Planet Microbe Semantic Web Application
Kai Blumberg, Alise J Ponsero, Bonnie L Hurwitz
Abstract
Tutorial for the use of the Planet Microbe Semantic Web Application, accompanying the PhD dissertation work of Kai Blumberg.
Steps
Home Page
Welcome, This is the protocol to accompany the use of the Planet Microbe Semantic Web API. This protocol was created as part of Kai Blumberg's PhD dissertation work. This work is contained within this github repository: https://github.com/hurwitzlab/planet-microbe-semantic-web-analysis.
This protocol is organized by the following sections:
- Home Page
- This table of contents.
- Introduction and System Overview
- Some basics about the system
- Getting Started
- A quick guide for how to get started using the Planet Microbe RDF web service.
- How to Navigate relevant OBO Ontologies
- A description of how to browse relevant ontologies to find terms of interest for queries.
- Create your own SPARQL Query
- A description of the available command line arguments in the python script that can be used to assemble and submit SPARQL queries to the Planet Microbe RDF database.
- Example SPARQL System Queries
- A "how to" examples guide showing how to use the system to query for all annotations of samples constrained by the three relevant ontologies.
- Tips for Analyzing Discovered Data
- Basic instructions on how to work with the provided example python and R code to process and analyze the query results delivered from the system.
- Appendix
-
- Table of environmental attributes (e.g., water temperature) available for use with the system
-
- Example RDF data structure
Introduction and System Overview
This protocol describers the use of the Planet Microbe RDF web service accessible through an open API. This web service can be used to retrieve data by which to ask and answer novel biological questions from the prokaryotic fraction of the Planet Microbe's metagenomic datasets.
This work created as part of Kai Blumberg's PhD dissertation integrates large-scale marine metagenomic datasets with community-driven life-science ontologies into a novel FAIR web service. This approach enables the retrieval of data discovered by intersecting the knowledge represented within ontologies against the functional genomic potential and taxonomic structure computed from marine sequencing data sourced from the Planet Microbe database.
This web service leverages several open source ontologies from the Open Biomedical and Biological Ontologies (OBO) Foundry and Library. These include Gene Ontology (GO) for representations of the biological processes and molecular functions of genes, the Environment Ontology (ENVO) for representations environment types and environmental parameters, as well as NCBITaxon, the ontology representation of the National Center for Biotechnological Information organismal taxonomy database.
The ontology searchable data products provided by this API are intended to be leveraged by future research efforts. I hope you do so with joy.
Getting Started
A quick guide for how to get started using the Planet Microbe RDF web service.
Requirements:
python3 packages:
- argparse
- sys
- os
- requests
- time
R (and optionally R studio)
Please make sure to install all python requirements prior to doing this tutorial.
The first step is to download the relevant software from this github repository. The code can be downloaded as a ZIP file or be cloned using github. Click on the code button in the top right hand corer of the hyper link. If you choose to download the ZIP file, make sure to unzip downloaded zip file.
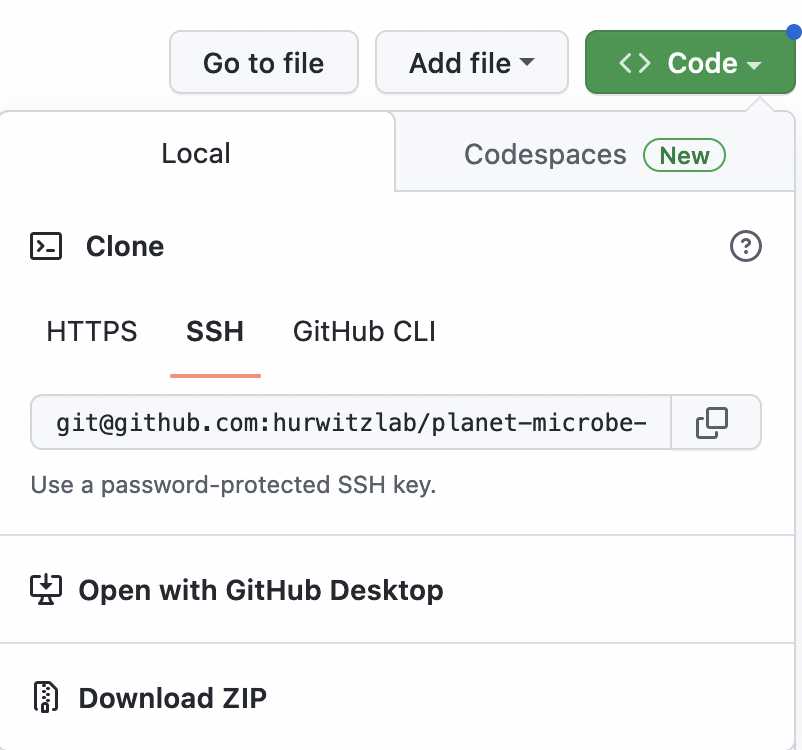
Navigate to the folder
planet-microbe-semantic-web-analysis/analysis/query
Create a directory called 'api_results' or similar for your analyses. If using a name other than 'api_results' make sure to change the name in any subsequent command line instructions.
Test that the query assembly script is working properly by running the following line:
python3 assemble_query.py -u base_metadata.rq -o api_results/base_metadata.csv
If this creates a file at the path "api_results/base_metadata.csv" then this was successful. If not make sure python3 is correctly installed, and you are in the right place within the downloaded code repository. Note the for the example questions used in the paper relative paths to the assemble_query.py script are used in the example commands (see section on Example R Analyses on Discovered Data)
Congratulations you are running the code correctly. To learn how to make your own custom query see the section about Creating your own SPARQL Query. However in order to do that you'll first need to be able to navigate and browse the relevant OBO foundry ontologies.
How to Navigate relevant OBO Ontologies
In order to discover ontology terms that can be used as inputs to queries to help answer natural language questions, we first need to learn to navigate the relevant ontologies. Although there are many ways this can be done, this tutorial recommends the use of the European Molecular Biology Laboratory (EMBL) European Bioinformatics Institute (EBI) Ontology Lookup Service.
Click here for the links to navigate the following ontologies:
Gene Ontology (GO)
Environment Ontology (ENVO)
NCBI taxonomy database ontology build (NCBITaxon)
Finally, extra terminology from the Planet Microbe Application Ontology which can also be used to query the API are list in Appendix I.
The following video shows an example of searching for a GO term and copying the CURIE from the OLS lookup page. OLS_lookup_tutorial.mp4
The important steps in the video are recounted here. The OLS ontology browser page can either be searched by typing a gene name into the text search box, e.g., typing "photosynthesis" will give you the following:
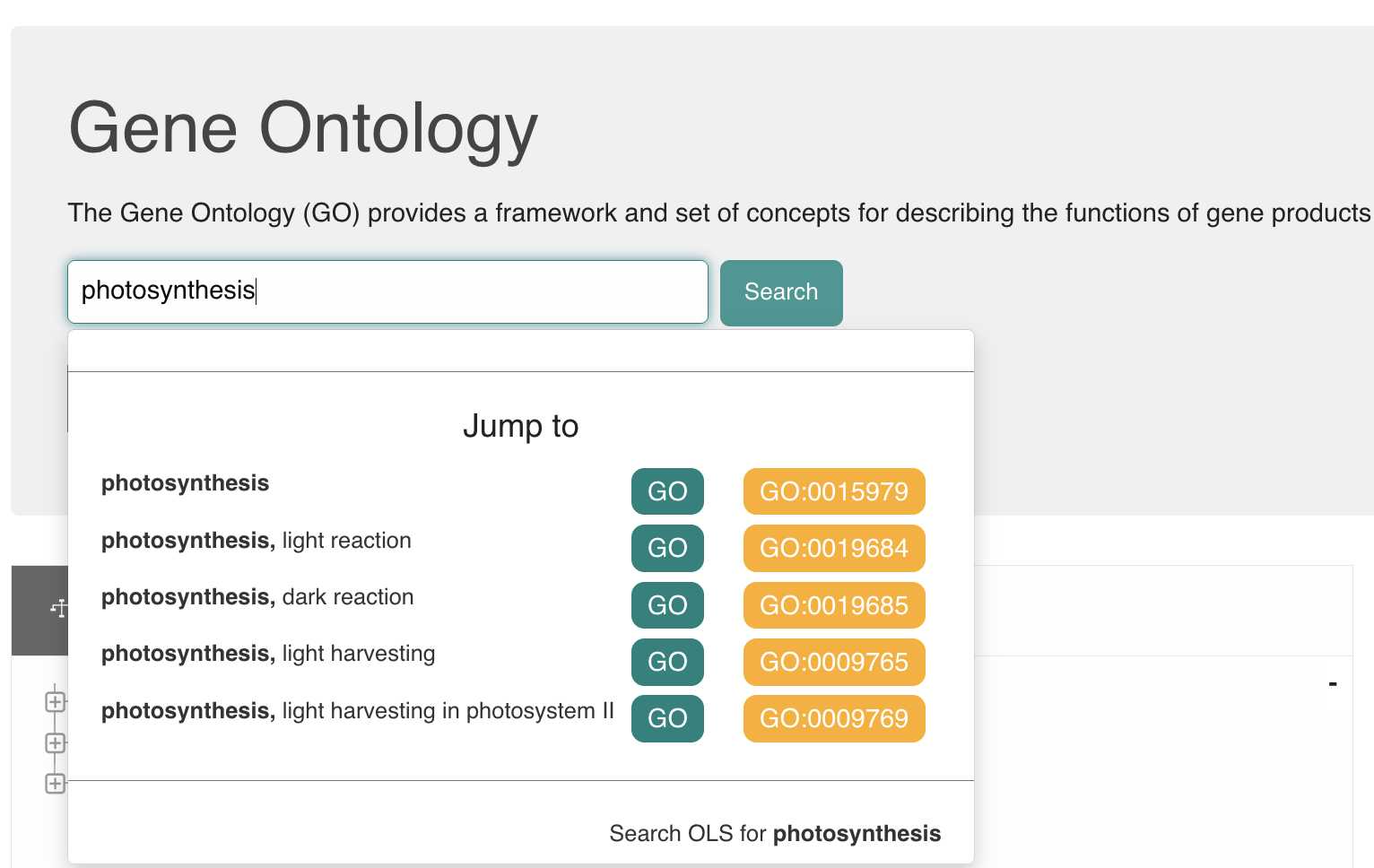
Or manually by clicking and expanding the + sign to expand any given term and view it's subclasses within the ontology.

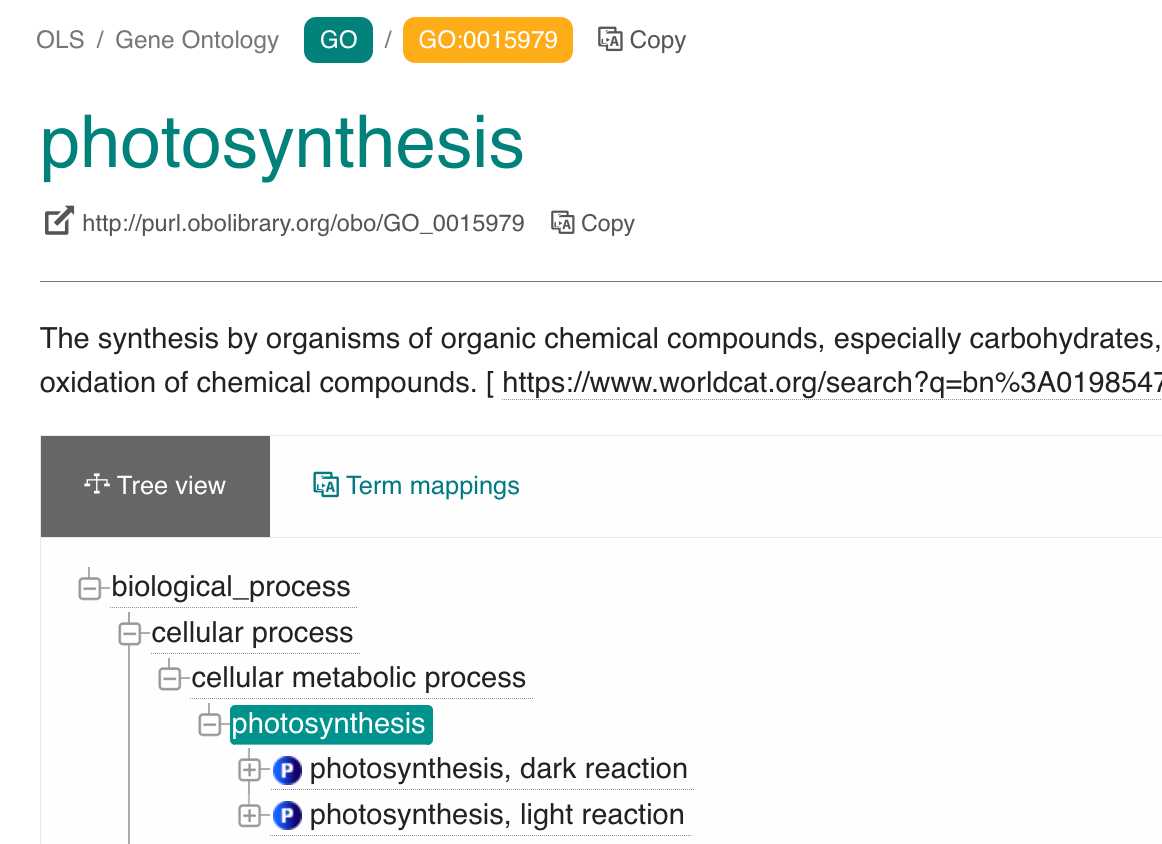
After selecting an ontology term you will be directed to a page like the above. To extract the "Compact URI" also known as "CURIE" version of an ontology term identifier, click on the Copy button which will copy the CURIE to clipboard.

Here for example the CURIE ID for the go "photosynthesis" term is GO:0015979, this will be needed later when creating queries to send against the Planet Microbe RDF web service.
Create your own SPARQL Query
This section describes use of the assemble_query.py python3 script which can be used to assemble and send off SPARQL queries to the Planet Microbe RDF web service. The next section provides examples of using the python script to create a query for various questions of interest.
The scripts usage is summarized as follows:
usage: assemble_query.py [-m str] [-b str] [-l str] [-g str] [-t str]
[-q str] [-ql str [str ...]] [-o str] [-p str]
[-u str] [-dmin int] [-dmax int]
Where the following are the table of flags that can optionally be added to a run command.
-m str, --env_medium
Environmental medium
Expects an ENVO CURIE from the environmental material hierarchy,
E.g., ENVO:00002149
-b str, --env_broad
Environment broad scale context
Expects an ENVO CURIE from the biome hierarchy
E.g., ENVO:00000447
-l str, --env_local
Environment local scale context
Expects an ENVO CURIE from the astronomical body part, or layer, hierarchies
E.g., ENVO:01000061
-g str, --go
Gene Ontology term
Expects a GO CURIE
E.g., GO:0015979*
*Note that the system will only search for terms from one of the three major GO hierarchies (biological process, cellular component, or molecular function ) at a time.
-t str, --taxon
NCBI Taxonomy ontology term
Expects a NCBITaxon CURIE from the Bacteria or Archaea lineages
E.g., NCBITaxon:1117
-q str, --quality
Query for subclasses of input quality argument
Experts a BFO, ENVO or PMO quality CURIE
E.g., BFO:0000019
See Appendix section for list of qualities
-ql str [str ...], --quality_list
Query for a list of input quality arguments
Experts BFO, ENVO or PMO quality CURIE see list in Appendix section for list
E.g., ENVO:09200014 ENVO:3100031
-o str, --output
Output file path to write tsv file of go term counts
Typical use would be `output/custom_file_name`
-p str, --project
Query for project name
E.g., "Amazon Plume Metagenomes"
*The list of available projects are as follows: "Amazon Plume Metagenomes", "Amazon River Metagenomes", "BATS Chisholm", "HOT 224-283", "HOT 144-166", or "Tara Oceans".
-u str, --universal
File path to input sparql query file with query for
basic metadata universal across samples
Default is: base_metadata.rq
Only needs to be run once but should be run at first to get metadata table
-dmin int, --depth_minimum int
Filter samples by depth with minimum value cutoff
Default: 0
E.g., 300
-dmax int, --depth_maximum
Filter samples by depth with maximum value cutoff
E.g., 400
Example SPARQL System Queries
This section provides examples of how to use the python query creation script to create a SPARQL query by which to data to answer a natural language questions.
Here we provide three examples demonstrating how the system can query for data leveraging 1) the Gene Ontology, 2) the NCBITaxonomy database ontology, and 3) the Environment Ontology.
The following examples are setup to be run from the following directory:
planet-microbe-semantic-web-analysis/analysis
Demonstration 1) querying using the Gene Ontology
Here we demonstrate an example usage of the script that assembles a query to search for data that can be used to ask the question:
What data do we have about metagenomes from the 'HOT 224-283' project, where we have observed occurrences of "cellular lipid metabolic process"(es), where there is also a recorded "temperature" value?
python3 query/assemble_query.py -o api_results/GO_0044255.csv -p "HOT 224-283" -ql ENVO:09200014 -g GO:0044255
Breaking this down by the various inputs we have the following:
The -o flag gives us a path where we are writing out our results (as a csv file).
The -p flag is specifying a particular project.
The -ql flag is specifying a list of additional attributes we want to constrain our query by (in this case just one) "temperature of water" expressed by the ontology CURIE "ENVO:09200014".
Finally the -g flag is specifying pre-computed gene ontology occurrence data, specifically to search for any type of "cellular lipid metabolic process" including the term itself as well as all of it's descendent terms within the GO hierarchy using the curie "GO:0044255".
The expected file downloaded form this query should be the following. GO_0044255.csv
Demonstration 2) querying using the NCBITaxonomy Ontology
Here we demonstrate an example usage of the script that assembles a query to search for data that can be used to ask the question:
What data do we have about metagenomes from the Amazon Plume project where we have observed occurrences of Prochlorococcus collected between the surface up to the depth of 300 meters?
python3 query/assemble_query.py -o api_results/NCBITaxon_1218.csv -p "Amazon Plume Metagenomes" -t NCBITaxon:1218 -dmin 0 -dmax 300
Breaking this down by the various inputs we have the following:
The -o flag (again) gives us a path where we are writing out our results (as a csv file).
The -p flag is again specifying a specific project.
The -t flag is specifying that we want to search for pre-computed taxonomic occurence data, specifically to search for any type of " Prochlorococcus" or descendent thereof within the NCBITaxon hierarchy using the curie "NCBITaxon:1218".
Finally, the -dmin 0 and -dmax 300 flags specify that we want to constrain the depth search from 0-300 meters (inclusively).
The expected file downloaded form this query should be the following. NCBITaxon_1218.csv
NOTE queries with some of top level taxonomic ranks e.g., Bacteria, or Proteobacteria are too large and WILL NOT WORK. Archaea has less representatives in the database therefore it will work, so too will some phyla e.g., Aquificae. If a top level taxonomic query fails due to too many representatives being included in the database, try specifying a finer level of taxonomic resolution.
Demonstration 3) querying using the Environment Ontology
Here we demonstrate an example usage of the script that assembles a query to search for data that can be used to ask the question:
What data do we have about metagenomes that were sampled from "sea water" collected from any type of "marine layer" from a "marine biome"?
python3 query/assemble_query.py -o api_results/context_constraint.csv -b ENVO:00000447 -l ENVO:01000295 -m ENVO:00002149
Breaking this down by the various inputs we have the following:
The -o flag (again) gives us a path where we are writing out our results (as a csv file).
The -b flag specifies an ENVO biome term in this case "marine biome" using the curie "ENVO:00000447".
The -l flag specifies an local scale environmental context term from ENVO, in this case "marine layer" using the curie "ENVO:01000295".
Finally, the -m flag specifies an environmental material term from ENVO environmental material hierarchy, in this case "sea water" using the curie "ENVO:00002149".
The expected file downloaded form this query should be the following. context_constraint.csv
Tips for Analyzing Discovered Data
The Planet Microbe Semantic Web API is designed to discover biological results based on user queries to the Planet Microbe RDF database API. As such this system can deliver FAIR data products that are annotated with various ontology terms. These data resulting from queries to the Planet Microbe RDF database API could be analyzed and or post-processed using any number of programs or packages (R, python, etc). Although the analyses conducted for the publication make use of R, this section gives general guidance on working with query scripts and analyzing the data. Please take the presented information into consideration regardless of what tools you choose to use for analysis. Please note that the python query script and RDF web-service is not meant to be run in parallel, thus one should only run one RDF query at a time. The data within the web-service are the summarized results of large-scale parallelized computations made available through an RDF query interface.
The example code and queries used in the paper are available from the planet-microbe-semantic-web-analysis github repository https://github.com/hurwitzlab/planet-microbe-semantic-web-analysis, see the directory:
planet-microbe-semantic-web-analysis/analysis/paper_questions
Within this directory you will find all the example queries and R code used in the manuscript to analyze the data and generate the figures. Note that each of the paper question directories have a api_results directory where the results are downloaded to as specified in the calls to the assemble_query.py python script. The command to create API results directory is included in each R file, along with the the assemble_query.py command relevant to that question. For example in the dissolved_inorganic_carbon_functional question directory the biosynthetic_process_glmnet_CLRT.r script has the following command included as an R comment.
python3 ../../query/assemble_query.py -o api_results/GO_0009058_DIC_30m.csv -dmax 30 -ql PMO:00000142 -g GO:0009058
To reuse these query scripts the user is asked to run such a command (without the pound symbol and trailing space "# ") in the command line within the appropriate directory.
Within the query scripts another query command is also included asking the user to retrieve the base metadata. For the same example above the user is asked to also run the following command in the shell in the appropriate directory:
python3 ../../query/assemble_query.py -u ../../query/base_metadata.rq -o api_results/base_metadata.csv
Note both of these commands have relative directory paths to the query script and base_metadata.rq file in the query directory. You may need to be adjust these depending on how you setup your file structure to do more queries. For example, one could make a new directory in the planet-microbe-semantic-web-analysis/analysis folder next to the paper_questions and query directories.
Another important note is that the analyses conducted in this work made use of Centered Log-Ratio (CLR) transformations on data in order to make comparisons across metagenomic projects. For more information on analyzing metagenomic data using CLR and similar methods, see the paper "Microbiome Datasets Are Compositional: And This Is Not Optional". Like the authors of that paper, we highly recommend the use of CLR transformation prior to analysis of discovered GO and NCBITaxon data using this system.
Finally, it should also be noted that the paper example R scripts make use of various packages including ggplot2, dplyr, tidyverse, glmnet as well as others. Make sure to download any and all appropriate packages and their dependencies to be able to run or re-purpose the existing source code. R studio may display a message asking to download the missing packages.
Appendix
Appendix I) Supplemental table of qualities available for use with the systems -ql and -q flags.
| A | B | C |
|---|---|---|
| 19'-butanoyloxyfucoxanthin concentration | PMO:00000156 | micromolar |
| 19'-hexanoyloxyfucoxanthin concentration | PMO:00000157 | micromolar |
| acidity of water | ENVO:3100030 | pH units |
| alkalinity of water | PMO:00000139 | milliequivalent per liter |
| alloxanthine concentration | ENVO:3100002 | micromolar |
| Adenosine 5-triphosphate concentration | ENVO:3100001 | micromolar |
| bacteriochlorophyll a concentration | ENVO:3100005 | microgram per liter |
| carbon dioxide concentration | PMO:00000174 | micromole per kilogram |
| carbonate concentration | PMO:00000175 | micromole per kilogram |
| carotene concentration | ENVO:3100007 | micromolar |
| chlorophyll a concentration | ENVO:3100008 | microgram per liter |
| chlorophyll b concentration | ENVO:3100009 | microgram per liter |
| chlorophyllide a concentration | ENVO:3100010 | microgram per liter |
| conductivity | ENVO:09200018 | milisiemens per centimeter |
| density of water | PMO:00000191 | kilogram per cubic meter |
| depth of water | ENVO:3100031 | meter |
| dioxygen concentration | ENVO:3100011 | micromole per kilogram |
| dissolved inorganic carbon concentration | PMO:00000142 | micromole per kilogram |
| dissolved organic carbon concentration | PMO:00000102 | microgram per liter |
| divinyl chlorophyll a concentration | ENVO:3100012 | microgram per liter |
| divinyl chlorophyll b concentration | ENVO:3100013 | microgram per liter |
| filter max cutoff | PMO:00000023 | micrometer |
| filter min cutoff | PMO:00000022 | micrometer |
| fucoxanthin concentration | ENVO:3100014 | micromolar |
| heterotrophic prokaryote count | PMO:00000162 | cells per milliliter |
| hydrogencarbonate concentration | PMO:00000176 | micromole per kilogram |
| lutein concentration | ENVO:3100019 | micromolar |
| neoxanthin concentration | ENVO:3100021 | micromolar |
| nitrate concentration | ENVO:3100022 | micromolar |
| nitrite concentration | ENVO:3100023 | micromolar |
| Photosynthetically active electromagnetic radiation of liquid water (PAR) | PMO:00000015 | micromole per square meter per second |
| particulate carbon concentration | PMO:00000150 | micromole per kilogram |
| particulate nitrogen concentration | PMO:00000151 | micromole per kilogram |
| particulate phosphorus concentration | PMO:00000153 | nanomole per kilogram |
| particulate silica concentration | PMO:00000165 | nanomole per kilogram |
| peridinin concentration | ENVO:3100025 | micromolar |
| phosphate concentration | ENVO:3100026 | micromolar |
| picoeukaryote count | PMO:00000161 | cells per milliliter |
| Prochlorococcus count | PMO:00000159 | cells per milliliter |
| prokaryotic leucine production | PMO:00000189 | picomolar per hour |
| salinity of water | PMO:00000014 | parts per thousand |
| silicic acid concentration | ENVO:3100034 | micromolar |
| Synechococcus count | PMO:00000160 | cells per milliliter |
| temperature of water | ENVO:09200014 | degree Celsius |
| turbidity of water | PMO:00000121 | formazin turbidity unit |
| violaxanthin concentration | ENVO:3100028 | micromolar |
| zeaxanthin concentration | ENVO:3100029 | micromolar |
Appendix II) Example RDF data structure
This is purely for reference and interest and is not required to use the system.
Example 1: Environmental context and physicochemical factors
@prefix ENVO: <http://purl.obolibrary.org/obo/ENVO_> .
:SRR1204581 rdf:type :sample_run ;
:has-env-broad-scale _:b0 .
_:b0 rdf:type ENVO:00000447 .
:SRR1204581 rdf:type :sample_run ;
:has-env-local-scale _:b0 .
_:b0 rdf:type ENVO:02000049 .
:SRR1204581 rdf:type :sample_run ;
:has-env-medium _:b0 .
_:b0 rdf:type ENVO:00002149 .
:SRR1204581 rdf:type :sample_run ;
:has-quality _:b0 .
_:b0 rdf:type ENVO:09200014 ;
:has-quantity "28.99"^^xsd:float .
Example 2: Gene Ontology and NCIBI Taxonomy Ontology annotated functional and taxonomic occurrence data
@prefix GO: <http://purl.obolibrary.org/obo/GO_> .
@prefix NCBITaxon: <http://purl.obolibrary.org/obo/NCBITaxon_> .
:ERR315856 rdf:type :sample_run ;
:has-go-annotation _:b0 .
_:b0 rdf:type GO:0000030 ;
:has-quantity 3 .
...
:ERR315856 :has-ncbitaxon-annotation _:b191 .
_:b191 rdf:type NCBITaxon:718192 ;
:has-quantity 106 .
...