.
This MRI data protocol details
-
The MRI data required / desirable for patients and controls in the MELD focal epilepsies project
-
How to convert the MRI data from dicom to nifti
-
How to ensure all identifiable data is stripped from the header
-
How to deface the data
-
How to convert the data into BIDS format
-
How to send the anonymised data to the study coordinators
Please refer to protocol 1 for inclusion / exclusion criteria and retrieval of necessary anonymised demographic and clinical information.
PLEASE NOTE: To take part in the MELD Project each site will be required to 1) sign our memorandum of understanding and 2) provide a letter from the head of department or local R&D office confirming that they have obtained appropriate ethical approval to share the anonymised data with UCL.
Before start
Ensure that you have "cloned" the MELD file structure and scripts from github. (https://github.com/MELDProject/meld_focal_epilepsy)
To do this, in a terminal window cd into the location you wish to store the data.
cd <path>
Then "clone" the repository using the following command:
git clone https://github.com/MELDProject/meld_focal_epilepsy
This will download all of the scripts & template folder structure necessary for the MELD focal epilepsy project.
Steps
Initial setup
For this protocol you will need to have installed :
- Anaconda
- FSL 6.0
- The meld_focal_epilepsy package
- The mfe_env conda environment
Install Anaconda & FSL
To check if you already have anaconda installed, in your terminal window run:
conda --version
```It should return the version of your current conda installation e.g "conda 4.10.1"
If not, please refer to the following instructions to install anaconda on your computer :
[https://www.anaconda.com/products/individual-d](https://www.anaconda.com/products/individual-d)
To check if you already have **FSL** installed, in your terminal window run:
flirt --version
If not, or if you don't have the version 6, please refer to the following instructions to install FSL 6.0 on your computer :
[https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation)
Install meld_focal_epilepsy github & create environment:
- Open a terminal and move to the folder you want to saved the package in:
cd <path_to_meld_focal_epilepsy_github_folder>
```2) Copy from GitHub the package by pasting the following sentence :
git clone https://github.com/MELDProject/meld_focal_epilepsy.git
Check if you have a folder called meld_focal_epilespy in <path_to_meld_focal_epilepsies_folder>
3) Create the environment :
cd <path_to_meld_focal_epilepsy_github_folder>/meld_focal_epilepsy/scripts conda env create -f mfe_env.yml
conda activate mfe_env
e.g
<img src="https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.3byl4k8jjvo5/image.png" alt="" loading="lazy" title=""/>
Your meld_focal_epilepsy package is ready to be used !
**Note** : you will have to activate your environment every time you open a new terminal and you want to use the meld_focal_epilepsy package
Retrieval of MRI data
Prepare your data
Download the meld_focal_epilepsy_data folder from : https://figshare.com/s/763e50f4eb51a4f76f58
Keep note of where you save this folder as it will be your data folder : <path_to_meld_focal_epilepsy_data_folder>
You will need to unzip the folder :
cd <path_to_meld_focal_epilepsy_data_folder>
tar -xzvf meld-focal-epilepsy-data.tar.gz
Anonymous participant IDs
During Protocol 1 each participant should have been given an anonymous ID according to the following naming structure:
MELD_[site code]_[patient/control]_number
In the meld_focal_epilepsy_data/participants folder copy the template file structure called meld_template and rename the folder with the participants anonymous ID. This can either be done by right clicking on the folder clicking rename and typing in the anonymous ID OR in your terminal window:
cd <path_to_meld_focal_epilepsy_folder>/meld_focal_epilepsy/participants
mv meld_template <MELD_anonymous_ID>
```e.g. mv meld_template MELD_H1_P_0001
Further details about the naming structure:
[site code] = site identifier which will be provided to you e.g. H1 for Great Ormond Street Hospital
[patient/control] = P if patient, C if control.
_If participant was included in the FCD study and has an ID such as MELD_H1_15T_FCD_0001, the ID should be kept the same. I.e. Use the original FCD flag and scanner flag._
_If control was included in the FCD study and has an ID such as MELD_H1_3T_C_0001, the ID should be kept the same._
[number] = 0001, 0002 etc.
Examples of participant IDs:
MELD_H1_P_0001
MELD_H1_C_0002
MELD_H2_FCD_0042
Please make sure to securely keep a spreadsheet at your centre which links the anonymous IDs used in this study back to the IDENTIFIABLE patient data. THIS MUST NOT BE SHARED and should be kept securely!
The following MRI data should be retrieved off the hospital system / research database for each participant and stored in the relevant folder for each participant:
| A | B | C | D | E |
|---|---|---|---|---|
| MRI scan | Notes on data | Essential / where available | Folder to store data | Data formats accepted |
| T1-weighted preoperative | 3D 1.5T, 3T or 7T | Essential | t1 | dicom, nifti |
| T2 preoperative | 3D or 2D coronal | Where available | t2 | dicom, nifti |
| FLAIR preoperative | 3D or 2D coronal | Where available | flair | dicom, nifti |
| DWI preoperative (diffusion weighted data, bval and bvec) | 3D | Where available | dwi | dicom, nifti |
| T1-weighted postoperative | 3D 1.5T, 3T or 7T | Essential in operated patients | postop_t1 | dicom, nifti |
Please note: the MRI data must NOT have contrast (e.g. gadolinium contrast)
Within the meld_template are 1.5T, 3T and 7T folders and within these there are folders to store the MRI data. Please save the patient's MRI data in the correct folder.
If you have MRI data from multiple scanner field strengths for the same patient (e.g. 1.5T and 3T data for a patient) - it would be wonderful to have both, so we can compare how the classifier is able to perform on data from different scanner field strengths.
Here is an overview of the folder structure:
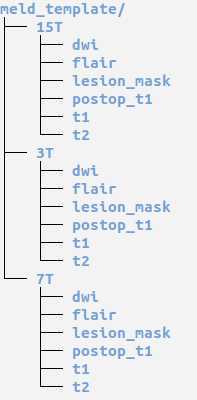
Diffusion Weighted Data:
Please note if you are providing the data in nifti format you need to provide the following files:
dwi.nii.gz
dwi.bval
dwi.bvec
If you are providing the data in dicom format, the script in the subsequent section will create these files.
Anonymising MRI data
Once you have retrieved all your patient / control MRI data and stored it in the MELD_focal_epilepsy file structure, you need to run the " meld_bidsify_data_step1.py " script. This script will convert any dicom data into nifti format and save them alongside dicoms
Note : you need to have your mfe_env environment activated : conda activate mfe_env
Please note that the meld_focal_epilepsy_github_folder contains the scripts and is different from the meld_focal_epilepsy_data_folder which should contains the data.
To run this script on all participants: Open a terminal window and type:
cd <path_to_meld_focal_epilepsy_github_folder>/meld_focal_epilepsy/scripts
python meld_bidsify_data_step1.py -d <path_to_meld_focal_epilepsy_data_folder>/meld_focal_epilepsy_data
Note :
This script also coregister FLAIR volumes to T1 if both available, and saves it in the 'flair' folder
You can now use the nifti T1 volume, and the additional coregistered FLAIR volume to create the lesion mask if not already provided.
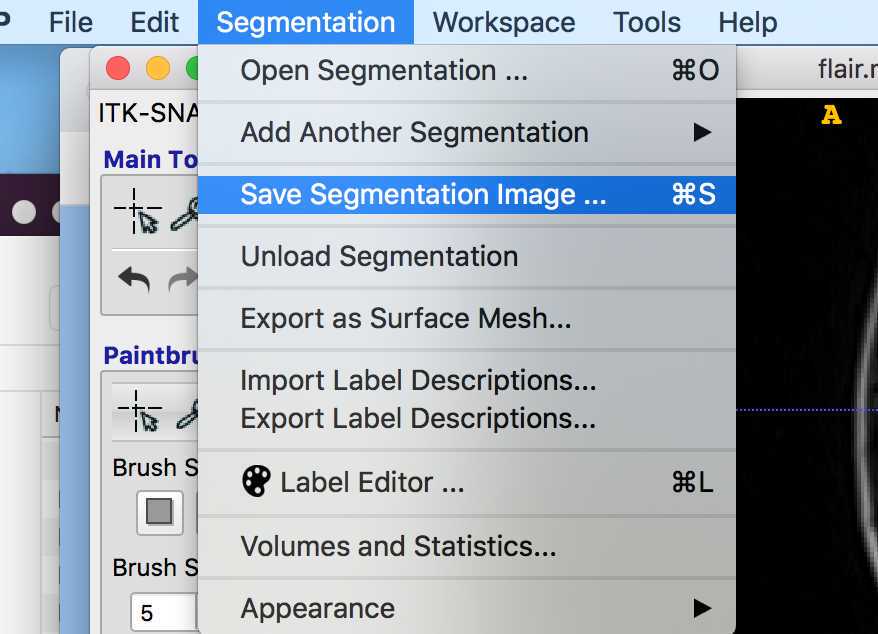
Lesion Masking
This section (Substeps 1 - 7) details how to create a lesion mask in ITK-SNAP and save it as a Nifti file.
Lesion masks should ideally be created for all patients included in the MELD Project EXCEPT patients with:
-
hippocampal sclerosis
-
or patients who are MRI negative
If your centre has an established method to create lesion masks / has already created lesion mask:
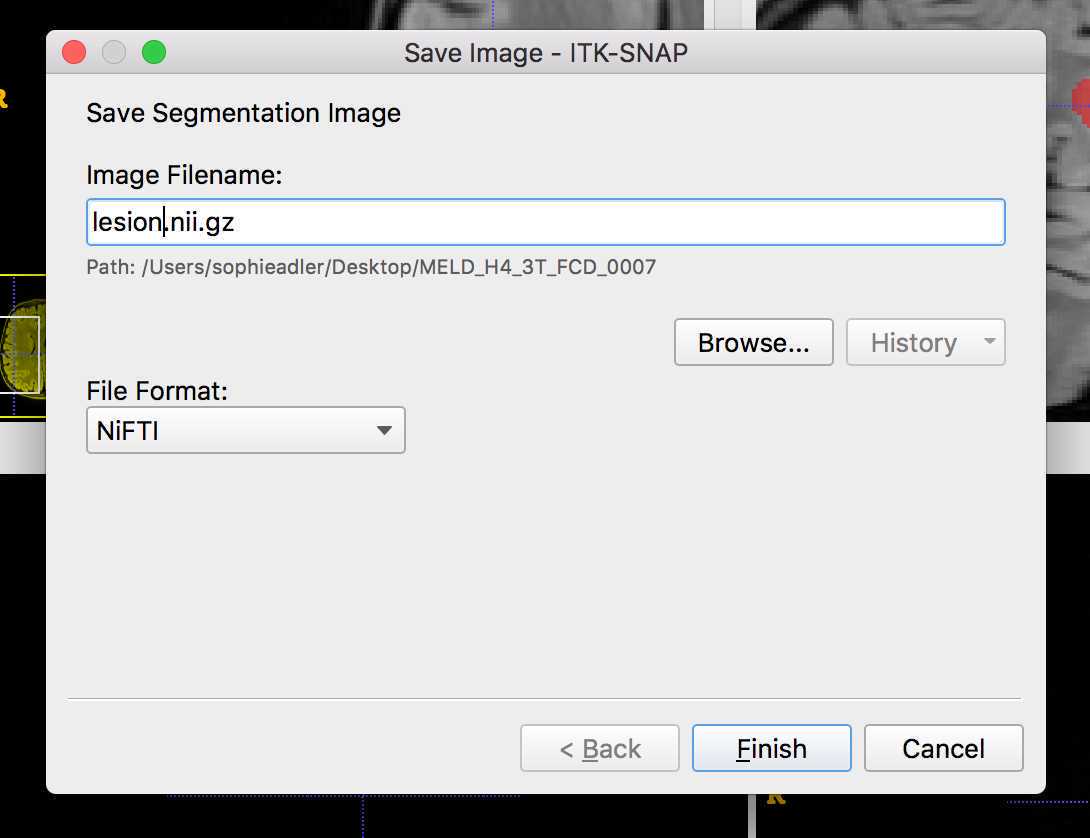
You do not need to redo them or change your method. The lesion masks will need to be converted to Nifti format. Please save the lesion mask as lesion.nii.gz or lesion.nii in the lesion_mask folder. If the lesion mask was created based on a 1.5T scan, please save in the 1.5T folder. If the lesion mask was created based on a 3T scan, please save in the 3T folder etc.
How to create a lesion mask:
The lesion mask should be done on the 3D T1-weighted scan but other scans e.g. FLAIR and the post-operative T1-weighted can be used to assist in defining the lesion. We provide the FLAIR coregistered (FLAIR_coreg.nii) which can be used to help delineate the lesion.
You may need assistance from a neuroradiologist at your centre for the subtle lesions.
If you have multiple T1-weighted scans at different field strengths - please create the lesion mask based on the T1-weighted scan you can visualise the lesion best on.
If the patient has hippocampal sclerosis OR is MRI negative there is no need to create a lesion mask.
If the patient is MRI-negative but with histological confirmation, the post-operative scan and other corroborative evidence such as EEG, PET, sEEG etc. can be used to help define where the lesion was on the preoperative scan in order to create the lesion mask.
If it is NOT possible to create a lesion mask. Please still include this patient in the study. In the RedCap Participant information survey under the question "Have you provided a lesion mask for this patient?" Click No
If you have any questions or run into problems, please feel free to contact the MELD project: (meld.study@gmail.com).
Downloading ITK-SNAP:
Click on the link below and download version 3.8.0 for windows / mac / linux depending on your workstation:
http://www.itksnap.org/pmwiki/pmwiki.php?n=Downloads.SNAP3
Follow the tutorial provided for the installation: http://www.itksnap.org/pmwiki/pmwiki.php?n=Documentation.TutorialSectionInstallation
Before starting manual lesion masking:
We advise watching part of the ITK-SNAP webinar:
If you start at 9minutes 57 seconds and watch until 12 minutes 5 seconds, you can watch the part of navigating an image in ITK-SNAP and manual segmentation. You may want to pause / rewatch the manual segmentation example as it is very fast but shows you 3 tools you can use to help create your lesion masks - polygon; paintbrush and smart brush
https://www.youtube.com/watch?v=Wob8beX88Ks
Here is a useful introductory guide on how to use ITK-SNAP:
http://www.itksnap.org/pmwiki/uploads/Train/RSNA2016-Manual-Guido-Final.pdf
Slides
11-14: Image navigation
16-22: Manual segmentation
24: Loading and saving manual segmentations
There are also many other useful youtube tutorials on how to manually segment using ITK-SNAP. Here is a longer (18 minute) but much more detailed webinar which is very useful. In this example they use segmentation of the hippocampal subfields as an example.
Example manual segmentation of a FCD Type IIB - Opening the MRI scans and navigating through them
Open ITK-SNAP and click open image
Then search for the MRI image (usually T1-weighted) of the patient who's lesion you are going to segment

In this example I have opened a 3T FLAIR image:
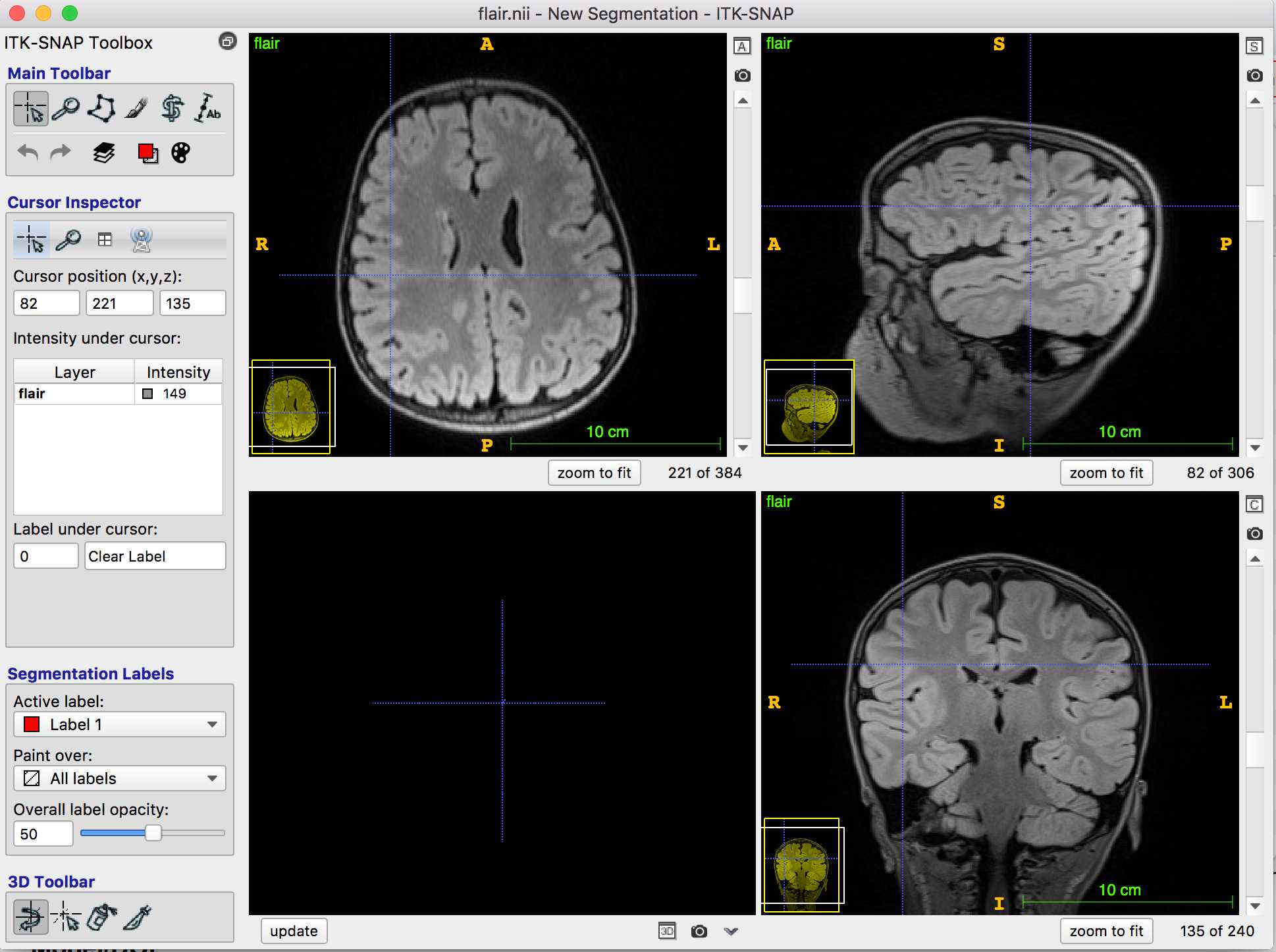
On the toolbar, the cursor is clicked, so I can navigate through the MRI to find the lesion.
If I want to add another MRI scan (e.g. the co-registered 3T T1-weighted), I can click file -> add another image
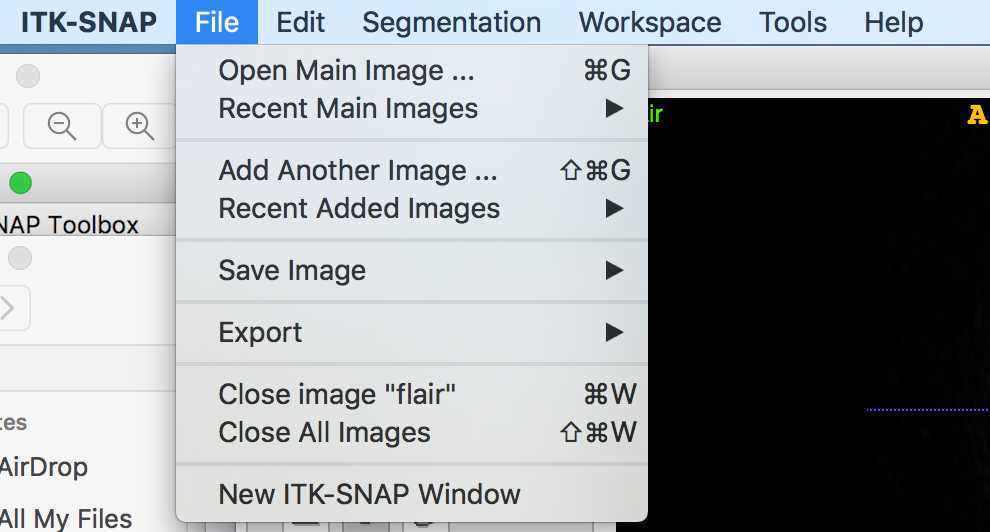
Example manual segmentation of a FCD Type IIB - Opening the MRI scans and navigating through them
Open ITK-SNAP and click open image
Then search for the MRI image (usually T1-weighted) of the patient who's lesion you are going to segment
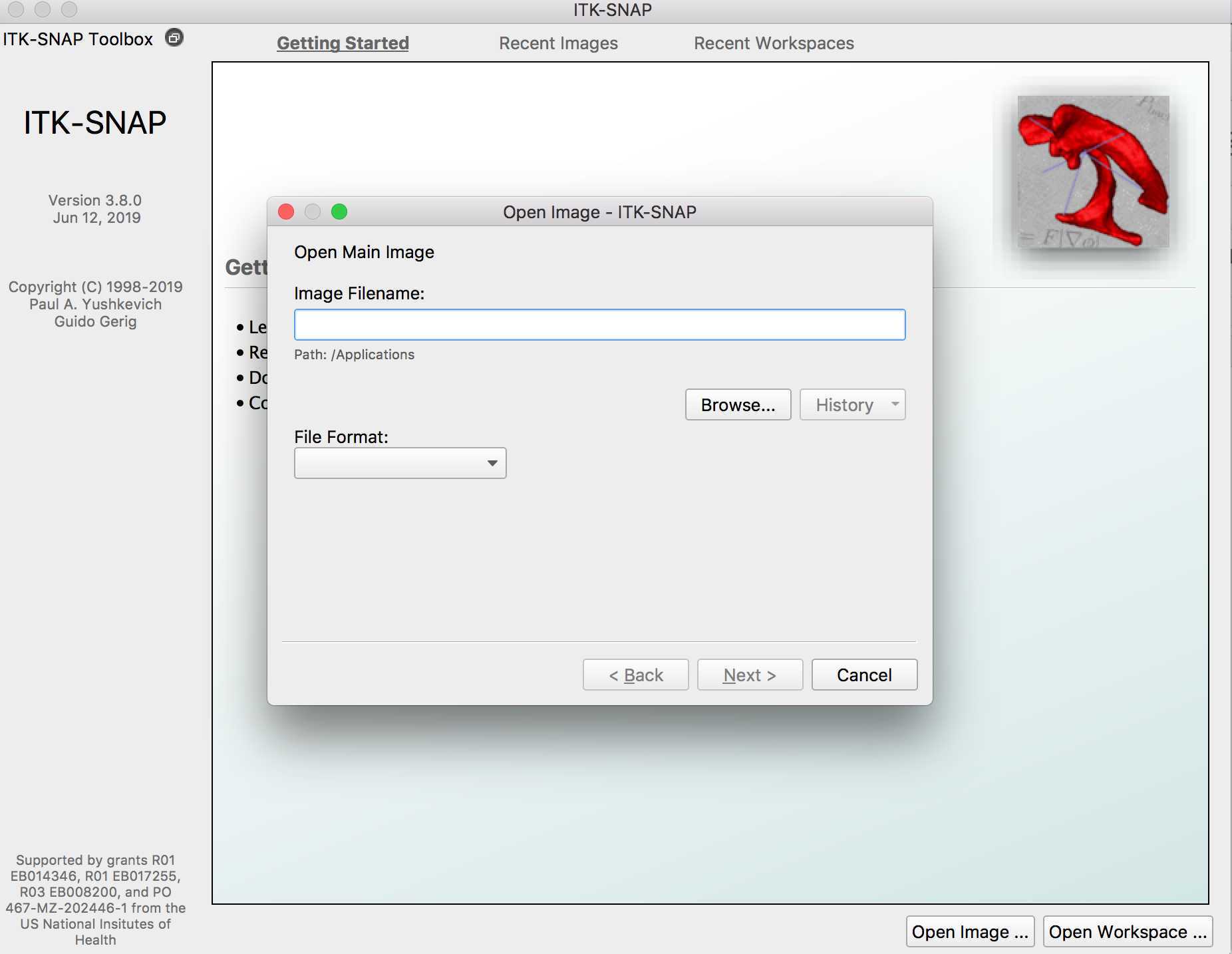
Example manual segmentation of a FCD Type IIB
Once you have identified the lesion, you may want to zoom in on it. Click the magnifying glass and increase the zoom until you can clearly see the lesion
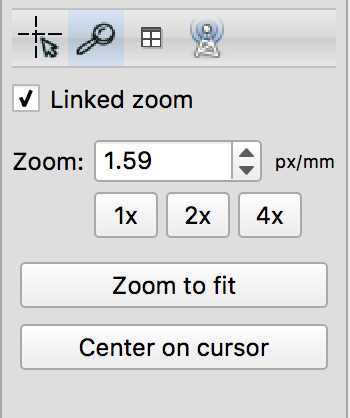
Then click the paintbrush and choose your brush style (I use the circle), and brush size. To create your lesion mask in 3D click 3D, to mask slice by slice, unclick the 3D button.
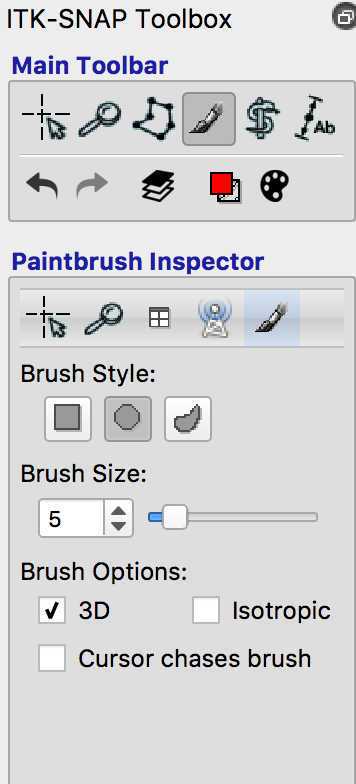
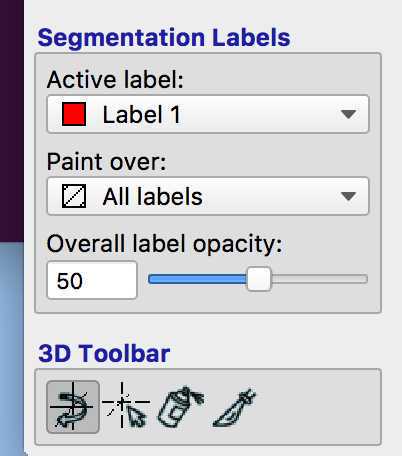
You can now start to mask your lesion. As you can see the "Active label" is 1 - this means that the voxels of the lesion will all have a value of 1. If you make a mistake and want to delete some of the voxels you have labelled, you can right click on them.
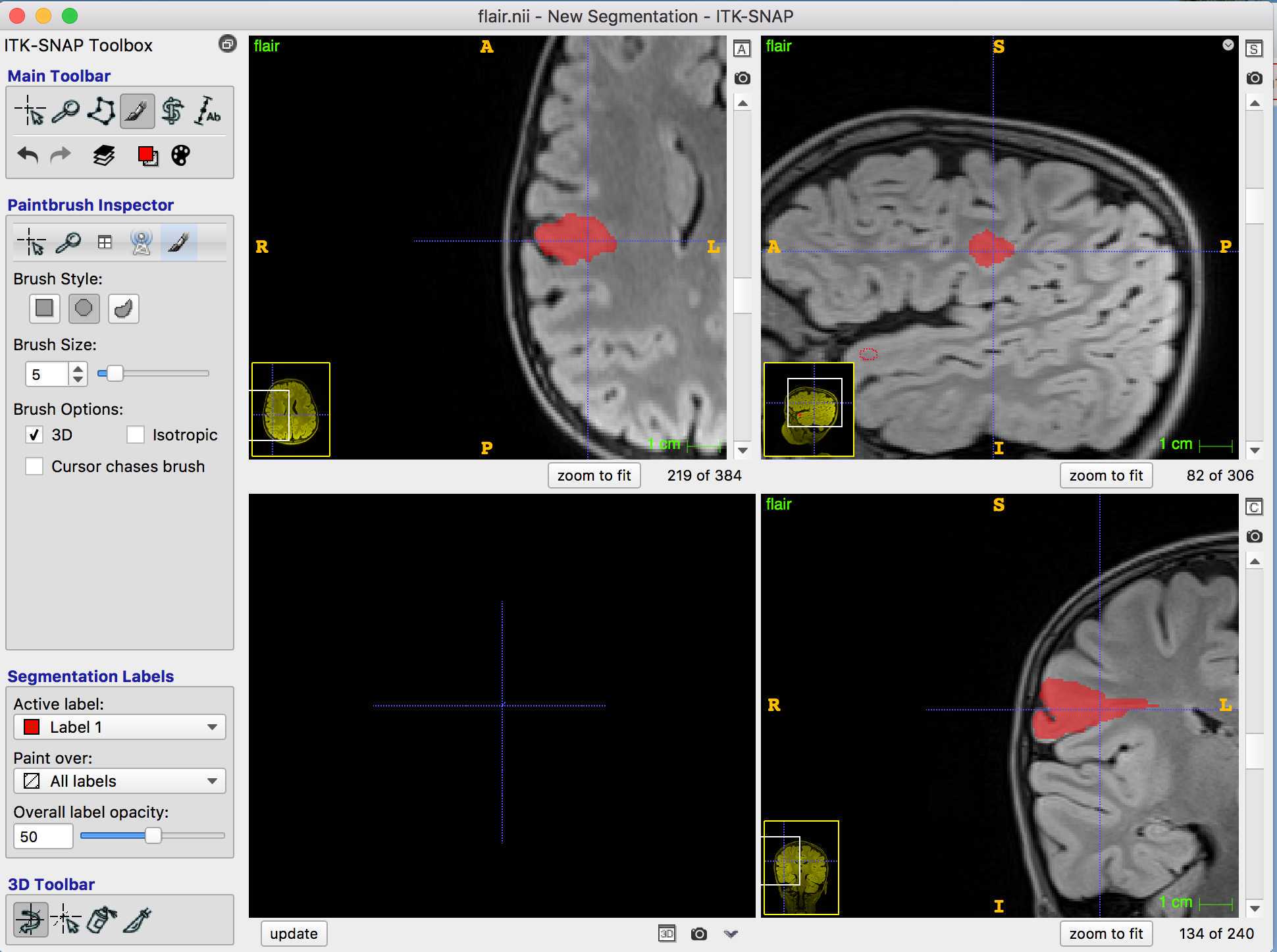
Creating the lesion label if there is more than 1 lesion e.g. FCD IIIA where you have a hippocampal lesion and a cortical lesion
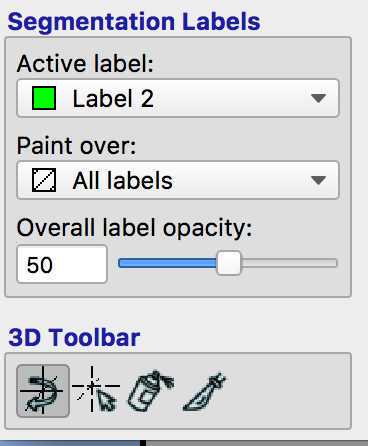
Mask one lesion using active label 1. Then change the label to 2 and mask the other lesion:
Save the lesions as before as "lesion.nii.gz". The first lesion mask will have values of 1 and the second will have values of 2.
Convert in BIDS and zip
Run meld_bidsify_data_step2.py to convert in BIDS format
Once you have placed the lesion nifti file into the right lesion_mask folder, you need to run " meld_bidsify_data_step2.py " script. This script will :
1. Reorganise MRI data into BIDS structure saved into meld_bids folder
2. Strip any identifiable header information from the scans
3. Deface the MRI data
4. Compress the meld_bids folder into different batchs
To run this script you need to provide a txt file with the list of participants you want to include in the batch.
To create the list of participants :
1) Open a terminal window
cd <path_to_meld_focal_epilepsy_data_folder>/meld_focal_epilepsy_data
nano list_participants_batch_<date_as_ddmmyyyy>.txt
``` Then type the MELD IDs of the participants you would like to run the script on. Press enter between each participant_id:
e.g.
MELD_H1_3T_P_0001
MELD_H1_3T_P_0002
MELD_H1_15T_P_0001
MELD_H1_15T_C_0002
File handling in nano
Ctrl+S Save current file
Ctrl+X Close buffer, exit from nano
Alternatively use any text editor to save your list of participant ids at list_participants_batch_<date_as_ddmmyyyy>.txt.
Once you have the participants file, type:
cd <path_to_meld_focal_epilepsy_github_folder>/meld_focal_epilepsy/scripts
python meld_bidsify_data_step2.py -d <path_to_meld_focal_epilepsy_data_folder>/meld_focal_epilepsy_data -ids <path_to_meld_focal_epilepsy_data_folder>/meld_focal_epilepsy_data/list_participants_batch_
You can find your new bids folder and its zipped batches at <path_to_meld_focal_epilepsy_data_folder>/meld_focal_epilepsy_data/meld_bids
The files to share begin with: "share_data_part"
**Note** : remember to keep track of the subjects you have already converted with the participants list file.
Sending MRI data
Note: Dropbox is now deprecated and files will be shared via File Request on OneDrive
Please contact meld.study@gmail.com and provide the email(s) of the person(s) who will be uploading the data. We will send you a file request to upload the data to a secure drop-only folder.
Upload each zipped batch
Please note – due to the file sizes, it may take a while to upload!
A member of the MELD team will confirm the reception of the files
REMINDER: ONLY SEND ANONYMISED DATA OVER