Proteome-Scale Drug-Target Interaction Predictions: Approaches and Applications
Stephen Scott MacKinnon, Stephen Scott MacKinnon, S. A. Madani Tonekaboni, S. A. Madani Tonekaboni, Andreas Windemuth, Andreas Windemuth
Abstract
Drug-Target interaction predictions are an important cornerstone of computer-aided drug discovery. While predictive methods around individual targets have a long history, the application of proteome-scale models is relatively recent. In this overview, we will provide the context required to understand advances in this emerging field within computational drug discovery, evaluate emerging technologies for suitability to given tasks, and provide guidelines for the design and implementation of new drug-target interaction prediction models. We will discuss the validation approaches used, and propose a set of key criteria that should be applied to evaluate their validity. We note that we find widespread deficiencies in the existing literature, making it difficult to judge the practical effectiveness of some of the techniques proposed from their publications alone. We hope that this review may help remedy this situation and increase awareness of several sources of bias that may enter into commonly used cross-validation methods. © 2021 Cyclica Inc. Current Protocols published by Wiley Periodicals LLC.
INTRODUCTION
Drug discovery prediction models are typically binned into two groups: ligand-based or structure-based. Ligand-based drug discovery approaches build predictive machine learning models from datasets that pair multiple compounds with a specific bioactivity of interest, which is often the activation, inhibition, or binding of a target protein. Ligand-based machine learning models are also known as quantitative structure activity relationship (QSAR) and date back to the 1960s (Craig, 1984). QSAR models do not take protein structures into account, and can only predict bioactivities for which there is sufficient experimental data, including both active and inactive compounds. Structure-based drug discovery approaches rely on knowledge of a target protein's 3D-structure. Molecular docking simulations first predict the 3D binding pose of a protein-ligand pair (Kuntz, Blaney, Oatley, Langridge, & Ferrin, 1982), and the pose is then scored to predict the likelihood of physical binding using empirical or knowledge-based force fields. Molecular docking force fields are mathematical functions that express protein-ligand binding propensity as a function of molecular coordinates. Empirical force fields implement functions whose primary terms model physical contributions to binding energy, such as bond rotations or hydrogen bonds, with select term weights or coefficients empirically fit to solved structures/complexes via regression (Eldridge, Murray, Auton, Paolini, & Mee, 1997). In contrast, the primary terms of knowledge-based force fields model non-physics-based statistical properties, such as geometric propensities, interatomic distance propensities for varied atom types, or functional group contact propensities consistent with other known protein-ligand binding systems (Gohlke, Hendlich, & Klebe, 2000; Mitchell, Laskowski, Alex, & Thornton, 1999; Muegge & Martin, 1999). In both cases, molecular docking force fields represent a hybrid between machine learning and expert systems.
Both ligand-based and structure-based approaches to virtual screening can be used to screen chemical libraries separately or in combination, scoring thousands to millions of compounds for molecules likely to exhibit desirable bioactivities (Drwal & Griffith, 2013). Together, they have long been staples of computational drug discovery, complementing one another with their benefits and limitations. Ligand-based models can be very accurate with sufficient data, but require many known actives and inactives to develop initial models. Structure-based methods can work without such data, but require detailed knowledge of the 3D structure of the target. They are also computationally expensive and have wide fluctuations in performance across different protein systems, and the underlying biophysically motivated model does not benefit from existing experimental data available for the drug/target of interest. Widespread use of structure-based approaches has, however, led to successful drug discovery programs, emphasizing the importance of the biophysical insights provided by protein structure. One notable advantage of structure-based systems is their generalizability. Generalizability is the property of a predictive model to correctly extrapolate beyond their original design, such as training data in the case of machine learning. Structure-based simulation (molecular docking, FEP, etc.) offers a great amount of generalizability, simply because there is no well-defined training set and predictions are made on the basis of assumed fundamental principles embodied by the force field. Structure-based approaches can be applied to targets without experimental bioactivity measurements, but do not inherently improve when there are known bioactivity measurements.
Over the past decade, the increased abundance of relevant data and rapid progress of machine learning methodologies have motivated new predictive solutions and consequently applications for Drug-Target Interaction (DTI) predictions. In particular, widespread accessibility of flexible deep learning software toolkits such as MXNet, PyTorch, and TensorFlow (Chen et al., 2015; TensorFlow Developers, 2021; Paszke et al., 2019) have provided computational scientists with the opportunity to model numerous sub-problems related to drug-target interaction prediction. For example, deep learning models trained on bound protein-ligand co-crystal structures are designed to learn atom-level binding patterns (Ragoza, Hochuli, Idrobo, Sunseri, & Koes, 2017). These models are used to score predicted poses from docking simulations to predict correct binding modes, or to perform virtual screening tasks. In another example, one-shot-learning models use neural networks to derive a universal metric for compound-compound similarity, which in turn can be used situationally for structure activity modeling on datasets that are otherwise too small for a conventional QSAR (Altae-Tran, Ramsundar, Pappu, & Pande, 2017; Baskin, 2019).
Moreover, improvements in the speed and accuracy of protein-ligand machine learning models have renewed interest in alternative applications outside of virtual screening. Specifically, accurate and generalizable models could be used to screen one or more compounds against a collection of proteins in search of on- or off-target interactions in living systems with potential applications in toxicology, drug repurposing, or phenotypic deconvolution. These tasks, collectively referred to herein as off-target profiling , were initially performed using molecular docking, under a range of different names including: inverse screening, reverse screening, ligand profiling, and target fishing (Hui-fang, Qing, Jian, & Wei, 2010; Rognan, 2010; Chen & Zhi, 2001; Meslamani et al., 2012; Paul, Kellenberger, Bret, Müller, & Rognan, 2004). In a 2013 review of proteome screening methods, Rognan notes that ligand-based approaches are faster and more effective when relevant data is available, but structure-based approaches provide a broader scope of coverage (Rognan, 2013). Structure-based approaches and ligand-based approaches are subject to separate target coverage restrictions, which also vary from algorithm to algorithm. In particular, data requirements for ligand-based approaches can have a big impact on target coverage. A recent analysis of DTI data points captured by the STITCH 5 database (Szklarczyk et al., 2016) revealed that 65% of known human proteins can be linked back to one or more ligands, while less than 2% of all human proteins have 50+ high-quality interactions (Somody, MacKinnon, & Windemuth, 2017). QSAR models aimed at individual protein activity generally require tens to thousands of known activities to yield predictive models. For example, Rifaioglu and colleagues were able to create 523 individual protein models for each human protein with more than 100 known active ligands (bioactivity values ≤10 μM) using the ChEMBL database (Mendez et al., 2019), in their development of their deep learning based DEEPScreen models (Rifaioglu et al. 2020). The most comprehensive such effort to date generated more than 1000 individual datasets from ChEMBL by systematically aggregating measurements of compatible assays, which include specific target-based activities (Mayr et al., 2018). The QSAR panel approach is particularly well suited for high-data systems, such as liability targets commonly screened in drug discovery, as the authors of the study further indicate that predictive performance on many of the virtual assays is comparable to in vitro experiments. However, the approach limits target space to high-data systems, constraining its application toward new drug targets or in off-target profiling tasks.
Drug-Target Interaction models are a class of drug discovery tools that base predictions on large bioactivity datasets, represented as paired protein-ligand activity measurements (Yamanishi, Araki, Gutteridge, Honda, & Kanehisa, 2008). Relative to standard ligand-based approaches, DTI models are trained on bioactivity datasets encompassing multiple different proteins, in order to extrapolate known binding data to new proteins. These approaches combine virtual representations of proteins and ligands, then train a machine learning model to identify interacting pairs. DTI models can be framed as a classification problem, aimed at predicting binding and non-binding pairs, or as a regression problem, aimed at predicting quantitative bioactivity measurements.
Past reviews on DTI prediction tools have provided historical context, summarized available datasets, described similarity versus machine-learning based approaches and provided lists of available tools (Bagherian et al., 2021; Chen et al., 2016; Ding, Takigawa, Mamitsuka, & Zhu, 2014). While the term Drug-Target Interactions (DTI) was originally coined to describe network-based predictions that operate on pharmaceutically relevant datasets (Cheng et al., 2012; Yamanishi et al., 2008), it has since been applied to small-molecule compounds and proteins not otherwise considered therapeutic targets. Alternatively, Compound Protein Interactions (CPI) is occasionally used to describe applications to broader datasets (Cheng, Zhou, Li, Liu, & Tang, 2012; Liu, Sun, Guan, Zheng, & Zhou, 2015), and Drug Target Affinity (DTA) models can refer to regression variants of DTI models (Zhao, Xiao, Yang, Li, & Wang, 2019). For simplicity, we will refer to these methodologies collectively as DTI predictions for the remainder of this article.
Here, we examine DTI prediction tools based on deep learning, with an emphasis on understanding how data selection, benchmarking, machine learning algorithms, and representations impact utility and application of the respective models. We will consider the suitability of these systems for class detection, polypharmacological profiling, or proteome screening applications, where the predictive power of deep learning can help generalize DTI datasets to low-data or dataless targets. We also examine the experimental designs introduced by DTI prediction studies, such as scientifically informed leave-out sets and ablation studies, with an emphasis on learned concepts that can be readily applied to new DTI models.
DEEP LEARNING MODELS FOR DRUG-TARGET INTERACTION PREDICTIONS
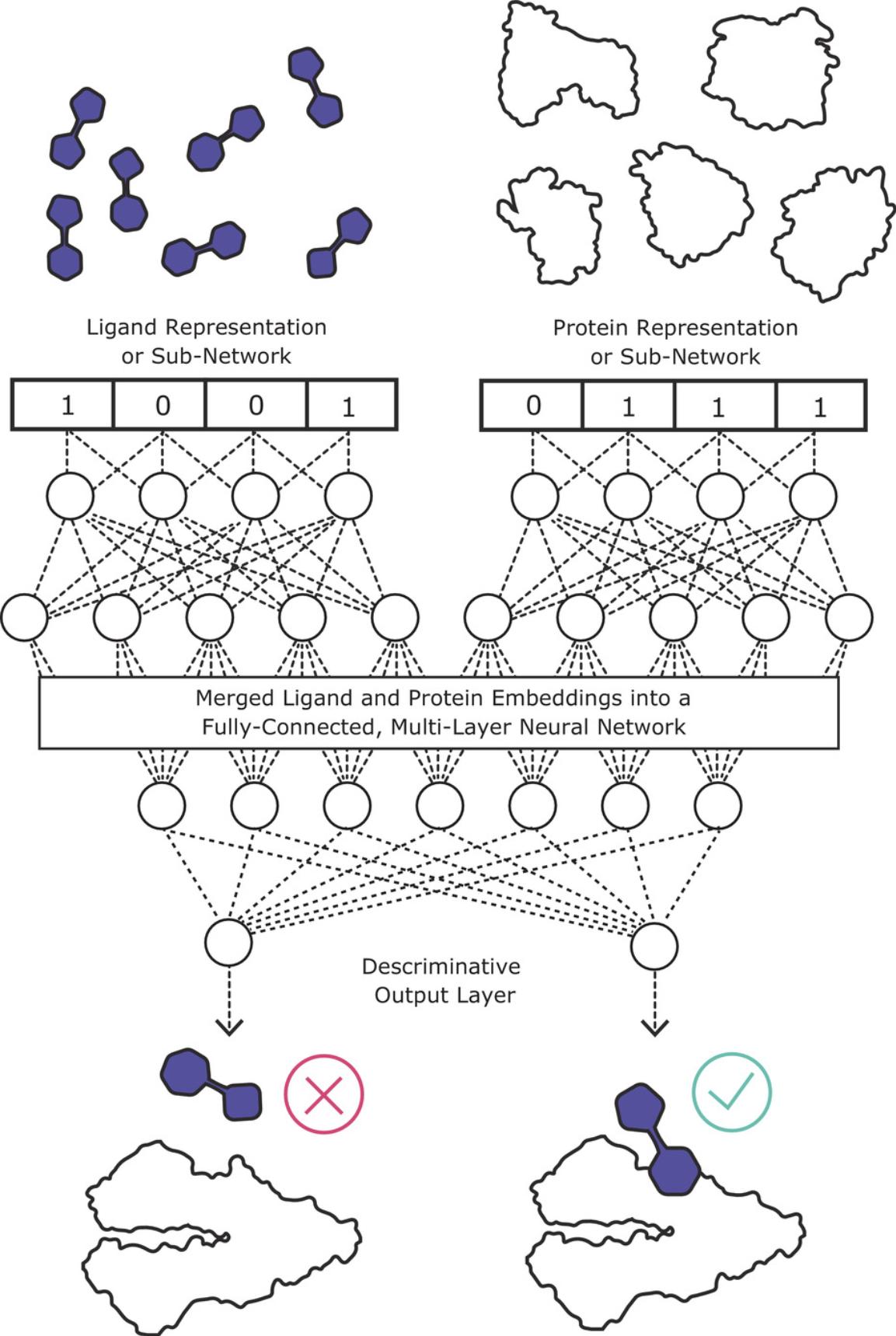
Deep learning approaches frame DTI predictions as discriminative supervised learning problems. In each case, the neural network accepts separate representations of a protein and a ligand, and ultimately fuses the representations into a numerical vector representing the paired association. The largest variation among deep learning architectures for DTI models is in the treatment of the individual ligand and protein representation prior to fusing their representation in the discriminative network. Once fused, most models implement a multi-layer, fully connected neural network with 2-4 layers (Hie, Cho, & Berger, 2018; Lee, Keum, & Nam, 2019; Nguyen et al., 2021), with some minor variations like the use of dropouts (Abdel-Basset, Hawash, Elhoseny, Chakrabortty, & Ryan, 2020; Öztürk, Özgür, & Ozkirimli, 2018). An output layer then evaluates a categorical binding or non-binding label (classification) or a numerical activity prediction, such as binding affinity (regression). Conceptually, this dual-input architecture allows a neural network to pair properties of a ligand that are compatible with its target protein and vice versa. The selection of protein and ligand representations, as well as the algorithmic approach that transforms these representations into machine-learnable data, are intertwined components of any deep learning DTI model. Figure 1 shows a schematic representation of a DTI model. Table 1 shows the diversity in protein and ligand representations among 12 recently-released DTI models.
| Name | Learning task | Negative data | Protein representation | Ligand representation | Applicability | Datasets | Largest dataset | Validations |
|---|---|---|---|---|---|---|---|---|
|
DeepDTA (Öztürk et al., 2018) |
Regression | Experimental | Protein Sequence CNN | SMILES String CNN | Kinases | Davis and KIBA Kinase Datasets | 118,254 Affinities |
Random Splits on Interactions; Ensures each test set protein is represented in the training data |
|
GraphDTA (Nguyen et al., 2021) |
Regression | Experimental | Protein Sequence CNN | Four variants of a Graph Neural Networks | Kinases | Davis and KIBA Kinase Datasets | 118,254 Affinities |
Random Splits on Interactions; Ensures each test set protein is represented in the training data |
|
DeepH-DTA (Abdel-Basset et al., 2020) |
Regression | Experimental | Residue K-mer Compositions |
Bidirectional ConvLSTM, SMILES string autoencoder Molecular Graphs |
Kinases | Davis and KIBA Kinase Datasets | 118,254 Affinities |
Random Splits on Interactions; Ensures each test set protein is represented in the training data |
|
DeepPurpose (Huang et al., 2021) |
Regression or Classification | Experimental |
Residue K-mer Compositions Sequence Neural Networks |
SMILES Encoders Chemical Fingerprints and Molecular Graphs |
Kinases | Davis and KIBA Kinase Datasets | 118,254 Affinities |
Random Splits on Interactions; Ensures each test set protein is represented in the training data |
|
MFDR (Hu et al., 2016) |
Classification | 1:1 Assumed |
Multi-Scale Feature Vectors, Derived from Primary Sequence |
Pubchem Fingerprints | Separate Models per Protein Class |
Yamanishi et al. Curated Dataset |
2926 Positives | Random Splits on Interactions |
|
AutoDTI++ (Sajadi et al., 2021) |
Unsupervised Imputation | N/A | N/A | Substructure-based fingerprint | Separate Models per Protein Class |
Yamanishi et al. Curated Dataset |
2926 Positives |
Random Splits on Interactions New Compound Test New Protein Test |
|
DeepDTIs (Wen et al., 2017) |
Classification |
1:1 Assumed (10 Replicates) |
Residue K-mer Compositions | ECFP Fingerprints | Multiple Drug Target Classes | Drugbank with EDTPs | 7352 Positives | Random Splits on Interactions |
|
DeepConv-DTI (Lee et al., 2019) |
Classification | 1:1 Assumed | Protein Sequence CNN | Morgan/Circular Fingerprints | Multiple Drug Target Classes | Drugbank, KEGG, IUPHAR, Matador | 32,568 Positives |
Random Splits on Interactions New Compound Test New Protein Test New Compound + Protein Test |
|
DL-DTI (Zhao et al., 2020) |
Classification | 1:1 Assumed | Residue K-mer Compositions | Morgan/Circular Fingerprints | Multiple Drug Target Classes | DrugBank Human Targets | 904 Drugs with 614 Targets | Randomly Sampled |
|
SecureDTI (Hie et al., 2018) |
Classification |
1:1 Assumed 1:10 Assumed 1:1 Shuffled |
PFam Domains | ECFP Fingerprint | General Protein-Ligand Binding | Stitch | 969,817 Positives |
Random Splits on Interactions New Compounds |
|
Gao et al. (Gao et al., 2018) |
Classification | Experimental |
GO Terms Protein Sequence RNN |
Graph CNN | General Protein-Ligand Binding | BindingDB | 33,777 Positives |
Random Splits on Interactions New Compound Test New Protein Test New Compound + Protein Test |
|
MatchMaker (Redka et al., 2020; Sugiyama et al., 2021) |
Classification |
1:19 Shuffled |
Functional Annotations 3D Structure Annotations |
Molecular Descriptions Molecular Fingerprints |
General Protein-Ligand Binding | ChEMBL, Stitch, BindingDB, GoStar | 1.5 Million Positives |
Random Splits on Interactions New Compound Test New Scaffold Test New Compound + Protein Test |
- aRatios listed in the negative data column represent positive to negative data balance selected in the generation of randomized negative data.
Protein and ligand representations within a DTI model provide the basis for learning and extrapolating patterns into unknown chemical space or new proteins. The source DTI datasets themselves represent proteins and ligands as database identifiers, such as PubChem IDs (Kim et al., 2021) or UniProt accessions (UniProt Consortium 2021). In principle, DTI predictions can be made directly on source database identifiers without any added context. For example, Cobanoglu et al. used Probabilistic Matrix Factorization (PMF) to predict DTIs based solely on the connectivity of a bipartite graph connecting proteins and ligands in the absence of ligand and protein features or similarity metrics (Cobanoglu, Liu, Hu, Oltvai, & Bahar, 2013). In contrast, earlier approaches to graph-based DTI predictions, including Yamanishi et al. (2008) and the Kernelized Bayesian Matrix Factorization (KBMF ; Gönen, 2012 ) factor in precompiled all-by-all chemical structure similarity and protein sequence similarity matrices to help model instances where similar compounds interact with the same protein and vice versa. The use of ligand or protein similarity matrices does, however, constrain the maximum dataset size that can be used for training. As noted by Cobanoglu et al. (2013), these similarity-based approaches would require ∼1012 protein similarity comparisons and ∼1010 ligand similarity comparisons to model larger DTI databases such as STITCH (Kuhn et al., 2012).
Deep learning based DTI models use vector-based representations of ligands and proteins instead of similarity matrices to generalize DTI training data onto new compounds and proteins. Selection of protein or ligand representation directly impacts predictivity, allowing the network to readily learn salient relationships. The process of transforming protein or ligand representations into numerical vectors used to train or evaluate a machine learning model is known as encoding. In this section, we will explore how different ligand and protein representations have been used in recently published DTI models.
Ligand Representations
SMILES strings are often the starting point for chemical representations. A ligand's SMILES representation is a 1-dimensional string of characters that convey the molecule's atoms, as well as their bonding connectivity and stereochemistry (Weininger, 1988). SMILES representations are then used to derive molecular graphs, descriptors, pharmacophores, or fingerprints as input features for machine learning models. The topic of ligand representations has been central to computational drug discovery tasks dating back to the earliest bioactivity models based on expert-defined molecular descriptors (Craig, 1984). Outside the domain of DTI models, the relationship between ligand representations and performance of predictive tasks has been thoroughly explored, benchmarked, and reviewed (Banegas-Luna, Cerón-Carrasco, & Pérez-Sánchez, 2018; Brereton et al., 2020; Jiang et al., 2021; Qing et al., 2014; Riniker & Landrum, 2013; Stepišnik, Škrlj, Wicker, & Kocev, 2021), generally noting that different bioactivity modeling and similarity problems benefit from different forms of chemical representation. Notably, the modeling toolkit DeepChem 2.5.0 offers over 40 different molecule featurizers that can be evaluated in different combinations with machine learning algorithms to identify optimal bioactivity models for specific tasks (Ramsundar et al., 2019). Among DTI prediction tools, DeepPurpose similarly implements a collection of eight ligand encoders to combine with alternative protein representations and architectures to derive new models (Huang et al., 2021); however, the resource has not yet been used for a thorough benchmarking of ligand representations.
Stepišnik et al. (2021) classify ligand features into expert-based representations, which include descriptors and fingerprints constructed with expert knowledge, and learnable representations, which include neural network−based ligand encoders, trained to interpret other than numerical vectors, such as SMILES strings or molecular graphs (Stepišnik et al., 2021). Learnable representations are subdivided into task-independent representations, in which the ligand encoding network is developed separately from a predictive task, and task-specific representations, where ligand encoding and the discriminative task are simultaneously trained in the same network. Within the context of ligand-based structure-activity models, the authors also noted that optimal ligand representations varied from task to task. Additionally, expert-based representations and task-independent, learned representations achieved comparable performance while the task-specific, learnable representations were more computationally demanding and had no clear predictive advantage. At this time, we are unaware of any similar study that benchmarks multiple diverse ligand representations in DTI models. Nonetheless, many different representations, including learned representations, have been explored in separate studies.
Three of the DTI prediction tools reviewed in Table 1 use automatic text-based encoders to train models directly from SMILES string inputs. As a task-specific, learned representation, the added challenge of simultaneously interpreting SMILES strings while performing a classification task requires slower, more complex network architectures. In general, multiple alternative ligand representations should be considered alongside the SMILES-based encoder to optimize predictive and runtime performance. Moreover, synonymous SMILES may pose a secondary challenge and practical constraints for SMILES-based encoders. For example, “CC(=O)NC1=CC=C(C=C1)O”, “CC(=O)NC1=CC=C(O)C=C1”, and “CC(=O)Nc1ccc(O)cc1” each encode acetaminophen, a popular pain relief medication. This long-standing issue is addressed in many cheminformatics software toolkits, such as RDKIT (Landrum, 2014), through a process known as canonicalization. Canonicalization is a pre-processing step, where each molecule in the dataset is converted into a graph representation, then back into a SMILES string using a consistent algorithm. Canonicalization is specific to each individual software package. Omitting a canonicalization step may introduce a data origin bias, while including a canonicalization will require each subsequent use of the model to equally pre-process new query molecules through the same algorithmic process as used for training molecules. It is currently unclear what impact the effects of synonymous SMILES have on DTI prediction models. Of the DTI models reviewed in this study, DeepH-DTI does perform a canonicalization step to eliminate stereochemistry from input training data (Abdel-Basset et al., 2020); however, none of the other DTI models that use string-based SMILES encoders have reported canonicalization steps in their methodology. Moreover, none of the studies to date have reported differences in DTI prediction performance associated with synonymous SMILES strings.
Molecular graph representations provide an alternative learnable representation to SMILES strings. GraphDTA compares graph-based neural network architectures encoding the ligand side of a DTI model to the SMILES-based encoder implemented in DeepDTA (Öztürk et al., 2018). This benchmark otherwise examines comparable network architectures on the same dataset for a head-to-head comparison of two task-specific, learned representations. In GraphDTA, molecules are represented as graphs where atoms are nodes and bonds are edges, capturing overall connectivity of a molecule with the notable exception of chirality. The authors report modest gains in predictive performance relative to SMILES-based encoders consistent with the application of graph-based neural networks in other molecular prediction problems (Hirohara, Saito, Koda, Sato, & Sakakibara, 2018; Kearnes, McCloskey, Berndl, Pande, & Riley, 2016; Liu et al., 2019; Torng & Altman, 2019). The authors further attribute these gains to the explicit structural information contained within the input molecular graphs relative to the SMILES-based encoder.
Protein Representations
Among the deep learning DTI models, proteins are mainly represented by global attributes, such as primary sequence, or other attributes that can be derived from source protein databases. For example, SecureDTI (Hie et al., 2018) introduces binary features to represent the presence or absence of Pfam domains (El-Gebali et al., 2019) in the protein, while Gao and colleagues use primary sequence features in addition to Gene Ontology (Ashburner et al., 2000; Gao et al., 2018). Conceptually, these features allow the network to match domains to relevant substrate ligand properties; e.g., an esterase domain may be linked to a ligand feature representing an ester group. More often, however, the network pairing operates at a higher level in the neural network, instead representing relationships between mathematically inferred features, known as latent variables. In the context of a supervised neural network model, latent variables correspond to intermediate values calculated in the middle layers between the input features and the model output layer (e.g., layers of circular nodes illustrated in Fig. 1). The use of protein representations beyond sequence features can however constrain the scope of usable data. In the example above, proteins with undocumented Pfam domains or Pfam domains that are not in the training dataset may not be sufficiently recognized by the DTI model. Selection of representations is therefore a tradeoff between augmenting DTI data with useful information versus limiting the scope of drugs or proteins in the dataset.
Instead, most DTI models reviewed in this study operate directly on the target's protein sequence, which is readily available for most proteins encountered in DTI datasets. DeepPurpose is a software library designed to let end users develop custom DTI models for their own datasets, using a broad range of different algorithms and representations (Huang et al., 2021). It provides seven separate protein encoders, which can be considered as two separate categories, (1) expert-designed algorithms or (2) neural networks designed for text processing. Among the algorithmic encoders, the ACC encoder creates a vector of 8420 elements, describing the frequency of all amino acid k-mers for k values up to 3 (Reczko & Bohr, 1994), while the conjoint triad encoder provides a 3-mer frequency count using a reduced amino acid alphabet (Shen et al., 2007). In contrast, neural network encoders operate directly on sequence. The DeepPurpose Convolutional Neural Network (CNN) encoder converts each amino acid into a numerical value as a fixed-length 1D array, then uses a convolutional neural network (Krizhevsky, Sutskever, & Hinton, 2017) to learn spatial information from the sequence (i.e., local amino acid neighborhoods) that may be relevant for the DTI binding model.
While the technical nature of primary sequence encoders differ, they all aim to represent proteins as a function of their local subsequences. Since proteins are much larger than small-molecule drugs, only a small portion of any protein sequence may be directly involved in a DTI interaction. Eukaryotic proteins average 472 amino acids (Tiessen, Pérez-Rodríguez, & Delaye-Arredondo, 2012), which is approximately 100×[ larger than a standard small-molecule drug with a molecular weight of 500 Da. To provide a specific example, we examine a typical drug-target complex, the structure of which is found in the PDB: 4HJO, a co-crystal structure of EGFR (a protein target) bound to erlotinib (a drug) (Park, Liu, Lemmon, & Radhakrishnan, 2012). Using the RCSB PDB web portal (Berman et al., 2000), we see that the erlotinib binding site is made up of residues 702, 719, 721, 764, 766-773, 820, 830, and 831. Only 15 of the 1210 amino acids from the full-length EGFR sequence are directly involved in ligand binding, and many are non-sequential. Some of the articles reviewed in this study acknowledge this limitation of sequence-based representation. In particular, Nguyen and colleagues hypothesize that the predictability of their GraphDTI model may improve by introducing graph-based representations of protein 3D structure (Nguyen et al., 2021). On the other hand, Lee and colleagues anticipate that the use of structure would narrow the domain of applicability of DTI models, as protein structure databases have fewer total records relative to sequence databases (Lee et al., 2019).
MatchMaker is a commercial DTI model that addresses the lack of structural coverage by systematically mapping DTIs to observed or inferred ligand-binding sites from experimentally determined protein structures and homology models (Redka et al., 2020; Sugiyama et al., 2021). Instead of matching targets directly with ligands, MatchMaker matches pockets with ligands, with each target having one or more pockets. Because the pockets are mapped onto the 3D structure, pocket features can be computed with full knowledge of which residues are relevant for binding. Models are then trained using the assumed pocket features to represent each protein, while simultaneously addressing uncertainties in data quality and pocket mapping with Filtered Transfer Learning (Tonekaboni et al., 2020). This combination of ligand- and structure- based data is believed to provide the DTI model with the ability to generalize from first principles in biophysics as well as from patterns in the experimental data.
Training and Evaluation Time
Few DTI prediction resources listed in Table 1 report model training times. Model training times impose an upper bound on training data, and consequently model performance and generalizability. Moreover, faster training times afford broader benchmarking options, including multiple splits or generalizability thresholds. Relative training times are reported for DeepH-DTA and four reference DTI algorithms trained on an unspecified GPU system (Abdel-Basset et al., 2020). DeepH-DTA requires 7.1 min per epoch to train on a 30,056 interaction dataset and 25.9 min per epoch on a 118,254 compound dataset, indicating roughly linear scaling with dataset size. The KIBA test set model running 200 epochs would therefore require 3.6 days per model. While these model training times are permissive for the benchmarked datasets, ChEMBL 28 reports 17,276,334 activity measurements (Mendez et al., 2019), implying that individual models may require GPU-year training times. Moreover, practical DTI model development could require dozens to hundreds of experimental models to explore hyper-parameters, feature refinements, multiple validation split replicas to evaluate averaged performance metrics, and alternate benchmarking tests to evaluate generalizability.
Currently, reaching compute scales capable of training DTI models with all publicly available training data remains a challenging task. Secure-DTI (Hie et al., 2018) trains on the STITCH 5.0 (Szklarczyk et al., 2016) dataset of nearly 1 million molecules and has arguably the simplest architecture, consisting of a standard multi-layer perceptron with two hidden layers, each with 250 neurons. Abdel-Basset et al. (2020) also note that simpler deep learning frameworks are more time efficient, and attribute large execution times to complex portions of the network linked to learning the protein and ligand representations. This observation suggests a speed advantage with respect to DTI prediction methods where protein and ligand embeddings can be performed independently and recycled for multiple interaction pairs. Independent embeddings may include the use of algorithmically defined features as well as separate ligand/protein unsupervised representation identifiers like autoencoders. As an example, DLDTI uses a stacked auto-encoder to compress large molecular fingerprint arrays into smaller, low-dimensional feature vectors independently of model training (Zhao et al., 2020).
Model evaluation time also impacts possible applications. MatchMaker technical documentation indicates a top speed of 789 DTI pair evaluations per CPU-second (Cyclica Inc. 2021). In contrast, DeepConv-DTI was benchmarked at 7.17 DTI pairs per CPU-second. This scaling potential allows for broader 2D all-by-all cross screening for select molecule datasets (Redka et al., 2020) or single-target screening on large on-demand molecule libraries, such as the Enamine REAL database (Grygorenko et al., 2020).
DRUG-TARGET INTERACTION DATASETS
Drug-target interaction databases define the problem scope for DTI predictions. At their core, DTI databases provide a ligand identifier, a protein identifier, and some experimental measure about the given association. Additional annotation fields vary between individual resources. These annotations can provide added context on the nature of the specific bioactivity, experimental details, experimental conditions, or measurement confidence. Table 1 describes the datasets used by the twelve reviewed DTI models, including dataset activity counts, scope, and testing framework. Reported source datasets used by DTI tools include: the Davis Kinase dataset (Davis et al., 2011), the KIBA Kinase dataset (Tang et al., 2014), Drugbank (Wishart et al., 2006), KEGG (Kanehisa et al., 2006), Brenda (Schomburg et al., 2004), Supertargets and Matador (Günther et al., 2008), IUPHAR (Southan et al., 2016), BindingDB (Gilson et al., 2016), ChEMBL (Mendez et al., 2019), and Stitch (Szklarczyk et al., 2016).
DTI Dataset Density
DTI data density is one attribute that impacts a model's capabilities and domain of applicability. Large DTI datasets have sparse coverage on specific protein-ligand pairs within the dataset. For example, ChEMBL 28 reports 17,276,334 activity measurements, covering 14,347 distinct targets and 2,086,898 compounds, for an estimated density of 0.05% (Mendez et al., 2019). Higher-density datasets are often preferred, particularly by regression models focused on binding affinity predictions. Specialized datasets do, however, exist with higher data density, but restricted in target coverage. For example, the Davis benchmark is a fully populated, all-by-all dataset of 68 drugs by 442 kinase targets (Davis et al., 2011), i.e., a DTI dataset with 100% density, made possible by systematic experimental assay panels. Another kinase-specific dataset, KIBA, offers a middle ground between density and coverage, with 246,088 total interactions between 52,498 ligands and 467 kinase targets, at an average density of 1% (Tang et al., 2014). In developing SimBoostQuant, a gradient-boosting DTI model, He and colleagues further restricted the KIBA dataset to only 2126 drugs and 229 targets in order to reach an average data density of 24% (He, Heidemeyer, Ban, Cherkasov, & Ester, 2017). The authors demonstrated that confidence in their drug-target affinity predictions was dependent on the number of observations attributed to each ligand in the dataset. KIBA was also used to train DeepDTA (Öztürk et al., 2018), a deep learning DTI model specializing in kinase affinities. Both DeepDTA and SimBoost also benchmark their respective tools with the Davis set, but reported higher performance on the KIBA benchmark. Predictive improvements introduced by the DeepDTA deep learning model are also more apparent in the KIBA dataset. Öztürk et al. attribute these findings to the 4× larger number of DTI pairs in the KIBA benchmark, stating that deep learning architectures are better able to capture information from larger datasets. Alternatively, greater chemical diversity may be a contributing factor, as the KIBA dataset has 31× as many distinct compounds. However, based on the materials presented in these studies, it is unclear what impact the dataset selections have on ligand generalizability and new scaffold discovery. Nonetheless, both tools restrict the domain of applicability to the human kinome in exchange for higher data density, capable of modeling binding affinities.
Some DTI models supplement small datasets with low data density by applying unsupervised pre-training approaches (Hu, Chan, & You, 2016; Wen et al., 2017). In unsupervised pre-training, a neural network is trained to recognize and recapitulate salient features of inputs in training data, without specifically training to predict drug-target interactions. For this task, the network can train using all pairwise combinations of proteins and ligands in the DTI dataset, even those lacking experimental measurements. Once the unsupervised model is built, it is used to generate feature vector representation for all other DTIs, which are subsequently used as input features to train a separate DTI prediction model.
Activity Calls and Negatives
DTI data is not all equal. One major caveat of DTI datasets is their lack of real negative data, as non-interacting pairs are generally considered uninteresting and are not reported. In the absence of true negative interactions, randomly selected protein-ligand pairs are selected as negative training examples. Assumed negatives are not explicitly known to be non-binders, but the likelihood of any protein interacting with an artificially generated molecule is considered sufficiently low to use in training or benchmarking DTI models. However, DTI data coverage biases related specific drug target families such as GPCRs or kinases, which increase the likelihood of a randomly sampled protein-ligand pair forming a real interaction. Corrections are however possible. Liu and colleagues introduce dissimilarity metrics to the selection of random protein-ligand pairs used as negative samples for machine learning DTI models (Liu et al., 2015).
Molecular decoy datasets, such as the DUD-E benchmark (Mysinger, Carchia, Irwin, & Shoichet, 2012), represent a separate class of assumed negatives designed to benchmark molecular docking engines. Decoys simulate negative pairs by generating realistic compounds that share similar chemical properties to known positives, but diverge in topology. While providing a useful negative control to molecular docking experiments, the use of molecular decoys in machine learning, however, can introduce unintended decoy biases (Chen et al., 2019). Chen et al. demonstrated that machine learning models could distinguish between real molecules and procedurally-generated molecular decoys in the absence of protein-specific information. Since decoys are restricted to negative data examples, complex deep learning models could be interpreting intrinsic properties of the decoy ligand as being associated with a non-binding outcome, rather than the intended properties of the interacting pair. This is called negative selection bias , and it can lead to an overestimation of prediction performance if not controlled for.
Biases that stem from intrinsic properties of ligands are not limited to molecular decoys. Any systematic differences between the proteins or ligands found in positive examples relative to negative examples could incur negative selection bias. For example, if a specific ligand is observed forming exclusively positive interactions in the dataset during training, the model may subsequently predict that the ligand will interact with any protein during future uses. This is particularly a risk when using real negative DTI data, but may also be the case with random negative selection. One clever solution to this bias trains a model exclusively on randomized negative data that has been shuffled in a manner to ensure equal representation of each ligand or protein among positive and negative labels (Hie et al., 2018).
What is particularly concerning is that very few of the DTI models listed in Table 1 control for such selection bias, which can lead to exaggerated performance measurements. If only the classification of positive versus negative DTIs is tested, a recognition of general “negativeness” of molecules will be measured as a success, even if it is not based on any properties of the target at all (Chen et al., 2019). One way to control for this is to test for the ability of the model to enrich actual targets for a molecule near the top of a ranked list of proteins. This approach is described by Zhou and colleagues in the description of a similarity-based DTI prediction platform, Dr. Prodis (Zhou, Gao, & Skolnick, 2015).
While regression models do not have explicit positive or negative binding pairs, a similar bias may arise if the distribution of affinity measurements for any given ligand or protein in the training data is not representative of systems where the model is subsequently applied. For example, if a protein only appears with a strong, nano-molar binder, it may predict all subsequent ligands within the same affinity range. When the KIBA dataset was first used for affinity predictions in SimBoostQuant, the authors explicitly removed all drugs and targets with fewer than 10 observations from the initial dataset (He et al., 2017). While classification tasks can rely on shuffled negatives to counter intrinsic ligand- or protein-specific biases, there are no obvious similar solutions for regression tasks. This suggests that near 100% density data sets are required for affinity prediction.
MODEL TESTING
The process of testing a machine learning model is itself a scientific experiment. A machine learning model provides a mathematical approximation to a complex function based on a finite number of observations. Any expectation concerning a model's behavior to explain a new phenomenon (make predictions) is inherently a scientific hypothesis that can be accepted or rejected based on suitable experiments. The standard approach to test machine learning models involves cross-validation, i.e., removing some observations from the source dataset prior to training, and later using the removed observations to evaluate how the model does predicting data points not seen before. If removal is random, such tests inform on the model's expected predictive performance when inputs are representative of the distribution of training data. In the case of DTI predictions, representative indicates proteins and ligands observed in the training data. Postulating that a DTI model can generalize beyond the scope of its training data is itself a separate hypothesis in need of a dedicated experiment. In this section, we examine and discuss how adaptations to testing protocols inform on the ligand or protein generalizability of DTI models.
Ligand Exclusion
Generalizing predictive performance to unseen ligands determines a model's suitability for drug design applications involving novel molecules. The simplest form of evaluating ligand-based generalizability involves excluding test set ligands from the associated model training data. Implementations of ligand exclusion experiments vary. For example, Secure-DTI performs a Divided Chemicals test by first performing a 70-30 split of all compounds present in the DTI dataset set, then building their test and training sets upon the corresponding DTIs associated with testing ligands and training ligands (Hie et al., 2018). In performing their ligand exclusion experiment, the authors documented a drop in ROC-AUC values from 0.98 to 0.95 relative to the otherwise randomly sampled DTI splits.
The ligand exclusion approach should also be subjected to canonicalization to avoid the unintended distribution of synonymous SMILES between testing and training datasets (see Ligand Representations). Moreover, ligand exclusions may require more stringent definitions of molecular identity. Some elements of molecular identity, such as isomers or the use of implicit versus explicit hydrogen atoms, may also escape canonicalization. Defining compound identity itself can be a challenging task. For example, the PubChem web service lists five separate types of molecular identity, including same CID , same stereochemistry , same isotopes , same connectivity , and same parent (see Internet Resources: PubChem Identifier Exchange Service). Instead, ligand identity exclusions are best handled with molecular fingerprinting approaches and by applying a similarity metric threshold.
Protein Exclusion
Generalizing predictive performance to unseen proteins determines a model's suitability for performing virtual screens on novel drug targets or characterizing a compound's off-target interactions via proteome screening. In a similar manner to ligand exclusion experiments, a DTI model's protein-dependent generalizability can be estimated via protein-exclusion experiments. However, most DTI models reviewed in Table 1 do not perform protein exclusions experiments. Moreover, in developing the kinase-specific benchmark used to validate SimBoost and subsequently used by deep learning counterpart DeepDTA, He and colleagues explicitly designed cross-validation splits to ensure that all interactions in the test set have corresponding protein representation in the training splits (He et al., 2017).
More recent models estimate protein target generalization by explicitly excluding targets used for validation from the training set. For example DeepConv-DTI separately reports model performance for all testing data, testing data consisting of unseen compounds, testing data consisting of unseen proteins, and testing data where both compounds and proteins are not seen in the training dataset (Lee et al., 2019). In their model, unseen proteins have the largest impact on predictive performance. AutoDTI++ also reports performance of ligand- and protein- exclusion studies (Sajadi, Chahooki, Gharaghani, & Abbasi, 2021), but notice a more significant loss in predictive power in the ligand exclusion test. AutoDTI++, however, differs from most other DTI prediction tools reviewed in this study in its lack of protein features and by framing DTI predictions as an unsupervised imputation task. In this specific case, the impact of ligand exclusion correlates with the sparsity of the four datasets evaluated in the study, indicating that inference is largely governed exclusively by ligand similarity in their model. Overall, the impact of ligand-exclusion versus protein-exclusion experiments may be dataset- or methodology-specific.
In one additional example, Gao and colleagues perform a similar set of ligand- and protein-exclusion experiments for their deep learning model trained on protein GO terms, amino acid sequence embeddings, and graph representations of compounds (Gao et al., 2018). Gao et al. (2018) also provide these benchmark tests for three alternate approaches, including a matrix factorization algorithm (Koren, Bell, & Volinsky, 2009), a similarity-based method (Fokoue, Sadoghi, Hassanzadeh, & Zhang, 2016), and an alternative deep learning method (Wen et al., 2017). For the most part, the ligand exclusion experiment caused a minor loss in predictive performance, while the protein exclusion experiment led to a more noticeable drop. Unexpectedly, the models proposed by Gao et al. appear to increase in performance when testing unseen proteins and unseen ligands, which is fundamentally the most difficult of the four tasks. Higher predictive power in the most difficult tasks could be a sign of data leakage, mapping errors, datasets that are too small for statistical inference, or a systemic bias in test-set DTIs belonging to each of the four exclusion tests. Such biases could be addressed by holding a fixed test set and applying the four exclusion criteria to the training data, yielding four separate models tested on the same benchmark. A fixed test set approach could also provide the basis for broader generalizability benchmarking, whereby increasingly stringent thresholds of ligand or protein similarity exclude more training data, making the training task increasingly difficult.
Measuring Extended Generalizability
Ligand and protein exclusions provide performance metrics to evaluate a model's performance on unseen proteins and unseen ligands. However, these naive leave-out tests, based entirely on ligand or protein identity matching, may not adequately represent real-world applications. In pharmaceutical drug design programs, it is not uncommon to screen a series of related compounds (analogs) of active leads in search of variations or derivatives with the optimal binding activity. This can lead to many near-identical bioactivities, bridging the test and the training data and resulting in apparent “generalization” that is really just memorization with a tolerance for very small changes. This is called compound series bias , and it renders ligand exclusion experiments non-representative of novel drug scaffolds. Most machine learning studies in drug discovery fail to account for compound series bias (Sheridan, 2013), including most DTI prediction models cited in this review.
DTI models may retain some predictive capacity outside of training target classes and scaffolds (generalizability), but such applications require separate validation benchmarks. Cluster cross-validation addresses compound series bias by grouping related molecules during cross-validation (Mayr, Klambauer, Unterthiner, & Hochreiter, 2016, 2018). A similar approach was applied to benchmark matchmaker, whereby following the selection of a fixed test set, all ligands with 0.4 tanimoto similarity to a test-set ligand were excluded from model training (morgan fingerprints with a 3 bond radius) (Cyclica Inc. 2021).
DISCUSSION
Pharmaceutical applications of DTI predictions largely require predictive models capable of addressing entirely new ligands or data-less protein targets. Early graph-based machine learning approaches to DTI prediction were inherently governed by pairwise compound-compound and protein-protein similarity metrics, and were bound to smaller training datasets due to constraints on algorithmic complexity (Gönen, 2012; Yamanishi et al., 2008). Feature-based machine learning approaches can extend beyond similarity metrics for predictive applications. Cao et al. (2012) reformulated the DTI prediction problem as an extension of conventional, ligand-based structure-activity classification models trained on ligand features. Rather than developing separate ligand-based models for each target bioactivity, a support vector machine was trained to model general bioactivity as a function of combined ligand and protein features. Recognizing that feature-based approaches suffer from predictive issues associated with high dimensionality, Hu et al. (2016) first introduced deep learning as a means to reduce the size of the combined ligand and protein feature vector through a stacked auto-encoder architecture, followed by a support vector machine for bioactivity predictions. This approach outperformed prior graph-based machine learning models as well as the Cao et al. approach on the standard Yamanishi benchmark.
Subsequent DTI models combine the ligand and protein vector embedding process with the discriminative task in a single neural network architecture, as depicted in Figure 1, each providing additional contributions in network architectures or feature representations. Notably, DeepDTIs implemented a Deep Belief Network (DBN) for DTI prediction, which combined the role of autoencoder and classifiers from Hu et al. mentioned above into a single deep learning network with a pre-training step and a supervised learning step (Wen et al., 2017). Gao et al. (2018) introduce more complex ligand and protein embedding solutions all into one end-to-end deep learning network. SecureDTI addresses compute scale in a DTI framework, training on datasets approaching 1 million interactions with the potential to securely combine private training data from multiple parties (Hie et al., 2018). DeepConv-DTI improves protein representations in DTI models by introducing CNN models that operate directly on protein sequences (Lee et al., 2019). In addition to standard ligand and protein feature vectors, DL-DTI encodes additional features to represent local drug-target network topology, including ligand and protein similarity matrices (Zhao et al., 2020). Lastly, MatchMaker introduces structural representations of protein pockets based on assumed binding sites (Redka et al., 2020; Sugiyama et al., 2021).
As new scientific advancements continue to progress beyond problem framing, deep learning model architecture, and feature representations, DTI models will inevitably transition from conceptual methodology studies to practical tools applied to pharmaceutical applications. We anticipate that this shift will be coupled with advances in training and testing methods that accurately gauge applicability to specific tasks and generalizability to novel compound drug discovery. In developing or evaluating DTI prediction models that are intended for real-world pharmaceutical applications, we propose asking the following questions to best evaluate a model's domain of applicability:
- Are validation experiments performed with ligand and protein exclusions?
- Are ligand and protein exclusions naive or based on a distance measure?
- Do validation experiments control for compound series bias?
- Do validation experiments control for negative selection bias?
- Are test sets comparable across different proposed models or exclusion tests?
- Are the training and testing datasets representative of proposed use?
- How do variations in input data, such as SMILES strings, impact model behavior?
While the absence of answers to these questions does not necessarily invalidate a given usage, they can help understand the risk profile associated with a proposed application.
AUTHOR CONTRIBUTIONS
Stephen Scott MacKinnon : investigation, methodology, writing original draft, writing review and editing; Seyed Ali Madani Tonekaboni : formal analysis, writing review and editing; Andreas Windemuth : investigation, methodology, writing original draft, writing review and editing.
CONFLICT OF INTEREST
S.S.M.K., S.A.M.T., and A.W. are employees of Cyclica, and may own stock in Cyclica Inc. Cyclica develops MatchMaker, a commercial DTI prediction model.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
LITERATURE CITED
- Abdel-Basset, M., Hawash, H., Elhoseny, M., Chakrabortty, R. K., & Ryan, M. (2020). DeepH-DTA: Deep learning for predicting drug-target interactions: A case study of COVID-19 drug repurposing. IEEE Access , 8, 170433–51. doi: 10.1109/ACCESS.2020.3024238.
- Altae-Tran, H., Ramsundar, B., Pappu, A. S., & Pande, V. (2017). Low data drug discovery with one-shot learning. ACS Central Science , 3(4), 283–293. doi: 10.1021/acscentsci.6b00367.
- Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Michael Cherry, J., … Sherlock, G. (2000). Gene ontology: Tool for the unification of biology. Nature Genetics , 25, 25–29. doi: 10.1038/75556.
- Bagherian, M., Sabeti, E., Wang, K., Sartor, M. A., Nikolovska-Coleska, Z., & Najarian, K. (2021). Machine learning approaches and databases for prediction of drug-target interaction: A survey paper. Briefings in Bioinformatics , 22(1), 247–269. doi: 10.1093/bib/bbz157.
- Banegas-Luna, A.-J., Cerón-Carrasco, J. P., & Pérez-Sánchez, H. (2018). A review of ligand-based virtual screening web tools and screening algorithms in large molecular databases in the age of big data. Future Medicinal Chemistry , 10(22), 2641–2658. doi: 10.4155/fmc-2018-0076.
- Baskin, I. I. (2019). Is one-shot learning a viable option in drug discovery? Expert Opinion on Drug Discovery , 14(7), 601–603. doi: 10.1080/17460441.2019.1593368.
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., … Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research , 28(1), 235–242. doi: 10.1093/nar/28.1.235.
- Brereton, A. E., MacKinnon, S., Safikhani, Z., Reeves, S., Alwash, S., Shahani, V., & Windemuth, A. (2020). Predicting drug properties with parameter-free machine learning: Pareto-optimal embedded modeling (POEM). Machine Learning: Science and Technology , 1(2), 025008.
- Cao, D.-S., Liu, S., Xu, Q.-S., Lu, H.-M., Huang, J.-H., Hu, Q.-N., & Liang, Y.-Z. (2012). Large-scale prediction of drug-target interactions using protein sequences and drug topological structures. Analytica Chimica Acta , 752(November), 1–10. doi: 10.1016/j.aca.2012.09.021.
- Cheng, F., Liu, C., Jiang, J., Lu, W., Li, W., Liu, G., … Tang, Y. (2012). Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Computational Biology , 8(5), e1002503. doi: 10.1371/journal.pcbi.1002503.
- Cheng, F., Zhou, Y., Li, W., Liu, G., & Tang, Y. (2012). Prediction of chemical-protein interactions network with weighted network-based inference method. PloS One , 7(7), e41064. doi: 10.1371/journal.pone.0041064.
- Chen, L., Cruz, A., Ramsey, S., Dickson, C. J., Duca, J. S., Hornak, V., … Kurtzman, T. (2019). Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PloS One , 14(8), e0220113. doi: 10.1371/journal.pone.0220113.
- Chen, T., Li, M., Li, Y., Lin, M., Wang, N., Wang, M., … Zhang, Z. (2015). MXNet: A flexible and efficient machine learning library for heterogeneous distributed systems. arXiv. Available at http://arxiv.org/abs/1512.01274.
- Chen, X., Yan, C. C., Zhang, X., Zhang, X., Dai, F., Yin, J., & Zhang, Y. (2016). Drug-target interaction prediction: Databases, web servers and computational models. Briefings in Bioinformatics , 17(4), 696–712. doi: 10.1093/bib/bbv066.
- Chen, Y. Z., & Zhi, D. G. (2001). Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins , 43(2), 217–226. doi: 10.1002/1097-0134(20010501)43:2%3c217::AID-PROT1032%3e3.0.CO;2-G.
- Cobanoglu, M. C., Liu, C., Hu, F., Oltvai, Z. N., & Bahar, I. (2013). Predicting drug-target interactions using probabilistic matrix factorization. Journal of Chemical Information and Modeling , 53(12), 3399–3409. doi: 10.1021/ci400219z.
- Craig, P. N. (1984). QSAR—origins and present status: A historical perspective. Drug Information Journal , 18(2), 123–130. doi: 10.1177/009286158401800203.
- Cyclica Inc. (2021). Comparison of MatchMaker to DeepConv-DTI reveals superior performance and compute efficiency for predicting drug-target interactions. Cyclica. June 29, 2021. Available at: https://www.cyclicarx.com/case-studies/comparison-of-matchmaker-to.
- Davis, M. I., Hunt, J. P., Herrgard, S., Ciceri, P., Wodicka, L. M., Pallares, G., … Zarrinkar, P. P. (2011). Comprehensive analysis of kinase inhibitor selectivity. Nature Biotechnology , 29(11), 1046–1051. doi: 10.1038/nbt.1990.
- Ding, H., Takigawa, I., Mamitsuka, H., & Zhu, S. (2014). Similarity-based machine learning methods for predicting drug-target interactions: A brief review. Briefings in Bioinformatics , 15(5), 734–747. doi: 10.1093/bib/bbt056.
- Drwal, M. N., & Griffith, R. (2013). Combination of ligand- and structure-based methods in virtual screening. Drug Discovery Today. Technologies , 10(3), e395–401. doi: 10.1016/j.ddtec.2013.02.002.
- Eldridge, M. D., Murray, C. W., Auton, T. R., Paolini, G. V., & Mee, R. P. (1997). Empirical scoring functions: I. the development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. Journal of Computer-Aided Molecular Design , 11(5), 425–445. doi: 10.1023/A:1007996124545.
- El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., … Finn, R. D. (2019). The pfam protein families database in 2019. Nucleic Acids Research , 47(D1), D427–32. doi: 10.1093/nar/gky995.
- Fokoue, A., Sadoghi, M., Hassanzadeh, O., & Zhang, P. (2016). Predicting drug-drug interactions through large-scale similarity-based link prediction. In The semantic web. Latest advances and new domains (pp. 774–789). Lecture Notes in Computer Science. Cham, Germany: Springer International Publishing.
- Gao, K. Y., Fokoue, A., Luo, H., Iyengar, A., Dey, S., & Zhang, P. (2018). Interpretable Drug Target Prediction Using Deep Neural Representation. In Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence (pp. 3371–3377). California: International Joint Conferences on Artificial Intelligence Organization.
- Gilson, M. K., Liu, T., Baitaluk, M., Nicola, G., Hwang, L., & Chong, J. (2016). BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Research , 44(D1), D1045–53. doi: 10.1093/nar/gkv1072.
- Gohlke, H., Hendlich, M., & Klebe, G. (2000). Knowledge-based scoring function to predict protein-ligand interactions. Journal of Molecular Biology , 295(2), 337–356. doi: 10.1006/jmbi.1999.3371.
- Gönen, M. (2012). Predicting drug-target interactions from chemical and genomic kernels using bayesian matrix factorization. Bioinformatics , 28(18), 2304–2310. doi: 10.1093/bioinformatics/bts360.
- Grygorenko, O. O., Radchenko, D. S., Dziuba, I., Chuprina, A., Gubina, K. E., & Moroz, Y. S. (2020). Generating multibillion chemical space of readily accessible screening compounds. iScience , 23(11), 101681. doi: 10.1016/j.isci.2020.101681.
- Günther, S., Kuhn, M., Dunkel, M., Campillos, M., Senger, C., Petsalaki, E., … Preissner, R. (2008). Supertarget and matador: Resources for exploring drug-target relationships. Nucleic Acids Research , 36(Database issue), D919–22.
- He, T., Heidemeyer, M., Ban, F., Cherkasov, A., & Ester, M. (2017). SimBoost: A read-across approach for predicting drug-target binding affinities using gradient boosting machines. Journal of Cheminformatics , 9(1), 24. doi: 10.1186/s13321-017-0209-z.
- Hie, B., Cho, H., & Berger, B. (2018). Realizing private and practical pharmacological collaboration. Science , 362(6412), 347–350. doi: 10.1126/science.aat4807.
- Hirohara, M., Saito, Y., Koda, Y., Sato, K., & Sakakibara, Y. (2018). Convolutional neural network based on SMILES representation of compounds for detecting chemical motif. BMC Bioinformatics , 19(Suppl 19), 526. doi: 10.1186/s12859-018-2523-5.
- Huang, K., Fu, T., Glass, L. M., Zitnik, M., Xiao, C., & Sun, J. (2021). DeepPurpose: A deep learning library for drug-target interaction prediction. Bioinformatics , 36(22-23), 5545–5547. doi: 10.1093/bioinformatics/btaa1005.
- Hui-fang, L., Qing, S., Jian, Z., & Wei, F. (2010). Evaluation of various inverse docking schemes in multiple targets identification. Journal of Molecular Graphics & Modelling, 29(3), 326–330.
- Hu, P.-W., Chan, K. C. C., & You, Z.-H. (2016). Large-scale prediction of drug-target interactions from deep representations. In 2016 International Joint Conference on Neural Networks (IJCNN) , 1236–1243.
- Jiang, D., Wu, Z., Hsieh, C.-Y., Chen, G., Liao, B., Wang, Z., … Hou, T. (2021). Could graph neural networks learn better molecular representation for drug discovery? A comparison study of descriptor-based and graph-based models. Journal of Cheminformatics , 13(1), 12. doi: 10.1186/s13321-020-00479-8.
- Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S., … Hirakawa, M. (2006). From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Research , 34(Database issue), D354–57. doi: 10.1093/nar/gkj102.
- Kearnes, S., McCloskey, K., Berndl, M., Pande, V., & Riley, P. (2016). Molecular graph convolutions: Moving beyond fingerprints. Journal of Computer-Aided Molecular Design , 30(8), 595–608. doi: 10.1007/s10822-016-9938-8.
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., … Bolton, E. E. (2021). PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Research , 49(D1), D1388–95. doi: 10.1093/nar/gkaa971.
- Koren, Y., Bell, R., & Volinsky, C. (2009). Matrix factorization techniques for recommender systems. Computer , 42(8), 30–37. doi: 10.1109/MC.2009.263.
- Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2017). ImageNet classification with deep convolutional neural networks. Communications of the ACM , 60(6), 84–90. doi: 10.1145/3065386.
- Kuhn, M., Szklarczyk, D., Franceschini, A., von Mering, C., Jensen, L. J., & Bork, P. (2012). STITCH 3: Zooming in on protein-chemical interactions. Nucleic Acids Research , 40(Database issue), D876–80. doi: 10.1093/nar/gkr1011.
- Kuntz, I. D., Blaney, J. M., Oatley, S. J., Langridge, R., & Ferrin, T. E. (1982). A geometric approach to macromolecule-ligand interactions. Journal of Molecular Biology , 161(2), 269–288. doi: 10.1016/0022-2836(82)90153-X.
- Landrum, G. (2014). RDKit: Open-Source Cheminformatics. Release 2014.03.1. doi: 10.5281/zenodo.10398.
- Lee, I., Keum, J., & Nam, H. (2019). DeepConv-DTI: Prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS Computational Biology , 15(6), e1007129. doi: 10.1371/journal.pcbi.1007129.
- Liu, H., Sun, J., Guan, J., Zheng, J., & Zhou, S. (2015). Improving compound-protein interaction prediction by building up highly credible negative samples. Bioinformatics , 31(12), i221–29. doi: 10.1093/bioinformatics/btv256.
- Liu, K., Sun, X., Jia, L., Ma, J., Xing, H., Wu, J., … Fan, J. (2019). Chemi-Net: A molecular graph convolutional network for accurate drug property prediction. International Journal of Molecular Sciences , 20(14), 3389. doi: 10.3390/ijms20143389.
- TensorFlow Developers. (2021). TensorFlow. Zenodo. doi: 10.5281/ZENODO.4724125.
- Tonekaboni, M., Ali, S., Brereton, A. E., Safikhani, Z., Windemuth, A., Haibe-Kains, B., & MacKinnon, S. (2020). Learning across label confidence distributions using filtered transfer learning. In 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA) , 1117–1123.
- Mayr, A., Klambauer, G., Unterthiner, T., & Hochreiter, S. (2016). DeepTox: Toxicity prediction using deep learning. Frontiers of Environmental Science & Engineering in China, 3(February). doi: 10.3389/fenvs.2015.00080.
- Mayr, A., Klambauer, G., Unterthiner, T., Steijaert, M., Wegner, J. K., Ceulemans, H., … Hochreiter, S. (2018). Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chemical Science , 9(24), 5441–5451. doi: 10.1039/C8SC00148K.
- Mendez, D., Gaulton, A., Bento, A. P., Chambers, J., De Veij, M., Félix, E., … Leach, A. R. (2019). ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Research , 47(D1), D930–40. doi: 10.1093/nar/gky1075.
- Meslamani, J., Li, J., Sutter, J., Stevens, A., Bertrand, H.-O., & Rognan, D. (2012). Protein-ligand-based pharmacophores: Generation and utility assessment in computational ligand profiling. Journal of Chemical Information and Modeling , 52(4), 943–955. doi: 10.1021/ci300083r.
- Mitchell, J. B. O., Laskowski, R. A., Alex, A., & Thornton, J. M. (1999). An improved method of potential of mean force for protein-protein interactions: I. Generating potential. Journal of Computational Chemistry , 20(11), 1165–1176. doi: 10.1002/(SICI)1096-987X(199908)20:11%3c1165::AID-JCC7%3e3.0.CO;2-A.
- Muegge, I., & Martin, Y. C. (1999). A general and fast scoring function for protein-ligand interactions: A simplified potential approach. Journal of Medicinal Chemistry , 42(5), 791–804. doi: 10.1021/jm980536j.
- Mysinger, M. M., Carchia, M., Irwin, J. J., & Shoichet, B. K. (2012). Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. Journal of Medicinal Chemistry , 55(14), 6582–6594. doi: 10.1021/jm300687e.
- Nguyen, T., Le, H., Quinn, T. P., Nguyen, T., Le, T. D., & Venkatesh, S. (2021). GraphDTA: Predicting drug-target binding affinity with graph neural networks. Bioinformatics , 37(8), 1140–1147. doi: 10.1093/bioinformatics/btaa921.
- Öztürk, H., Özgür, A., & Ozkirimli, E. (2018). DeepDTA: Deep drug-target binding affinity prediction. Bioinformatics , 34(17), i821–29. doi: 10.1093/bioinformatics/bty593.
- Park, J. H., Liu, Y., Lemmon, M. A., & Radhakrishnan, R. (2012). Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochemical Journal , 448(3), 417–423. doi: 10.1042/BJ20121513.
- Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., … Chintala, S. (2019). PyTorch: An imperative style, high-performance deep learning library. arXiv. Available at: http://arxiv.org/abs/1912.01703.
- Paul, N., Kellenberger, E., Bret, G., Müller, P., & Rognan, D. (2004). Recovering the true targets of specific ligands by virtual screening of the protein data bank. Proteins , 54(4), 671–680. doi: 10.1002/prot.10625.
- Qing, X., Lee, X. Y., De Raeymaecker, J., Tame, J. R. H., Zhang, K. Y. J., De Maeyer, M., & Voet, A. R. D. (2014). Pharmacophore modeling: Advances, limitations, and current utility in drug discovery. Journal of Receptor, Ligand and Channel Research , 7(November), 81–92.
- Ragoza, M., Hochuli, J., Idrobo, E., Sunseri, J., & Koes, D. R. (2017). Protein-ligand scoring with convolutional neural networks. Journal of Chemical Information and Modeling , 57(4), 942–957. doi: 10.1021/acs.jcim.6b00740.
- Ramsundar, B., Eastman, P., Walters, P., Pande, V., Leswing, K., & Wu, Z. (2019). Deep learning for the life sciences. Sebastopol, CA: O'Reilly Media.
- Reczko, M., & Bohr, H. (1994). The DEF data base of sequence based protein fold class predictions. Nucleic Acids Research , 22(17), 3616–3619.
- Redka, D. S., MacKinnon, S. S., Landon, M., Windemuth, A., Kurji, N., & Shahani, V. (2020). PolypharmDB, a deep learning-based resource, quickly identifies repurposed drug candidates for COVID-19. ChemRxiv. doi: 10.26434/chemrxiv.12071271.v1.
- Riniker, S., & Landrum, G. A. (2013). Open-source platform to benchmark fingerprints for ligand-based virtual screening. Journal of Cheminformatics , 5(1), 26. doi: 10.1186/1758-2946-5-26.
- Rognan, D. (2010). Structure-based approaches to target fishing and ligand profiling. Molecular Informatics , 29(3), 176–187. doi: 10.1002/minf.200900081.
- Rognan, D. (2013). Proteome-scale docking: Myth and reality. Drug Discovery Today. Technologies , 10(3), e403–9. doi: 10.1016/j.ddtec.2013.01.003.
- Sajadi, S. Z., Chahooki, M. A. Z., Gharaghani, S., & Abbasi, K. (2021). AutoDTI++: Deep unsupervised learning for DTI prediction by autoencoders. BMC Bioinformatics , 22(1), 204. doi: 10.1186/s12859-021-04127-2.
- Schomburg, I., Chang, A., Ebeling, C., Gremse, M., Heldt, C., Huhn, G., & Schomburg, D. (2004). BRENDA, the enzyme database: Updates and major new developments. Nucleic Acids Research , 32(Database issue), D431–33. doi: 10.1093/nar/gkh081.
- Shen, J., Zhang, J., Luo, X., Zhu, W., Yu, K., Chen, K., … Jiang, H. (2007). Predicting protein-protein interactions based only on sequences information. Proceedings of the National Academy of Sciences of the United States of America , 104(11), 4337–4341. doi: 10.1073/pnas.0607879104.
- Sheridan, R. P. (2013). Time-split cross-validation as a method for estimating the goodness of prospective prediction. Journal of Chemical Information and Modeling , 53(4), 783–790. doi: 10.1021/ci400084k.
- Somody, J. C., MacKinnon, S. S., & Windemuth, A. (2017). Structural coverage of the proteome for pharmaceutical applications. Drug Discovery Today , 22(12), 1792–1799. doi: 10.1016/j.drudis.2017.08.004.
- Southan, C., Sharman, J. L., Benson, H. E., Faccenda, E., Pawson, A. J., Alexander, S. P. H., … NC-IUPHAR (2016). The IUPHAR/BPS guide to PHARMACOLOGY in 2016: Towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Research , 44(D1), D1054–68. doi: 10.1093/nar/gkv1037.
- Stepišnik, T., Škrlj, B., Wicker, J., & Kocev, D. (2021). A comprehensive comparison of molecular feature representations for use in predictive modeling. Computers in Biology and Medicine , 130(March), 104197. doi: 10.1016/j.compbiomed.2020.104197.
- Sugiyama, M. G., Cui, H., Redka, D. S., Karimzadeh, M., Rujas, E., Maan, H., … Antonescu, C. N. (2021). Multiscale interactome analysis coupled with off-target drug predictions reveals drug repurposing candidates for human coronavirus disease. bioRxiv. doi: 10.1101/2021.04.13.439274 bioRxiv.
- Szklarczyk, D., Santos, A., von Mering, C., Jensen, L. J., Bork, P., & Kuhn, M. (2016). STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research , 44(D1), D380–84. doi: 10.1093/nar/gkv1277.
- Tang, J., Szwajda, A., Shakyawar, S., Xu, T., Hintsanen, P., Wennerberg, K., & Aittokallio, T. (2014). Making sense of large-scale kinase inhibitor bioactivity data sets: A comparative and integrative analysis. Journal of Chemical Information and Modeling , 54(3), 735–743. doi: 10.1021/ci400709d.
- Tiessen, A., Pérez-Rodríguez, P., & Delaye-Arredondo, L. J. (2012). Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Research Notes , 5(February), 85. doi: 10.1186/1756-0500-5-85.
- Torng, W., & Altman, R. B. (2019). Graph convolutional neural networks for predicting drug-target interactions. Journal of Chemical Information and Modeling , 59(10), 4131–4149. doi: 10.1021/acs.jcim.9b00628.
- UniProt Consortium. (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research , 49(D1), D480–89. doi: 10.1093/nar/gkaa1100.
- Weininger, D. (1988). SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. Journal of Chemical Information and Computer Sciences , 28(1), 31–36.
- Wen, M., Zhang, Z., Niu, S., Sha, H., Yang, R., Yun, Y., & Lu, H. (2017). Deep-learning-based drug-target interaction prediction. Journal of Proteome Research , 16(4), 1401–1409. doi: 10.1021/acs.jproteome.6b00618.
- Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., … Woolsey, J. (2006). DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Research , 34(Database issue), D668–72. doi: 10.1093/nar/gkj067.
- Yamanishi, Y., Araki, M., Gutteridge, A., Honda, W., & Kanehisa, M. (2008). Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics , 24(13), i232–40. doi: 10.1093/bioinformatics/btn162.
- Zhao, Q., Xiao, F., Yang, M., Li, Y., & Wang, J. (2019). AttentionDTA: Prediction of drug–target binding affinity using attention model. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) , 64–69.
- Zhao, Y., Zheng, K., Guan, B., Guo, M., Song, L., Gao, J., … Zhang, Y. (2020). DLDTI: A learning-based framework for drug-target interaction identification using neural networks and network representation. Journal of Translational Medicine , 18(1), 434. doi: 10.1186/s12967-020-02602-7.
- Zhou, H., Gao, M., & Skolnick, J. (2015). Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Scientific Reports , 5(June), 11090. doi: 10.1038/srep11090.
INTERNET RESOURCES
PubChem Identifier Exchange Service. n.d. Accessed June 15, 2021.
Citing Literature
Number of times cited according to CrossRef: 4
- Shiqiang Zhu, Ting Yu, Tao Xu, Hongyang Chen, Schahram Dustdar, Sylvain Gigan, Deniz Gunduz, Ekram Hossain, Yaochu Jin, Feng Lin, Bo Liu, Zhiguo Wan, Ji Zhang, Zhifeng Zhao, Wentao Zhu, Zuoning Chen, Tariq S. Durrani, Huaimin Wang, Jiangxing Wu, Tongyi Zhang, Yunhe Pan, Intelligent Computing: The Latest Advances, Challenges, and Future, Intelligent Computing, 10.34133/icomputing.0006, 2 , (2023).
- S. Morteza Hashemi, Arash Zabihian, Mohsen Hooshmand, Sajjad Gharaghani, DRaW: prediction of COVID-19 antivirals by deep learning—an objection on using matrix factorization, BMC Bioinformatics, 10.1186/s12859-023-05181-8, 24 , 1, (2023).
- Irini Doytchinova, Drug Design—Past, Present, Future, Molecules, 10.3390/molecules27051496, 27 , 5, (1496), (2022).
- K. Nandhini, P. Mayilvahanan, Molecular Sub-structural Pattern Discovery using Transformer-CNN-OMBO Algorithm for Drug-target interaction (DTI) Prediction, 2022 6th International Conference on Trends in Electronics and Informatics (ICOEI), 10.1109/ICOEI53556.2022.9776666, (241-247), (2022).

