OT-2 Counter-Selection
Ana Mariya Anhel, Lorea Alejaldre, Ángel Goñi-Moreno
Abstract
This protocol is meant to select the samples from 1 source plate that has been growing in different conditions and has different values for these different conditions, one condition gives higher values and the other lower values. From this selected samples this protocol create final plates with 2 types of reactives
The instructions for running this protocol are a python script for an Opentrons 2 robot and need a csv file with several variables setted making the protocol modular to reactives, volumes of transfer, type of plates, etc.
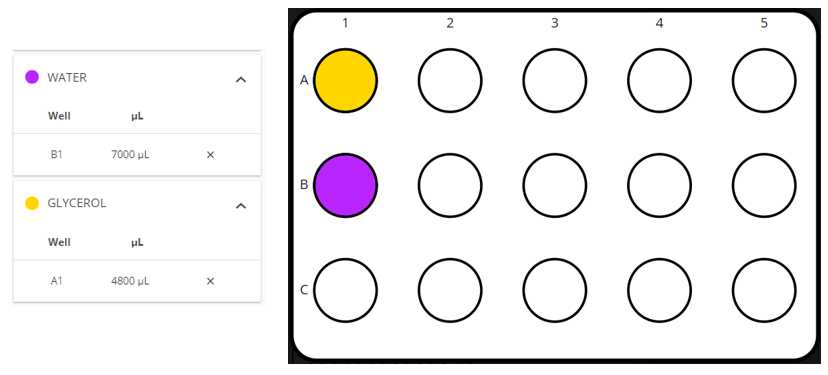
In our lab we are selecting samples by their after night OD in 2 different antibiotics and create PCR plates (with water) and stock plates (with glycerol), but it can be used with other selection characteristics as expression of GFP and with other types of reactives.
Before start
Note that this protocol can be done for one 96-well source plate at a time. In addition, it can only be performed with two selection variables (2 files with 1 condition per file in which one should be higher than a value and the other lower than this same value ).
Steps
Files Preparation
Preparing Customized Template
Preparing the template (a .csv) with the specific variables for each experiment.
Here we attach one excel with several sheets:
- Template to use in protocol
- Explanation of each variable
- Example
Fill the template with the corresponding values
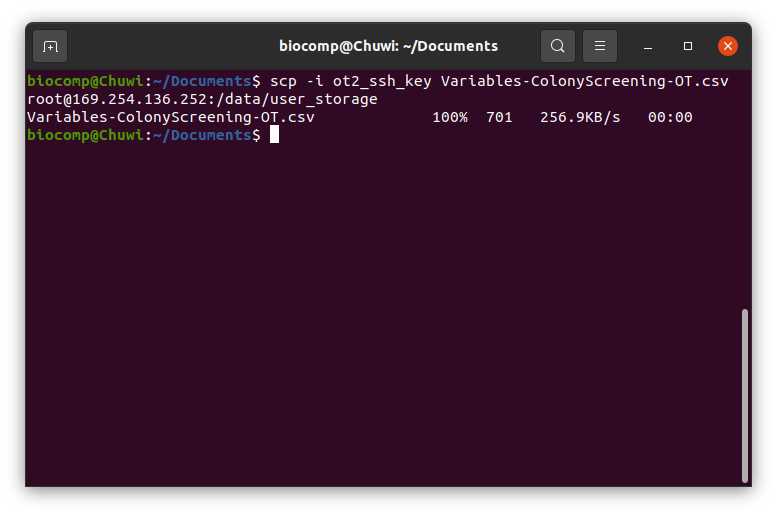
Store it as a comma-separated values file (csv) with the name Variables-ColonieScreening-OT.csv
Transferring csv to Robot
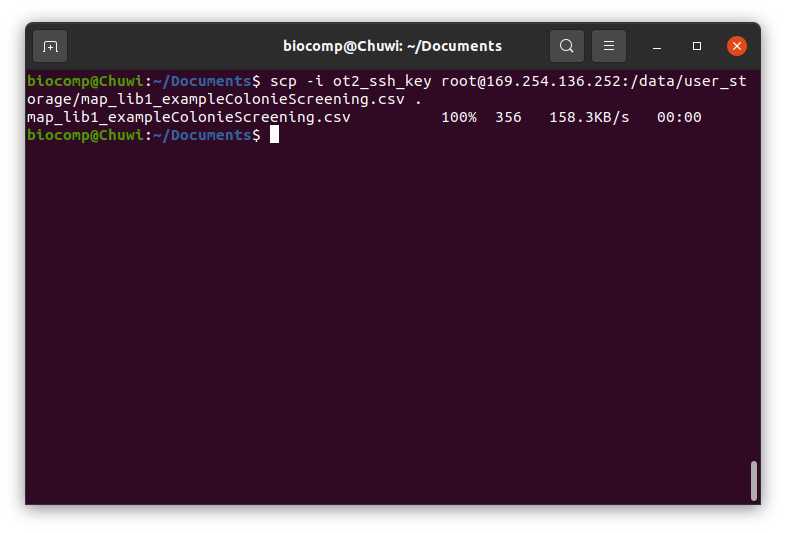
Transfer the Variables-ColonieScreening-OT.csv to the directory /data/user_storage of the OT system that we are going to use to perform the protocol.
Here we present a summary of how to transfer the files in 3 OS: Windows, Mac and Ubuntu (applicable to most Linux)
Mac/Linux
We will use the command line with scp to transfer the file Variables-ColonieScreening-OT.csv to the OT system.
We need to perform the following line:
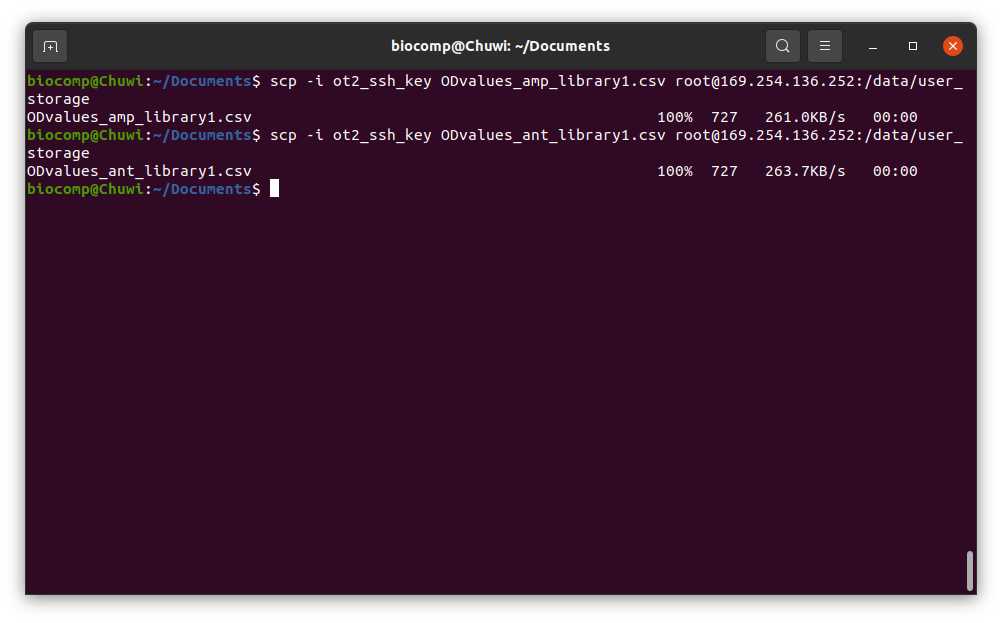
#Passing Files to OT (Ubuntu)
scp -i [ot_key] [file] root@[IP_OT]:/data/user_storage
Windows
There are several ways to pass files from a windows to a linux (for example with a virtual machine).
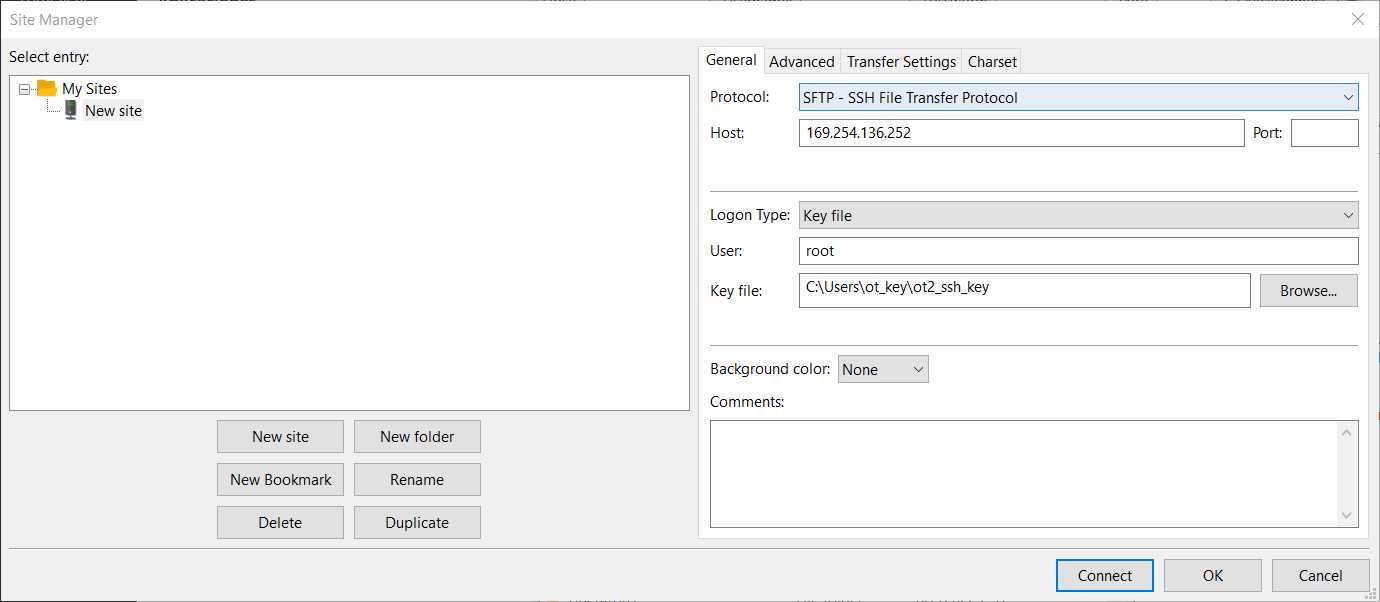
Here we will use FileZilla (https://filezilla-project.org/download.php?type=client).
Go to File > Site Manager > New Site > Change Protocol to SFTP . Then introduce in Host the OT IP, change the Logon Type to key file, change the user name to root and give the directory were the ot key is. Should look something like this
Then press connect and we will have a conection between our computer and the robot.
After this connection, we should be able to move out Variables-ColonieScreening-OT.csv (in our computer) to the directory /data/user_storage in the robot.
Adding the custom labware
Creation of .json file
The description file can be obtained by describing the labware dimensions in https://labware.opentrons.com/create/
Creating description folder
We need to create for our custom labware a folder with the API name containing the description file (.json) called 1.json
Transfer the description folder to the OT robot
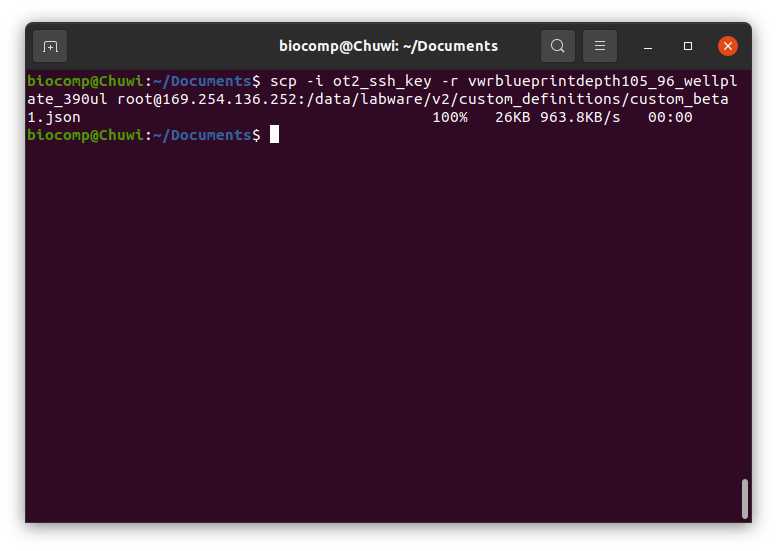
We need to transfer the directory or directories created in Step 3.2
For that we need to introduce the following command (in Linux), for Windows process
#Transfering directory to OT (Linux)
scp -i [ot_key] -r [directory_custom_labware] root@[IP_OT]:/data/labware/v2/custom_definitions/custom_beta
Simulating and getting the output (user instructions)
For more information in how to setup the ssh visit the official OT support blog (https://support.opentrons.com/s/article/Setting-up-SSH-access-to-your-OT-2)
Transferring python script
To make sure that the protocol will work with the variables we have introduced we need to run the python script in the robot with the OT simulator.
The last script version can be found at https://github.com/Biocomputation-CBGP/OT2/tree/main/ColonySelection (name of this file is user's choice)
For that we can but instead of passing the variables file, we pass the python script. In this case we can transfer it to the directory /data/user_storage or we can directly pass it to the directory /root (recommended)
Simulating
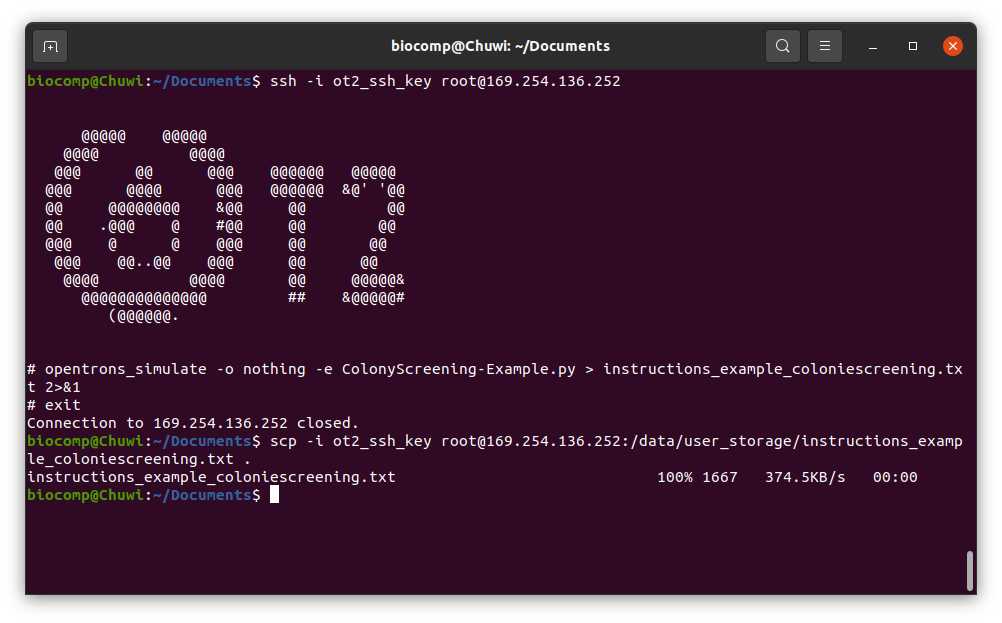
We should connect to the robot via ssh
#Connect to Linux based OT via ssh
ssh -i [ot_key] root@[Robot_IP]
Then we would move to the directory where the python script is with the cd command
Finally we are going to perform the simulation of the opentrons
#Simulate OT protocol (Linux)
opentrons_simulate -o nothing -e [name_script].py > [name_instruction_file].txt 2>&1
In case that you want to see the instruction file before the next step, you can see the instruction file with cat
#Show content file cmd (Linux)
cat [name_instruction_file].txt
Retrieving output file (user instructions)
To retrieve the output from the OT system you can , replicate it but transfering the file from the OT to your computer
In this file we have 3 or more sections:
- (Optional) Warnings
- General Information: General Information: pieces of information that are extracted from variables in the variables file and are only remainders
- Deck Labware Position: this information will be provided by the OT as well
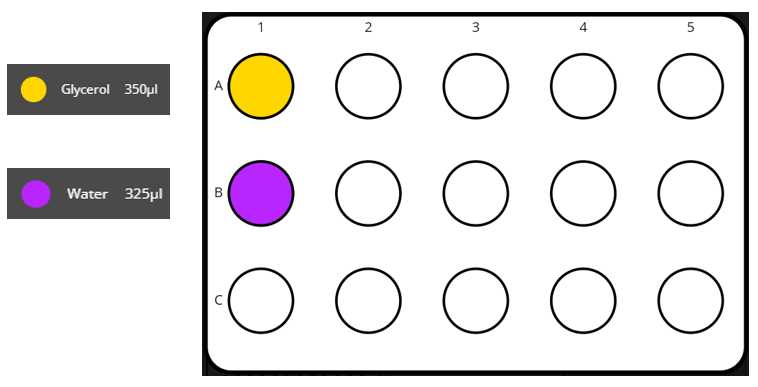
- 15mL Tuberack Reactive Positions: for each tuberack that is needed (with 15 positions) we have a table with the reactives and their respective volumes
In the case that something went wrong, the variables are not consistents or there is not enough deck place to perform the experiments the instruction file will be sorter and will have a single section, Errors .
Running Protocol
Setting Labware
Wipe the surface of the deck with 70% ethanol to clean and desinfect the surfaces
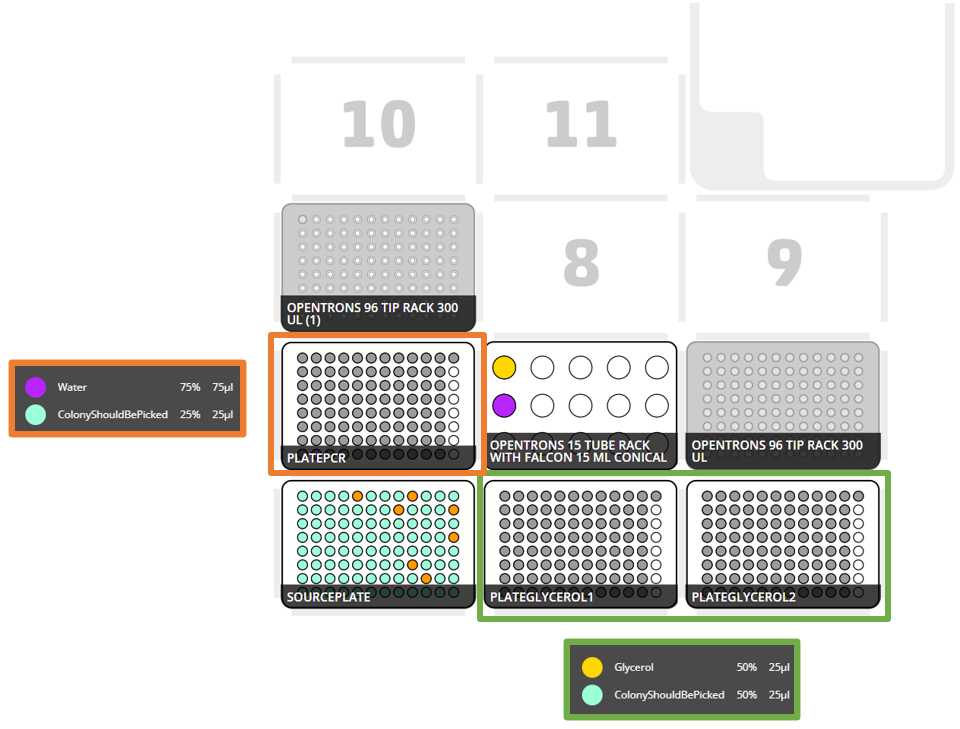
Set the respective labwares in the slots as the OT App and the instruction file set
Set the different reactives in their respective coldblock positions, as stated in the instruction file
Load script in OT-App
Now that we have made sure that the protocol is going to work we import the python script in the OT-App.
Load the script
Protocols > Import > Drag the python script
Select Robot to Perform Script
Click in the protocol > Run Protocol > Choose the OT where the file Variables-ColonieScreening-OT.csv is > Proceed To Setup Variables-ColonieScreening-OT.csv is > Proceed To Setup
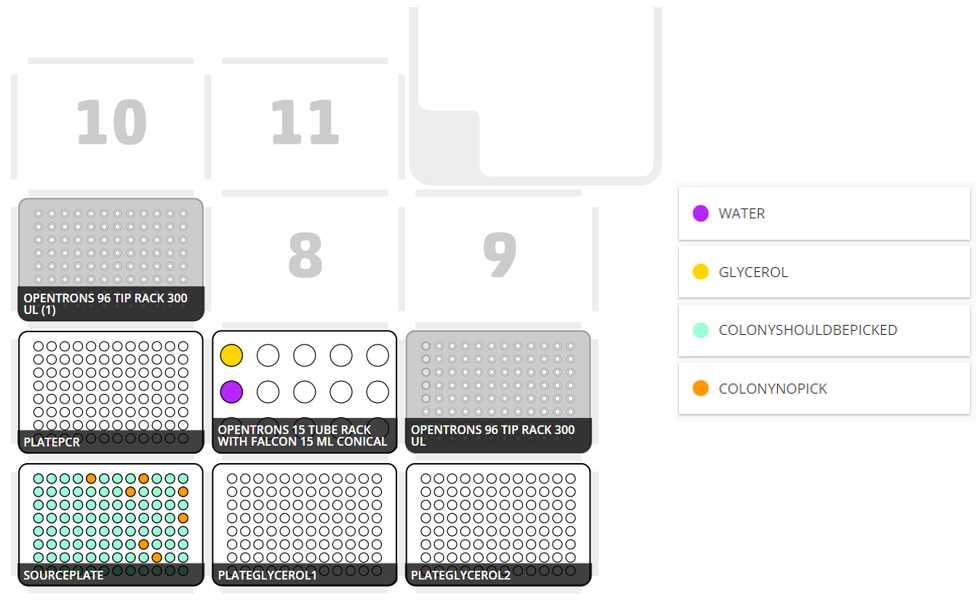
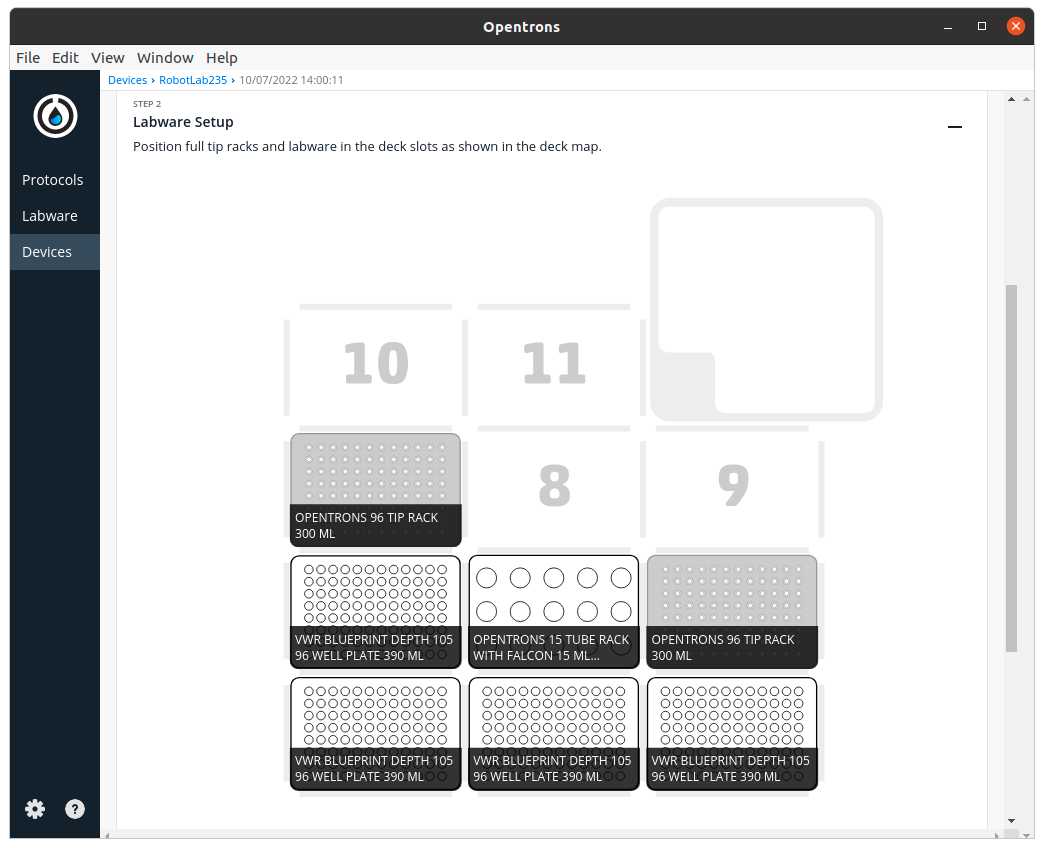
After clicking in Proceed to Setup you should obtain the setup of the labware as instructed in the Deck Labware Positions section of the instructions file.
The Labware Setup tab should look similar to the following image
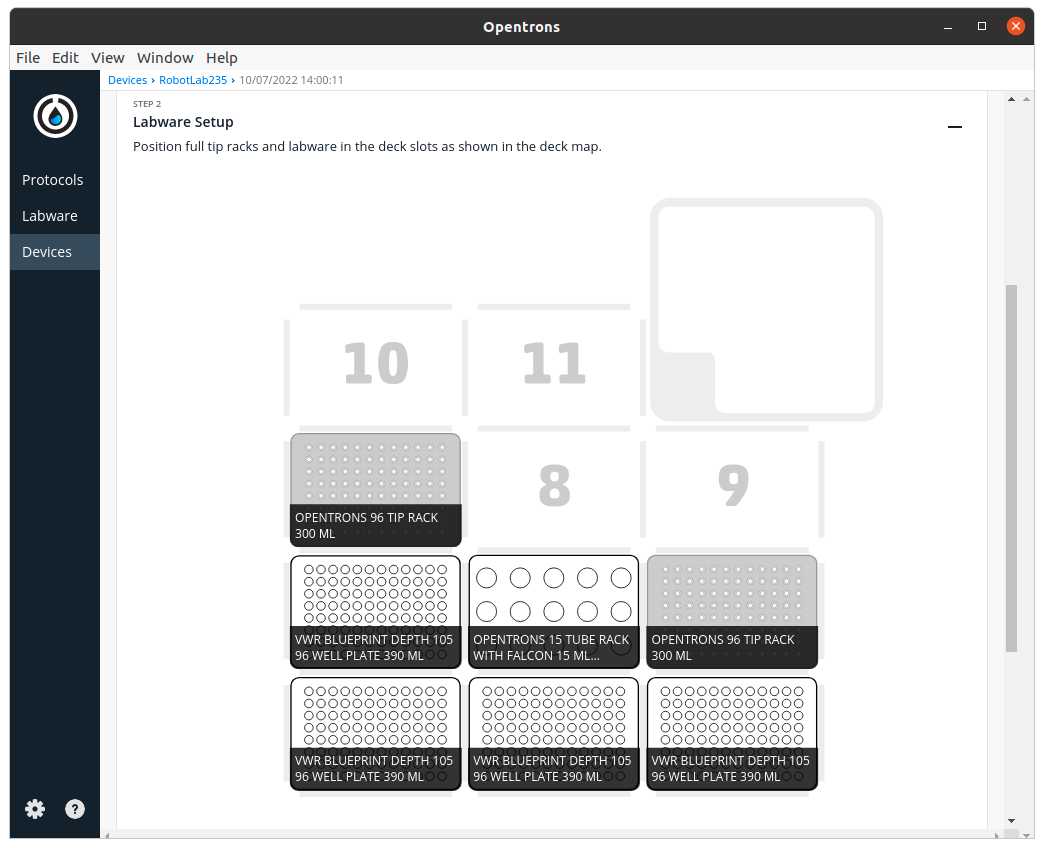
Run Protocol in OT
Make sure the needed calibrations are done
Labware position check is performed (if needed)
Start Run
The procedure that the robot is going to do is mainly divided in 3 parts:
- Select colonies that comply with the selected parameters
- Distribute reactives to respective plates
- Distribute to all plates the selected culture samples
- Generate identity maps (to be exported in following steps)
After-Running
Retrieve labware from the OT
Importing maps from robot
This can be done if in Step 1 the variable Optional Map was set to True
There will be as many maps as source plates with the following structure of names: VariableNameMap.csv
To retrieve we can and reproduce it with transfering the files to the computer.
They will be in the directory /data/user_storage
Example
We want to select the samples that have an OD600nm higher than 0.07 in the Antibiotic Transposition genome file 600nm higher than 0.07 in the Antibiotic Transposition genome file ( ODvalues_ant_library1.csv ) and have an OD600nm lower than 0.07 in the Antibiotic ampicillin plasmid file 600nm lower than 0.07 in the Antibiotic ampicillin plasmid file ( ODvalues_amp_library1.csv ). This corresponds to the example in the Template excel file in
Excel temple filled and exported to csv with the name Variables-ColonyScreening-OT.csv
Retrieve labwares from the OT
Pass the scipt that I have downloaded from https://github.com/Biocomputation-CBGP/OT2/tree/main/ColonySelection (I name it ColonyScreening-Example.py )

Turn HEPA module on and wipe OT with 70% ethanol
Load protocol labware as instructed in Step 10.6