Plant material collection protocol for eastern hemlock (Tsuga canadensis) to assess chemical and expression profiles associated with HWA resistance
Jill Wegrzyn, victoriaburton, Vidya S Vuruputoor, karlfetter
Abstract
This protocol was developed by the PCG team for sampling needle and branch tissue from eastern hemlock ( Tsuga canadensis ) for terpenoid profiling, as well as transcriptomic assessment (RNA-Seq). This study is a time-course assessment comparing known HWA-resistant (lingering) individuals from the New Jersey bullet-proof population (NJ3 and NJ4) to likely HWA-susceptible individuals. The plots assessed include those described in Kinahan et al. 2020 (https://www.mdpi.com/1999-4907/11/3/312). Sampling occurs once per month and includes deeper sampling (more bulletproof population members) at the Arnold Arboretum. Non-targeted LC/MS and GC/MS assessment is underway with Tim Cernak and Roland Kersten at the University of Michigan. RNA-Seq will be conducted at UConn on a subset of the individuals collected. This protocol describes the collection of three tubes worth of needle/stem samples for each individual (two for terpenoids and one for RNA-Seq), to be conducted on a monthly basis across all plots.
Steps
TreeSnap Observation
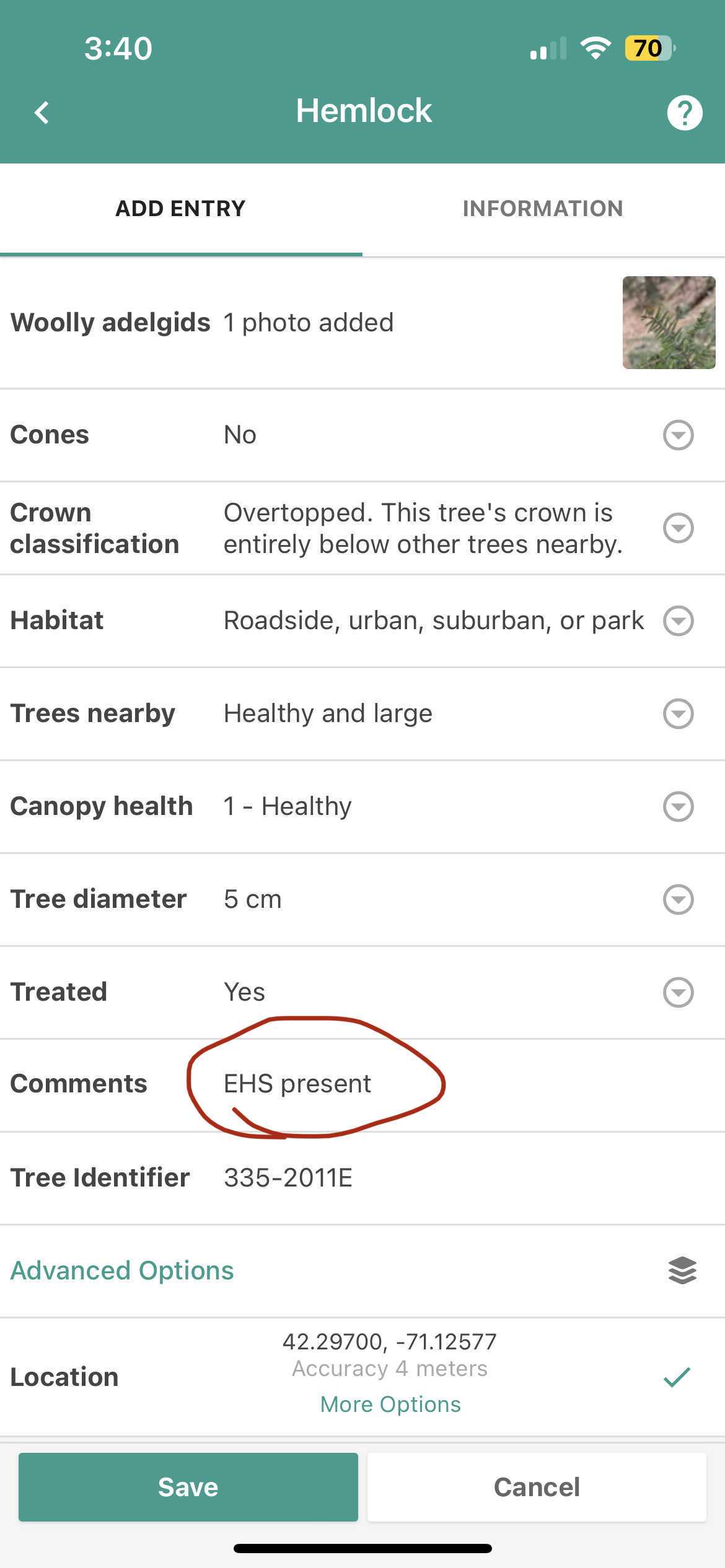
TreeSnap is a mobile application that can run on both Apple OS and Android that utilizes the phone’s GPS to collect location information and present a set of standardized questions to the collector on basic traits of the tree, including image data. This is the application of choice for this project to record this information, including local identifiers associated with the tree. The application will record the coordinates even if cell service is limited or absent in that region.
Take at least 3 photos with TreeSnap. This includes at least one of the trees itself, one photo of the tree tag if that is present, and one photo of the collection vial with the label as well.
Geolocation will be recorded automatically but TreeSnap can also connect with a more precise external GPS device.
Note DBH in appropriate column
Note any other observation you may have- including plant condition (Excellent/Good/Poor), and placement of individual (lower canopy, full sun, etc.)
Prepare sampling sheet
The sample sheet would look like this:- Link to spreadsheet
Collection
You will collect two sets of samples from each tree. One for RNA extraction for transcriptomics and one for metabolomics. The first needs less material so we will use the 5ml tubes. The latter needs more material and will use the 10ml tubes (you'll do this for two tubes). The first should include two branches that are approximately 5 cm in length. The second should include four branches, ideally taken from different locations around the tree.
Metabolomics- You would use 2 10ml vials. You will be able to fit 4 branches (5cm) in each tube- so sample from one direction of the tree, then collect the three other branches from the other sides of the tree (you would ideally try to sample around the individual).
RNA- You would use one 5ml vial. For this, 2 branches would be more than sufficient to fit into the vial. Ideally, you would want to sample from 2 different locations of the individual.
In total- THREE vials per individual. TWO for terpenoids/metabolomics, and ONE for RNA.
Placing in Liquid Nitrogen
Close cryovials, and place the vial slowly into the LN dewar. Be wary of LN splashing out, and wear appropriate gear, and stay at a distance.
Cleaning before sampling from new individual
Clean trimmers between trees with ethanol (spray bottle).
Use a clipboard to manually record notes on a pre-printed spreadsheet with sample names.
After returning from the field, ensure that notes are consistent with TreeSnap entries. These entries can be amended after collection as needed.
Post-sampling and Pre-shipping
Remove all vials from the dewar (cross-check with the sample sheet). Samples must be stored at -80C or should be on dry ice during transport.
Cross-check all samples at this stage with your sample sheet.
Shipping
Shipping instructions will be provided via email. Shipments must be sent on dry ice that is contained inside a styrofoam box insert. You will be shipping separate parcels for the Metabolomics and RNA samples.
Log all data and samples in a Google sheet, or send over sample sheet so that we log your data